Abstract
It was the work of Jan Anderson, together with Keith Boardman, that showed it was possible to physically separate photosystem I (PSI) from photosystem II (PSII), and it was Jan Anderson who realized the importance of this work in terms of the fluid-mosaic model as applied to the thylakoid membrane. Since then, there has been a steady progress in the development of biochemical procedures to isolate PSII and PSI both for physical and structural studies. Dodecylmaltoside (DM) has emerged as an effective mild detergent for this purpose. DM is a glucoside-based surfactant with a bulky hydrophilic head group composed of two sugar rings and a non-charged alkyl glycoside chain. Two isomers of this molecule exist, differing only in the configuration of the alkyl chain around the anomeric centre of the carbohydrate head group, axial in α-DM and equatorial in β-DM. We have compared the use of α-DM and β-DM for the isolation of supramolecular complexes of PSII by a single-step solubilization of stacked thylakoid membranes isolated from peas. As a result, we have optimized conditions to obtain homogeneous preparations of the C2S2M2 and C2S2 supercomplexes following the nomenclature of Dekker & Boekema (2005 Biochim. Biophys. Acta 1706, 12–39). These PSII–LHCII supercomplexes were subjected to biochemical and structural analyses.
Keywords: photosynthesis, photosystem II, PSII–LHCII supercomplex, thylakoids, n-dodecyl-α-d-maltoside, n-dodecyl-β-d-maltoside
1. Introduction
Photosystem II (PSII) uses sunlight to split water into its elemental constituents: dioxygen is released, providing our planet with an aerobic atmosphere, and a protective ozone layer, whereas the ‘hydrogen’ supplies reducing equivalents, which, with additional input of light energy absorbed by photosystem I (PSI), are used to convert carbon dioxide into organic molecules that constitute almost all the global biosphere. The names PSII and PSI were coined as photoreactions II and I by Duysens et al. [1] soon after the concept of the Z-scheme was postulated by Hill & Bendall [2]. Experiments at that time indicated that there was efficient redox coupling between the two photosystems [3] and it seemed inconceivable that they could exist as separate biochemical and structural entities. However, it was the work of Jan Anderson, together with Keith Boardman, that showed this to be the case [4], and it was Jan Anderson who realized the importance of this work in terms of the fluid-mosaic model as applied to the thylakoid membrane [5]. Since then, there has been a steady progress in the development of biochemical procedures to isolate both PSII and PSI. In the case of higher plants, the biochemical procedures developed have built on the recognition that PSII and PSI are laterally separated between granal and stromal regions as demonstrated by Andersson & Anderson [6]. Perhaps the most widely employed exploitation of this lateral separation was in the preparation of Berthold Babcock and Yocum (BBY) particles using Triton X-100 [7]. These particles, derived from the grana partition lamellae, are highly enriched in PSII and therefore have often been the starting point for PSII preparation used for detailed biochemical and structural studies.
The first direct structural information about PSII was derived from electron microscopy (EM) via freeze fracture studies on thylakoid membranes, as reviewed by Staehelin [8], followed by the imaging of isolated PSII particles, as reviewed by Hankamer et al. [9]. However, it was the application of electron crystallography that revealed the position and helix organization of the major subunits of the higher plant PSII dimeric reaction centre core [10–13] and its outer light-harvesting complexes (LHCs) containing chlorophyll (Chl) a and b. In 2001, the electron crystallographic studies of the higher plant PSII core dimer were complemented by X-ray crystallography conducted on PSII isolated from the cyanobacterium, Thermosynechococcus elongatus [14]. X-ray crystallography then provided a slightly more detailed model of PSII from Thermosynechococcus vulcanus [15], followed by a fully refined crystal structure of the T. elongatus PSII at 3.5 Å [16] and later at 3.0 Å/2.9 Å [17,18]. These atomic structures revealed the positioning of all the protein subunits of cyanobacterial PSII and allowed the assignment of all the transmembrane helices, the major cofactors and their protein environments, giving information also about the location of lipids and detergent molecules within the membrane-spanning portion of the dimeric complex. More recently, the crystal structure of PSII isolated from T. vulcanus was refined to 1.9 Å giving very valuable details about the positioning of water molecules and providing more information about the oxygen-evolving centre (OEC) [19]. Complementing these achievements has been the determination of the crystal structures of higher plant PSII proteins in their isolated forms: LHCII at 2.7 Å [20] and 2.5 Å [21], CP29, a LHC-like Chl a/b-binding protein [22], and the extrinsic proteins PsbP [23] and PsbQ [24,25].
The challenge is to extend this degree of structural knowledge to obtain a high-resolution model of the complete PSII complex of higher plants. Indeed, there are a number of significant differences between the PSII of cyanobacteria and that of higher plants and green algae. In higher plant PSII, there are at least two small subunits that are not found in cyanobacteria, the intrinsic PsbW and the extrinsic PsbR. In the case of the OEC, plant PSII has the extrinsic protein PsbO complemented with PsbP and PsbQ [26] instead of PsbU and PsbV as in cyanobacteria. Other differences concern the outer antenna systems. In higher plants, the PSII reaction centre core complex is serviced by intrinsic LHC-binding Chl a and Chl b, whereas in cyanobacteria, this function is carried out by extrinsic phycobilisomes attached to the stromal surface of PSII. This difference in the outer antenna systems suggests significant differences in binding sites for these additional pigment-containing proteins.
Our understanding of how LHC and LHC-like proteins bind to the PSII dimeric core in higher plants and green algae comes mainly from combining single particle analysis of electron microscopic images with appropriate crystallographic data of individual components for interpreting two-dimensional projection maps (see [27] for review) or constructing three-dimensional models [28,29]. Two-dimensional projection maps from negatively stained particles have provided models of a range of PSII particles of which two supercomplexes maintain a twofold symmetry, C2S2M2 and C2S2 as named by Dekker & Boekema [27]. Three-dimensional reconstructions of non-stained particles recorded in vitreous ice have provided a more detailed model of the C2S2 supercomplex [30] using the crystal structure of the cyanobacterial core dimer [16] and the isolated higher plant LHCII [20], PsbP [23] and PsbQ [24]. Clearly, such interpretations and models have serious limitations and uncertainties. Therefore, to fully address the architecture of the PSII in higher plants and to complement functional, biochemical and molecular biological studies of PSII and its OEC proteins, structural information at high resolution is required.
There have been reports of plant PSII preparations that form three-dimensional crystals [31–33], but unfortunately, they did not give X-ray diffractions of sufficient quality for high-resolution analysis. To make progress with this challenge, it is necessary to isolate homogeneous preparations with well-defined protein composition. Several methods have been used to prepare PSII reaction centre cores from higher plants that include multiple steps of membrane solubilization with rather long exposure to different non-ionic detergents (i.e. Triton X-100, n-octyl beta-d-glucopyranoside, β-DM, digitonin) followed by a final isolation step through sucrose density centrifugation [34–38], column chromatography [39–41] or a combination of these two methods [42]. A few of these preparations start from digitonin-solubilized chloroplasts, whereas the majority rely on BBY particles [7]. Such isolated reaction centre cores are usually depleted of LHC proteins. They have provided a minimal system for the study of primary photochemistry and oxygen evolution as well as for structural studies by EM (see [43] for review). Milder detergent treatment with α-DM and β-DM directly on isolated thylakoids was reported by Eshaghi et al. [44], Dekker et al. [45] and Bumba et al. [46] yielding a variety of different oligomeric forms of PSII: monomeric PSII and different-sized supramolecular LHCII–PSII structures, including the C2S2M2 and C2S2 supercomplexes. More recently, Caffarri et al. [47] have used α-DM to solubilize BBY preparations from wild-type and LHC mutants of Arabidopsis to obtain a wide range of PSII–LHCII supercomplexes separated by sucrose density centrifugation. Among this range were the twofold symmetrical C2S2M2 and C2S2 supercomplexes.
