Abstract
The Salmonella enterica species includes about 2600 diverse serotypes, most of which cause a wide range of food- and water-borne diseases ranging from self-limiting gastroenteritis to typhoid fever in both humans and animals. Moreover, some serotypes are restricted to a few animal species, whereas other serotypes are able to infect plants as well as cold- and warm-blooded animals. An essential feature of the pathogenicity of Salmonella is its capacity to cross a number of barriers requiring invasion of a large variety of phagocytic and nonphagocytic cells. The aim of this review is to describe the different entry pathways used by Salmonella serotypes to enter different nonphagocytic cell types. Until recently, it was accepted that Salmonella invasion of eukaryotic cells required only the type III secretion system (T3SS) encoded by the Salmonella pathogenicity island-1. However, recent evidence shows that Salmonella can cause infection in a T3SS-1-independent manner. Currently, two outer membrane proteins Rck and PagN have been clearly identified as Salmonella invasins. As Rck mediates a Zipper-like entry mechanism, Salmonella is therefore the first bacterium shown to be able to induce both Zipper and Trigger mechanisms to invade host cells. In addition to these known entry pathways, recent data have shown that unknown entry routes could be used according to the serotype, the host and the cell type considered, inducing either Zipper-like or Trigger-like entry processes. The new paradigm presented here should change our classic view of Salmonella pathogenicity. It could also modify our understanding of the mechanisms leading to the different Salmonella-induced diseases and to Salmonella-host specificity.
Keywords: Adhesion, invasin, invasion, Salmonella, Trigger, type III secretion system, Zipper
Salmonella and Salmonelloses
The bacteria and diseases
Salmonella is a member of the Enterobacteriaceae family, a large group of Gram-negative, facultative anaerobic and nonspore-forming bacilli. The genus Salmonella consists of only two species, Salmonella bongori and Salmonella enterica, and the latter is divided into six subspecies: enterica, salamae, arizonae, diarizonae, houtenae, and indica (Guibourdenche et al. 2010). The agglutinating properties of the somatic O, flagellar H, and capsular Vi antigens are used to differentiate more than 2600 serologically distinct Salmonella (Guibourdenche et al. 2010). Strains belonging to S. enterica subsp. enterica cause approximately 99% of Salmonella infections in humans and warm-blooded animals (McClelland et al. 2001). Moreover, this subspecies is able to infect plants and numerous international outbreaks of S. enterica have been linked to plant contamination (Pezzoli et al. 2007; Nygard et al. 2008). Serotypes in other subspecies are usually isolated from cold-blooded animals and the environment, but rarely from humans (Uzzau et al. 2000). Salmonella nomenclature is now based on the name of serotypes belonging to subspecies. For example, Salmonella enterica subsp. enterica serotype Typhimurium is shortened to Salmonella Typhimurium (Brenner et al. 2000).
From a clinical perspective, Salmonella serotypes may be broadly grouped on the basis of host range and disease outcomes (Uzzau et al. 2000). Host-specific serotypes are associated with severe systemic disease in adults of a single species, which may seldom involve diarrhea (e.g., Salmonella Typhi, Salmonella Gallinarum). For example, S. Typhi is a human-specific pathogen causing a septicaemic typhoid syndrome (enteric fever). S. Gallinarum has a host range restricted to birds and causes a severe systemic disease called fowl typhoid (Shivaprasad 2000). Host-restricted serotypes are primarily associated with systemic disease in few hosts (e.g., Salmonella Dublin in cattle, Salmonella Choleraesuis in pigs and humans), but may cause disease in a limited number of other species (Chiu et al. 2004). Broad host range serotypes, such as S. Typhimurium and S. Enteritidis cause the majority of human gastrointestinal salmonelloses (Velge et al. 2005). They are able to infect, among many other animal species, domestic livestock and fowl worldwide, resulting in a spectrum of outcomes ranging from severe systemic disease to asymptomatic carriage. For example, in contrast to S. Gallinarum, which induces a systemic infection only in fowl, S. Enteritidis and S. Typhimurium generate a subclinical intestinal infection in poultry after a short systemic infection, and infected hens can become chronic carriers and lay contaminated eggs (Shivaprasad 2000; Velge et al. 2005). In humans and cattle, infection with these broad host range serotypes manifests as enterocolitis, usually limited to the gastrointestinal tract and rarely spreads to systemic organs (Stevens et al. 2009). In susceptible mice, these serotypes cross the gut epithelium efficiently, colonize the spleen and the liver, and are consequently responsible for a typhoid-like disease (Santos et al. 2001). Moreover, even within the same serotype, different strains isolated from animals or humans exhibited different virulence levels (Heithoff et al. 2008), and host-restricted variants exist, such as pigeon-associated S. Typhimurium definitive type (DT)-2 and DT99 strains (Rabsch et al. 2002). Finally, for the same host, the different Salmonella serotypes can induce different pathologies. For example, oral inoculation of weaned calves with S. Dublin produces severe systemic infection, whereas S. Gallinarum is avirulent and S. Typhimurium elicits acute enteritis (Paulin et al. 2002). Pathogenesis is not only influenced by the dose and route of inoculation but also by the genetic background and immune status of the host (Calenge et al. 2009, 2010). In summary, no other bacterial pathogen belonging to a single species shows such a remarkable ability to infect different hosts and induce so many different diseases.
Host invasion pathways
Despite the availability of complete genome sequences of isolates representing several serotypes, the molecular mechanisms underlying Salmonella colonization, pathogenesis, and transmission have been described mainly in rodents, and thus little is known about these mechanisms in farm animals and humans.
Host infections are usually initiated by ingestion of contaminated food or water followed by the passage of the bacteria from the stomach to the intestine. There, the bacteria adhere to and enter the cells lining the intestinal epithelium. Passage of the bacteria through the intestinal wall is believed to be initiated by transcytosis, that is, invasion of either enterocytes or M cells at the apical side, migration to the basolateral side, and exocytosis into the interstitial space of the lamina propria (Takeuchi 1967; Clark et al. 1994; Muller et al. 2012). Direct capture by CD18+ phagocytes and CD11b+, CD11c+, CX3CR1high phagocytes has also been observed (Vazquez-Torres et al. 1999; Muller et al. 2012). Within the lamina propria, S. Typhimurium is taken up randomly by the different phagocytes (macrophages, dendritic cells, and polymorphonuclear cells) and disseminates rapidly through efferent lymph in mesenteric lymph nodes and through the blood stream in spleen and liver (Salcedo et al. 2001). However, different behaviors have been observed depending on the serotype and the host. In cattle, S. Dublin transits rapidly through the epithelial layer and associates with MHC class II-positive cells in the lamina propria. These bacteria are predominantly extracellular within efferent lymph, but it remains unclear how they arrive in draining nodes or escape into an extracellular niche in this organ (Pullinger et al. 2007). S. Typhi, like S. Typhimurium, is capable of entering the murine intestinal epithelium via M cells. However, unlike S. Typhimurium, it does not destroy the epithelium and is cleared from the Peyer's patches soon after M-cell entry (Pascopella et al. 1995).
Cell invasion pathways
The reasons why some Salmonella serotypes are confined to the intestine while others translocate to distal organs remain unclear. An essential feature of the pathogenicity of Salmonella is its interaction with phagocytic and nonphagocytic cells, and Salmonella entry into host cells is known to be critical for bacterial survival and establishment of disease in a host. In general, intracellular bacterial pathogens enter nonphagocytic eukaryotic cells via two mechanisms, which are initially differentiated according to morphological criteria based on membrane remodeling. The “Trigger” mechanism involves dramatic cytoskeletal rearrangements known as “membrane ruffles” (Fig. 1A and B). In contrast, in the “Zipper” mechanism, or “receptor-mediated entry,” the invading bacteria are tightly bound to the host cell membrane, and only minor cytoskeletal protein rearrangements are initiated by specific contact between bacterial ligands (invasin) and host cell surface receptors (Fig. 1C and D). An important mechanistic difference between the Trigger and Zipper modes of entry is that the former is triggered from “inside” via the action of bacterial effectors delivered by secretion systems, whereas the latter is promoted from “outside” through activation of host cell receptors. However, in both cases, bacteria hijack the cell's physiological processes through the modulation of existing cell signaling cascades. It has recently been reported that Salmonella is the first bacteria shown to be able to enter cells using both these mechanisms (Rosselin et al. 2010). An emerging idea is that Salmonella strains can enter nonphagocytic cells by multiple pathways involving a Trigger or a Zipper mechanism. This is in contrast to the prevailing paradigm of Salmonella pathogenesis asserting that the Salmonella pathogenicity island-1 type III secretion system (SPI-1 T3SS or T3SS-1) is essential for bacterial invasion of host cells. The T3SS-1 and other cell invasion mechanisms are described below.
Figure 1.
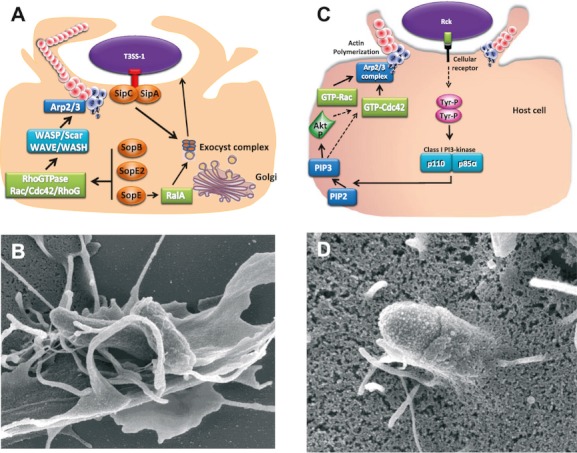
Trigger and Zipper mechanisms used by Salmonella to enter cells. (A) Schematic representation of the Trigger mechanism: Using a type III secretion system (T3SS), Salmonella bacterial effectors (SipA, SipC, SopB, SopE, SopE2) are directly injected into eukaryotic cells. SopE, SopE2, and SopB activate the RhoGTPases Rac/Cdc42/RhoG to allow actin cytoskeleton remodeling via cellular proteins, such as WASP/Scar/WAVE/WASH, which activate the Arp2/3 complex. In contrast, SipA and SipC bind directly to actin. To induce the formation of membrane ruffles and internalization, the recruitment of the exocyst complex is required and is manipulated by SipC and by SopE via the Ras-related protein RalA. (B) Scanning electron microscopy of Salmonella entering into cells via a Trigger mechanism, which is characterized by the apparition of large membrane ruffles at the bacterial entry site. (C) Schematic representation of the Zipper mechanism: the Rck invasin expressed on Salmonella outer membrane interacts with its receptor on the host cell membrane, leading to phosphorylation of at least one tyrosine kinase. Activation of the class I PI 3-kinase induces PI (3,4,5)P3 formation, participating to Akt activation. The activation of the GTPase Rac1, downstream of the Akt/PI 3-kinase activation, and the GTPase Cdc42 trigger actin polymerization via the Arp2/3 nucleator complex. The mechanism controlling Cdc42 during Rck-induced signaling pathway is still unknown. Dotted arrows represent possible signaling events and/or interactions. (D) Scanning electron microscopy of Salmonella entering into cells via a Zipper mechanism, which is characterized by weak membrane rearrangements.
