Abstract
A substantial part of the human genome is derived from transposable elements; remnants of ancient retroviral infections. Conservative estimates set the percentage of human endogenous retroviruses (HERVs) in the genome at 8%. For the most part, the interplay between mutations, epigenetic mechanisms and posttranscriptional regulations silence HERVs in somatic cells. We first highlight mechanisms by which activation of members of several HERV families may be associated with tumor development before discussing the arising chances for both diagnosis and therapy. It has been shown that at least in some cases, tumor cells expressing HERV open reading frames (ORFs) thus gain tumor-promoting functions. However, since these proteins are not expressed in healthy tissues, they become prime target structures. Of potential pharmacological interest are the prevention of HERV transposition, the inhibition of HERV-encoded protein expression and the interference with these proteins’ activities. Evidence from recent studies unequivocally proves that HERV ORFs represent a very interesting source of novel tumor-specific antigens with even the potential to surpass entity boundaries. The development of new tumor (immune-) therapies is a very active field and true tumor-specific targets are of outstanding interest since they minimize the risk of autoimmunity and could reduce side effects. Finally, we postulate on main future research streams in order to stimulate discussion on this hot topic.
Keywords: Human endogenous retroviruses, Gastrointestinal cancer, Therapeutic targets, Tumor-specific antigens, Tumorigenesis
HUMAN ENDOGENOUS RETROVIRUSES: AN INTRODUCTION
Human endogenous retroviruses (HERVs) are remnants of ancient retroviral infections. Many insertions into the genome have taken place tens of millions of years ago[1,2]. Since the first set of data on the human genome project has been published in 2001, it is well established that about 8% of the genome consists of HERVs and their total number is approximately 3 × 105 copies[3]. Generally, HERVs are classified into three groups: class I [gamma (like) retroviruses], class II [beta (like) retroviruses] and class III [spuma (like) retroviruses][1]. The most common nomenclature utilizes the single-letter amino acid code corresponding to the tRNA primer that is used for reverse transcription of the HERV genome[4]. For the most part, their canonical structure of a single open reading frame (ORF) consists of the gag, pol and env genes flanked by 5’ and 3’ long terminal repeats (LTR)[2,5]. The latter features are what endogenous and exogenous retroviruses have in common.
ENDOGENIZATION
Following a retroviral infection, the fate of the host depends on the pathogenicity of the infectious virus. Highly pathogenic ones will kill the host whereas ones with only weak pathogenicity may manage to infect many different cell types, including reproductive tissue cells. Subsequently, an endogenous retrovirus will establish if virus and host proceed to fixation of the virus’ sequences in the host genome. This process has been termed endogenization or molecular domestication[6]. In most cases, HERV activities have been silenced by a variety of mechanisms. Mutational inactivation includes deletions as well as point mutations and probably has been triggered by specific regulatory proteins such as APOBEC[7,8]. Furthermore, epigenetic mechanisms including methylation and histone modification contributed to HERV inactivation[9]. Besides directly silencing the expression, posttranscriptional regulation further protects the host’s genome[10]. It has been argued that the presence of such a high number of HERV copies must be advantageous for the host, too[11]. An amazing idea suggests that over time of evolution, retroelements like HERVs actively contribute to the development of novel physiological capacities[12,13]. It is for example easily imaginable that a de novo ORF which basically encodes a membrane protein may give rise to a protein with novel functions when mutated. If such a protein is beneficial to the host, it will be fixed. Thus, HERVs are together with other mobile genetic elements drivers of the (human) evolution by providing material for genomic evolution, variation and natural selection[6,12,14]. This argumentation adds another level of complexity to the relationship of humans (and all other vertebrates) and retroviruses[15]. Consequently, HERVs must not be considered as parasites but as true symbionts - on the population level. However, the individual risk for de novo insertions is rather low with estimated rates of only 1 in 100 births[16].
PRESERVED FUNCTIONS
Although the vast majority of HERV sequences have been inactivated over time as outlined above, there are some examples of HERVs with potentially useful functional modules; comparable to the proviruses of their exogenous counterparts. Among the cellular functions influenced by HERVs are enhancement and promotion of gene expression. In a study on primate evolution ERV-9 LTR sequences were found in higher primates and humans. In the latter, tissue specific enhancer activity could be detected in hematopoietic cells and even stronger in embryonic cells[17]. HERV-E LTR functions as enhancer for endothelin B receptor and apolipoprotein C-I genes in humans[18]. Furthermore, HERV sequences also give rise to novel or alternative splicing and polyadenylation sites[19]. They also can be involved in membrane fusion, with Syncytin in the placenta being the most prominent representative here of[20].