In this paper, we describe an efficient one-step method, avoiding a BBY intermediate preparation, to obtain in relatively high yields homogenous preparations of the C2S2M2 and C2S2 supercomplexes, as a starting point for high-resolution structural studies. Isolated thylakoid membranes of peas grown under controlled conditions were subjected to a single very short and mild solubilization step, using low concentration of either α-DM or β-DM in combination with sucrose density gradient centrifugation. The spectral and biochemical properties of both supercomplexes have been investigated, and the initial characterizations of their structures conducted by single particle analysis of negatively stained particles.
2. Material and methods
(a). Plant growth conditions and thylakoid membranes isolation
Before sowing, pea (Pisum sativum L., var. Palladio nano) seeds were treated as described in Pagliano et al. [48]. Germinated seedlings were transferred to pots and grown hydroponically in Long Ashton nutrient solution [49] in a growth chamber with 8 h daylight, 20°C, 60 per cent humidity and 150 µmol m−2 s−1 photons. Leaves from plants grown for three weeks were harvested and thylakoid membranes isolated according to Pagliano et al. [50] and finally stored in 25 mM 2-(N-morpholino)ethanesulfonic acid (MES), pH 6.0, 10 mM NaCl, 5 mM MgCl2 and 2 M glycine betaine (buffer A) at a Chl concentration in excess of 1 mg ml−1.
(b). Isolation of PSII–LHCII supercomplexes
Thylakoid membranes having a Chl concentration of 1 mg ml−1 in buffer A were treated with 50 mM α-DM or β-DM for 1 min at 4°C in the dark. Phenylmethylsulphonylfluoride (500 μM) was present during the solubilization to inhibit protease activity. After a short centrifugation at 21 000g for 10 min at 4°C, 700 μl of the supernatants was added to the top of linear sucrose gradients, prepared by freezing and thawing ultracentrifuge tubes filled with a buffer made of 0.65 M sucrose, 25 mM MES pH 5.7, 10 mM NaCl, 5 mM CaCl2 and 0.03 per cent α-DM or β-DM. Centrifugation was carried out at 100 000g for 12 h at 4°C (Surespin 630 rotor, Thermo Scientific). Sucrose bands, containing PSII-LHCII particles, were carefully removed using a syringe and, if necessary, concentrated by membrane filtration with Amicon Ultra 100 kDa cut-off devices (Millipore) and then stored at −80°C.
(c). Spectroscopic analyses
The Chl concentration was determined after extraction in 80 per cent acetone according to Arnon [51]. Absorption spectra were recorded, using a Lambda25 spectrophotometer (Perkin Elmer). When dilution was necessary, the same buffers as for the gradients were used. Low temperature (77 K) fluorescence emission spectra were registered by a FL55 spectrofluorometer (Perkin Elmer), equipped with a red sensitive photomultiplier and a low temperature cuvette holder. Samples were excited at 436 nm. The spectral bandwidth was 7.5 nm (excitation) and 5.5 nm (emission). The Chl concentration was approximately 0.5 μg ml−1 in 90 per cent (v/v) glycerol/sucrose gradient buffers.
(d). PSII–LHCII supercomplexes biochemical characterization
Sodium dodecylsulfate–polyacrylamide gel electrophoresis (SDS–PAGE) was carried out using either the Laemmli's system [52] with a 12.5 per cent acrylamide resolving gel containing 5 M urea or the Tris–Tricine system [53] with a 16 per cent acrylamide resolving gel containing 6 M urea. Pre-stained protein size markers (Precision plus, Bio-Rad) were used for the estimation of apparent size of PSII–LHCII components. The separated proteins were either stained by Coomassie brilliant blue R-250 or transferred onto nitro-cellulose membrane and immuno-detected with specific antisera against LHCII subunits and PsbO, PsbP, PsbR and PsbW polypeptides, by using the alkaline phosphatase conjugate method, with 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium as chromogenic substrates (Sigma).
The purity of the PSII-LHCII preparations was analysed by size-exclusion chromatography on a Jasco high performance liquid chromatography (HPLC) system with a BioSep-SEC-S 3000 (Phenomenex) column. The 20-μl samples injected contained 6 µg of Chl and profiles were monitored at 280 nm. The mobile phase consisted of 20 mM MES pH 6.5, 10 mM MgCl2, 30 mM CaCl2, 0.5 M mannitol, 0.03 per cent α-DM or β-DM and was passed through the column at a flow rate of 1 ml min−1.
(e). Oxygen evolution measurements
The oxygen evolution was measured at 20°C using a Clark-type oxygen electrode (Hansatech) under saturating light intensity (≈1000 and 5000 μmol m−2 s−1 photons, respectively, for thylakoids and PSII-LHCII particles). Chl of thylakoids (10 μg) and 25 or 20 μg Chl of PSII-LHCII particles isolated, respectively, with α-DM or β-DM were added to 1 ml of a medium made of 25 mM MES pH 6.5, 2 M glycine betaine, 10 mM NaHCO3, 10 mM NaCl, in the presence or absence of 25 mM CaCl2. A mixture of 1 mM potassium ferricyanide (K3Fe(CN)6) and 200 μM 2,6-dichlorobenzoquinone (DCBQ) or 500 μM DCBQ alone were used as electron acceptors for thylakoids and PSII–LHCII particles, respectively.
(f). Electron microscopy and image analyses
Samples from sucrose gradient bands were diluted 10-fold in a buffer composed of 10 mM HEPES pH 7.5 and stained with 2 per cent uranyl acetate on glow discharged carbon-coated copper grids. EM was performed with a Phillips CM200FEG electron microscope operated at 200 kV. The images were recorded at 50 000× on a TVIPS F415 4Kx4K CCD camera leading to a final pixel size of 1.76 Å at specimen level. Single particle analysis was performed using EMAN2 [54] and IMAGIC [55] softwares, including initial reference-free alignment, followed by multireference alignment and multivariate statistical analysis and classification, as described in van Heel et al. [56].
The modelling studies of the two-dimensional projection maps were conducted with X-ray structures of the PSII core of cyanobacteria at 1.9 Å [19], the LHCII complex of pea at 2.5 Å [21] and the CP29 of spinach at 2.8 Å [22] (PDB accession numbers: 3ARC, 2BHW and 3PL9, respectively) using UCSF Chimera [57].
3. Results
(a). Isolation and characterization of differently sized PSII–LHCII supercomplexes
To obtain homogenous preparation of highly active PSII–LHCII supercomplexes, we optimized the conditions for direct solubilization of thylakoid membranes in the presence of salts. Previously, we had found that using a wide range of Chl/DM ratios to access the partition regions of stacked pea thylakoids resulted in PSII particles of different complexities that could be separated on a sucrose density gradient at low pH containing salts [50]. From this work, we identified a short-time single step with α-DM and β-DM, using a Chl/DM ratio of 1 : 50 as being effective for isolating complexes containing LHCII and PSII proteins.
