Abstract
Despite the well-documented influence of genetics on susceptibility to cardiovascular diseases, delineation of the full spectrum of the risk alleles had to await the development of modern Next Generation Sequencing technologies. The techniques provide unbiased approaches for identification of the DNA sequence variants (DSVs) in the entire genome (Whole Genome Sequencing) or the protein-coding exons (Whole Exome Sequencing). Each genome contains approximately 4 million DSVs and each exome about 13,000 single nucleotide variants (SNVs). The challenge facing the researchers and clinicians alike is to decipher biological and clinical significance of these variants and harness the information for the practice of medicine. The common DSVs typically exert modest effect sizes, as evidenced by the results of Genome-Wide association Studies (GWAS), and hence, modest or negligible clinical implications. The focus is on the rare variants with large effect sizes, which are expected to have stronger clinical implications, as in single gene disorders with Mendelian patterns of inheritance. However, the clinical implications of the rare variants for common complex cardiovascular diseases remain to be established. In our view, the most important contribution of the WES or WGS is in delineation of the novel molecular pathways involved in the pathogenesis of the phenotype, which would be expected to provide for preventive and therapeutic opportunities.
Introduction
The advent of massively parallel nucleic acid sequencing or next generation sequencing (NGS) technologies has had an enabling impact in delineating the genetic make up of each individual. NGS has raised considerable interest in potential applications of the information embedded in the DNA sequence of each genome into the practice of medicine. The NGS platforms have afforded the opportunity to sequence the entire genome of an individual (whole genome sequencing or WGS), which is composed of 3.2 billion nucleotides, within a week. Likewise, it has enabled sequencing of the entire protein coding regions, referred to as exome, which is comprised of about 180,000 exons in ~ 23,500 genes and 30 million nucleotides in dozens of individuals within a few days (whole exome sequencing or WES). The enormity of this progress is remarkable, as the draft sequence of the human genome (The Human Genome Project), which was completed about a decade ago, took about 11 years and cost approximately 3 billion dollars (Lander et al., 2001). In contrast, the modern NGS instruments can generate up to 600 gigabases (Gb) output per sequencing run, in less than 2 weeks and at the cost of several thousand dollars. The output is sufficient to sequence several human genomes and several dozen exomes at an excellent average coverage per read. The exciting advances along with improvements in bioinformatics tools, which provide for the analysis of massive amount of sequence reads generated by the sequencing instruments, have led to dawning of the genetic-based practice of medicine. Despite these advances, however, a number of important obstacles have to be resolved before WGS or WES is routinely utilized in the practice of medicine.
Genetic etiology of susceptibility to cardiovascular diseases
Clinical phenotypes are the consequences of complex, non-linear and often stochastic interactions among various etiological determinants, including genetics factors. It is probably accurate to state that there is no phenotype that does not have a genetic etiological component. It is equally important to realize that there is no phenotype; Mendelian or otherwise, that is not influenced by the non-genetic factors. The magnitude of contribution of the genetic factors to the clinical phenotypes typically follows a gradient that ranges from minimal to large (Marian, 2009). It is best illustrated in familial segregation, which is indicative of the heritable component of the phenotype. Heritability, which is the fraction of observed variability in the phenotype that is attributed to genetic differences among the population with the trait, is best estimated through twin and family studies. It is strongest for Mendelian diseases, wherein the presence of the risk allele is sufficient to cause the phenotype, even though the phenotype is also influenced by a number of other genetic and non-genetic factors. In complex traits, however, the risk allele is neither sufficient nor necessary for the disease to manifest. Nevertheless, heritability estimates of the complex cardiovascular diseases, estimated from monozygotic and dizygotic twins studies, are quite variable and typically range from 30 to 80% (Marian, 2012). The well-documented heritability of the cardiovascular phenotypes is the basis for intense interest in Genome-Wide Association Studies (GWAS), WGS or WES to delineate the genetic basis for susceptibility to disease, response to therapy and the clinical outcomes. GWAS, which by design analyzes the common variants, have led to identification of over 200 susceptibility loci for cardiovascular diseases (http://www.genome.gov/gwastudies/). However, alleles identified through GWAS account only for a small fraction of heritability of the complex cardiovascular diseases. Hence, the emphasis has shifted toward identification and analysis of the rare variants through WES/WGS approaches, which might have larger effect sizes.