HERV AND CANCER
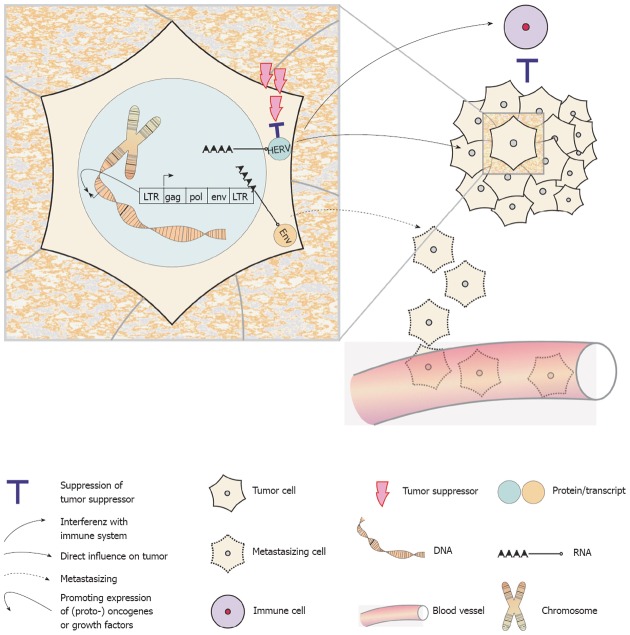
A variety of oncogenic mechanisms have been attributed to animal oncogenic retroviruses[21,22]. Moreover, it has been suggested, that failures and errors in single somatic cells’ efficiency to control HERV activity potentially results in genome damage and may thus contribute to the formation of cancer[14]. The possible oncogenic mechanisms of HERVs include (Figure 1): (1) the general or more specific (re)activation of HERV sequences by hypomethylation[23-25]; (2) the expression of HERV encoded oncogenes such as Rec and NP9[26]; (3) the inactivation of tumor suppressor genes by de novo insertion or translocation of retroelements within the genome[26]; (4) the regulation of nearby (proto-) oncogenes or growth factors by the regulatory sequences of LTRs[27,28]; and (5) the potential of Env proteins to induce cell fusions, which may contribute to tumor progression or even aid in metastasizing processes[29]. Far from being complete, this is already a quite impressive list. An additional aspect comes from the observation that Env proteins of the mouse leukemia virus, the Mason-Pfizer monkey virus and also of HERV-K have strong immunosuppressive properties and may thus help tumor cells evade an anti-tumoral immune response[26,30,31].
Figure 1.
Possible mechanisms by which human endogenous retroviruses contribute to oncogenesis. Human endogenous retroviruses (HERVs) transcripts or proteins may directly have tumor promoting properties. The long terminal repeat (LTR) elements can function as promotors or enhancers for nearby (proto-) oncogenes or growth factors. Especially Env proteins might attract regulatory immune cells and thus provide an immunosuppressive microenvironment. And finally, the Env proteins may be directly involved in the metastasizing process.
Methylation
As a general rule, all human regulatory genomic sequences become methylated unless specific factors prevent methylation[23]. In addition, methylated sites are more prone to mutations[23] and by this means, virus inactivation is further strengthened. Demethylation of regulatory regions is possible in the context of normal physiological processes by strong transcriptional activators. Re-expression of methylated sites is also possible during cell stress dependent on chromatin remodeling as a reaction to this stress[32]. Obviously, the maintenance of methylation patterns and status must play a central role in HERV transcriptional control. In healthy somatic and mature germ cells HERV sequences are generally (hyper-) methylated. Thus, HERV transcriptional activity is mainly restricted to germ cell development or the desensitization of check-point activation in meiotic cells. This mechanism may also be responsible for a high(er) retroelement expression in germ cell tumors[23]. In somatic cells, severe global hypomethylation leads to apoptosis induction mediated by TP53 and other tumor suppressive factors[33]. Premalignant and malignant cells are typically insensitive to apoptosis induction[34] and aberrant expression from normally methylated promoters is a main oncogenic force. In line with this, a general hypomethylation of HERV sequences can be found in the cancer cells of different entities, including testicular germ cell cancer, teratocarcinomas, colorectal, breast and ovarian cancer[35-39]. However, methylation analyses are biased by a lack of accuracy of the bisulfide sequencing technique[40]. When not highly standardized, this may account for a number of false positive or negative results in methylation analyses. Thus, it is always recommended to combine methylation analyses together with an investigation of mRNA or superior protein expression.
HERV ORF expression
Of interest, Syncytin-1 is the only expressed HERV sequence with a presumable physiological function. Syncytin-1 expression takes place in the placenta in the context of syncytiotrophoblast generation by cellular fusion of precursor cells, the cytotrophoblasts[41,42]. This expression follows after a general hypomethylation of a HERV-W env sequence and the Env protein is considered to contribute to this cellular fusion process. It may be a coincidence, but for many tumor entities, naturally occurring cellular fusions have been described[43-45] and this may hint towards the expression of similar HERV Env proteins. Expression of HERV sequences has been described for several tumor entities including melanoma, breast, ovarian, prostate and colon cancer[37-39,46,47]. Active retrotranspositions cause DNA strand-breaks and will thus lead to an activation of check-point signaling, e.g., TP53. Thus, transpositions as another mechanism for HERV re-expression may consequently occur especially in tumors with defect check-points and TP53 mutations[23].
Tumor induction/promotion
Beside sheer tumor specific expression, HERVs have repeatedly been discussed to induce or promote tumorigenesis. Potential mechanisms have been outlined in the preceding paragraphs. Here we want to gather the bits of evidence that have been obtained so far.
Several groups could show the production of HERV-derived proteins or even of viral particles in tumor cells[48-54]. In a mouse study, Howard and coworkers could directly link genome hypomethylation to ERV up-regulation[55]. Further research could make the connection between hypomethylation of (H)ERVs and chromosomal instability; it is by mediating ectopic recombination[56]. These HERV-induced recombination events have been found to produce large scale chromosomal anomalies[57], a hallmark of most tumors[34]. Finally, Lamprecht and colleagues could link the deregulated expression of the colony-stimulating factor 1 receptor (CSF1R) in B cell-derived Hodgkin’s lymphoma cells to hypomethylation of an up-stream HERV-derived LTR, which promotes ectopic expression of the CSF1R proto-oncogene[58]. However, the question if the reactivation of a (pro-) virus could actively promote cellular transformation or at least contribute to tumor progression is formally unsolved for human cancer. Similarly, it is unknown, if HERV activation is an early or a late step in tumor formation. Still, when considering the above listed bits of evidence, it seems reasonable to conclude that HERVs’ contribution to the multi-step process of tumor development in humans is very likely[14].