Role of T3SS-1 in Cell Invasion
The SPI-1 encodes the T3SS-1
Pathogenicity islands (PAIs) are genetic elements integrated into the core genome of bacteria, which confer a virulence phenotype on the bacteria that have acquired them (Groisman and Ochman 1996; Shames et al. 2009). Currently, 21 different Salmonella pathogenicity islands (SPIs) have been identified in Salmonella (Blondel et al. 2009). However, many of the identified PAI-encoded genes have only predicted putative functions with no clear role in Salmonella pathogenesis (Blondel et al. 2010). The SPI-1 locus is a 40-kb chromosomal island, which carries among others all the genes required for the biosynthesis of a functional T3SS apparatus, a number of effector proteins and their chaperones, and some regulatory proteins (Galan and Curtiss 1989; Galan and Collmer 1999). A T3SS is a multi-subunit protein complex capable of injecting effectors directly from bacterial cytoplasm into the host cell cytosol. These effectors modulate cellular processes to the benefit of the pathogen. The T3SS consists of a needle complex and an export apparatus allowing the secreted proteins to pass through the bacterial inner and outer membranes, and of a translocon which creates a pore in the host cell membrane. The T3SS-1, encoded by SPI-1, is among the best characterized of all Salmonella virulence factors and triggers entry of Salmonella in a wide range of eukaryotic cells.
Of all SPIs reported in Salmonella only SPI-1, SPI-4, and SPI-9 are present in both Salmonella bongori and S. enterica species suggesting that they were acquired by Salmonella at the beginning of the evolution of Salmonella and prior to speciation. Moreover, 10 of the 15 known T3SS-1 translocated effectors are almost entirely conserved in S. enterica and S. bongori. Genes encoding effectors are even found at the same genomic loci in S. bongori as they are in S. enterica, that is, carried on SPI-1 itself or on SPI-5 or at identical sites in the chromosomal backbone (Fookes et al. 2011). The use of microarrays to identify the virulence gene profiles in 24 Salmonella strains of different serotypes from food and/or food animal environment showed that nearly 58% of the virulence-associated genes tested were present in all Salmonella strains tested. In general, genes belonging to inv, prg, sic, sip, or spa families were detected in more than 90% of the isolates, whereas the iacP, avrA, invH, sopB, or sopE genes were detected in 40–80% of the isolates (Zou et al. 2011). Moreover, an epidemiological analysis suggested that sopE, which is encoded by a temperate bacteriophage, appears to be associated with epidemic strains (Hopkins and Threlfall 2004), whereas sopE2 is present in all Salmonella strains tested (Bakshi et al. 2000).
Role of T3SS-1 in cell invasion
Inside the host, after entering the lumen of the small intestine, Salmonella sense the environment (pH, oxygen tension, osmolarity, etc.), enabling T3SS-1 genes to be expressed and subsequently the secretion apparatus to be assembled at the bacterial membrane (Galan and Collmer 1999; Rosselin et al. 2012). Host cell invasion is initiated by pathogen binding to the host cell surface, which activates the insertion of the translocon into the host cell membrane through its affinity for cholesterol (Hayward et al. 2005). This bacteria-cell contact allows translocation of effectors into the host cell. This translocation is precisely coordinated ensuring that bacterial proteins engage in a coherent order through a cytoplasmic sorting platform, which ensures secretion of the translocases (SipB, SipC, and SipD) before the effectors. The sequential loading on this platform may be facilitated by the different affinities of the T3SS-chaperones, ensuring the hierarchy in type III effector secretion (Lara-Tejero et al. 2011). Cell entry is characterized by profuse rearrangements of the actin cytoskeleton at the site of bacteria–host-cell contact, which envelop external bacteria and internalize them into membrane-bound vacuoles. Currently, it seems difficult to know whether the entry process mediated by the T3SS-1 is the same for all cell types (phagocytic and nonphagocytic cells), mainly because no experiment has been designed to compare the entry processes between cell lines. Only one experiment was performed to compare the transcriptome of a S. Typhimurium strain within a phagocytic and a nonphagocytic cell line (Hautefort et al. 2008). This study especially showed that SPI-1- and flagella-related genes were expressed inside epithelial cells at later stages of the infection than in macrophage-like cells. At least 15 effectors can be translocated into the host cell by T3SS-1 to induce bacterial entry (reviewed in McGhie et al. 2009). Among them, five major Salmonella effectors, each being able to manipulate the cytoskeletal machinery within the host, are known to drive engulfment (SopE, SopE2, SopB, SipA, and SipC) (Fig. 1A).
SopE, SopE2, and SopB target the RhoGTPase switch and promote activation of the Rho family members Cdc42 and Rac, leading to activation of N-WASP- and Scar/WAVE-Arp2/Arp3 (Arp2/3) complexes which trigger actin remodeling. Despite their differing biochemical activities, these effectors have key redundant roles during internalization. Indeed, Salmonella strains lacking just one of these effectors display only a modest reduction in entry, whereas a triple ΔsopE/E2/B mutant is completely abrogated for cytoskeletal remodeling and entry (Zhou et al. 2001). SopE and SopE2 function as guanine exchange factors (GEFs) and directly catalyze GTPase activation. SopB is an inositol phosphatase that acts on host cell membrane phospholipids and thus functions indirectly by activating endogenous eukaryotic SH-3-containing GEF (SGEF), an exchange factor for the Rho-family GTPase: RhoG (Patel and Galan 2006). Recent data also showed that SopB constitutes an important regulator of an additional entry pathway which activates RhoA, the Rho kinase, and myosin II leading to stress fiber formation and contractility (Hanisch et al. 2011).
SipA and SipC can engage actin directly controlling and localizing actin polymerization at bacterial attachment site. SipC possesses distinct C- and N-terminal domains which are able to nucleate filamentous actin (F-actin) and promote F-actin bundling (Myeni and Zhou 2010). Chang et al. (2005) demonstrated that the effector translocation function of SipC is dissociable from the actin-nucleating function. In vitro, SipA stimulates these SipC activities and can stabilize F-actin by directly antagonizing the action within the cell of depolymerizing factors (Zhou et al. 1999), such as ADF/cofilin and gelsolin (McGhie et al. 2004). However, recent single molecule imaging studies have questioned the latter finding (Popp et al. 2008). SipA also plays an important role in induction of invasion-competent membrane ruffles in synergy with the other major effectors (Perrett and Jepson 2009). Invasion efficiency is also increased by promoting localized membrane expansion directly through SipC-dependent recruitment of the exocyst and indirectly via SopE-dependent activation of RalA (Braun and Brumell 2010; Nichols and Casanova 2010). Nichols and Casanova (2010) have proposed a role for the exocyst in delivering vesicles to the site of bacterial entry to provide additional membranes to allow the extension and ruffling of the plasma membrane necessary to promote invasion.
The actin remodeling events initiated by these five major effectors which undergo profuse membrane ruffling after 10–30 min of contact are transient and typically reversed 2–3 h postentry to display a normal actin cytoskeleton, despite the presence of a large number of intracellular bacteria (Kubori and Galan 2003). Remarkably, Salmonella actively helps the host cell to regain its normal cellular architecture through the action of SptP, a SPI-1 effector with GAP activity that returns Cdc42 and Rac1 to the nonactivated state. The interplay between the bacterial effectors acting as GEFs and GAPs (SopE/SptP) is based on different half-lives of these proteins following translocation (Kubori and Galan 2003). SptP shows a higher resistance to the ubiquitin-proteasome system, and downregulates Cdc42 and Rac1 once SopE is degraded.
These effectors involved in T3SS-1-dependent entry also have profound effects on later processes, such as membrane trafficking, cell division, apoptosis, bacterial killing, cytokine and chemokine production, and antigen presentation (reviewed in Santos et al. 2009). It is noteworthy that several T3SS-1 effectors also mediate the disruption of tight junctions and thus impair intestinal barrier integrity (Boyle et al. 2006). After bacterial entry, SopB also plays a role in Salmonella-containing vacuole (SCV) biogenesis, sealing, and trafficking. Recently, it has been reported that ubiquitination downregulates SopB activity at the plasma membrane, but prolongs retention of SopB on the induced vesicles enriched in phospho-inositide (3)P (Knodler et al. 2009; Patel et al. 2009). The SCV initially acquires the early endosome markers, which are sequentially replaced by the late endosome and lysosome markers. However, the SCV does not fuse directly with the lysosomes, thereby avoiding Salmonella destruction, partly due to the phospho-inositide phosphatase activity of SopB (Rudge et al. 2004). By reducing the local concentration of PI(4,5)P2, SopB cooperates with SopD to destabilize cytoskeleton-plasma membrane interactions and to reduce membrane rigidity, promoting the fission and the sealing of the future SCV (Bakowski et al. 2007).
As infection progresses, Salmonella uses a second type III secretion system, the T3SS-2, encoded by the SPI-2 which delivers additional effector proteins through the SCV membrane allowing bacterial survival and replication (reviewed in Malik-Kale et al. 2011). Nevertheless, it has been demonstrated that SPI-1 T3SS effectors may be involved in vacuole biogenesis and intracellular survival, functions which were previously attributed solely to the actions of SPI-2 T3SS effectors (Steele-Mortimer et al. 2002).
Role of T3SS-1 in Salmonella Pathogenesis
Involvement of the T3SS-1 in host infection
Based on studies on the cellular and molecular mechanisms of S. Typhimurium infection, SPI-1 is considered to be essential for the invasion of animal cells by Salmonella, and SPI-2 is required for intracellular proliferation and survival. In vivo experiments have been performed in different animal species to investigate the role of SPI-1 T3SS during the course of Salmonella infection.
In a mouse model of systemic lethal infection, it was observed that S. Typhimurium mutants unable to assemble functional T3SS-1 were recovered from intestinal contents and systemic sites at a lower level than the wild-type strain after oral but not after intraperitoneal inoculation (Galan and Curtiss 1989). The T3SS-1 is also involved in a mouse model of Salmonella-induced colitis based on the pretreatment of animals with streptomycin. It was demonstrated that the SPI-1 effectors SipA, SopE, and SopE2, in addition to flagella and chemotaxis, were required to induce intestinal inflammation and significant histopathological changes (Hapfelmeier et al. 2004; Stecher et al. 2004).
Within minutes of injecting Salmonella into ligated ileal loops in calves, Salmonella can be seen to invade both M cells and enterocytes (Frost et al. 1997). This is in marked contrast to what is seen in mice where the major portal of entry appears to be through M cells (Jones et al. 1994). Different in vivo studies have demonstrated that mutations in T3SS-1 decrease enterocyte invasion and abolish induction of fluid secretion and the recruitment of polymorphonuclear cells in bovine ligated ileal loops (Galyov et al. 1997).