Abundance of DNA sequence variants in the human genome
The human genome is comprised of 3.2 billion pairs of nucleotides of which approximately 4 million nucleotides are polymorphic in a given individual, i.e., has a different nucleotide at a given position than the reference sequence. However, in a population of about 10,000 -15,000 individuals, approximately one in every 20 nucleotides is polymorphic (about 5% of the population genome) and the vast majority of the polymorphic alleles are exceedingly rare (Keinan and Clark, 2012; Nelson et al., 2012). With the exception of monozygotic twins, no two individuals are genetically, i.e., at the DNA sequence level, identical. In average, each human genome contains approximately 4 million DNA sequence variants (DSVs), of which about 3.5 million are single nucleotide variants (SNVs)(Levy et al., 2007; Ng et al., 2008; Pennisi, 2010; Wang et al., 2008; Wheeler et al., 2008). In addition, the genome contains thousands of small insertion/deletions (indels) and large segments of DNA duplications, insertions, deletions and rearrangements, which are collectively referred to as structural variations (SVs)(Kidd et al., 2008; Korbel et al., 2007). Structural variations that increase or decrease the two copies of the genes or chromosomal segments are referred to as Copy Number Variants (CNVs). The vast majority of the DSVs in the human genome are SNVs, and a considerable number of them are unique to an individual (Table 1). Almost half of the genes in each individual genome is polymorphic, i.e., the two copies of the genes are not precisely identical at the DNA sequence level (Levy et al., 2007). In addition, each genome/exome contains about 10,000 non–synonymous variants (nsSNVs) that – by definition – affect the amino acid sequence in the encoded proteins and hence, might exert biological effects (Levy et al., 2007; Ng et al., 2008; Wang et al., 2008; Wheeler et al., 2008). The commonly used bioinformatics tools for predicting the functional consequences of the DSVs, outside of nonsense, frameshift and splice junction variants, often do not offer concordant results (Tennessen et al., 2012). On average 2.3% of 13,595 SNVs identified in each exome are predicted to affect protein function by multiple bioinformatics tools (Tennessen et al., 2012). Thus, each exome carries several hundred to several thousand SNVs that are considered potentially damaging. Likewise, each genome contains approximately 100–120 loss-of-function (LoF) variants of which about 20 are homozygous and hence, inactivate the corresponding gene (MacArthur et al., 2012). Approximately 25 to 35 heterozygous and 2–3 homozygous variants affect stop codons and lead to inactivation of one or two copies of the corresponding gene. Approximately 50 to 100 variants in each genome are known to be associated with inherited disorders and about 30 variants in each genome are de novo, i.e., absent in the parents. The plethora of DSVs including putatively functional DSVs highlights the complexity of the genetic diversity of the humans, which is accentuated by the recent accelerate expansion of the human population and introduction of a very large number of new alleles in each generation. The newly introduced alleles are, as expected, rare, often private and expected to be more deleterious because of an adequate filtering by natural selection. This complexity complicates clinical applications of the DNA sequencing data.
TABLE 1.