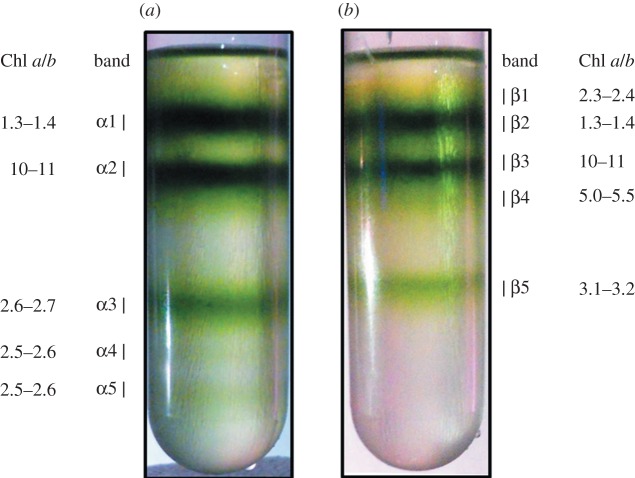
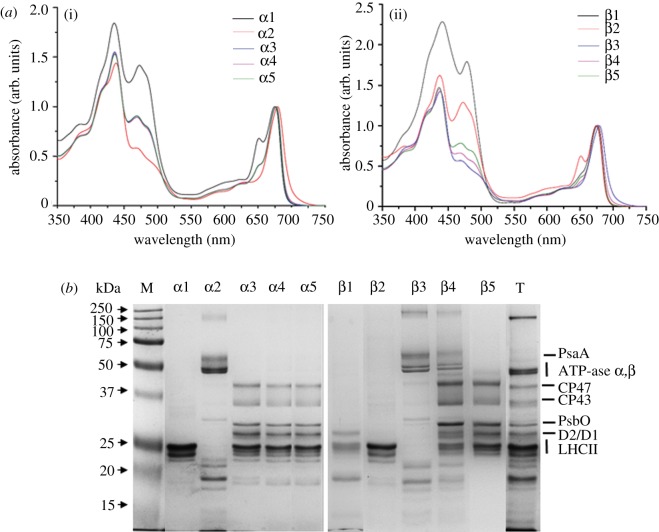
Figure 1 shows the sucrose density profile after treating isolated pea thylakoids with α-DM and β-DM at a Chl/DM ratio of 1 : 50 followed by a relatively quick (12 h) ultracentrifugation. Five different green bands were separated either with α-DM (figure 1a) or with β-DM (figure 1b). All bands were characterized by their Chl a/b ratio (figure 1), absorption spectra (figure 2a) and protein composition (figure 2b). These results indicated that: (i) bands α1 and β2 contained mainly trimeric LHCII subunits; (ii) bands α2 and β3 were composed of PSI–LHCI complexes co-migrating with ATP-ase; (iii) a monomeric LHC/Chl b-enriched fraction was separated only by using β-DM, corresponding to band β1; (iv) lower bands (α3–5 and β4–5) contained PSII and LHCII proteins. Chls b, bound to Lhcb (polypeptides of LHCII), show two main peaks around 470 and 650 nm, whereas Chls a in the Lhcb and PSII proteins are responsible for the absorption around 435 and 675 nm. The relatively high intensity of the absorption in the Chl b region in bands α3–5 and β5 together with the Chl a/b ratios ranging between 2.5 and 3.2 is consistent with previous studies conducted on PSII–LHCII supercomplexes isolated either from BBYs [37,47] or directly from thylakoid membranes [44]. Therefore, we conclude that these bands are likely to contain PSII supercomplexes with a higher level of Chl b in those derived with α-DM (Chl a/b ratio of 2.5–2.6) compared with β-DM (3.1–3.2). Instead, in band β4, there was a high level of PSII cores subunits, but its LHCII content was lower than in band β5, and there was a contamination by PSI and LHCI polypeptides, as detected by SDS–PAGE (figure 2b), accounting for its higher Chl a/b ratio (5.0–5.5).
Figure 1.
Separation of pigment-binding complexes by sucrose gradient ultracentrifugation of thylakoids solubilized with (a) α-DM and (b) β-DM.
Figure 2.
(a) Absorption spectra of sucrose gradient bands α1–α5 obtained from thylakoids solubilized with α-DM (i) and bands β1–β5 obtained from thylakoids solubilized with β-DM (ii). The spectra are normalized to the maximum in the red region. α3, α4 and α5 are almost superimposed. The Chl b content, which is proportional to the antenna content, is deducible from the intensity of the bands at 470 and 650 nm. (b) Protein composition of sucrose gradient bands α1–α5 isolated from thylakoids solubilized with α-DM, of bands β1–β5 isolated from thylakoids solubilized with β-DM and of thylakoid membranes (lane T). Chl (4 µg) was loaded on each lane. On the left-hand side, pre-stained protein markers with indicated apparent molecular weight (kDa).
(b). Biochemical characterization of two predominant forms of PSII–LHCII supercomplexes
The aim of our work was to develop a (reproducible) method of isolating abundant, highly pure and intact PSII–LHCII supercomplexes for structural studies. For this reason, we focused our biochemical characterization only on the two strongest sucrose gradient bands, α3 and β5, referred hereafter to as α and β, respectively.
Absorption spectra and 77 K fluorescence emission spectra of bands α and β are presented respectively in figure 3a,b. The α particles showed absorption peaks at 677 and 436 nm and a single fluorescence emission peak at 684 nm, whereas the β particle had its maxima absorption peaks at 676 and 436 nm and a single fluorescence emission peak at 682 nm. From a comparison of the absorption spectra in the region around 650 nm, it was evident that the particles isolated with α-DM have a higher Lhcb content compared with β particles, which is in accordance with the Chl a/b ratios given in figure 1. The absence of any 77 K fluorescence peaks at 735 nm, corresponding to emission of long-wavelength PSI antennae, in both emission spectra (figure 3b) confirmed the absence of any PSI contamination in the PSII–LHCII supercomplexes isolated with α- and β-DM.
Figure 3.
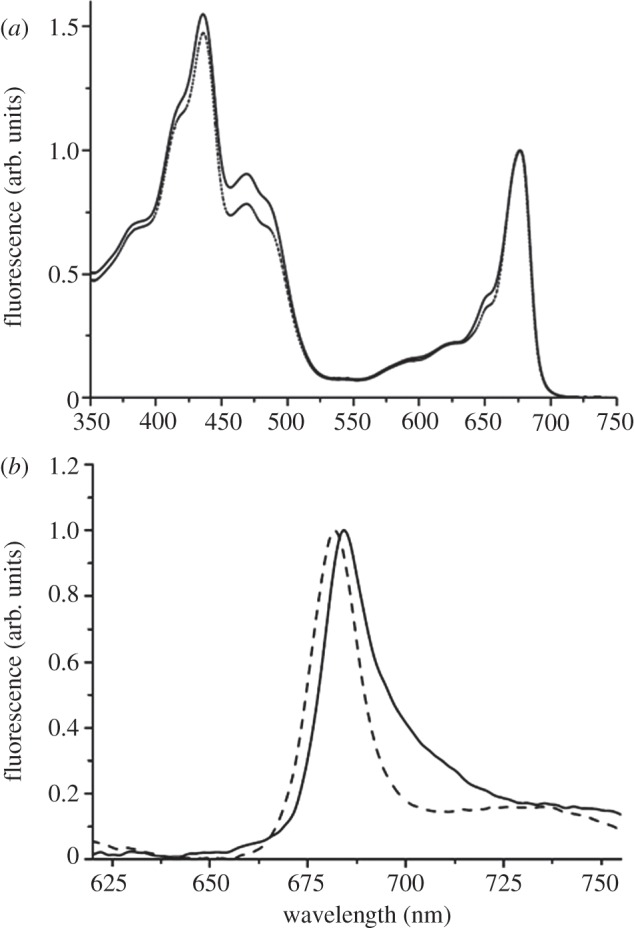
(a) Absorption spectra of α particles (solid line) and β particles (dashed line). The spectra are normalized to the maximum in the red region. (b) Low temperature (77 K) fluorescence emission spectra of α particles (solid line) and β particles (dashed line). Spectra are normalized to the maximum value.
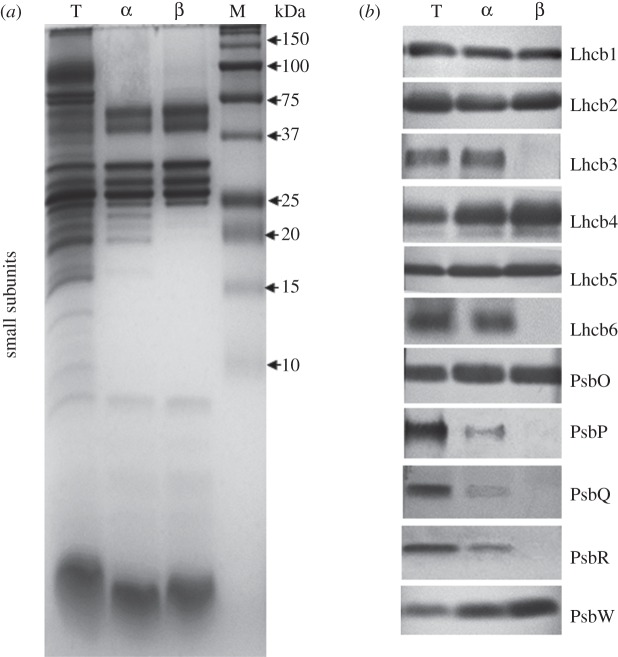
The overall purity, as well as the complete protein composition of the two particles, was further explored by using the Tris–Tricine SDS–PAGE system [53] loading the same amount of total Chl for each sample on the gel (figure 4a). In addition to the presence of a number of small PSII core subunits in the region below 10 kDa common to both samples, this electrophoretic system revealed clearly additional polypeptides in the region between 25 and 15 kDa only in the α particle. Western blot analyses based on the same Chl loading (figure 4b) showed that: (i) both particles displayed almost the same quantity of the antennae polypeptides Lhcb1 and Lhcb5 and of the extrinsic subunit PsbO; (ii) the amount of Lhcb2 was slightly higher in the particle isolated with β-DM and the same was even more evident in the case of Lhcb4; (iii) the Lhcb3 and Lhcb6 polypeptides were present and in high amounts only in the α particle; (iv) low amounts of the extrinsic subunits PsbP, PsbQ and PsbR were detected only in the particle isolated with α-DM; and (v) the small subunit PsbW, present in both α and β particles, was more abundant in the particle isolated with β-DM, on an equal Chl basis.