Similarly, S. Typhimurium SPI-1 mutants are impaired in their ability to colonize the porcine gut in a ligated intestinal loop model (Boyen et al. 2006). The involvement of SPI-1 and not of other SPIs in proinflammatory signaling and heterophil infiltration in the intestine has also been demonstrated in chicks infected with S. Enteritidis (Rychlik et al. 2009). In this model, the role of SPI-1 and SPI-2 in the liver and spleen colonization has also been shown. Mutants lacking all the five major pathogenicity islands (SPI 1-5), but expressing only SPI-1 or SPI-2 have a medium virulence compared with the wild-type S. Enteritidis strain, whereas mutants bearing both SPI-1 and SPI-2 are almost as virulent as the wild-type strain (Rychlik et al. 2009). The role of SPI-1 T3SS has also been shown with sipD and iacP (invasion associated ACP) mutants which were unable to colonize spleens, whereas the wild-type S. Enteritidis strain could, three days postinfection in chickens infected subcutaneously (Parker and Guard-Petter 2001). However, numerous other data have shown that the role of the T3SS-1 is controversial in chicken and especially for the intestinal translocation (see the section “In vivo evidences for a non-essential role of T3SS-1”).
Until recently, little was known about the infection mechanisms of Salmonella in the plant kingdom. Contaminated plants are nonetheless responsible for 25% of food poisoning outbreaks in the United States (Rangel et al. 2005). Generally, it was believed that Salmonella could survive on leaves. However, a growing body of evidence points to an active process in which Salmonella infects plant organs and uses them as a viable host or vector between animals (Golberg et al. 2011). This invasion process uses both SPI-1 and SPI-2-encoded apparatus because the proliferation rate in plant of invA and prgH (encoded by SPI-1) mutants and ssaJ and ssaV (encoded by SPI-2) mutants are lower than the wild-type S. Typhimurium strain (Schikora et al. 2011). Moreover, Salmonella actively suppresses plant defense mechanisms using the SPI-1 T3SS. Mutants defective in virulence factors induced, indeed, disease symptoms (tissue damage, oxidative burst, and pH changes) contrary to the wild-type S. Typhimurium strain (Schikora et al. 2011; Shirron and Yaron 2011). Involvement of the T3SS appears to depend on the Salmonella serotype, plant species, and even the cultivar. Barak et al. (2011) showed, for example, diverse resistant/susceptible phenotypes to Salmonella in different tomato cultivars.
In vivo evidences for a non-essential role of T3SS-1
Although numerous studies demonstrate a role of T3SS-1 in host infection, recent evidence from bovine, chicken, and murine models suggests that S. Typhimurium and other serotypes can also cause infection in a SPI-1-independent manner (Coombes et al. 2005; Hapfelmeier et al. 2005; Desin et al. 2009). For example, Morgan et al. (2004) concluded that S. Typhimurium uses different strategies to colonize calves and chicks. They observed that T3SS-1 and T3SS-2 are required for efficient colonization of cattle, whereas disruption of these secretion systems only caused a minor defect in S. Typhimurium colonization of chicks. The role of SPI-1 T3SS in chicken infection remains unclear. Desin et al. (2009) showed, for example, that a S. Enteritidis ΔSPI-1 mutant was not impaired in the caecal colonization of 1-week-old chicks, whereas the deletion of this region caused a delay in systemic infection. Similar results were observed by Rychlik et al. (2009), who showed that SPI-1 from S. Enteritidis was poorly involved in intestinal chick colonization, but was involved in internal organ colonization. Another argument for a nonessential role of the T3SS-1 in vivo was reported by Murray and Lee (2000) in a murine model of infection. They found that a S. Typhimurium strain, in which SPI-1 was entirely deleted, was recovered from spleens of infected mice at a frequency similar to that of its parental wild-type strain, indicating that S. Typhimurium did not require T3SS-1 genes to cross the intestinal epithelium and infect systemic tissues. Moreover, it has been reported that nonfunctional SPI-1 mutants, which render them severely deficient for invasion of polarized epithelial cells, retain their ability to invade M cells in a murine gut loop model, implying that intestinal invasion may involve other factors (Clark et al. 1996).
The large majority of the in vivo studies on T3SS-1 have been performed with broad host range serotypes (S. Typhimurium and S. Enteritidis), and only a few analyses have involved host-specific serotypes. The translocation mechanism of typhoidal serotypes from the gut to other organs in farm animals, thus remains elusive. For S. Gallinarum, which induces a typhoid-like disease, the molecular and cellular mechanisms of fowl typhoid are relatively poorly understood, but the 85-kb S. Gallinarum plasmid has been shown to be essential for virulence, whereas a functional SPI-1 T3SS is not required. In addition, unidentified gene(s) under the control of ppGpp has (have) been shown to be involved in internalization (Jones et al. 2001; Jeong et al. 2008).
Overall, numerous articles have reported that Salmonella strains impaired in their capacities to use their T3SS-1 can still colonize the intestinal tissue and induce different pathologies in different animals, whereas others demonstrate an in vivo role of this T3SS. Discrepancies that exist between studies could, in part, be due to the type of infection induced (systemic/enteric), the time analyzed postinoculation, the serotype used, the host resistance, and the immune status of the host. The differences could also be due to the mutant used. For example, Murray and Lee (2000) using a ΔSPI-1 mutant demonstrated that SPI-1 is not essential, but they also found that S. Typhimurium strains containing a mutation in hilA or invG, which are encoded by SPI-1, were recovered from the intestinal tissues and internal organs of mice at a lower frequency than their parental wild-type strain. One explanation for such results is that deletion of SPI-1 genes suppresses the hilA infection defect and allows Salmonella to colonize and infect their hosts in a SPI-1-independent manner (Murray and Lee 2000).
All these in vivo experimental studies support clinical observations. Indeed, some Salmonella Senftenberg and Salmonella Litchfield isolates carrying a deletion encompassing a vast segment of SPI-1 have been identified (Ginocchio et al. 1997), and some of them have been responsible for food-borne disease outbreaks, indicating that SPI-1 is not required for enteropathogenesis in humans (Hu et al. 2008; Li et al. 2011).
Existence of Other Entry Mechanisms
To infect different hosts, and cause various diseases ranging from typhoid fever to gastroenteritis or to an asymptomatic carrier state, Salmonella needs to cross several barriers. Crossing these barriers and multiplying within the host require invasion of a large variety of phagocytic and nonphagocytic cells. Research on the invasion mechanisms has until recently focused on SPI-1 and SPI-2 due to their key roles in entry and intracellular multiplication within different cell types, in particular enterocyte cell lines (Agbor and McCormick 2011). However, the role of outer membrane proteins (OMP) as possible adhesion molecules and virulence factors has been demonstrated in different pathogenic bacteria. In Salmonella, the association of OMP with host cells is known to trigger a variety of biological events that include induction of innate and adaptative immune response (Galdiero et al. 2003). More recently, some Salmonella OMP, namely Rck and PagN, have been shown to be able to induce cell invasion (Heffernan et al. 1994; Rosselin et al. 2010).
Role of the OMP Rck
Rck is a 19-kDa OMP encoded by the rck gene located on the large virulence plasmid which contributes to the expression of virulence genes like the spvRABCD (Salmonella plasmid virulence), pef (plasmid-encoded fimbriae), srgA (SdiA-regulated gene, putative disulphide bond oxidoreductase), or mig-5 (macrophage-inducible gene coding for putative carbonic anhydrase) genes (Rychlik et al. 2006). Among the different serotypes harboring a virulence plasmid (i.e., Typhimurium, Enteritidis, Gallinarum, Pullorum, Dublin, Abortus-ovis, and Choleraesuis), only a few carries the rck gene (Buisan et al. 1994). It is highly conserved in most isolates of S. Enteritidis and S. Typhimurium (Futagawa-Saito et al. 2010), found in some isolates of S. Dublin, but was not detected on the virulence plasmid of S. Choleraesuis, S. Gallinarum, and S. Pullorum (Chu et al. 1999; Rychlik et al. 2006). This gene is therefore present in the serovars which are frequently associated with infections of both humans and farm animals, and which have the wider range of hosts.
This protein belongs to a family of five homologous OMP characterized in Salmonella (Rck and PagC), Escherichia coli (Lom), Yersinia enterocolitica (Ail), and Enterobacter cloacae (OmpX) (Heffernan et al. 1992). A role in complement resistance, cell attachment and invasion has been attributed to individual members of this family. Molecular analysis has shown that Rck is homologous to PagC and Ail with 53% and 42% identity, respectively, but both complement resistance and invasion phenotypes have been attributed to only Rck and Ail. However, these proteins do not exhibit homologous regions that could support this role (Heffernan et al. 1992).
Rck alone is able to promote adhesion and internalization of coated beads (Rosselin et al. 2010) or of noninvasive E. coli strains (Heffernan et al. 1994). Fourty-six amino acids of Rck were identified as being necessary and sufficient in this process. Their binding to the cell surface is inhibited by soluble Rck and induces discrete membrane rearrangements due to reprogramming of cell signaling (Rosselin et al. 2010). These findings demonstrate that Rck induces a Zipper-like entry mechanism supporting the fact that Salmonella is the first bacterium to be described as able to induce both Zipper and Trigger mechanisms for host cell invasion. This Zipper entry process requires protein tyrosine kinase activation and class I PI3 kinase, which activate Akt and the small GTPase Rac1 and Cdc42 (but not Rho), leading to activation of the Arp2/3 complex and actin polymerization (Mijouin et al. 2012). The cellular receptor of Rck required in this process, however, remains unknown.
In spite of the absence of Rck expression in usual laboratory culture conditions, its role in Salmonella invasion has been demonstrated in vitro after growing Salmonella in swarming culture conditions (Rosselin et al. 2010). However, its role in Salmonella pathogenesis is still poorly understood. The fact that Rck expression is dependent on SdiA, a LuxR homolog which is a quorum sensing regulator, suggests an intestinal role of this invasin (Ahmer et al. 1998; Michael et al. 2001). However, Salmonella cannot synthesize the acyl homoserine lactones (AHL) that allow SdiA activation, but can detect AHL signaling molecules of other microbes using SdiA. Surprisingly, AHL have not been found in the intestinal tract of healthy mammals, with the exception of the bovine rumen (Erickson et al. 2002). SdiA from Salmonella could be activated in mice whose intestinal flora contained the AHL-producing Yersinia enterocolitica strain (Dyszel et al. 2010) or in turtle carrying Aeromonas hydrophila (Smith et al. 2008). After co-infection of mice with two S. Typhimurium strains engineered to produce AHL, a sdiA mutant and a sdiA+ strain, it was shown that the constant activation of SdiA conferred a selective advantage to Salmonella (Dyszel et al. 2010). However, under physiological conditions, SdiA activation did not confer a fitness advantage for intestinal colonization, suggesting that even if SdiA activation is achieved, it is not always sufficient to induce the expression of the Rck regulon. This hypothesis is supported by the fact that rck is also regulated by an unidentified SdiA-independent system (Smith and Ahmer 2003). Moreover, in relation to its role in resistance to the complement-dependent bactericidal action, it is conceivable that Rck also plays a role in systemic infection.