DNA Sequence Variants in the Human Genome
| Nucleotides (base pairs) | 3.2 × 109 |
| Protein-coding genes | 23,500 |
| Number of exons | 180,000 |
| Size of exome (base pairs) | 30 × 106 |
| DNA Sequence variants (DSVs) | 4 ×106 |
| Single nucleotide polymorphisms (SNPs) | 3.5 ×106 |
| Structural variants (SVs)/Copy number variants (CNVs) | 104 – 105 |
| Non-synonymous SNPs (nsSNPs) | 10,000–12,000 |
| NsSNP potentially damaging | 100s-1,000s |
| Loss-of-function (loF) variants | 120 |
| Homozygous LoF variants | 20 |
| Variants known to be associated with inherited diseases | 50–100 |
| Stop-codon variants | 25–35 |
| Homozygous stop codon variants | 2–3 |
| De novo variants | 30 |
Medical DNA Sequencing
The strength of the NGS in offering an unbiased approach to identification of DSVs in an individual genome or exome, in terms of medical applications of the findings, has to be considered in the context of the enormous genetic diversity of the humans and the presence of a very large number of DSVs in each genome or exome. Further complicating the clinical implications is the unknown contributions of these variants to the phenotype. In considering the clinical utility of NGS data, the effect sizes of the DSVs, which follow a gradient, have major implications (Marian, 2009). By and large, rare DSVs exert larger effect sizes and hence, are more likely to be pathogenic, as in diseases with Mendelian patterns of inheritance. In contrast, common DSVs typically exert modest effect sizes and hence, have modest, if any clinical utility at in individual level, even though their population attributable risk might be greater, simply because of their abundance (Marian and Belmont, 2011). For common complex diseases, WES/WGS, in the best-case scenario, might lead to identification of a clinically meaningful risk allele for at least one disease (Roberts et al., 2012). However, the majority of the individuals are likely to have a negative WES/WGS results but the negative predictive value of such test would be relatively small for common complex diseases (Roberts et al., 2012). Overall, the direct clinical impact of NGS is expected to be greater for single gene diseases, while the indirect impact – through delineation of the responsible mechanisms for the pathogenesis of the phenotype – is equally important for single gene and complex polygenic traits. The complexity of applying the NGS data to medical practice has two major components, which are imperfectness of the NGS technologies and difficulty in identifying the clinically significant alleles.
Technical challenges
At the technical level, the error rate of allele calling, i.e., false positives, is probably the most important aspect of NGS, as even a small error rate leads to an inflated number of false positives in the genome, simply because of the size of human genome. The lowest sequencing error rate of 10−4 per nucleotide with the best available platforms is sufficient to introduce a large number of false positive calls. At the best circumstances, the false positive rate is about 5%, which means that of about 4 million variants detected in each genome, 200,000 allele calls would be erroneous. This would complicate clinical implications of the discoveries. Perhaps, the simplest way to reduce the number of false positives in WES/WGS is to increase the number of reads per each nucleotide or read, i.e., the coverage. Typically there is an inverse relationship between the average coverage rate of reads and the number of false positive alleles, albeit within a limit, as some errors might simply get amplified with increasing average. Likewise, certain genomic regions might be more prone to false positive calls and hence, re-sequencing alone might not be sufficient to eliminate such calls. In addition, increasing the coverage or re-sequencing, at least with the present instruments, adds to the cost of DNA sequencing as well the complexity of managing the massive amount of data and the bioinformatics analysis. Nevertheless, a coverage rate, which offers the maximum confidence in accurate allele calling, is essential in medical sequencing. Given the size of the whole genome, a higher mean nucleotide coverage rate in WGS would be necessary to reduce the technical difficulties in accurate allele caling. The mean coverage rate for WES is typically much higher because of the smaller size of the exome as opposed to genome. Also relevant to medical sequencing is inadequate capture of 1,000 to 2,000 genes for variant detection, as the current WES offers adequate capture for ~ 80 to 90 % of the exons. Hence, this imperfect sensitivity of the NGS is another source for potential missed diagnosis (Kiezun et al., 2012).
From the population genetic aspects, the issue of discerning the false positive calls is further complicated by fact that about 75% of alleles in the population genome are singleton or doubleton, which are subject to a high false positive rate (Keinan and Clark, 2012; Nelson et al., 2012). Eliminating singleton and doubleton is expected to reduce the number of false positive calls and yet it could also eliminate potentially important causal variants with large effect sizes. In addition, the false positive rate or the confidence in accurate allele calling is not evenly distributed to eliminate the false positives by re-sequencing the whole exome or the genome. Thus, false positive allele calls is expected to remain a major limitation in medical DNA sequencing, necessitating an alternative method for validation of the variants identified by WES or WGS in an individuals.