Figure 4.
(a) Protein composition of α particles (α) and β particles (β) and of pea thylakoids (lane T). Chl (8 µg) was loaded on each lane. Lane M, pre-stained protein markers with their apparent molecular weight (kDa) indicated. (b) Western blot with antibodies against antenna polypeptides Lhcb1–6, OEC subunits PsbO, PsbP, PsbQ and PsbR and PsbW of α particles (α) and β particles (β) and of pea thylakoids (lane T). Chl (1 µg) was loaded on each lane.
The purity and relative molecular masses of the α and β particles were checked by size-exclusion chromatography. From the HPLC profiles shown in figure 5, a single distinct peak for both types of particles was detected, suggesting homogeneous preparations in both cases. The longer eluting time displayed by the β particle (5.6 min) with respect to the α (5.3 min) indicates that the latter is larger than the former.
Figure 5.
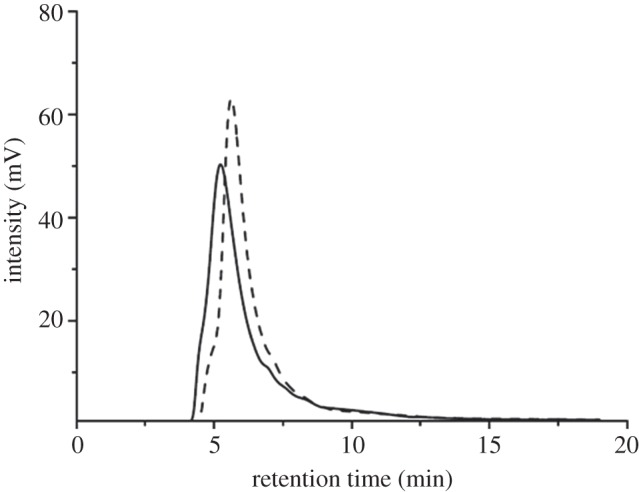
Size-exclusion profiles recorded at 280 nm of α particles (solid line) and β particles (dashed line). Chl (6 µg) was applied for injection of each sample.
(c). Activity measurements
Both α and β particles were assayed for their ability to evolve oxygen. Triplicate measurements of oxygen evolution in the presence of an optimal concentration of Ca2+ and Cl− (25 mM CaCl2) showed that the particle isolated with α-DM gave rates of 478 ± 6 µmol O2 mg Chl−1 h−1, whereas its counterpart isolated with β-DM reached values of 591 ± 20 µmol O2 mg Chl−1 h−1 (table 1). When compared on a Chl basis, the higher rates displayed by the β particle can reflect its higher amount of reaction centre with respect to the α particle, which instead is characterized by a larger-sized antenna system.
Table 1.
Oxygen evolution measurements on the α and β particles and the starting thylakoid membranes performed in the presence and absence of 25 mM CaCl2 in the assay medium. The oxygen evolution rates are average ± s.d. of triplicate measurements of at least three independent experiments.
| µmol O2 mg Chl−1 h−1 |
||
|---|---|---|
| +CaCl2 | −CaCl2 | |
| thylakoids | 189 ± 8 | 175 ± 12 |
| α particle | 478 ± 6 | 475 ± 12 |
| β particle | 591 ± 20 | 437 ± 16 |
It is known that the reduction in oxygen-evolving capacity of PSII, due to the removal of the PsbP extrinsic subunit, can be restored by addition of calcium [58,59] and chloride ions [60]. The complex isolated with α-DM, which was found to bind a certain amount of the extrinsic subunit PsbP, did not significantly change its rate of oxygen evolution in the absence of CaCl2, whereas its counterpart isolated with β-DM in the same ionic conditions showed a reduction of about 26 per cent (table 1). Thus, in the case of the α particle, the absence of a significant stimulation by addition of 25 mM CaCl2 suggests that the PsbP subunit is present in a functionally active manner and that the OEC is in a more intact state than in the β particle.
(d). Determination of the supramolecular structure of the PSII–LHCII supercomplexes
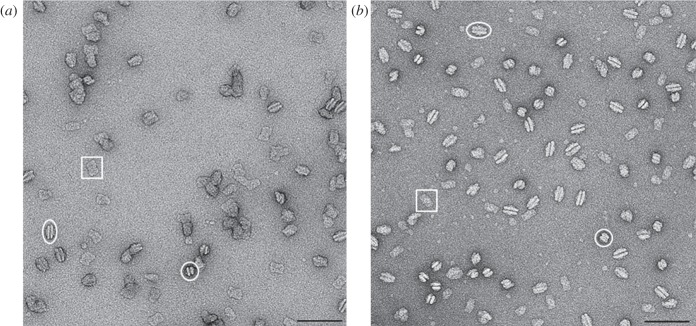
Electron micrographs of the negatively stained α and β particles (figure 6) revealed the high purity of the two preparations and confirmed that the isolated complexes are LHCII-PSII supercomplexes. From a total of 600 images for each preparation, a set of 15 563 projections for the α particles and 26 340 for the β particles were analysed, respectively. From these analyses, it was evident that for both particles a preferential orientation in side-view projections (as paired PSII–LHCII supercomplexes) and in top-view projections was present. Moreover, a higher number of side-view projections characterized the β particles, whereas similar amounts of projections in slightly tilted oblique views were observed in both particles.
Figure 6.
Electron micrographs of (a) α particles and (b) β particles obtained by sucrose density gradient, negatively stained with 2% uranyl acetate. Top-view, side-view and end-view projections of the α and β particles are indicated by white rectangles, ovals and circles, respectively. Scale bar, 100 nm.