Role of the OMP PagN
PagN is a 26-kDa OMP, which displays similarities with the Hek and Tia adhesins/invasins of pathogenic E. coli. pagN is widely conserved in the Salmonella genus. All the subspecies of S. enterica and two strains of S. bongori represented in the Salmonella reference collection C (SARC) carry the pagN gene (Boyd et al. 1996). This gene was originally identified in a TnphoA random-mutagenesis screen of phoP-activated S. Typhimurium genes (Belden and Miller 1994) and through the use of in vivo expression technology performed in BALB/c mice (Heithoff et al. 1997). These studies showed that pagN is a PhoP-activated gene and thus is not expressed in S. Typhimurium strains grown under typical laboratory culture conditions, but is maximally expressed intracellularly as observed using gene fusion (Conner et al. 1998) or microarray analysis (Eriksson et al. 2003). The function of PagN as an invasin is supported by the fact that PagN overexpressed in a noninvasive E. coli strain induced cell invasion (Lambert and Smith 2008). However, PagN-defective bacteria displayed a consistent two- to fivefold reduction in cell invasion when compared to the wild-type strain only after overnight culture in pH 5.8 minimal media, suggesting that PagN is induced in the intracellular compartment. To correlate the intracellular expression pattern of PagN to its role as an invasin, Lambert and Smith (2008) postulated that Salmonella exiting from epithelial cells or macrophages might have an optimal level of PagN expression. Thus, PagN might facilitate interactions between Salmonella and mammalian cells in specific conditions that do not allow SPI-1 expression (Lambert and Smith 2008). This hypothesis is supported by the low T3SS-1 expression detected inside macrophages at 4 h postinfection (Eriksson et al. 2003) but not by the more recent data showing that SPI-1 genes remained expressed or were even upregulated few hours after invasion into different cell types (Drecktrah et al. 2005; Hautefort et al. 2008).
The PagN protein interacts with cell surface heparin sulfate proteoglycans to invade the mammalian cell line CHO-K1 (Lambert and Smith 2009). However, because proteoglycans cannot transduce a signaling cascade, they might act as co-receptors for invasion and not as the receptor per se. More studies are necessary to identify the PagN receptor at the molecular level.
Role of the hemolysin HlyE
HlyE is a pore-forming hemolysin that is encoded by SPI-18, a small 2.3 kb genomic island missing in S. Typhimurium, but present in S. Typhi and S. Paratyphi A. HlyE shares more than 90% identity with the 34 kDa E. coli HlyE (ClyA) hemolysin. S. Typhi hlyE mutants are impaired in their ability to invade HEp-2 cells, compared with their wild-type parental strain. Moreover, the heterologous expression of HlyE in S. Typhimurium improves the colonization of deep organs in mice, demonstrating that HlyE is a new virulence determinant (Fuentes et al. 2008). This result is consistent with those from other laboratories showing that pore-forming hemolysins play critical roles in invasion of eukaryotic cells (Strauss et al. 1997; Doran et al. 2002). However, the precise mechanism by which hemolysins enhance invasion of intracellular pathogens remains unknown. It is possible that these hemolysins are invasins, but they could also modulate the entry of bacteria by inducing changes in calcium flux as described for Listeria monocytogenes (Dramsi and Cossart 2003).
Existence of unknown factors involved in Salmonella invasion
The different data mentioned above show that Salmonella has developed different strategies to invade cells. Currently, three invasion pathways have been described for the broad host range serotypes (T3SS-1, Rck, PagN). However, recent data have shown that other unknown entry routes may be used depending on the serotype, the host and the cell-type considered.
Rosselin et al. (2011) have demonstrated that a S. Enteritidis strain which does not express Rck, PagN, and the T3SS-1 is still able to invade fibroblasts, epithelial, and endothelial cells significantly. The relative degree of the unknown entry processes depends on the cell type and the cell line. Among the cell types tested, 3T3 fibroblasts and MA104 kidney epithelial cells are the most permissive to these mechanisms, allowing one bacterium out of three (33% of the invasion) to enter independently of T3SS-1, PagN, and Rck. In contrast, nonpolarized HT29 human enterocytes are not prone to the unknown entry mechanisms, as less than 4% of the internalized bacteria invade cells in the absence of these known invasion processes. This heterogeneity in invasion profiles demonstrates a cell specificity of the T3SS-1, Rck, PagN-independent mechanisms. Other results reinforce the idea that nonidentified invasion factors are involved during the entry of S. Typhimurium. Indeed, Aiastui et al. (2010) and Sorge et al. (2011) have shown that nonidentified invasion factors are involved during entry of S. Typhimurium strains lacking the T3SS-1 into rat and mouse fibroblasts or into human brain microvascular endothelial cells. The entry mechanism used by Salmonella into immortalized human foreskin fibroblasts seems different from those described for Rck and PagN, as none of the GTPases tested (Rac1, Cdc42, RhoA, or RhoG) was essential for invasion of these cells (Aiastui et al. 2010). Moreover, the Salmonella SPI-1 T3SS is not required to invade intestinal cells grown in three dimensions as an invA S. Typhimurium mutant unable to express the T3SS-1, invades 3-D HT-29 cells at similar levels to the wild-type strain (Radtke et al. 2010). To our knowledge Rck and PagN are not expressed in bacterial culture conditions used in these studies; therefore, these results also highlight the fact that Salmonella possesses noncharacterized invasion factors.
Certain indications about the nature of the unknown invasion mechanisms have been obtained. Indeed, a Salmonella mutant expressing none of the known invasion factors displayed both local and massive actin accumulations, as well as discrete and intense membrane rearrangements. These observations obtained using different cell types show that invasion factors other than PagN, Rck, and the T3SS-1 apparatus are able to induce either a Zipper or a Trigger mechanism (Rosselin et al. 2011). Moreover, Zipper-like entry processes have been observed with fibroblast, epithelial, and endothelial cells (Aiastui et al. 2010; Rosselin et al. 2011; Sorge et al. 2011).
Overall and contrary to the prevailing theory, Salmonella can enter cells through a Zipper-like mechanism mediated by Rck and other unknown invasins, in addition to the Trigger mechanism mediated by its T3SS-1 apparatus and also other unknown determinants. These observations thus open new avenues for the identification of new invasion factors.
Possible invasion factors
The cell specificity and consequently the organs or hosts targeted by Salmonella serotypes could be determined by the cells targeted by the different entry processes, in particular, by the Zipper mechanisms which involve a cell receptor. However, in addition to these different entry processes, the role of adhesins in cell specificity and invasion should also be taken into account. Fimbriae and/or nonfimbrial adhesins may indeed mediate attachment to cell surfaces, and thus could mediate part of the cell specificity. Moreover, some “typical” adhesins in some species are known to be involved, in other species, in entry in professional phagocytes or in nonprofessional phagocytes. This has been clearly described for E. coli where FimH mediates not only bacterial adherence but also invasion of numerous epithelial cells (Martinez et al. 2000).
Depending on the serotype, Salmonella can express a wide range of adhesion factors. Salmonella gene clusters encode more than 13 different fimbrial adhesins, such as Fim (type I fimbriae), Lpf (long polar fimbriae), Tafi (thin aggregative fimbriae, formerly SEF17), or the type IV pili of serotype Typhi (reviewed in Wagner and Hensel 2011). In addition, auto-transporter adhesins, such as ShdA, MisL, SadA, the type I secreted large repetitive adhesins SiiE, and BapA have been identified. Although the functions of various adhesins are not well understood, different studies have shown how they act in concert with other virulence determinants. For example, type 1 fimbrial adhesin FimH mediates binding to epithelial cells and it also helps to induce actin-dependent uptake in the absence of T3SS-1 in murine dendritic (Guo et al. 2007) or HeLa cells (Horiuchi et al. 1992; Lara-Tejero and Galan 2009). Similarly, Hensel's group has shown that SiiE mediates intimate contact of Salmonella with polarized cell surface allowing entry through the T3SS-1 (Gerlach et al. 2008). Mutant strains lacking SiiE fail to invade polarized cells. This giant nonfimbrial adhesin SiiE is secreted by a T1SS both encoded by the SPI-4. SPI-4 seems to be involved in calves but not in chickens or pigs challenged with S. Typhimurium (Morgan et al. 2004, 2007). The role of SPI-4 in virulence has also been demonstrated in mice following the oral challenge of BALB/c mice but not following the intraperitoneal infection of Nrampr mice (Morgan et al. 2004; Lawley et al. 2006).
These adhesion factors could also be involved in cell specificity. To invade epithelial cells, Salmonella first binds to the cell surface. For instance, it is well known that some fimbrial adhesins play a role in targeting in vivo S. Typhimurium to a particular cell lineage in the host. For example, in an intestinal-organ culture model, the pef fimbrial operon mediates attachment to the murine villous small intestine, whereas selective adhesion of S. Typhimurium to murine ileal Peyer's patches is mediated by the lpf fimbrial operon. Similarly, MisL and ShdA are outer membrane fibronectin-binding proteins that are expressed in the intestine and could be involved in carrier state in mice (Kingsley et al. 2002; Dorsey et al. 2005). This synergy between adhesion and entry has been described for fimbriae and T3SS-1-independent internalization (Guo et al. 2007) and also for adhesins and T3SS-1 (Gerlach et al. 2008; Lara-Tejero and Galan 2009; Misselwitz et al. 2011).
The ability of some bacteria to invade human intestinal epithelial cells is also linked to outer membrane vesicles (OMV). It has been suggested that OMV carry effector proteins into the host cell resulting in the uptake of bacteria. In support of this hypothesis, purified Treponema denticola vesicles have been shown to harbor the chymotrypsin-like protease, dentilisin (PrtP), a protease that allows T. denticola to penetrate the epithelial barrier and Hep2 cells (Chi et al. 2003). In addition, a nonadherent-invasive E. coli mutant was discovered to have a defect in OMV formation (Rolhion et al. 2005). Recently, Kitagawa et al. (2010) have demonstrated that the OMP PagC is a major constituent of Salmonella OMV. However, until now, there has been no evidence of the role of these OMV in cell invasion by Salmonella because they could also function as a delivery system for virulence-related proteins from the SCV to the cytoplasm of macrophage cells (Kitagawa et al. 2010).
The T6SS represents a new paradigm of protein secretion that is crucial for the pathogenesis of many gram-negative bacteria. T6SS has been linked to a wide variety of functions ranging from inter-bacterial relationships, biofilm formation, cytotoxicity, survival in phagocytic cells, and also host cell invasion (Schwarz et al. 2010). Bioinformatic studies in different S. enterica serotypes have revealed four gene clusters encoding T6SS, acquired by independent lateral transfer events. These T6SS loci are located in different genomic islands: SPI-6, SPI-19, SPI-20, and SPI-21. The T6SS encoded in the SPI-19 is present in at least four serotypes: Dublin, Agona, Gallinarum, and Enteritidis. Interestingly, whereas S. Gallinarum appears to encode a complete T6SS, S. Enteritidis has a degenerate genetic element lacking most of the T6SS-related components (Blondel et al. 2009). SPI-19 contributes to the efficient colonization of the intestinal tract and systemic sites in chicks infected by S. Gallinarum strain 287/91 (Blondel et al. 2010). However, the transfer of a complete T6SS locus from S. Gallinarum to S. Enteritidis impaired the ability of this bacterium to efficiently colonize chicken tissues. The T6SS associated with SPI-6, formerly known as the S. enterica centisome seven islands (Sci), is present in at least 16 serotypes including Typhimurium and Typhi. Complete deletion of the Sci genomic island resulted in a decreased ability of S. Typhimurium to enter host cells (Folkesson et al. 2002). Similarly, increased levels of a dominant negative variant of S. Typhimurium ClpV strongly reduced the ability of this bacterium to invade epithelial cells (Schlieker et al. 2005). These results differ from the data obtained with a sciS (icmF-like) transposon mutant (Parsons and Heffron 2005). In that case, SciS limited intracellular growth in macrophages at late stages of infection, and attenuated the lethality of S. Typhimurium in a murine host. However, it is difficult to compare these studies due to the cell lines used (epithelial vs. macrophage). The function and characteristics of the T6SS are far from being understood, but this secretion system appears as a novel key player in bacterial pathogenesis and bacteria–host interaction.