Challenges in linking the DSVs to clinical phenotypes
Perhaps even a bigger challenge than the problem of false positives is the difficulty in identifying the true causal allele from the vast number of relatively innocuous alleles, as no gene or protein is perfect and each carries a number of non-pathogenic variants, including non-SNVs. Evidently, the larger the gene the greater likelihood of identifying nsSNVs in the gene, some of which might be the causal variants. For example, TTN gene encoding sarcomere protein Titin is one of the largest genes in the genome. It carries a very large number of nsSNVs, of which only a fraction might be true causal variants despite the well-established causal role of TTN variants in cardiomyopathies (Herman et al., 2012). In a recent study, Seidman and colleagues only considered the truncating variants, i.e., those that shortened the length of the Titin protein, as the causal variants for dilated cardiomyopathy and did not include the nsSNVs in the initial analysis, partly because of their abundance in this large gene (Herman et al., 2012). Moreover, the challenge is to identify the clinically significant alleles from those might be functional but clinically not significant. In population genetics, various strategies might be employed to reduce the chance of a random association, such as setting a proper threshold for statistical significance, corrections for multiple hypothesis testing by Bonferroni’s methods or permutation analysis (Kiezun et al., 2012). Such measures apply to population data and do not apply to a single individual. Thus, in a given individual one can only estimate the risk based on the population genetic data and hence, might not be sufficiently accurate.
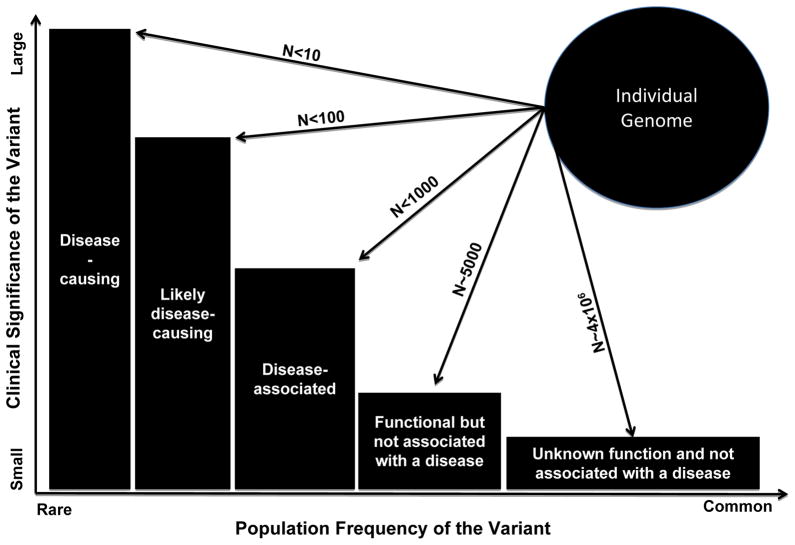
To better appreciate the clinical and biological significance of the DSVs in the genome, the variants might be categorized, in order of strength of the evidence for causality, into five classes (Figure 1) as follows: 1. Disease-causing; 2. Likely disease-causing; 3. Disease-associated; 4. Functional but not associated with a disease; and 5. Unknown biological function (Marian and Belmont, 2011). The clinical significance follows a gradient being the highest for disease-causing variants (category 1) and negligible for variants with unknown biological function (category 5). As would be expected the frequency of the alleles, in general, follows the opposite gradient, being rare for disease-causing variants and common for the variants that are not know to carry biological significance. Thus, the practical approach would be to focus on those variants that already have been implicated as the causal variants in the pathogenesis of phenotype, such as non-sense or missense mutations previously identified in patients with cardiovascular phenotypes or in a gene previously shown to be a causal gene for a Mendelian disorder. There are probably a handful of such variants in each genome, which could provide for an early detection of those at risk prior to development of the disease (pre-clinical). The genetic information might be exploited for close monitoring and follow up of the mutation carriers and even interventions to prevent the evolving phenotype. Nevertheless, like any medical diagnostic test, the results must not be over-interpreted because of the presence of considerable phenotype variability as well as plasticity (Klassen et al., 2011).
Figure 1. Plot showing population frequency of the DNA sequence variants (DSVs), abundance in an individual genome and the clinical significance (effect size) of the variants.