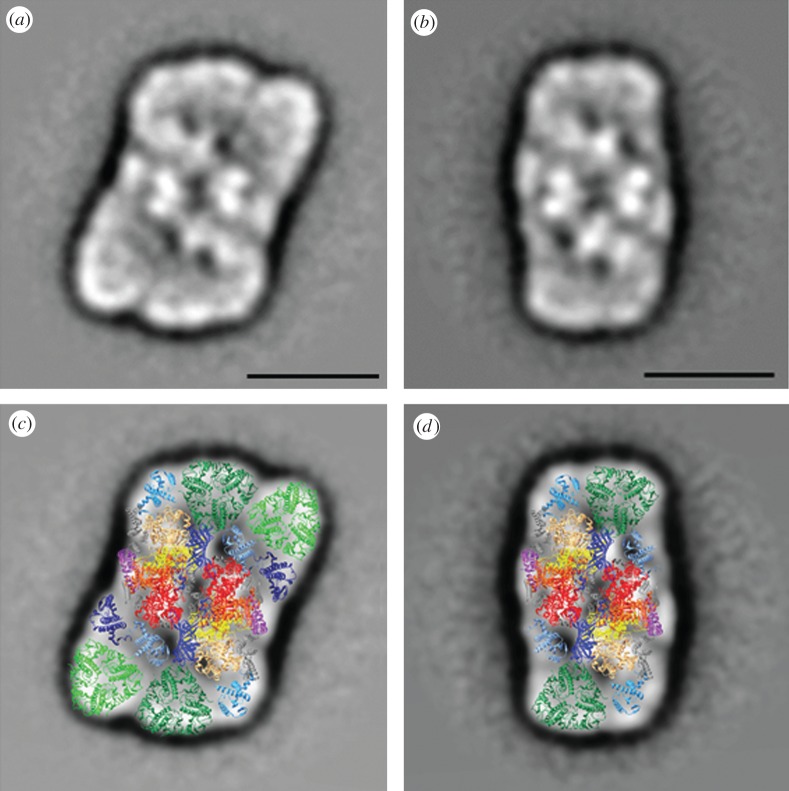
In order to investigate the supramolecular organization of PSII and its associated light-harvesting antenna, a total number of 6197 single particle top-views from the α dataset and 6985 top-views from the β dataset were extracted from the EM images. The two datasets were separately processed and final results after alignment, multivariate statistical analysis and classification showed that the α particle is mainly composed of C2S2M2 supercomplexes, and the β particle is mainly composed of C2S2 supercomplexes. As the most abundant PSII–LHCII supercomplexes in the α and β preparations showed respectively a C2S2M2 and a C2S2 organization, homogeneous classes of their corresponding top-view projections (1049 for α and 1765 for β) were summed to give the final two-dimensional projection maps shown in figure 7a,b. Despite uncertainties in these projection maps, the assignment of the main subunits of the supercomplexes can be made by overlaying the projections of X-ray structures of individual components as previously shown [30,47]. Here, we have used the cyanobacterial PSII core at 1.9 Å [19], the trimeric LHCII complex of pea at 2.5 Å [21] and the monomeric Lhcb protein CP29 of spinach at 2.8 Å [22]. The latter has been used as a template for CP24 and CP26, as well as for CP29. The fitting of the X-ray data into the electron density of the projection maps of the α and β particles was essentially the same as that adopted by Caffarri et al. [47], with the exception that CP24 and CP26 were modelled at a slightly different orientation, based on density distribution. The final projection maps of the C2S2M2 and the C2S2 supercomplexes with the assignment of their main PSII core and antenna subunits are shown in figure 7c,d.
Figure 7.
(a) Final projection maps of the C2S2M2 supercomplex isolated with α-DM and (b) the C2S2 supercomplex isolated with β-DM. The scale bar equals 150 Å. (c) Assignment of the subunits in the C2S2M2 and (d) C2S2 supercomplexes by fitting the high-resolution structures of the PSII core [19] (subunits D1, D2, CP47, CP43 and PsbO are in yellow, orange, red, sandy brown and blue, respectively), the LHCII [21] (S-trimer is in dark green and M-trimer in light green) and the monomeric Lhcb [22] (CP29, CP26 and CP24 are in steel blue, turquoise and dark blue, respectively).
4. Discussion
Progress in the determination of the structure of PSII isolated from cyanobacteria has advanced in a most striking way over the past 10 years culminating in an X-ray-derived model at 1.9 Å [19]. This advance has not been mirrored for PSII isolated from higher plants. Despite reports that crystals of higher plant PSII core preparations have been grown, they showed poor X-ray diffraction [31–33]. However, there is every reason to believe that most of the details revealed by the cyanobacterial crystal structures will be the same for the PSII core of higher plants. For example, all the key amino acids of the D1, D2 and CP43 proteins, which are intimate with the Mn4Ca-cluster of the OEC, are conserved between higher plant and cyanobacterial PSII [61]. Similarly, the amino acids that act as ligands for cofactors involved in energy capture and charge separation are conserved. The differences between the two systems relate to the OEC extrinsic proteins, the presence of additional core proteins in higher plant PSII and in their outer light-harvesting systems. In the latter case, the phycobiliproteins serve as an extrinsic located outer light-harvesting system for cyanobacteria, whereas the Chl a/b-binding Lhcb proteins are an intrinsic outer antenna system for higher plant PSII. In both cases, there is no high-resolution data to reveal how these outer antenna systems structurally interact with the PSII core. In the case of plants, this information is vital for the interpretation of long-range energy-transfer processes and, in particular, to give a detailed explanation to the phenomenon of non-photochemical quenching [62]. For this purpose alone, a high-resolution structure of higher plant PSII is urgently needed. Such a challenge requires careful biochemistry to obtain homogeneous and stable preparations of PSII–LHCII supercomplexes.
Previous studies [37,44,46,47,63] have identified DM as an effective mild detergent for obtaining preparations of discrete PSII–LHCII supercomplexes. DM is a glucoside-based surfactant with a bulky hydrophilic head group composed of two sugar rings and a non-charged alkyl glycoside chain. Two isomers of this molecule exist, differing only in the configuration of the alkyl chain around the anomeric centre of the carbohydrate head group, axial in α-DM and equatorial in β-DM. We have compared the use of α-DM and β-DM for the isolation of supramolecular complexes of PSII by a single-step solubilization of stacked thylakoid membranes isolated from peas. We chose: (i) to solubilize directly thylakoids instead of PSII membranes (BBYs), in order to minimize the possibility of detergent-induced artefacts; (ii) to use stacked membranes in order to avoid destabilization of the interaction of LHCII with the PSII core. As a result, we optimized conditions to obtain homogeneous preparations of the C2S2 and C2S2M2 supercomplexes as the main product. In agreement with previous works [64,65], our biochemical analyses of these two types of LHCII-PSII supercomplexes showed that unlike C2S2, the C2S2M2 supercomplex contained both the Lhcb3 and Lhcb6 antennae subunits, crucial polypeptides for the assembly and the macro organization of this supercomplex in the thylakoid membrane of higher plants [47]. Moreover, the C2S2M2 supercomplex contained detectable levels of PsbP, PsbQ and PsbR compared with the C2S2, although both contained the extrinsic PsbO polypeptide and the small intrinsic subunit PsbW. The reason for this is almost certainly due to the milder nature of α-DM compared with β-DM and is unlikely to have any physiological significance. However, Caffarri et al. [47] concluded that PsbQ can bind to PSII cores only in the presence of peripheral antennae subunits (at least the domain composed of CP26/LHCII S-trimer) and PsbP binds to the supercomplex only in the presence of CP29/CP24/LHCII M-trimer and is absent from PSII cores. This last conclusion has been challenged by Pagliano et al. [66].
In all other respects, the protein compositions of the two supercomplexes are the same as expected, given that C2S2 forms the inner PSII–LHCII core to C2S2M2. This structural relationship was suggested previously [67] and confirmed by our analyses of α and β particles by negative stain EM and interpretation of density within the top-view projections using currently available X-ray data.
We identify density for two types of LHCII trimers designated as S and M, where S, common to both supercomplexes, consists of Lhcb1 and Lhcb2 proteins, whereas M, exclusive to the α particle, is a LHCII trimer containing the Lhcb3 protein. These results are in accordance with previous findings suggesting that Lhcb3 is a subunit of the M-trimer [68] on the basis that it is present in the C2S2M1–2 supercomplexes but absent in the C2S2 supercomplex [37]. Additionally, Lhcb3 is frequently found in a Lhcb6–Lhcb4–LHCII trimer subcomplex [69,70]. Moreover, it had previously been shown that the C2S2 supercomplex contained the Lhcb4 and Lhcb5 and not the Lhcb6 subunit [28,30]. By taking this information into account, we interpret the projection maps of the α and β particles in the same way as Dekker & Boekema [27] and conclude that Lhcb4 is likely to be located close to the M-trimer in C2S2M2 and linked to it via Lhcb6.
In modelling the organization of the subunits within the supercomplex, we used the latest X-ray data for the PSII dimeric core [19], LHCII trimer [21] and CP29 [22], where the latter was also used for modelling CP26 and CP24. As we observed similar features in the projections as Caffarri et al. [47], we followed essentially the same fitting of the crystal structures into the electron density as they did, except for small rotational adjustments of CP24 and CP26, although the reliability of the fitting must be treated with caution: the protein is stained with uranyl acetate, the spatial resolution is limiting and the problem of fitting three-dimensional crystal structures into a two-dimensional projection map is always somewhat tentative.