In contrast, some bacterial invasion factors remain unidentified, whereas their cell receptors have been identified. Pier et al. found that human epithelial cells expressing wild-type CFTR (cystic fibrosis transmembrane conductance regulator) are significantly more infected by S. Typhi than cells expressing ΔF508 Cftr. Moreover, a 15-amino-acid peptide derived from the first CFTR extracellular domain inhibited S. Typhi invasion of T84 cells, whereas a scrambled synthetic peptide of these residues did not. These results indicate that CFTR is a major epithelial-cell receptor for S. Typhi internalization (Pier et al. 1998). In line with in vitro results, translocation of S. Typhi into the intestinal submucosa of ΔF508 Cftr heterozygotes was 86% less effective than in wild-type mice, and in homozygotes it was almost completely abrogated. In contrast, there was no significant difference between wild-type, heterozygous, or homozygous ΔF508 Cftr mice in intestinal barrier translocation of S. Typhimurium, demonstrating the specificity of the CFTR receptor for S. Typhi internalization (Pier et al. 1998).
Conclusion
The precise traits associated with the zoonotic and epidemic potential of Salmonella strains remain unknown. This is particularly true with the recent identification of human clinical cases associated with SPI-1 deficient Salmonella strains indicating that SPI-1 T3SS is not required to cause entero-pathogenesis (Hu et al. 2008; Li et al. 2011). Identification of such traits is vital to assess the risk posed to humans by S. enterica strains found in animals or plants and for effective targeting of intervention strategies. Moreover, the reasons why some Salmonella serotypes are confined to the intestines while others translocate to internal organs remain unclear. A common feature of pathogens associated with enteric fevers is their use of different strategies to evade detection or subvert host innate immunity. Until recently, it was accepted that Salmonella entered cells only via its T3SS-1. However, new evidence has shown that Salmonella is able to use other pathways to enter phagocytic and nonphagocytic cells. Moreover, these T3SS-1-independent processes could mediate several Trigger and Zipper entry processes. We can thus speculate that these different entry mechanisms are efficient for particular cell types or species and could thus be involved in particular diseases induced by the different Salmonella serotypes. The coexistence of several invasion processes also raises the interesting possibility that synergy might exist between these different bacterial entry proteins or between the two invasion pathways (Zipper vs. Trigger) used by this pathogen. We can hypothesize that the receptor-ligand interaction mediated by the Zipper mechanism improves bacteria-cell contact and starts a process which facilitates the injection and the effects of bacterial effectors injected via the Trigger mechanism.
Several studies performed in different models suggest that the T3SS-1-independent entry processes might enhance bacterial entry into some cell lines or cell types of one or several species. They could also mediate invasion into polarized epithelial cells. Thus, particular invasins or entry processes may contribute to the invasion of specific host cells, and be involved in cell and tissue tropism as well as host specificity in both animals and plants. However, the number of identified adhesins or invasins which are only expressed in vivo is continually increasing, thus making it difficult to unravel the function of these new virulence factors. The use of more sophisticated in vivo studies, such as noninvasive in vivo imaging methods should help to understand the role of the entry factors in the interplay between bacteria and their hosts.
The new paradigm presented here stating that Salmonella strains are able to enter nonphagocytic cells by various routes should modify our view of the mechanisms that lead to the different Salmonella-induced diseases and should encourage us to revisit the host specificity bases. Future studies will undoubtedly focus on whether cell entry mechanisms are different according to the host and the serotype, and whether there is a link between host specificity, cell tropism, cell entry mechanism, cell response, and disease outcomes.
Acknowledgments
We apologize to all whose work could not be cited because of space limitations. Work on the new invasion routes has been funded by the Région Centre. N. Abed holds a Post-Doctoral fellowship funded and performed within the “SAVIRE” project, funded by the Délégation Régionale à la Recherche et à la Technologie du Centre (FEDER) number 1634-32245 and by the Région Centre number 2008-00036085. F. Namdari holds a Doctoral fellowship funded by the Région Centre. A. Rossignol holds a Doctoral fellowship funded by the Région Centre and the Institut National de la Recherche Agronomique. Z. Boumart holds a Doctoral fellowship funded by the Institut National de la Recherche Agronomique and the Agence Nationale de Sécurité Sanitaire de l'alimentation, de l'environnement et du travail.
Conflict of Interest
All authors certify that they have no commercial associations (e.g., consultancies, stock ownership, equity interests, patent license arrangement, etc.) that might pose a conflict of interest in connection with the submitted article.
References
- Agbor TA, McCormick BA. Salmonella effectors: important players modulating host cell function during infection. Cell Microbiol. 2011;13:1858–1869. doi: 10.1111/j.1462-5822.2011.01701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmer BMM, Vanreeuwijk J, Timmers CD, Valentine PJ, Heffron F. Salmonella Typhimurium encodes an SdiA homolog, a putative quorum sensor of the luxR family, that regulates genes on the virulence plasmid. J. Bacteriol. 1998;180:1185–1193. doi: 10.1128/jb.180.5.1185-1193.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiastui A, Pucciarelli MG, Garcia-del Portillo F. Salmonella enterica serovar Typhimurium invades fibroblasts by multiple routes differing from the entry into epithelial cells. Infect. Immun. 2010;78:2700–2713. doi: 10.1128/IAI.01389-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakowski MA, Cirulis JT, Brown NF, Finlay BB, Brumell JH. SopD acts cooperatively with SopB during Salmonella enterica serovar Typhimurium invasion. Cell Microbiol. 2007;9:2839–2855. doi: 10.1111/j.1462-5822.2007.01000.x. [DOI] [PubMed] [Google Scholar]
- Bakshi CS, Singh VP, Wood MW, Jones PW, Wallis TS, Galyov EE. Identification of SopE2, a Salmonella secreted protein which is highly homologous to SopE and involved in bacterial invasion of epithelial cells. J. Bacteriol. 2000;182:2341–2344. doi: 10.1128/jb.182.8.2341-2344.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak JD, Kramer LC, Hao LY. Colonization of tomato plants by Salmonella enterica is cultivar dependent, and type 1 trichomes are preferred colonization sites. Appl. Environ. Microbiol. 2011;77:498–504. doi: 10.1128/AEM.01661-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belden WJ, Miller SI. Further characterization of the phoP regulon: identification of new phoP-activated virulence loci. Infect. Immun. 1994;62:5095–5101. doi: 10.1128/iai.62.11.5095-5101.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel CJ, Jimenez JC, Contreras I, Santiviago CA. Comparative genomic analysis uncovers 3 novel loci encoding type six secretion systems differentially distributed in Salmonella serotypes. BMC Genomics. 2009;10:354. doi: 10.1186/1471-2164-10-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel CJ, Yang HJ, Castro B, Chiang S, Toro CS, Zaldivar M, et al. Contribution of the type VI secretion system encoded in SPI-19 to chicken colonization by Salmonella enterica serotypes Gallinarum and Enteritidis. PLoS ONE. 2010;5:e11724. doi: 10.1371/journal.pone.0011724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd EF, Wang FS, Whittam TS, Selander RK. Molecular genetic relationships of the Salmonellae. Appl. Environ. Microbiol. 1996;62:804–808. doi: 10.1128/aem.62.3.804-808.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyen F, Pasmans F, Van Immerseel F, Morgan E, Adriaensen C, Hernalsteens JP, et al. Salmonella Typhimurium SPI-1 genes promote intestinal but not tonsillar colonization in pigs. Microbes Infect. 2006;8:2899–2907. doi: 10.1016/j.micinf.2006.09.008. [DOI] [PubMed] [Google Scholar]
- Boyle EC, Brown NF, Finlay BB. Salmonella enterica serovar Typhimurium effectors SopB, SopE, SopE2 and SipA disrupt tight junction structure and function. Cell Microbiol. 2006;8:1946–1957. doi: 10.1111/j.1462-5822.2006.00762.x. [DOI] [PubMed] [Google Scholar]
- Braun V, Brumell JH. Bacterial invasion: entry through the exocyst door. Curr. Biol. 2010;20:R677–R679. doi: 10.1016/j.cub.2010.06.047. [DOI] [PubMed] [Google Scholar]
- Brenner FW, Villar RG, Angulo FJ, Tauxe R, Swaminathan B. Salmonella nomenclature. J. Clin. Microbiol. 2000;38:2465–2467. doi: 10.1128/jcm.38.7.2465-2467.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buisan M, Rodriguez-Pena JM, Rotger R. Restriction map of the Salmonella Enteritidis virulence plasmid and its homology with the plasmid of Salmonella Typhimurium. Microb. Pathog. 1994;16:165–169. doi: 10.1006/mpat.1994.1017. [DOI] [PubMed] [Google Scholar]
- Calenge F, Lecerf F, Demars J, Feve K, Vignoles F, Pitel F, et al. QTL for resistance to Salmonella carrier state confirmed in both experimental and commercial chicken lines. Anim. Genet. 2009;40:590–597. doi: 10.1111/j.1365-2052.2009.01884.x. [DOI] [PubMed] [Google Scholar]
- Calenge F, Kaiser P, Vignal A, Beaumont C. Genetic control of resistance to salmonellosis and to Salmonella carrier-state in fowl: a review. Genet. Sel. Evol. 2010;42:11. doi: 10.1186/1297-9686-42-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J, Chen J, Zhou D. Delineation and characterization of the actin nucleation and effector translocation activities of Salmonella SipC. Mol. Microbiol. 2005;55:1379–1389. doi: 10.1111/j.1365-2958.2004.04480.x. [DOI] [PubMed] [Google Scholar]