Immune responses towards HERV sequences
The human immune system’s capability to recognize HERV sequences has so far only scarcely been analyzed. However, some examples can be found in the literature. In patients with kidney cancer, cytotoxic T lymphocytes (CTLs) reactive to a HERV-E sequence encoded on chromosome 6q were found[59]. Serological responses and CTLs reactive to HERV-K sequences were detected in melanoma patients[60,61]. Anti-Env antibodies for HERV-K, -E and ERV3 were present in sera of patients with ovarian cancer[38] and in male patients with germ cell tumors[62]. Similarly, in breast cancer patients, anti-HERV-K serum antibodies were detected together with HERV-K-specific CTLs[63]. The orchestrated activation of both arms of the adaptive immune system in the latter cases is a strong indicator of HERV sequences’ high immunogenicity. Consequently, one may conclude that at least no strong tolerance towards HERV encoded sequences is induced during lymphocyte development. Future studies will have to analyze whether the immunological recognition of HERV sequences is executed by highly avid or only by intermediate avid T cells and antibodies. Also, it must be carefully analyzed, which HERV sequences give rise to strong immune responses when aberrantly expressed in tumor cells. We would like to state that immune recognition is a strong indicator for endogenous expression of a given HERV protein, as has been shown for other tumor antigens[64]. Of note, T cell reactions against HERVs, such as HERV-K, HERV-L and HERV-H, were associated with successful control of human immunodeficiency virus (HIV) in a subset of HIV patients[65]. It can be anticipated that this association of successful HIV control by HERV specific immune reactions will be translated into the tumor field. One of the major questions with clinical relevance is whether HERV-specific immune signatures can be associated with better prognosis or not.
THERAPEUTIC STRATEGIES
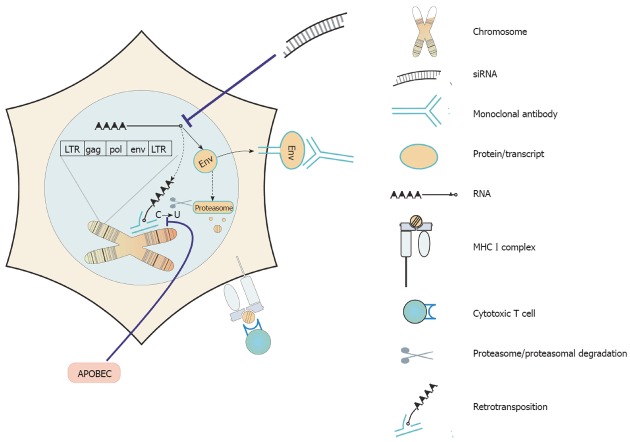
Assuming that in the normal physiology of adult tissues, HERVs do not play a vital role and following the line of evidence that HERV sequences are of significance in tumor formation, development and metastasis, HERVs recommend themselves as prime targets for tumor therapy. Several targeting strategies have been suggested (Figure 2 for an overview) and first experimental results can be found in the literature.
Figure 2.
Therapeutic possibilities to target tumor cells with active human endogenous retroviruses. Expressed Env proteins may be targeted by therapeutic monoclonal antibodies. (Re-) activation of retroelement-controlling proteins may help to reduce human endogenous retrovirus (HERV) activities. Small molecule inhibitors of HERV proteins or inhibitory targeting of expressed HERV sequences potentially will prevent oncogenic properties of HERV and harm or kill tumor cells with activated HERV oncogenesis. HERV proteins with tumor-specific antigen properties can be targeted by specific T cells. LTR: Long terminal repeat; MHC: Major histocompatibility complex.
Inhibition
In the light of the tremendous success in HIV control with infected people treated by antiretroviral combination therapies, it would make sense to simply reverse the expression of HERV sequences in human tumor cells. The group of Carlini analyzed the effect of a reverse transcription inhibitor (Abcavir) on prostate cancer cell lines[66]. It showed a strong anti-proliferative capacity and even triggered senescence in the cancer cells. Interestingly, the authors found an up-regulation of transcripts from LINE elements in the treated cells but unfortunately, they did not analyze HERV expression[66].
A direct targeting of HERV proteins by small molecular inhibitors or via RNA interference would also be worth trying. However, this has not yet been done. Therapeutical use of natural inhibitors of retroviruses such as APOBEC[67,68] or TRIM5[69] would be another possible future option. First, detail knowledge on how and when such retroviral restriction elements act on HERVs must be build up.
Passive immune therapy
Only very recently, Wang-Johanning and coworkers designed a monoclonal antibody (mAb) recognizing a HERV-K Env protein. They described that HERV-K Env protein expression was substantially higher in malignant breast cancer cell lines than in non-malignant breast cells. Furthermore, HERV-K expression was detected in 148 (66%) of 223 primary breast tumors. And a higher rate of lymph node metastasis was associated with HERV-K-positive tumors. Anti-HERV-K-specific mAbs inhibited tumor growth and induced apoptosis of breast cancer cells in vitro. Mice treated with these mAbs showed significantly reduced growth of xenograft tumors. In vitro, this treatment resulted in an over-expression of several proteins involved in the apoptotic signaling pathways in malignant breast cells[70]. In principle, targeting HERV Env proteins by therapeutical antibodies should be exploitable to all individual tumors expressing HERV Env. Moreover, passive immune therapies may well be applied in combination with active immune therapies.