In terms of chlorophyll content, we can conclude that C2S2M2 binds approximately a total of 316 Chls per OEC (214 Chl a and 102 Chl b, giving a Chl a/b ratio of 2.1), whereas C2S2 contains a total of 206 Chls per OEC (150 Chl a and 56 Chl b, giving a Chl a/b ratio of 2.7). With these assumptions, the oxygen evolution rates for the two types of supercomplexes can be compared on an equal Chl basis. The maximum rates with CaCl2 added were 478 ± 6 and 591 ± 20 μmol O2 mg Chl−1 h−1 for the C2S2M2 and C2S2 particles, respectively, giving a ratio of 1.27 in favour of C2S2 supercomplexes. However, because C2S2M2 contains approximately 110 more Chls per OEC than C2S2, the ratio of the maximum rates per OEC become 1.24 in favour of C2S2M2. From this difference and the fact that the C2S2M2 preparation showed only a slight dependence of the addition of CaCl2 and contained detectable levels of PsbP, PsbQ and PsbR, we can conclude that this supercomplex is more catalytically competent than the smaller C2S2 supercomplex. Nevertheless, both preparations offer an opportunity for further structural analyses by cryo-EM and open up a new direction for crystallography.
Acknowledgements
The authors kindly thank Dr Tomas Morosinotto (University of Padova, Italy) for supplying the antibody against Lhcb6 and Taha Shahid (Imperial College London, UK) for the initial technical support to C.P. and T.P. with the softwares EMAN2 and IMAGIC. This work was supported by the European Commission (SOLHYDROMICS project (227192), FP-7-Energy-2008-FET).
References
- 1.Duysens L. N. M., Amesz J., Kamp B. M. 1961. Two photochemical systems in photosynthesis. Nature 190, 510–511 10.1038/190510a0 (doi:10.1038/190510a0) [DOI] [PubMed] [Google Scholar]
- 2.Hill R., Bendall F. 1960. Function of the two cytochrome components in chloroplasts: a working hypothesis. Nature 186, 136–137 10.1038/186136a0 (doi:10.1038/186136a0) [DOI] [Google Scholar]
- 3.Duysens L. N. M., Amesz J. 1962. Function and identification of two photochemical systems in photosynthesis. Biochim. Biophys. Acta 64, 243–260 10.1016/0006-3002(62)90735-7 (doi:10.1016/0006-3002(62)90735-7) [DOI] [Google Scholar]
- 4.Boardman N. K., Anderson J. M. 1964. Isolation from spinach chloroplasts of particles containing different proportions of chlorophyll a and chlorophyll b and their possible role in the light reactions of photosynthesis. Nature 203, 166–167 10.1038/203166a0 (doi:10.1038/203166a0) [DOI] [Google Scholar]
- 5.Anderson J. M. 1975. The molecular organization of chloroplast thylakoids. Biochim. Biophys. Acta 416, 191–235 10.1016/0304-4173(75)90007-5 (doi:10.1016/0304-4173(75)90007-5) [DOI] [PubMed] [Google Scholar]
- 6.Andersson B., Anderson J. M. 1980. Lateral heterogeneity in the distribution of chlorophyll-protein complexes of the thylakoid membranes of spinach chloroplasts. Biochim. Biophys. Acta 593, 427–440 10.1016/0005-2728(80)90078-X (doi:10.1016/0005-2728(80)90078-X) [DOI] [PubMed] [Google Scholar]
- 7.Berthold D. A., Babcock G. T., Yocum C. F. 1981. A highly resolved, oxygen evolving photosystem II preparation from spinach thylakoid membranes. FEBS Lett. 134, 231–234 10.1016/0014-5793(81)80608-4 (doi:10.1016/0014-5793(81)80608-4) [DOI] [Google Scholar]
- 8.Staehelin A. 2003. Chloroplast structure: from chlorophyll granules to supra-molecular architecture of thylakoid membranes. Photosynth. Res. 76, 185–196 10.1023/A:1024994525586 (doi:10.1023/A:1024994525586) [DOI] [PubMed] [Google Scholar]
- 9.Hankamer B., Barber J., Boekema E. J. 1997. Structure and membrane organization of photosystem II in green plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 641–671 10.1146/annurev.arplant.48.1.641 (doi:10.1146/annurev.arplant.48.1.641) [DOI] [PubMed] [Google Scholar]
- 10.Rhee K. H., Morris E. P., Zheleva D., Hankamer B., Kühlbrandt W., Barber J. 1997. Two-dimensional structure of plant photosystem II at 8 Å resolution. Nature 389, 522–526 10.1038/39103 (doi:10.1038/39103) [DOI] [PubMed] [Google Scholar]
- 11.Rhee K. H., Morris E. P., Barber J., Kühlbrandt W. 1998. Three-dimensional structure of the plant photosystem II reaction centre at 8 Å resolution. Nature 396, 283–286 10.1038/24421 (doi:10.1038/24421) [DOI] [PubMed] [Google Scholar]
- 12.Hankamer B., Morris E. P., Barber J. 1999. Revealing the structure of the oxygen evolving core dimer of photosystem II by cryoelectron crystallography. Nat. Struct. Biol. 6, 560–564 10.1038/9341 (doi:10.1038/9341) [DOI] [PubMed] [Google Scholar]
- 13.Hankamer B., Morris E. P., Nield J., Gerle C., Barber J. 2001. Three dimensional structure of the photosystem II core dimer of higher plants determined by electron microscopy. J. Struct. Biol. 135, 262–269 10.1006/jsbi.2001.4405 (doi:10.1006/jsbi.2001.4405) [DOI] [PubMed] [Google Scholar]
- 14.Zouni A., Witt H. T., Kern J., Fromme P., Krauss N., Saenger W., Orth P. 2001. Crystal structure of photosystem II from Synechococcus elongatus at 3.8 Å resolution. Nature 409, 739–743 10.1038/35055589 (doi:10.1038/35055589) [DOI] [PubMed] [Google Scholar]
- 15.Kamiya N., Shen J. R. 2003. Crystal structure of oxygen-evolving photosystem II from Thermosynechococcus vulcanus at 3.7 Å resolution. Proc. Natl Acad. Sci. USA 100, 98–103 10.1073/pnas.0135651100 (doi:10.1073/pnas.0135651100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferreira K. N., Iverson T. M., Maghlaoui K., Barber J., Iwata S. 2004. Architecture of the photosynthetic oxygen evolving center. Science 303, 1831–1838 10.1126/science.1093087 (doi:10.1126/science.1093087) [DOI] [PubMed] [Google Scholar]
- 17.Loll B., Kern J., Saenger W., Zouni A., Biesiadka J. 2005. Towards complete cofactor arrangement in the 3.0 Å resolution structure of photosystem II. Nature 438, 1040–1044 10.1038/nature04224 (doi:10.1038/nature04224) [DOI] [PubMed] [Google Scholar]
- 18.Guskov A., Kern J., Gabdulkhakov A., Broser M., Zouni A., Saenger W. 2009. Cyanobacterial photosystem II at 2.9 Å resolution and the role of quinones, lipids, channels and chloride. Nat. Struct. Mol. Biol. 16, 334–342 10.1038/nsmb.1559 (doi:10.1038/nsmb.1559) [DOI] [PubMed] [Google Scholar]
- 19.Umena Y., Kawakami K., Shen J. R., Kamiya N. 2011. Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature 473, 55–60 10.1038/nature09913 (doi:10.1038/nature09913) [DOI] [PubMed] [Google Scholar]
- 20.Liu Z. F., Yan H. C., Wang K. B., Kuang T. Y., Zhang J. P., Gui L. L., An X. M., Chang W. R. 2004. Crystal structure of spinach major light-harvesting complex at 2.72 Å resolution. Nature 428, 287–292 10.1038/nature02373 (doi:10.1038/nature02373) [DOI] [PubMed] [Google Scholar]