- Chi B, Qi M, Kuramitsu HK. Role of dentilisin in Treponema denticola epithelial cell layer penetration. Res. Microbiol. 2003;154:637–643. doi: 10.1016/j.resmic.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Chiu CH, Su LH, Chu C. Salmonella enterica serotype Choleraesuis: epidemiology, pathogenesis, clinical disease, and treatment. Clin. Microbiol. Rev. 2004;17:311–322. doi: 10.1128/CMR.17.2.311-322.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CS, Hong SF, Tsai CJ, Lin WS, Liu TP, Ou JT. Comparative physical and genetic maps of the virulence plasmids of Salmonella enterica serovars Typhimurium, Enteritidis, Choleraesuis, and Dublin. Infect. Immun. 1999;67:2611–2614. doi: 10.1128/iai.67.5.2611-2614.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MA, Jepson MA, Simmons NL, Hirst BH. Preferential interaction of Salmonella typhimurium with mouse peyer's patch m cells. Res. Microbiol. 1994;145:543–552. doi: 10.1016/0923-2508(94)90031-0. [DOI] [PubMed] [Google Scholar]
- Clark MA, Reed KA, Lodge J, Stephen J, Hirst BH, Jepson MA. Invasion of murine intestinal M cells by Salmonella typhimurium inv mutants severely deficient for invasion of cultured cells. Infect. Immun. 1996;64:4363–4368. doi: 10.1128/iai.64.10.4363-4368.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conner CP, Heithoff DM, Julio SM, Sinsheimer RL, Mahan MJ. Differential patterns of acquired virulence genes distinguish Salmonella strains. Proc. Natl Acad. Sci. USA. 1998;95:4641–4645. doi: 10.1073/pnas.95.8.4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombes BK, Coburn BA, Potter AA, Gomis S, Mirakhur K, Li Y, et al. Analysis of the contribution of Salmonella pathogenicity islands 1 and 2 to enteric disease progression using a novel bovine ileal loop model and a murine model of infectious enterocolitis. Infect. Immun. 2005;73:7161–7169. doi: 10.1128/IAI.73.11.7161-7169.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desin TS, Lam PK, Koch B, Mickael C, Berberov E, Wisner AL, et al. Salmonella enterica serovar Enteritidis pathogenicity island 1 is not essential for but facilitates rapid systemic spread in chickens. Infect. Immun. 2009;77:2866–2875. doi: 10.1128/IAI.00039-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doran KS, Chang JC, Benoit VM, Eckmann L, Nizet V. Group B streptococcal beta-hemolysin/cytolysin promotes invasion of human lung epithelial cells and the release of interleukin-8. J. Infect. Dis. 2002;185:196–203. doi: 10.1086/338475. [DOI] [PubMed] [Google Scholar]
- Dorsey CW, Laarakker MC, Humphries AD, Weening EH, Baumler AJ. Salmonella enterica serotype Typhimurium MisL is an intestinal colonization factor that binds fibronectin. Mol. Microbiol. 2005;57:196–211. doi: 10.1111/j.1365-2958.2005.04666.x. [DOI] [PubMed] [Google Scholar]
- Dramsi S, Cossart P. Listeriolysin O-mediated calcium influx potentiates entry of Listeria monocytogenes into the human Hep-2 epithelial cell line. Infect. Immun. 2003;71:3614–3618. doi: 10.1128/IAI.71.6.3614-3618.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drecktrah D, Knodler LA, Galbraith K, Steele-Mortimer O. The Salmonella SPI1 effector SopB stimulates nitric oxide production long after invasion. Cell Microbiol. 2005;7:105–113. doi: 10.1111/j.1462-5822.2004.00436.x. [DOI] [PubMed] [Google Scholar]
- Dyszel JL, Smith JN, Lucas DE, Soares JA, Swearingen MC, Vross MA, et al. Salmonella enterica serovar Typhimurium can detect acyl homoserine lactone production by Yersinia enterocolitica in mice. J. Bacteriol. 2010;192:29–37. doi: 10.1128/JB.01139-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson DL, Nsereko VL, Morgavi DP, Selinger LB, Rode LM, Beauchemin KA. Evidence of quorum sensing in the rumen ecosystem: detection of N-acyl homoserine lactone autoinducers in ruminal contents. Can. J. Microbiol. 2002;48:374–378. doi: 10.1139/w02-022. [DOI] [PubMed] [Google Scholar]
- Eriksson S, Lucchini S, Thompson A, Rhen M, Hinton JC. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol. Microbiol. 2003;47:103–118. doi: 10.1046/j.1365-2958.2003.03313.x. [DOI] [PubMed] [Google Scholar]
- Folkesson A, Lofdahl S, Normark S. The Salmonella enterica subspecies I specific centisome 7 genomic island encodes novel protein families present in bacteria living in close contact with eukaryotic cells. Res. Microbiol. 2002;153:537–545. doi: 10.1016/s0923-2508(02)01348-7. [DOI] [PubMed] [Google Scholar]
- Fookes M, Schroeder GN, Langridge GC, Blondel CJ, Mammina C, Connor TR, et al. Salmonella bongori provides insights into the evolution of the Salmonellae. PLoS Pathog. 2011;7:e1002191. doi: 10.1371/journal.ppat.1002191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost AJ, Bland AP, Wallis TS. The early dynamic response of the calf ileal epithelium to Salmonella typhimurium. Vet. Pathol. 1997;34:369–386. doi: 10.1177/030098589703400501. [DOI] [PubMed] [Google Scholar]
- Fuentes JA, Villagra N, Castillo-Ruiz M, Mora GC. The Salmonella Typhi hlyE gene plays a role in invasion of cultured epithelial cells and its functional transfer to S. typhimurium promotes deep organ infection in mice. Res. Microbiol. 2008;159:279–287. doi: 10.1016/j.resmic.2008.02.006. [DOI] [PubMed] [Google Scholar]
- Futagawa-Saito K, Okatani AT, Sakurai-Komada N, Ba-Thein W, Fukuyasu T. Epidemiological characteristics of Salmonella enterica serovar Typhimurium from healthy pigs in Japan. J. Vet. Med. Sci. 2010;72:61–66. doi: 10.1292/jvms.09-0041. [DOI] [PubMed] [Google Scholar]
- Galan JE, Collmer A. Type III secretion machines: bacterial devices for protein delivery into host cells. Science. 1999;284:1322–1328. doi: 10.1126/science.284.5418.1322. [DOI] [PubMed] [Google Scholar]
- Galan JE, Curtiss R., III Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc. Natl Acad. Sci. USA. 1989;86:6383–6387. doi: 10.1073/pnas.86.16.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galdiero M, Pisciotta MG, Galdiero E, Carratelli CR. Porins and lipopolysaccharide from Salmonella Typhimurium regulate the expression of CD80 and CD86 molecules on B cells and macrophages but not CD28 and CD152 on T cells. Clin. Microbiol. Infect. 2003;9:1104–1111. doi: 10.1046/j.1469-0691.2003.00728.x. [DOI] [PubMed] [Google Scholar]
- Galyov EE, Wood MW, Rosqvist R, Mullan PB, Watson PR, Hedges S, et al. A secreted effector protein of Salmonella dublin is translocated into eukaryotic cells and mediates inflammation and fluid secretion in infected ileal mucosa. Mol. Microbiol. 1997;25:903–912. doi: 10.1111/j.1365-2958.1997.mmi525.x. [DOI] [PubMed] [Google Scholar]
- Gerlach RG, Claudio N, Rohde M, Jackel D, Wagner C, Hensel M. Cooperation of Salmonella pathogenicity islands 1 and 4 is required to breach epithelial barriers. Cell Microbiol. 2008;10:2364–2376. doi: 10.1111/j.1462-5822.2008.01218.x. [DOI] [PubMed] [Google Scholar]
- Ginocchio CC, Rahn K, Clarke RC, Galan JE. Naturally occurring deletions in the centisome 63 pathogenicity island of environmental isolates of Salmonella spp. Infect. Immun. 1997;65:1267–1272. doi: 10.1128/iai.65.4.1267-1272.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golberg D, Kroupitski Y, Belausov E, Pinto R, Sela S. Salmonella Typhimurium internalization is variable in leafy vegetables and fresh herbs. Int. J. Food Microbiol. 2011;145:250–257. doi: 10.1016/j.ijfoodmicro.2010.12.031. [DOI] [PubMed] [Google Scholar]
- Groisman EA, Ochman H. Pathogenicity islands: bacterial evolution in quantum leaps. Cell. 1996;87:791–794. doi: 10.1016/s0092-8674(00)81985-6. [DOI] [PubMed] [Google Scholar]
- Guibourdenche M, Roggentin P, Mikoleit M, Fields PI, Bockemuhl J, Grimont PA, et al. Supplement 2003-2007 (No. 47) to the White-Kauffmann-Le Minor scheme. Res. Microbiol. 2010;161:26–29. doi: 10.1016/j.resmic.2009.10.002. [DOI] [PubMed] [Google Scholar]
- Guo A, Lasaro MA, Sirard JC, Kraehenbuhl JP, Schifferli DM. Adhesin-dependent binding and uptake of Salmonella enterica serovar Typhimurium by dendritic cells. Microbiology. 2007;153:1059–1069. doi: 10.1099/mic.0.2006/000331-0. [DOI] [PubMed] [Google Scholar]
- Hanisch J, Kolm R, Wozniczka M, Bumann D, Rottner K, Stradal TE. Activation of a RhoA/myosin II-dependent but Arp2/3 complex-independent pathway facilitates Salmonella invasion. Cell Host Microbe. 2011;9:273–285. doi: 10.1016/j.chom.2011.03.009. [DOI] [PubMed] [Google Scholar]
- Hapfelmeier S, Ehrbar K, Stecher B, Barthel M, Kremer M, Hardt WD. Role of the Salmonella pathogenicity island 1 effector proteins SipA, SopB, SopE, and SopE2 in Salmonella enterica subspecies 1 serovar Typhimurium colitis in streptomycin-pretreated mice. Infect. Immun. 2004;72:795–809. doi: 10.1128/IAI.72.2.795-809.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hapfelmeier S, Stecher B, Barthel M, Kremer M, Muller AJ, Heikenwalder M, et al. The Salmonella pathogenicity island (SPI)-2 and SPI-1 type III secretion systems allow Salmonella serovar Typhimurium to trigger colitis via MyD88-dependent and MyD88-independent mechanisms. J. Immunol. 2005;174:1675–1685. doi: 10.4049/jimmunol.174.3.1675. [DOI] [PubMed] [Google Scholar]
- Hautefort I, Thompson A, Eriksson-Ygberg S, Parker ML, Lucchini S, Danino V, et al. During infection of epithelial cells Salmonella enterica serovar Typhimurium undergoes a time-dependent transcriptional adaptation that results in simultaneous expression of three type 3 secretion systems. Cell Microbiol. 2008;10:958–984. doi: 10.1111/j.1462-5822.2007.01099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward RD, Cain RJ, McGhie EJ, Phillips N, Garner MJ, Koronakis V. Cholesterol binding by the bacterial type III translocon is essential for virulence effector delivery into mammalian cells. Mol. Microbiol. 2005;56:590–603. doi: 10.1111/j.1365-2958.2005.04568.x. [DOI] [PubMed] [Google Scholar]