Rare alleles impart larger effect sizes and in general, have a greater clinical significance than common alleles, which have modest effect sizes and typically not clinically significant. Each genome contains a few disease-causing variants, a category for which there is strong genetic evidence of causality, typically through studies in large pedigrees with single gene-disorders. Each genome also contains a number rare variants with large effect sizes that are considered “Likely disease-causing variants” as the genetic evidence for their causal role is not sufficiently conclusive. This group typically includes rare variants that are identified in the affected individuals, wherein co-segregation with the phenotype cannot be robustly established because or small size of the families or incomplete penetrance. Disease-associated variants are relatively more common and are those that in large –scale studies, such as GWAS have been associated with the phenotype. However, they are neither sufficient to cause the disease nor necessary but rather increase the risk of the disease. Each genome/exome has about 7,000 non-synonymous putatively functional variants. The most abundant variants encompasses about 4 million DSV that neither have a known biological function or are associated with a phenotype.
It would be expected that putatively functional nsSNVs in each genome also to contribute to phenotypic expression of the diseases. However, the clinical utility of these relatively common functional variants in a given individual is relatively modest and negligible for most instances. Accordingly, medical applications of the NGS are largely restricted to about hundred variants that are either known to causal inherited diseases or impart major effects on gene structure and function, such as the LoF variants. Astute clinical phenotypic characterization and periodic follow up of those who carry these typically rare and functionally important variants is necessary. Likewise, close monitoring of those who do not exhibit any discernible clinical phenotype is necessary to detect any evolving disease early and prior to full-blown manifestations. The goal is to intervene early to prevent development of the disease. Nevertheless, despite the plausibility of the genetic-based medical practice, the potential utility of the genetic information on early diagnosis and prevention of cardiovascular diseases awaits to be tested and validated.
Concluding remarks
WES affords an unbiased discovery of rare and common variants in the protein-coding regions of the genomes and hence, has the potential for clinical applications. However, the imperfectness of variant detection by NGS, which reduces the sensitivity of the approach to 80–90% and the specificity to 90 to 95%, poses major challenges. While the sensitivity and specificity of NGS in variant detection and calling are within the realm of clinical testing, the potential implications of a false or a missed diagnosis are greater – both from medical point of view as well as because of the psychological stress that such mis-diagnosis might generate. In addition, clinical application of the information content of DSVs is partially restricted by the phenotypic variability and plasticity, and hence, the lack of a one-to-one correlation. Thus, experienced clinicians who are trained in medical genetics should carefully discuss the results of WES with those involved in order to reduce and hopefully eliminate the potential for providing false information.
In addition to direct medical implications, WES/WGS is a also robust approach for various clinical applications including identification of the causal genes for cardiovascular disorders; particularly single gene disorders (Herman et al., 2012), genetic testing in probands and family members with cardiovascular diseases with a Mendelian pattern of inheritance, (Meder et al., 2011) and in defining the genetic architecture of a disease in a given individual/family. Establishing the causal role of the rare variants for complex phenotypes in populations is quite challenging, because of the need for a very large sample size of study population, undefined effect size of the rare alleles, multi-directionality of the effects, and population heterogeneity, among others (Kiezun et al., 2012). Perhaps, the most important contribution of WES/WGS is in providing insights into the molecular pathogenesis of the phenotype through delineating its genetic etiology. The latter, namely, elucidation of the molecular genetic basis of cardiovascular diseases has the potential to provide for new therapeutic targets to prevent the disease and reverse the established phenotype.