- 21.Standfuss J., van Scheltinga A. C. T., Lamborghini M., Kühlbrandt W. 2005. Mechanisms of photoprotection and nonphotochemical quenching in pea light harvesting complex at 2.5 Å resolution. EMBO J. 24, 918–928 10.1038/sj.emboj.7600585 (doi:10.1038/sj.emboj.7600585) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pan X., Li M., Wan T., Wang L., Jia C., Hou Z., Zhao X., Zhang J., Chang W. 2011. Structural insights into energy regulation of light-harvesting complex CP29 from spinach. Nat. Struct. Mol. Biol. 18, 309–316 10.1038/nsmb.2008 (doi:10.1038/nsmb.2008) [DOI] [PubMed] [Google Scholar]
- 23.Ifuku K., Nakatsu T., Kato H., Sato F. 2004. Crystal structure of the PsbP protein of photosystem II from Nicotiana tabacum. EMBO Rep. 5, 362–367 10.1038/sj.embor.7400113 (doi:10.1038/sj.embor.7400113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calderone V., Trabucco M., Vujicic A., Battistutta R., Giacometti G. M., Andreucci F., Barbato R., Zanotti G. 2003. Crystal structure of the PsbQ protein of photosystem II from higher plants. EMBO Rep. 4, 900–905 10.1038/sj.embor.embor923 (doi:10.1038/sj.embor.embor923) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Balsera M., Arellano J. B., Revuelta J. L., De Las Rivas J., Hermoso J. A. 2005. The 1.49 Å resolution crystal structure of PsbQ from photosystem II of Spinacia oleracea reveals a PPII structure in the N-terminal region. J. Mol. Biol. 350, 1051–1060 10.1016/j.jmb.2005.05.044 (doi:10.1016/j.jmb.2005.05.044) [DOI] [PubMed] [Google Scholar]
- 26.Bricker T. M., Burnap R. L. 2005. The extrinsic proteins of photosystem II. In Photosystem II: the water/plastoquinone oxido-reductase of photosynthesis (eds Wydrzynski T., Satoh K.), pp. 95–120 Dordrecht, The Netherlands: Springer [Google Scholar]
- 27.Dekker J. P., Boekema E. J. 2005. Supramolecular organization of thylakoid membrane proteins in green plants. Biochim. Biophys. Acta 1706, 12–39 10.1016/j.bbabio.2004.09.009 (doi:10.1016/j.bbabio.2004.09.009) [DOI] [PubMed] [Google Scholar]
- 28.Nield J., Orlova E. V., Morris E. P., Gowen B., van Heel M., Barber J. 2000. 3D map of the plant photosystem II supercomplex obtained by cryoelectron microscopy and single particle analysis. Nat. Struct. Biol. 7, 44–47 10.1038/71242 (doi:10.1038/71242) [DOI] [PubMed] [Google Scholar]
- 29.Nield J., Balsera M., De Las Rivas J., Barber J. 2002. Three-dimensional electron cryo-microscopy study of the extrinsic domains of the oxygen-evolving complex of spinach: assignment of the PsbO protein. J. Biol. Chem. 277, 15 006–15 012 10.1074/jbc.M110549200 (doi:10.1074/jbc.M110549200) [DOI] [PubMed] [Google Scholar]
- 30.Nield J., Barber J. 2006. Refinement of the structural model for the photosystem II supercomplex of higher plants. Biochim. Biophys. Acta 1757, 353–361 10.1016/j.bbabio.2006.03.019 (doi:10.1016/j.bbabio.2006.03.019) [DOI] [PubMed] [Google Scholar]
- 31.Adir N., Okamura M. Y., Feher G. 1992. Crystallization of the PSII-reaction centre. In Research in photosynthesis, vol. 2 (ed. Murata N.), pp. 5195–5198 Dordrecht, The Netherlands: Kluwer Academic Publishers [Google Scholar]
- 32.Fotinou C., Kokkinidis M., Fritzsch G., Haase W., Michel H., Ghanotakis D. 1993. Characterization of a photosystem II core and its three-dimensional crystals. Photosynth. Res. 37, 41–48 10.1007/BF02185437 (doi:10.1007/BF02185437) [DOI] [PubMed] [Google Scholar]
- 33.Piano D., Alaoui S. E., Korza H. J., Filipek R., Sabala I., Haniewicz P., Büchel C., De Sanctis D., Bochtler M. 2010. Crystallization of the Photosystem II core complex and its chlorophyll binding subunit CP43 from transplastomic plants of Nicotiana tabacum. Photosynth. Res. 106, 221–226 10.1007/s11120-010-9597-x (doi:10.1007/s11120-010-9597-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ikeuchi M., Yuasa M., Inoue Y. 1985. Simple and discrete isolation of an O2-evolving PSII reaction center complex retaining Mn and the extrinsic 33 kDa protein. FEBS Lett. 185, 316–322 10.1016/0014-5793(85)80930-3 (doi:10.1016/0014-5793(85)80930-3) [DOI] [Google Scholar]
- 35.Bricker T. M., Pakrasi H. B., Sherman L. A. 1985. Characterization of a spinach photosystem II core preparation isolated by a simplified method. Arch. Biochem. Biophys. 237, 170–176 10.1016/0003-9861(85)90266-8 (doi:10.1016/0003-9861(85)90266-8) [DOI] [PubMed] [Google Scholar]
- 36.Haag E., Irrgang K. D., Boekema E. J., Renger G. 1990. Functional and structural analysis of photosystem II core complex from spinach with high oxygen evolution capacity. Eur. J. Biochem. 189, 47–53 10.1111/j.1432-1033.1990.tb15458.x (doi:10.1111/j.1432-1033.1990.tb15458.x) [DOI] [PubMed] [Google Scholar]
- 37.Hankamer B., Nield J., Zheleva D., Boekema E., Jansson S., Barber J. 1997. Isolation and biochemical characterization of monomeric and dimeric photosystem II complexes from spinach and their relevance to the organization of photosystem II in vivo. Eur. J. Biochem. 243, 422–429 10.1111/j.1432-1033.1997.0422a.x (doi:10.1111/j.1432-1033.1997.0422a.x) [DOI] [PubMed] [Google Scholar]
- 38.Wang Z. G., Xu T. H., Liu C., Yang C. H. 2010. Fast isolation of highly active Photosystem II core complexes from spinach. J. Int. Plant Biol. 52, 793–800 10.1111/j.1744-7909.2010.00971.x (doi:10.1111/j.1744-7909.2010.00971.x) [DOI] [PubMed] [Google Scholar]
- 39.Tang X. S., Satoh K. 1985. The oxygen-evolving photosystem II core complex. FEBS Lett. 179, 60–64 10.1016/0014-5793(85)80191-5 (doi:10.1016/0014-5793(85)80191-5) [DOI] [Google Scholar]
- 40.Ghanotakis D. F., Demetriou D. M., Yocum C. F. 1987. Isolation and characterization of an oxygen evolving photosystem II reaction center core and a 28 kDa Chl a-binding protein. Biochim. Biophys. Acta 891, 15–21 10.1016/0005-2728(87)90078-8 (doi:10.1016/0005-2728(87)90078-8) [DOI] [Google Scholar]
- 41.Leeuwen P. J., Nieveen M. C., van de Meent E. J., Dekker J. P., van Gorkom H. J. 1991. Rapid and simple isolation of pure photosystem II core reaction center particles from spinach. Photosynth. Res. 28, 149–153 10.1007/BF00054128 (doi:10.1007/BF00054128) [DOI] [PubMed] [Google Scholar]
- 42.Satoh K., Butler W. L. 1978. Low temperature spectral properties of subchloroplast fractions purified from spinach. Plant Physiol. 61, 373–379 10.1104/pp.61.3.373 (doi:10.1104/pp.61.3.373) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barber J. 2003. Photosystem II: the engine of life. Biophys. Q. Rev. 36, 71–89 10.1017/S0033583502003839 (doi:10.1017/S0033583502003839) [DOI] [PubMed] [Google Scholar]
- 44.Eshaghi S., Andersson B., Barber J. 1999. Isolation of a highly active PSII–LHCII supercomplex from thylakoid membranes by a direct method. FEBS Lett. 446, 23–26 10.1016/S0014-5793(99)00149-0 (doi:10.1016/S0014-5793(99)00149-0) [DOI] [PubMed] [Google Scholar]
- 45.Dekker J. P., Germano M., van Roon H., Boekema E. J. 2002. Photosystem II solubilizes as a monomer by mild detergent treatment of unstacked thylakoid membranes. Photosynth. Res. 72, 203–210 10.1023/A:1016188818591 (doi:10.1023/A:1016188818591) [DOI] [PubMed] [Google Scholar]