Active immune therapy
The ideal cancer therapeutic agent should be able to discriminate between cancer and normal cells (i.e., specificity) and be potent enough to kill small or large numbers of tumor cells (i.e., sensitivity). A feature that makes immunotherapies unique is that an ideal cancer immunotherapy should be able to prevent recurrence of the tumor (i.e., durability). In the last decades it became increasingly apparent that this durability in prevention of tumor recurrences is due to persistent recognition of tumor antigens by lymphocytes.
Researchers distinguish between tumor-associated antigens (TAAs) and tumor-specific antigens (TSAs). TAAs are antigens that are expressed in normal tissue but to a much higher extent in malignant cells. Contrary to this, TSA are truly specifically expressed in tumor cells alone. Beside specific point[71] or frameshift mutations[72], proteins from tumor-inducing viruses[73] for the most part form this class of tumor antigens. Most of the features an ideal TSA should possess have been assigned to HERV encoded proteins. This being beside exclusivity also the necessity of expression for maintenance of the cancer cells’ transformed state. Thus, immune-escape by simple down-regulation of expression is prevented[74]. Moreover, to ease therapy development, ideal TSA expression should be not only present in single tumors but shared between individual tumors of a given entity or even superior between tumors of different entities[75,76]. Finally, the immune system should be able to mount both a cellular and a humoral response[63]. When summing up these desired properties of TSAs attributable to (at least some) HERV-encoded proteins, one may conclude that they might indeed be ideal targets for tumor immunotherapy. Because of the multitude of HERV-encoded sequences one can even expect that the development of a polyvalent (i.e., containing many epitopes) vaccine basing only on HERV epitopes may be possible. Even more visionary, actual bioinformatics approaches will allow the identification of immunogenic core epitopes shared between different HERV copy ORFs active in different tumor entities in order to design a universal HERV-based vaccine. As a first step in that direction, we recently described two CD8+ T cell epitopes encoded by a HERV-H copy located on Xp22.3[77].
FUTURE PERSPECTIVES
At the moment, in the field of HERVs more questions are open than answered. Are there human (tumor-) cells producing virus particles? If so, are those particles infectious? Further analyses on expression of HERV sequences and proteins - and in especially of Env proteins - would add to the full picture and understanding of the relationship of tumors and endogenous retroviruses. In a first step the mutual interaction between HERV Env and the immune system, also in a suppressive manner, has been addressed[26,30,31]. These analyses should be expanded. Especially a broader knowledge on tumor infiltrating cells specific for HERV epitopes and their prognostic value would be interesting. Furthermore, it would be very beneficial to know if there is a correlation between tumor grade, stage, progression or outcome and the expression of HERV sequences.
CONCLUSION
Our understanding of HERVs has come a long way. They must be considered as domesticated retroviruses with even main functions in evolution. On the level of an individual human being, however, their activities most likely are tightly controlled. Heavy genetic disorders, as present in tumor cells, generally seem to be linked with HERV activities. The tumor-specific expression of HERV-encoded proteins opens the way to diagnostically and therapeutically interesting opportunities: (1) The targeting of HERV proteins either biochemically or immunologically as TSAs; (2) Immune recognition of tumor cells takes place already early in tumor development. HERV-encoded ORF-derived proteins are likely candidates of this early recognition. Consequently, they may be ideal for screening people at risk to develop cancer as we suggested for frameshift mutations in lynch syndrome[72,78]; (3) the recognition of expressed HERV sequences by the adaptive immune system is likely to result in a better prognosis for patients raising to-be-defined minimum levels of immune responses. Such HERV-specific responses may well be suited for prognostic purposes.
POSTULATES
We would like to take the chance and hypothesize on some of the open questions and obvious tasks in the HERV/tumor field: APOBEC and other retroelement controlling factors are likely to be inactivated in cancer cells with active HERV-driven oncogenesis. If this is frequently the case, they must be considered tumor suppressor genes and screening for their inactivation would possibly hint towards specific HERV activation.
HERV-encoded TSAs are released into the circulation[79] and thus screening of HERV-TSA blood levels will become an interesting field of investigation. Similarly, HERV-specific (immune-) therapies will be developed in the near future for several tumor entities. For these immunotherapies, beside knowledge about expression in different tumors, the level of tolerance towards HERV-TSAs will guide the decision on which candidates to investigate in clinical trials.