- Heffernan EJ, Harwood J, Fierer J, Guiney D. The Salmonella Typhimurium virulence plasmid complement resistance gene rck is homologous to a family of virulence-related outer membrane protein genes, including pagC and ail. J. Bacteriol. 1992;174:84–91. doi: 10.1128/jb.174.1.84-91.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffernan EJ, Wu L, Louie J, Okamoto S, Fierer J, Guiney DG. Specificity of the complement resistance and cell association phenotypes encoded by the outer membrane protein genes rck from Salmonella Typhimurium and ail from Yersinia enterocolitica. Infect. Immun. 1994;62:5183–5186. doi: 10.1128/iai.62.11.5183-5186.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heithoff DM, Conner CP, Hanna PC, Julio SM, Hentschel U, Mahan MJ. Bacterial infection as assessed by in vivo gene expression. Proc. Natl Acad. Sci. USA. 1997;94:934–939. doi: 10.1073/pnas.94.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heithoff DM, Shimp WR, Lau PW, Badie G, Enioutina EY, Daynes RA, et al. Human Salmonella clinical isolates distinct from those of animal origin. Appl. Environ. Microbiol. 2008;74:1757–1766. doi: 10.1128/AEM.02740-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins KL, Threlfall EJ. Frequency and polymorphism of sopE in isolates of Salmonella enterica belonging to the ten most prevalent serotypes in England and Wales. J. Med. Microbiol. 2004;53:539–543. doi: 10.1099/jmm.0.05510-0. [DOI] [PubMed] [Google Scholar]
- Horiuchi S, Inagaki Y, Okamura N, Nakaya R, Yamamoto N. Type 1 pili enhance the invasion of Salmonella braenderup and Salmonella typhimurium to HeLa cells. Microbiol. Immunol. 1992;36:593–602. doi: 10.1111/j.1348-0421.1992.tb02059.x. [DOI] [PubMed] [Google Scholar]
- Hu Q, Coburn B, Deng W, Li Y, Shi X, Lan Q, et al. Salmonella enterica serovar Senftenberg human clinical isolates lacking SPI-1. J. Clin. Microbiol. 2008;46:1330–1336. doi: 10.1128/JCM.01255-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong JH, Song M, Park SI, Cho KO, Rhee JH, Choy HE. Salmonella enterica serovar Gallinarum requires ppGpp for internalization and survival in animal cells. J. Bacteriol. 2008;190:6340–6350. doi: 10.1128/JB.00385-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones BD, Ghori N, Falkow S. Salmonella typhimurium initiates murine infection by penetrating and destroying the specialized epithelial M cells of the peyer's patches. J. Exp. Med. 1994;180:15–23. doi: 10.1084/jem.180.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MA, Wigley P, Page KL, Hulme SD, Barrow PA. Salmonella enterica serovar Gallinarum requires the Salmonella pathogenicity island 2 type III secretion system but not the Salmonella pathogenicity island 1 type III secretion system for virulence in chickens. Infect. Immun. 2001;69:5471–5476. doi: 10.1128/IAI.69.9.5471-5476.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsley RA, Santos RL, Keestra AM, Adams LG, Baumler AJ. Salmonella enterica serotype Typhimurium ShdA is an outer membrane fibronectin-binding protein that is expressed in the intestine. Mol. Microbiol. 2002;43:895–905. doi: 10.1046/j.1365-2958.2002.02805.x. [DOI] [PubMed] [Google Scholar]
- Kitagawa R, Takaya A, Ohya M, Mizunoe Y, Takade A, Yoshida S, et al. Biogenesis of Salmonella enterica serovar Typhimurium membrane vesicles provoked by induction of PagC. J. Bacteriol. 2010;192:5645–5656. doi: 10.1128/JB.00590-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knodler LA, Winfree S, Drecktrah D, Ireland R, Steele-Mortimer O. Ubiquitination of the bacterial inositol phosphatase, SopB, regulates its biological activity at the plasma membrane. Cell Microbiol. 2009;11:1652–1670. doi: 10.1111/j.1462-5822.2009.01356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubori T, Galan JE. Temporal regulation of Salmonella virulence effector function by proteasome-dependent protein degradation. Cell. 2003;115:333–342. doi: 10.1016/s0092-8674(03)00849-3. [DOI] [PubMed] [Google Scholar]
- Lambert MA, Smith SG. The PagN protein of Salmonella enterica serovar Typhimurium is an adhesin and invasin. BMC Microbiol. 2008;8:142. doi: 10.1186/1471-2180-8-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert MA, Smith SG. The PagN protein mediates invasion via interaction with proteoglycan. FEMS Microbiol. Lett. 2009;297:209–216. doi: 10.1111/j.1574-6968.2009.01666.x. [DOI] [PubMed] [Google Scholar]
- Lara-Tejero M, Galan JE. Salmonella enterica serovar Typhimurium pathogenicity island 1-encoded type III secretion system translocases mediate intimate attachment to nonphagocytic cells. Infect. Immun. 2009;77:2635–2642. doi: 10.1128/IAI.00077-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Tejero M, Kato J, Wagner S, Liu X, Galan JE. A sorting platform determines the order of protein secretion in bacterial type III systems. Science. 2011;331:1188–1191. doi: 10.1126/science.1201476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley TD, Chan K, Thompson LJ, Kim CC, Govoni GR, Monack DM. Genome-wide screen for Salmonella genes required for long-term systemic infection of the mouse. PLoS Pathog. 2006;2:e11. doi: 10.1371/journal.ppat.0020011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YH, Xu XB, Hu QH, Shi XL, Lin YM, Qiu YQ. [Characteristics of Salmonella enterica serovar Senftenberg lacking Salmonella pathogenicity island 1] Zhonghua Yu Fang Yi Xue Za Zhi. 2011;45:899–903. [PubMed] [Google Scholar]
- Malik-Kale P, Jolly CE, Lathrop S, Winfree S, Luterbach C, Steele-Mortimer O. Salmonella – at home in the host cell. Front. Microbiol. 2011;2:125. doi: 10.3389/fmicb.2011.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez JJ, Mulvey MA, Schilling JD, Pinkner JS, Hultgren SJ. Type 1 pilus-mediated bacterial invasion of bladder epithelial cells. EMBO J. 2000;19:2803–2812. doi: 10.1093/emboj/19.12.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M, Sanderson KE, Spieth J, Clifton SW, Latreille P, Courtney L, et al. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature. 2001;413:852–856. doi: 10.1038/35101614. [DOI] [PubMed] [Google Scholar]
- McGhie EJ, Hayward RD, Koronakis V. Control of actin turnover by a Salmonella invasion protein. Mol. Cell. 2004;13:497–510. doi: 10.1016/s1097-2765(04)00053-x. [DOI] [PubMed] [Google Scholar]
- McGhie EJ, Brawn LC, Hume PJ, Humphreys D, Koronakis V. Salmonella takes control: effector-driven manipulation of the host. Curr. Opin. Microbiol. 2009;12:117–124. doi: 10.1016/j.mib.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael B, Smith JN, Swift S, Heffron F, Ahmer BM. SdiA of Salmonella enterica is a LuxR homolog that detects mixed microbial communities. J. Bacteriol. 2001;183:5733–5742. doi: 10.1128/JB.183.19.5733-5742.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mijouin L, Rosselin M, Bottreau E, Pizarro-Cerda J, Cossart P, Velge P, et al. Salmonella Enteritidis Rck-mediated invasion requires activation of Rac1, which is dependent on the class I PI 3-kinases-Akt signaling pathway. FASEB J. 2012;26:1569–1581. doi: 10.1096/fj.11-189647. [DOI] [PubMed] [Google Scholar]
- Misselwitz B, Kreibich SK, Rout S, Stecher B, Periaswamy B, Hardt WD. Salmonella enterica serovar Typhimurium binds to HeLa cells via Fim-mediated reversible adhesion and irreversible type three secretion system 1-mediated docking. Infect. Immun. 2011;79:330–341. doi: 10.1128/IAI.00581-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan E, Campbell JD, Rowe SC, Bispham J, Stevens MP, Bowen AJ, et al. Identification of host-specific colonization factors of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 2004;54:994–1010. doi: 10.1111/j.1365-2958.2004.04323.x. [DOI] [PubMed] [Google Scholar]
- Morgan E, Bowen AJ, Carnell SC, Wallis TS, Stevens MP. SiiE is secreted by the Salmonella enterica serovar Typhimurium pathogenicity island 4-encoded secretion system and contributes to intestinal colonization in cattle. Infect. Immun. 2007;75:1524–1533. doi: 10.1128/IAI.01438-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller AJ, Kaiser P, Dittmar KE, Weber TC, Haueter S, Endt K, et al. Salmonella gut invasion involves TTSS-2-dependent epithelial traversal, basolateral exit, and uptake by epithelium-sampling lamina propria phagocytes. Cell Host Microbe. 2012;11:19–32. doi: 10.1016/j.chom.2011.11.013. [DOI] [PubMed] [Google Scholar]
- Murray RA, Lee CA. Invasion genes are not required for Salmonella enterica serovar Typhimurium to breach the intestinal epithelium: evidence that salmonella pathogenicity island 1 has alternative functions during infection. Infect. Immun. 2000;68:5050–5055. doi: 10.1128/iai.68.9.5050-5055.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myeni SK, Zhou D. The C terminus of SipC binds and bundles F-actin to promote Salmonella invasion. J. Biol. Chem. 2010;285:13357–13363. doi: 10.1074/jbc.M109.094045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols CD, Casanova JE. Salmonella-directed recruitment of new membrane to invasion foci via the host exocyst complex. Curr. Biol. 2010;20:1316–1320. doi: 10.1016/j.cub.2010.05.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nygard K, Lassen J, Vold L, Andersson Y, Fisher I, Lofdahl S, et al. Outbreak of Salmonella Thompson infections linked to imported rucola lettuce. Foodborne Pathog. Dis. 2008;5:165–173. doi: 10.1089/fpd.2007.0053. [DOI] [PubMed] [Google Scholar]
- Parker CT, Guard-Petter J. Contribution of flagella and invasion proteins to pathogenesis of Salmonella enterica serovar Enteritidis in chicks. FEMS Microbiol. Lett. 2001;204:287–291. doi: 10.1111/j.1574-6968.2001.tb10899.x. [DOI] [PubMed] [Google Scholar]
- Parsons DA, Heffron F. sciS, an icmF homolog in Salmonella enterica serovar Typhimurium, limits intracellular replication and decreases virulence. Infect. Immun. 2005;73:4338–4345. doi: 10.1128/IAI.73.7.4338-4345.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascopella L, Raupach B, Ghori N, Monack D, Falkow S, Small PL. Host restriction phenotypes of Salmonella Typhi and Salmonella Gallinarum. Infect. Immun. 1995;63:4329–4335. doi: 10.1128/iai.63.11.4329-4335.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel JC, Galan JE. Differential activation and function of Rho GTPases during Salmonella-host cell interactions. J. Cell Biol. 2006;175:453–463. doi: 10.1083/jcb.200605144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel JC, Hueffer K, Lam TT, Galan JE. Diversification of a Salmonella virulence protein function by ubiquitin-dependent differential localization. Cell. 2009;137:283–294. doi: 10.1016/j.cell.2009.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulin SM, Watson PR, Benmore AR, Stevens MP, Jones PW, Villarreal-Ramos B, et al. Analysis of Salmonella enterica serotype-host specificity in calves: avirulence of S. enterica serotype Gallinarum correlates with bacterial dissemination from mesenteric lymph nodes and persistence in vivo. Infect. Immun. 2002;70:6788–6797. doi: 10.1128/IAI.70.12.6788-6797.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrett CA, Jepson MA. Regulation of Salmonella-induced membrane ruffling by SipA differs in strains lacking other effectors. Cell Microbiol. 2009;11:475–487. doi: 10.1111/j.1462-5822.2008.01268.x. [DOI] [PubMed] [Google Scholar]