Finally, it is important to recognize that clinical phenotypes are the outcomes of complex intertwined, stochastic and typically non-linear interactions among genetic, genomics and the environmental factors. Hence, information embedded in the genome/exome should be utilized wisely to fulfill the promise of “primum non nocere”.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Herman DS, Lam L, Taylor MR, Wang L, Teekakirikul P, Christodoulou D, Conner L, DePalma SR, McDonough B, Sparks E, et al. Truncations of titin causing dilated cardiomyopathy. The New England Journal Of Medicine. 2012;366:619–628. doi: 10.1056/NEJMoa1110186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinan A, Clark AG. Recent explosive human population growth has resulted in an excess of rare genetic variants. Science. 2012;336:740–743. doi: 10.1126/science.1217283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd JM, Cooper GM, Donahue WF, Hayden HS, Sampas N, Graves T, Hansen N, Teague B, Alkan C, Antonacci F, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiezun A, Garimella K, Do R, Stitziel NO, Neale BM, McLaren PJ, Gupta N, Sklar P, Sullivan PF, Moran JL, et al. Exome sequencing and the genetic basis of complex traits. Nature Genetics. 2012;44:623–630. doi: 10.1038/ng.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klassen T, Davis C, Goldman A, Burgess D, Chen T, Wheeler D, McPherson J, Bourquin T, Lewis L, Villasana D, et al. Exome sequencing of ion channel genes reveals complex profiles confounding personal risk assessment in epilepsy. Cell. 2011;145:1036–1048. doi: 10.1016/j.cell.2011.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbel JO, Urban AE, Affourtit JP, Godwin B, Grubert F, Simons JF, Kim PM, Palejev D, Carriero NJ, Du L, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Levy S, Sutton G, Ng PC, Feuk L, Halpern AL, Walenz BP, Axelrod N, Huang J, Kirkness EF, Denisov G, et al. The Diploid Genome Sequence of an Individual Human. PLoSBiol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur DG, Balasubramanian S, Frankish A, Huang N, Morris J, Walter K, Jostins L, Habegger L, Pickrell JK, Montgomery SB, et al. A systematic survey of loss-of-function variants in human protein-coding genes. Science. 2012;335:823–828. doi: 10.1126/science.1215040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marian AJ. Nature’s genetic gradients and the clinical phenotype. Circ Cardiovasc Genet. 2009;2:537–539. doi: 10.1161/CIRCGENETICS.109.921940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marian AJ. Elements of missing heritability. Current Opinion in Cardiology. 2012;27 doi: 10.1097/HCO.0b013e328352707d. [DOI] [PubMed] [Google Scholar]
- Marian AJ, Belmont J. Strategic approaches to unraveling genetic causes of cardiovascular diseases. Circulation Research. 2011;108:1252–1269. doi: 10.1161/CIRCRESAHA.110.236067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meder B, Haas J, Keller A, Heid C, Just S, Borries A, Boisguerin V, Scharfenberger-Schmeer M, Stahler P, Beier M, et al. Targeted Next-Generation Sequencing for the Molecular Genetic Diagnostics of Cardiomyopathies. Circ Cardiovasc Genet. 2011;4:110–122. doi: 10.1161/CIRCGENETICS.110.958322. [DOI] [PubMed] [Google Scholar]
- Nelson MR, Wegmann D, Ehm MG, Kessner D, St Jean P, Verzilli C, Shen J, Tang Z, Bacanu SA, Fraser D, et al. An Abundance of Rare Functional Variants in 202 Drug Target Genes Sequenced in 14,002 People. Science. 2012;337:100–104. doi: 10.1126/science.1217876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Levy S, Huang J, Stockwell TB, Walenz BP, Li K, Axelrod N, Busam DA, Strausberg RL, Venter JC. Genetic variation in an individual human exome. PLoS Genet. 2008;4:e1000160. doi: 10.1371/journal.pgen.1000160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennisi E. Genomics. 1000 Genomes Project gives new map of genetic diversity. Science. 2010;330:574–575. doi: 10.1126/science.330.6004.574. [DOI] [PubMed] [Google Scholar]
- Roberts NJ, Vogelstein JT, Parmigiani G, Kinzler KW, Vogelstein B, Velculescu VE. The predictive capacity of personal genome sequencing. Sci Transl Med. 2012;4:133ra158. doi: 10.1126/scitranslmed.3003380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen JA, Bigham AW, O’Connor TD, Fu W, Kenny EE, Gravel S, McGee S, Do R, Liu X, Jun G, et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wang W, Li R, Li Y, Tian G, Goodman L, Fan W, Zhang J, Li J, Guo Y, et al. The diploid genome sequence of an Asian individual. Nature. 2008;456:60–65. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A, He W, Chen YJ, Makhijani V, Roth GT, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]