Footnotes
Supported by Grants from the State Mecklenburg-Vorpommern and from Deutsche Krebshilfe, No. 108446
Peer reviewers: Yujin Hoshida, MD, PhD, Cancer Program, Broad Institute, 7 Cambridge Center, Cambridge, MA 02142, United States; Atsushi Nakajima, Professor, Division of Gastroenterology, Yokohama City University Graduate School of Medicine, 3-9 Fuku-ura, Kanazawa-ku, Yokohama 236-0004, Japan
S- Editor Gou SX L- Editor A E- Editor Li JY
References
- 1.Blikstad V, Benachenhou F, Sperber GO, Blomberg J. Evolution of human endogenous retroviral sequences: a conceptual account. Cell Mol Life Sci. 2008;65:3348–3365. doi: 10.1007/s00018-008-8495-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bannert N, Kurth R. Retroelements and the human genome: new perspectives on an old relation. Proc Natl Acad Sci USA. 2004;101 Suppl 2:14572–14579. doi: 10.1073/pnas.0404838101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 4.Cohen M, Larsson E. Human endogenous retroviruses. Bioessays. 1988;9:191–196. doi: 10.1002/bies.950090603. [DOI] [PubMed] [Google Scholar]
- 5.Nelson PN, Carnegie PR, Martin J, Davari Ejtehadi H, Hooley P, Roden D, Rowland-Jones S, Warren P, Astley J, Murray PG. Demystified. Human endogenous retroviruses. Mol Pathol. 2003;56:11–18. doi: 10.1136/mp.56.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Volff JN. Turning junk into gold: domestication of transposable elements and the creation of new genes in eukaryotes. Bioessays. 2006;28:913–922. doi: 10.1002/bies.20452. [DOI] [PubMed] [Google Scholar]
- 7.Goff SP. Retrovirus restriction factors. Mol Cell. 2004;16:849–859. doi: 10.1016/j.molcel.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 8.Mbisa JL, Bu W, Pathak VK. APOBEC3F and APOBEC3G inhibit HIV-1 DNA integration by different mechanisms. J Virol. 2010;84:5250–5259. doi: 10.1128/JVI.02358-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maksakova IA, Mager DL, Reiss D. Keeping active endogenous retroviral-like elements in check: the epigenetic perspective. Cell Mol Life Sci. 2008;65:3329–3347. doi: 10.1007/s00018-008-8494-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim DS, Hahn Y. Human-specific antisense transcripts induced by the insertion of transposable element. Int J Mol Med. 2010;26:151–157. [PubMed] [Google Scholar]
- 11.Bannert N, Kurth R. The evolutionary dynamics of human endogenous retroviral families. Annu Rev Genomics Hum Genet. 2006;7:149–173. doi: 10.1146/annurev.genom.7.080505.115700. [DOI] [PubMed] [Google Scholar]
- 12.Hedges DJ, Batzer MA. From the margins of the genome: mobile elements shape primate evolution. Bioessays. 2005;27:785–794. doi: 10.1002/bies.20268. [DOI] [PubMed] [Google Scholar]
- 13.Kazazian HH. Mobile elements: drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- 14.Kozeretska IA, Demydov SV, Ostapchenko LI. Mobile genetic elements and cancer. From mutations to gene therapy. Exp Oncol. 2011;33:198–205. [PubMed] [Google Scholar]
- 15.Rowe HM, Trono D. Dynamic control of endogenous retroviruses during development. Virology. 2011;411:273–287. doi: 10.1016/j.virol.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 16.Deininger PL, Batzer MA. Mammalian retroelements. Genome Res. 2002;12:1455–1465. doi: 10.1101/gr.282402. [DOI] [PubMed] [Google Scholar]
- 17.Ling J, Pi W, Bollag R, Zeng S, Keskintepe M, Saliman H, Krantz S, Whitney B, Tuan D. The solitary long terminal repeats of ERV-9 endogenous retrovirus are conserved during primate evolution and possess enhancer activities in embryonic and hematopoietic cells. J Virol. 2002;76:2410–2423. doi: 10.1128/jvi.76.5.2410-2423.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Medstrand P, Landry JR, Mager DL. Long terminal repeats are used as alternative promoters for the endothelin B receptor and apolipoprotein C-I genes in humans. J Biol Chem. 2001;276:1896–1903. doi: 10.1074/jbc.M006557200. [DOI] [PubMed] [Google Scholar]
- 19.Kim DS, Hahn Y. Identification of human-specific transcript variants induced by DNA insertions in the human genome. Bioinformatics. 2011;27:14–21. doi: 10.1093/bioinformatics/btq612. [DOI] [PubMed] [Google Scholar]
- 20.Pötgens AJ, Drewlo S, Kokozidou M, Kaufmann P. Syncytin: the major regulator of trophoblast fusion? Recent developments and hypotheses on its action. Hum Reprod Update. 2004;10:487–496. doi: 10.1093/humupd/dmh039. [DOI] [PubMed] [Google Scholar]
- 21.Braoudaki M, Tzortzatou-Stathopoulou F. Tumorigenesis related to retroviral infections. J Infect Dev Ctries. 2011;5:751–758. doi: 10.3855/jidc.1773. [DOI] [PubMed] [Google Scholar]
- 22.Maeda N, Fan H, Yoshikai Y. Oncogenesis by retroviruses: old and new paradigms. Rev Med Virol. 2008;18:387–405. doi: 10.1002/rmv.592. [DOI] [PubMed] [Google Scholar]
- 23.Schulz WA, Steinhoff C, Florl AR. Methylation of endogenous human retroelements in health and disease. Curr Top Microbiol Immunol. 2006;310:211–250. doi: 10.1007/3-540-31181-5_11. [DOI] [PubMed] [Google Scholar]