- Pezzoli L, Elson R, Little C, Fisher I, Yip H, Peters T, et al. International outbreak of Salmonella Senftenberg in 2007. Euro. Surveill. 2007;12:E070614.3. doi: 10.2807/esw.12.24.03218-en. [DOI] [PubMed] [Google Scholar]
- Pier GB, Grout M, Zaidi T, Meluleni G, Mueschenborn SS, Banting G, et al. Salmonella Typhi uses CFTR to enter intestinal epithelial cells. Nature. 1998;393:79–82. doi: 10.1038/30006. [DOI] [PubMed] [Google Scholar]
- Popp D, Yamamoto A, Iwasa M, Nitanai Y, Maeda Y. Single molecule polymerization, annealing and bundling dynamics of SipA induced actin filaments. Cell Motil. Cytoskelet. 2008;65:165–177. doi: 10.1002/cm.20252. [DOI] [PubMed] [Google Scholar]
- Pullinger GD, Paulin SM, Charleston B, Watson PR, Bowen AJ, Dziva F, et al. Systemic translocation of Salmonella enterica serovar Dublin in cattle occurs predominantly via efferent lymphatics in a cell-free niche and requires type III secretion system 1 (T3SS-1) but not T3SS-2. Infect. Immun. 2007;75:5191–5199. doi: 10.1128/IAI.00784-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabsch W, Andrews HL, Kingsley RA, Prager R, Tschape H, Adams LG, et al. Salmonella enterica serotype Typhimurium and its host-adapted variants. Infect. Immun. 2002;70:2249–2255. doi: 10.1128/IAI.70.5.2249-2255.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radtke AL, Wilson JW, Sarker S, Nickerson CA. Analysis of interactions of Salmonella type three secretion mutants with 3-D intestinal epithelial cells. PLoS ONE. 2010;5:e15750. doi: 10.1371/journal.pone.0015750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangel JM, Sparling PH, Crowe C, Griffin PM, Swerdlow DL. Epidemiology of Escherichia coli O157:H7 outbreaks, United States, 1982-2002. Emerg. Infect. Dis. 2005;11:603–609. doi: 10.3201/eid1104.040739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolhion N, Barnich N, Claret L, Darfeuille-Michaud A. Strong decrease in invasive ability and outer membrane vesicle release in Crohn's disease-associated adherent-invasive Escherichia coli strain LF82 with the yfgL gene deleted. J. Bacteriol. 2005;187:2286–2296. doi: 10.1128/JB.187.7.2286-2296.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosselin M, Virlogeux-Payant I, Roy C, Bottreau E, Sizaret PY, Mijouin L, et al. Rck of Salmonella enterica, subspecies enterica serovar Enteritidis, mediates Zipper-like internalization. Cell Res. 2010;20:647–664. doi: 10.1038/cr.2010.45. [DOI] [PubMed] [Google Scholar]
- Rosselin M, Abed N, Virlogeux-Payant I, Bottreau E, Sizaret PY, Velge P, et al. Heterogeneity of type III secretion system (T3SS)-1-independent entry mechanisms used by Salmonella Enteritidis to invade different cell types. Microbiology. 2011;157:839–847. doi: 10.1099/mic.0.044941-0. [DOI] [PubMed] [Google Scholar]
- Rosselin M, Abed N, Namdari F, Virlogeux Payant I, Velge P, Wiedemann A. Salmonella/Book2. Rijeka, Croatia: In Tech; 2012. The different strategies used by Salmonella to invade host cells. ISBN 979-953-307-690-3. [Google Scholar]
- Rudge SA, Anderson DM, Emr SD. Vacuole size control: regulation of PtdIns(3,5)P2 levels by the vacuole-associated Vac14-Fig 4 complex, a PtdIns(3,5)P2-specific phosphatase. Mol. Biol. Cell. 2004;15:24–36. doi: 10.1091/mbc.E03-05-0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rychlik I, Gregorova D, Hradecka H. Distribution and function of plasmids in Salmonella enterica. Vet. Microbiol. 2006;112:1–10. doi: 10.1016/j.vetmic.2005.10.030. [DOI] [PubMed] [Google Scholar]
- Rychlik I, Karasova D, Sebkova A, Volf J, Sisak F, Havlickova H, et al. Virulence potential of five major pathogenicity islands (SPI-1 to SPI-5) of Salmonella enterica serovar Enteritidis for chickens. BMC Microbiol. 2009;9:268. doi: 10.1186/1471-2180-9-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salcedo SP, Noursadeghi M, Cohen J, Holden DW. Intracellular replication of Salmonella Typhimurium strains in specific subsets of splenic macrophages in vivo. Cell Microbiol. 2001;3:587–597. doi: 10.1046/j.1462-5822.2001.00137.x. [DOI] [PubMed] [Google Scholar]
- Santos RL, Zhang S, Tsolis RM, Kingsley RA, Adams LG, Baumler AJ. Animal models of Salmonella infections: enteritis versus typhoid fever. Microbes Infect. 2001;3:1335–1344. doi: 10.1016/s1286-4579(01)01495-2. [DOI] [PubMed] [Google Scholar]
- Santos RL, Raffatellu M, Bevins CL, Adams LG, Tukel C, Tsolis RM, et al. Life in the inflamed intestine, Salmonella style. Trends Microbiol. 2009;17:498–506. doi: 10.1016/j.tim.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schikora A, Virlogeux-Payant I, Bueso E, Garcia AV, Nilau T, Charrier A, et al. Conservation of Salmonella infection mechanisms in plants and animals. PLoS ONE. 2011;6:e24112. doi: 10.1371/journal.pone.0024112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlieker C, Zentgraf H, Dersch P, Mogk A. ClpV, a unique Hsp100/Clp member of pathogenic proteobacteria. Biol. Chem. 2005;386:1115–1127. doi: 10.1515/BC.2005.128. [DOI] [PubMed] [Google Scholar]
- Schwarz S, West TE, Boyer F, Chiang WC, Carl MA, Hood RD, et al. Burkholderia type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Pathog. 2010;6:e1001068. doi: 10.1371/journal.ppat.1001068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shames SR, Auweter SD, Finlay BB. Co-evolution and exploitation of host cell signaling pathways by bacterial pathogens. Int. J. Biochem. Cell Biol. 2009;41:380–389. doi: 10.1016/j.biocel.2008.08.013. [DOI] [PubMed] [Google Scholar]
- Shirron N, Yaron S. Active suppression of early immune response in tobacco by the human pathogen Salmonella Typhimurium. PLoS ONE. 2011;6:e18855. doi: 10.1371/journal.pone.0018855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaprasad HL. Fowl typhoid and pullorum disease. Rev. Sci. Tech. 2000;19:405–424. doi: 10.20506/rst.19.2.1222. [DOI] [PubMed] [Google Scholar]
- Smith JN, Ahmer BM. Detection of other microbial species by Salmonella: expression of the SdiA regulon. J. Bacteriol. 2003;185:1357–1366. doi: 10.1128/JB.185.4.1357-1366.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JN, Dyszel JL, Soares JA, Ellermeier CD, Altier C, Lawhon SD, et al. SdiA, an N-acylhomoserine lactone receptor, becomes active during the transit of Salmonella enterica through the gastrointestinal tract of turtles. PLoS ONE. 2008;3:e2826. doi: 10.1371/journal.pone.0002826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Sorge NM, Zialcita PA, Browne SH, Quach D, Guiney DG, Doran KS. Penetration and activation of brain endothelium by Salmonella enterica serovar Typhimurium. J. Infect. Dis. 2011;203:401–405. doi: 10.1093/infdis/jiq048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecher B, Hapfelmeier S, Muller C, Kremer M, Stallmach T, Hardt WD. Flagella and chemotaxis are required for efficient induction of Salmonella enterica serovar Typhimurium colitis in streptomycin-pretreated mice. Infect. Immun. 2004;72:4138–4150. doi: 10.1128/IAI.72.7.4138-4150.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele-Mortimer O, Brumell JH, Knodler LA, Meresse S, Lopez A, Finlay BB. The invasion-associated type III secretion system of Salmonella enterica serovar Typhimurium is necessary for intracellular proliferation and vacuole biogenesis in epithelial cells. Cell Microbiol. 2002;4:43–54. doi: 10.1046/j.1462-5822.2002.00170.x. [DOI] [PubMed] [Google Scholar]
- Stevens MP, Humphrey TJ, Maskell DJ. Molecular insights into farm animal and zoonotic Salmonella infections. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2009;364:2709–2723. doi: 10.1098/rstb.2009.0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss EJ, Ghori N, Falkow S. An Edwardsiella tarda strain containing a mutation in a gene with homology to shlB and hpmB is defective for entry into epithelial cells in culture. Infect. Immun. 1997;65:3924–3932. doi: 10.1128/iai.65.9.3924-3932.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi A. Electron microscope studies of experimental Salmonella infection. I. Penetration into the intestinal epithelium by Salmonella typhimurium. Am. J. Pathol. 1967;50:109–136. [PMC free article] [PubMed] [Google Scholar]
- Uzzau S, Brown DJ, Wallis T, Rubino S, Leori G, Bernard S, et al. Host adapted serotypes of Salmonella enterica. Epidemiol. Infect. 2000;125:229–255. doi: 10.1017/s0950268899004379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Torres A, Jones-Carson J, Baumler AJ, Falkow S, Valdivia R, Brown W, et al. Extraintestinal dissemination of Salmonella by CD18-expressing phagocytes. Nature. 1999;401:804–808. doi: 10.1038/44593. [DOI] [PubMed] [Google Scholar]
- Velge P, Cloeckaert A, Barrow P. Emergence of Salmonella epidemics: the problems related to Salmonella enterica serotype Enteritidis and multiple antibiotic resistance in other major serotypes. Vet. Res. 2005;36:267–288. doi: 10.1051/vetres:2005005. [DOI] [PubMed] [Google Scholar]
- Wagner C, Hensel M. Adhesive mechanisms of Salmonella enterica. Adv. Exp. Med. Biol. 2011;715:17–34. doi: 10.1007/978-94-007-0940-9_2. [DOI] [PubMed] [Google Scholar]
- Zhou D, Mooseker MS, Galan JE. Role of the S. typhimurium actin-binding protein Sipa in bacterial internalization. Science. 1999;283:2092–2095. doi: 10.1126/science.283.5410.2092. [DOI] [PubMed] [Google Scholar]
- Zhou D, Chen LM, Hernandez L, Shears SB, Galan JE. A Salmonella inositol polyphosphatase acts in conjunction with other bacterial effectors to promote host cell actin cytoskeleton rearrangements and bacterial internalization. Mol. Microbiol. 2001;39:248–260. doi: 10.1046/j.1365-2958.2001.02230.x. [DOI] [PubMed] [Google Scholar]
- Zou W, Al-Khaldi SF, Branham WS, Han T, Fuscoe JC, Han J, et al. Microarray analysis of virulence gene profiles in Salmonella serovars from food/food animal environment. J. Infect. Dev. Ctries. 2011;5:94–105. doi: 10.3855/jidc.1396. [DOI] [PubMed] [Google Scholar]