- 24.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 25.Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002;21:5400–5413. doi: 10.1038/sj.onc.1205651. [DOI] [PubMed] [Google Scholar]
- 26.Ruprecht K, Mayer J, Sauter M, Roemer K, Mueller-Lantzsch N. Endogenous retroviruses and cancer. Cell Mol Life Sci. 2008;65:3366–3382. doi: 10.1007/s00018-008-8496-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dunn CA, Medstrand P, Mager DL. An endogenous retroviral long terminal repeat is the dominant promoter for human beta1,3-galactosyltransferase 5 in the colon. Proc Natl Acad Sci USA. 2003;100:12841–12846. doi: 10.1073/pnas.2134464100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamprecht B, Walter K, Kreher S, Kumar R, Hummel M, Lenze D, Köchert K, Bouhlel MA, Richter J, Soler E, et al. Derepression of an endogenous long terminal repeat activates the CSF1R proto-oncogene in human lymphoma. Nat Med. 2010;16:571–579, 1p following 579. doi: 10.1038/nm.2129. [DOI] [PubMed] [Google Scholar]
- 29.Oricchio E, Sciamanna I, Beraldi R, Tolstonog GV, Schumann GG, Spadafora C. Distinct roles for LINE-1 and HERV-K retroelements in cell proliferation, differentiation and tumor progression. Oncogene. 2007;26:4226–4233. doi: 10.1038/sj.onc.1210214. [DOI] [PubMed] [Google Scholar]
- 30.Mangeney M, Heidmann T. Tumor cells expressing a retroviral envelope escape immune rejection in vivo. Proc Natl Acad Sci USA. 1998;95:14920–14925. doi: 10.1073/pnas.95.25.14920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blaise S, Mangeney M, Heidmann T. The envelope of Mason-Pfizer monkey virus has immunosuppressive properties. J Gen Virol. 2001;82:1597–1600. doi: 10.1099/0022-1317-82-7-1597. [DOI] [PubMed] [Google Scholar]
- 32.Kim C, Rubin CM, Schmid CW. Genome-wide chromatin remodeling modulates the Alu heat shock response. Gene. 2001;276:127–133. doi: 10.1016/s0378-1119(01)00639-4. [DOI] [PubMed] [Google Scholar]
- 33.Jackson-Grusby L, Beard C, Possemato R, Tudor M, Fambrough D, Csankovszki G, Dausman J, Lee P, Wilson C, Lander E, et al. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat Genet. 2001;27:31–39. doi: 10.1038/83730. [DOI] [PubMed] [Google Scholar]
- 34.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 35.Götzinger N, Sauter M, Roemer K, Mueller-Lantzsch N. Regulation of human endogenous retrovirus-K Gag expression in teratocarcinoma cell lines and human tumours. J Gen Virol. 1996;77(Pt 12):2983–2990. doi: 10.1099/0022-1317-77-12-2983. [DOI] [PubMed] [Google Scholar]
- 36.Boller K, König H, Sauter M, Mueller-Lantzsch N, Löwer R, Löwer J, Kurth R. Evidence that HERV-K is the endogenous retrovirus sequence that codes for the human teratocarcinoma-derived retrovirus HTDV. Virology. 1993;196:349–353. doi: 10.1006/viro.1993.1487. [DOI] [PubMed] [Google Scholar]
- 37.Menendez L, Benigno BB, McDonald JF. L1 and HERV-W retrotransposons are hypomethylated in human ovarian carcinomas. Mol Cancer. 2004;3:12. doi: 10.1186/1476-4598-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang-Johanning F, Liu J, Rycaj K, Huang M, Tsai K, Rosen DG, Chen DT, Lu DW, Barnhart KF, Johanning GL. Expression of multiple human endogenous retrovirus surface envelope proteins in ovarian cancer. Int J Cancer. 2007;120:81–90. doi: 10.1002/ijc.22256. [DOI] [PubMed] [Google Scholar]
- 39.Wentzensen N, Coy JF, Knaebel HP, Linnebacher M, Wilz B, Gebert J, von Knebel Doeberitz M. Expression of an endogenous retroviral sequence from the HERV-H group in gastrointestinal cancers. Int J Cancer. 2007;121:1417–1423. doi: 10.1002/ijc.22826. [DOI] [PubMed] [Google Scholar]
- 40.Krueger F, Kreck B, Franke A, Andrews SR. DNA methylome analysis using short bisulfite sequencing data. Nat Methods. 2012;9:145–151. doi: 10.1038/nmeth.1828. [DOI] [PubMed] [Google Scholar]
- 41.Mallet F, Bouton O, Prudhomme S, Cheynet V, Oriol G, Bonnaud B, Lucotte G, Duret L, Mandrand B. The endogenous retroviral locus ERVWE1 is a bona fide gene involved in hominoid placental physiology. Proc Natl Acad Sci USA. 2004;101:1731–1736. doi: 10.1073/pnas.0305763101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blond JL, Lavillette D, Cheynet V, Bouton O, Oriol G, Chapel-Fernandes S, Mandrand B, Mallet F, Cosset FL. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J Virol. 2000;74:3321–3329. doi: 10.1128/jvi.74.7.3321-3329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dittmar T, Schwitalla S, Seidel J, Haverkampf S, Reith G, Meyer-Staeckling S, Brandt BH, Niggemann B, Zänker KS. Characterization of hybrid cells derived from spontaneous fusion events between breast epithelial cells exhibiting stem-like characteristics and breast cancer cells. Clin Exp Metastasis. 2011;28:75–90. doi: 10.1007/s10585-010-9359-3. [DOI] [PubMed] [Google Scholar]
- 44.Rappa G, Mercapide J, Lorico A. Spontaneous formation of tumorigenic hybrids between breast cancer and multipotent stromal cells is a source of tumor heterogeneity. Am J Pathol. 2012;180:2504–2515. doi: 10.1016/j.ajpath.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lazova R, Chakraborty A, Pawelek JM. Leukocyte-cancer cell fusion: initiator of the warburg effect in malignancy? Adv Exp Med Biol. 2011;714:151–172. doi: 10.1007/978-94-007-0782-5_8. [DOI] [PubMed] [Google Scholar]
- 46.Sauter M, Schommer S, Kremmer E, Remberger K, Dölken G, Lemm I, Buck M, Best B, Neumann-Haefelin D, Mueller-Lantzsch N. Human endogenous retrovirus K10: expression of Gag protein and detection of antibodies in patients with seminomas. J Virol. 1995;69:414–421. doi: 10.1128/jvi.69.1.414-421.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ishida T, Obata Y, Ohara N, Matsushita H, Sato S, Uenaka A, Saika T, Miyamura T, Chayama K, Nakamura Y, et al. Identification of the HERV-K gag antigen in prostate cancer by SEREX using autologous patient serum and its immunogenicity. Cancer Immun. 2008;8:15. [PMC free article] [PubMed] [Google Scholar]
- 48.Contreras-Galindo R, Kaplan MH, Leissner P, Verjat T, Ferlenghi I, Bagnoli F, Giusti F, Dosik MH, Hayes DF, Gitlin SD, et al. Human endogenous retrovirus K (HML-2) elements in the plasma of people with lymphoma and breast cancer. J Virol. 2008;82:9329–9336. doi: 10.1128/JVI.00646-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Büscher K, Hahn S, Hofmann M, Trefzer U, Ozel M, Sterry W, Löwer J, Löwer R, Kurth R, Denner J. Expression of the human endogenous retrovirus-K transmembrane envelope, Rec and Np9 proteins in melanomas and melanoma cell lines. Melanoma Res. 2006;16:223–234. doi: 10.1097/01.cmr.0000215031.07941.ca. [DOI] [PubMed] [Google Scholar]
- 50.Löwer R, Boller K, Hasenmaier B, Korbmacher C, Müller-Lantzsch N, Löwer J, Kurth R. Identification of human endogenous retroviruses with complex mRNA expression and particle formation. Proc Natl Acad Sci USA. 1993;90:4480–4484. doi: 10.1073/pnas.90.10.4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Muster T, Waltenberger A, Grassauer A, Hirschl S, Caucig P, Romirer I, Födinger D, Seppele H, Schanab O, Magin-Lachmann C, et al. An endogenous retrovirus derived from human melanoma cells. Cancer Res. 2003;63:8735–8741. [PubMed] [Google Scholar]
- 52.Morgan D, Brodsky I. Human endogenous retrovirus (HERV-K) particles in megakaryocytes cultured from essential thrombocythemia peripheral blood stem cells. Exp Hematol. 2004;32:520–525. doi: 10.1016/j.exphem.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 53.Seifarth W, Skladny H, Krieg-Schneider F, Reichert A, Hehlmann R, Leib-Mösch C. Retrovirus-like particles released from the human breast cancer cell line T47-D display type B- and C-related endogenous retroviral sequences. J Virol. 1995;69:6408–6416. doi: 10.1128/jvi.69.10.6408-6416.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stocking C, Kozak CA. Murine endogenous retroviruses. Cell Mol Life Sci. 2008;65:3383–3398. doi: 10.1007/s00018-008-8497-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Howard G, Eiges R, Gaudet F, Jaenisch R, Eden A. Activation and transposition of endogenous retroviral elements in hypomethylation induced tumors in mice. Oncogene. 2008;27:404–408. doi: 10.1038/sj.onc.1210631. [DOI] [PubMed] [Google Scholar]
- 56.Romanish MT, Cohen CJ, Mager DL. Potential mechanisms of endogenous retroviral-mediated genomic instability in human cancer. Semin Cancer Biol. 2010;20:246–253. doi: 10.1016/j.semcancer.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 57.Sun C, Skaletsky H, Rozen S, Gromoll J, Nieschlag E, Oates R, Page DC. Deletion of azoospermia factor a (AZFa) region of human Y chromosome caused by recombination between HERV15 proviruses. Hum Mol Genet. 2000;9:2291–2296. doi: 10.1093/oxfordjournals.hmg.a018920. [DOI] [PubMed] [Google Scholar]
- 58.Lamprecht B, Bonifer C, Mathas S. Repeat-element driven activation of proto-oncogenes in human malignancies. Cell Cycle. 2010;9:4276–4281. doi: 10.4161/cc.9.21.13682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takahashi Y, Harashima N, Kajigaya S, Yokoyama H, Cherkasova E, McCoy JP, Hanada K, Mena O, Kurlander R, Tawab A, et al. Regression of human kidney cancer following allogeneic stem cell transplantation is associated with recognition of an HERV-E antigen by T cells. J Clin Invest. 2008;118:1099–1109. doi: 10.1172/JCI34409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schiavetti F, Thonnard J, Colau D, Boon T, Coulie PG. A human endogenous retroviral sequence encoding an antigen recognized on melanoma by cytolytic T lymphocytes. Cancer Res. 2002;62:5510–5516. [PubMed] [Google Scholar]
- 61.Hahn S, Ugurel S, Hanschmann KM, Strobel H, Tondera C, Schadendorf D, Löwer J, Löwer R. Serological response to human endogenous retrovirus K in melanoma patients correlates with survival probability. AIDS Res Hum Retroviruses. 2008;24:717–723. doi: 10.1089/aid.2007.0286. [DOI] [PubMed] [Google Scholar]
- 62.Boller K, Janssen O, Schuldes H, Tönjes RR, Kurth R. Characterization of the antibody response specific for the human endogenous retrovirus HTDV/HERV-K. J Virol. 1997;71:4581–4588. doi: 10.1128/jvi.71.6.4581-4588.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang-Johanning F, Radvanyi L, Rycaj K, Plummer JB, Yan P, Sastry KJ, Piyathilake CJ, Hunt KK, Johanning GL. Human endogenous retrovirus K triggers an antigen-specific immune response in breast cancer patients. Cancer Res. 2008;68:5869–5877. doi: 10.1158/0008-5472.CAN-07-6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ripberger E, Linnebacher M, Schwitalle Y, Gebert J, von Knebel Doeberitz M. Identification of an HLA-A0201-restricted CTL epitope generated by a tumor-specific frameshift mutation in a coding microsatellite of the OGT gene. J Clin Immunol. 2003;23:415–423. doi: 10.1023/a:1025329819121. [DOI] [PubMed] [Google Scholar]
- 65.SenGupta D, Tandon R, Vieira RG, Ndhlovu LC, Lown-Hecht R, Ormsby CE, Loh L, Jones RB, Garrison KE, Martin JN, et al. Strong human endogenous retrovirus-specific T cell responses are associated with control of HIV-1 in chronic infection. J Virol. 2011;85:6977–6985. doi: 10.1128/JVI.00179-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Carlini F, Ridolfi B, Molinari A, Parisi C, Bozzuto G, Toccacieli L, Formisano G, De Orsi D, Paradisi S, Grober OM, et al. The reverse transcription inhibitor abacavir shows anticancer activity in prostate cancer cell lines. PLoS One. 2010;5:e14221. doi: 10.1371/journal.pone.0014221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Harris RS. Enhancing immunity to HIV through APOBEC. Nat Biotechnol. 2008;26:1089–1090. doi: 10.1038/nbt1008-1089. [DOI] [PubMed] [Google Scholar]
- 68.Argyris EG, Acheampong E, Wang F, Huang J, Chen K, Mukhtar M, Zhang H. The interferon-induced expression of APOBEC3G in human blood-brain barrier exerts a potent intrinsic immunity to block HIV-1 entry to central nervous system. Virology. 2007;367:440–451. doi: 10.1016/j.virol.2007.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sawyer SL, Wu LI, Emerman M, Malik HS. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc Natl Acad Sci USA. 2005;102:2832–2837. doi: 10.1073/pnas.0409853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang-Johanning F, Rycaj K, Plummer JB, Li M, Yin B, Frerich K, Garza JG, Shen J, Lin K, Yan P, et al. Immunotherapeutic potential of anti-human endogenous retrovirus-K envelope protein antibodies in targeting breast tumors. J Natl Cancer Inst. 2012;104:189–210. doi: 10.1093/jnci/djr540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sensi M, Anichini A. Unique tumor antigens: evidence for immune control of genome integrity and immunogenic targets for T cell-mediated patient-specific immunotherapy. Clin Cancer Res. 2006;12:5023–5032. doi: 10.1158/1078-0432.CCR-05-2682. [DOI] [PubMed] [Google Scholar]
- 72.Linnebacher M, Gebert J, Rudy W, Woerner S, Yuan YP, Bork P, von Knebel Doeberitz M. Frameshift peptide-derived T-cell epitopes: a source of novel tumor-specific antigens. Int J Cancer. 2001;93:6–11. doi: 10.1002/ijc.1298. [DOI] [PubMed] [Google Scholar]
- 73.Mizuuchi M, Hirohashi Y, Torigoe T, Kuroda T, Yasuda K, Shimizu Y, Saito T, Sato N. Novel oligomannose liposome-DNA complex DNA vaccination efficiently evokes anti-HPV E6 and E7 CTL responses. Exp Mol Pathol. 2012;92:185–190. doi: 10.1016/j.yexmp.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 74.Serafino A, Balestrieri E, Pierimarchi P, Matteucci C, Moroni G, Oricchio E, Rasi G, Mastino A, Spadafora C, Garaci E, et al. The activation of human endogenous retrovirus K (HERV-K) is implicated in melanoma cell malignant transformation. Exp Cell Res. 2009;315:849–862. doi: 10.1016/j.yexcr.2008.12.023. [DOI] [PubMed] [Google Scholar]
- 75.Herbst H, Sauter M, Fuchs H, Kühler-Obbarius C, Löning T, Mueller-Lantzsch N. Gene products of human endogenous retrovirus (HERV)-K in germ cell and trophoblastic tumors. Verh Dtsch Ges Pathol. 1997;81:464–470. [PubMed] [Google Scholar]
- 76.de Parseval N, Diop G, Blaise S, Helle F, Vasilescu A, Matsuda F, Heidmann T. Comprehensive search for intra- and inter-specific sequence polymorphisms among coding envelope genes of retroviral origin found in the human genome: genes and pseudogenes. BMC Genomics. 2005;6:117. doi: 10.1186/1471-2164-6-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mullins CS, Linnebacher M. Endogenous retrovirus sequences as a novel class of tumor-specific antigens: an example of HERV-H env encoding strong CTL epitopes. Cancer Immunol Immunother. 2012;61:1093–1100. doi: 10.1007/s00262-011-1183-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Garbe Y, Maletzki C, Linnebacher M. An MSI tumor specific frameshift mutation in a coding microsatellite of MSH3 encodes for HLA-A0201-restricted CD8+ cytotoxic T cell epitopes. PLoS One. 2011;6:e26517. doi: 10.1371/journal.pone.0026517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Balaj L, Lessard R, Dai L, Cho YJ, Pomeroy SL, Breakefield XO, Skog J. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat Commun. 2011;2:180. doi: 10.1038/ncomms1180. [DOI] [PMC free article] [PubMed] [Google Scholar]