- 46.Bumba L., Husak M., Vacha F. 2004. Interaction of photosystem 2-LHC2 supercomplexes in adjacent layers of stacked chloroplast thylakoid membranes. Photosynthetica 42, 193–199 10.1023/B:PHOT.0000040590.10919.02 (doi:10.1023/B:PHOT.0000040590.10919.02) [DOI] [Google Scholar]
- 47.Caffarri S., Kouřil R., Kereïche S., Boekema E. J., Croce R. 2009. Functional architecture of higher plant photosystem II supercomplexes. EMBO J. 28, 3052–3063 10.1038/emboj.2009.232 (doi:10.1038/emboj.2009.232) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pagliano C., Raviolo M., Dalla Vecchia F., Gabbrielli R., Gonnelli C., Rascio N., Barbato R., La Rocca N. 2006. Evidence for PSII donor-side damage and photoinhibition induced by cadmium treatment on rice (Oryza sativa L.). J. Photochem. Photobiol. 84, 70–78 10.1016/j.jphotobiol.2006.01.012 (doi:10.1016/j.jphotobiol.2006.01.012) [DOI] [PubMed] [Google Scholar]
- 49.Hewitt E. J. 1966. Sand and water culture methods in the study of plant nutrition. In Technical Communication No. 22 (revised) Commonwealth Agricultural bureaux, 2nd edn. London, UK: Farnham [Google Scholar]
- 50.Pagliano C., Barera S., Chimirri F., Saracco G., Barber J. 2012. Comparison of the α and β isomeric forms of the detergent n-dodecyl-D-maltoside for solubilizing photosynthetic complexes from pea thylakoid membranes. Biochim. Biophys. Acta 1817, 1506–1515 10.1016/j.bbabio.2011.11.001 (doi:10.1016/j.bbabio.2011.11.001) [DOI] [PubMed] [Google Scholar]
- 51.Arnon D. J. 1949. Copper enzymes in isolated chloroplasts, polyphenoloxidase in Beta vulgaris. Plant Physiol. 24, 1–14 10.1104/pp.24.1.1 (doi:10.1104/pp.24.1.1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Laemmli U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 10.1038/227680a0 (doi:10.1038/227680a0) [DOI] [PubMed] [Google Scholar]
- 53.Schagger H. 2006. Tricine–SDS–PAGE. Nat. Protoc. 1, 16–22 10.1038/nprot.2006.4 (doi:10.1038/nprot.2006.4) [DOI] [PubMed] [Google Scholar]
- 54.Tang G., Peng L., Baldwin P. R., Mann D. S., Jiang W., Rees I., Ludtke S. J. 2007. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 10.1016/j.jsb.2006.05.009 (doi:10.1016/j.jsb.2006.05.009) [DOI] [PubMed] [Google Scholar]
- 55.van Heel M., Portugal R., Rohou A., Linnemayr C., Bebeacua C., Schmidt R., Grant T., Schatz M. 2011. Four-dimensional cryo electron microscopy at quasi atomic resolution: ‘IMAGIC 4D’. In International tables for crystallography. Vol. F: Crystallography of biological macromolecules (eds Rossmann M. G., Arnold E., Himmel D.), pp. 624–628 Chichester, UK: John Wiley and Sons [Google Scholar]
- 56.van Heel M., et al. 2000. Single-particle electron cryo-microscopy: towards atomic resolution. Q. Rev. Biophys. 33, 307–369 10.1017/S0033583500003644 (doi:10.1017/S0033583500003644) [DOI] [PubMed] [Google Scholar]
- 57.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. 2004. UCSF chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 10.1002/jcc.20084 (doi:10.1002/jcc.20084) [DOI] [PubMed] [Google Scholar]
- 58.Ghanotakis D. F., Topper J. N., Babcock G. T., Yocum C. F. 1984. Water soluble 17- and 23-kDa polypeptides restore oxygen evolution activity by creating a high affinity binding site for Ca2+ on the oxidizing side of photosystem II. FEBS Lett. 170, 169–173 10.1016/0014-5793(84)81393-9 (doi:10.1016/0014-5793(84)81393-9) [DOI] [Google Scholar]
- 59.Miyao M., Murata N. 1984. Calcium ions can be substituted for the 24-kDa polypeptide in photosynthetic oxygen evolution. FEBS Lett. 168, 118–120 10.1016/0014-5793(84)80218-5 (doi:10.1016/0014-5793(84)80218-5) [DOI] [Google Scholar]
- 60.Miyao M., Murata N. 1985. The Cl− effect on photosynthetic oxygen evolution: interaction of Cl− with 18-kDa, 24-kDa and 33-kDa proteins. FEBS Lett. 180, 303–308 10.1016/0014-5793(85)81091-7 (doi:10.1016/0014-5793(85)81091-7) [DOI] [Google Scholar]
- 61.Murray J. W. 2012. Sequence variation at the oxygen-evolving centre of photosystem II: a new class of ‘rogue’ cyanobacterial D1 proteins. Photosynth. Res. 110, 177–184 10.1007/s11120-011-9714-5 (doi:10.1007/s11120-011-9714-5) [DOI] [PubMed] [Google Scholar]
- 62.Horton P., Ruban A. V., Walters R. G. 1996. Regulation of light harvesting in green plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 655–684 10.1146/annurev.arplant.47.1.655 (doi:10.1146/annurev.arplant.47.1.655) [DOI] [PubMed] [Google Scholar]
- 63.Morosinotto T., Bassi R., Frigerio S., Finazzi G., Morris E., Barber J. 2006. Biochemical and structural analyses of a higher plant photosystem II supercomplex of a photosystem I-less mutant of barley. FEBS J. 273, 4616–4630 10.1111/j.1742-4658.2006.05465.x (doi:10.1111/j.1742-4658.2006.05465.x) [DOI] [PubMed] [Google Scholar]
- 64.Boekema E. J., van Roon H., Calkoen F., Bassi R., Dekker J. P. 1999. Multiple types of association of photosystem II and its light harvesting antenna in partially solubilized photosystem II membranes. Biochemistry 38, 2233–2239 10.1021/bi9827161 (doi:10.1021/bi9827161) [DOI] [PubMed] [Google Scholar]
- 65.Kovács L., Damkjær J., Kereïche S., Ilioaia C., Ruban A. V., Boekema E. J., Jansson S., Horton P. 2006. Lack of the light-harvesting complex CP24 affects the structure and function of the grana membranes of higher plant chloroplasts. Plant Cell 18, 3106–3120 10.1105/tpc.106.045641 (doi:10.1105/tpc.106.045641) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pagliano C., Chimirri F., Saracco G., Marsano F., Barber J. 2011. One-step isolation and biochemical characterization of a highly active plant PSII monomeric core. Photosynth. Res. 108, 33–46 10.1007/s11120-011-9650-4 (doi:10.1007/s11120-011-9650-4) [DOI] [PubMed] [Google Scholar]
- 67.Boekema E. J., van Roon H., Dekker J. P. 1998. Specific association of photosystem II and light-harvesting complex II in partially solubilized photosystem II membranes. FEBS Lett. 424, 95–99 10.1016/S0014-5793(98)00147-1 (doi:10.1016/S0014-5793(98)00147-1) [DOI] [PubMed] [Google Scholar]
- 68.Boekema E. J., van Roon H., van Breemen J. F. L., Dekker J. P. 1999. Supramolecular organization of photosystem II and its light-harvesting antenna in partially solubilized photosystem II membranes. Eur. J. Biochem. 266, 444–452 10.1046/j.1432-1327.1999.00876.x (doi:10.1046/j.1432-1327.1999.00876.x) [DOI] [PubMed] [Google Scholar]
- 69.Peter G. F., Thornber J. P. 1991. Biochemical composition and organization of higher plant photosystem II light-harvesting pigment-proteins. J. Biol. Chem. 266, 16 745–16 754 [PubMed] [Google Scholar]
- 70.Bassi R., Dainese P. 1992. A supramolecular light-harvesting complex from chloroplast photosystem-II membranes. Eur. J. Biochem. 204, 317–326 10.1111/j.1432-1033.1992.tb16640.x (doi:10.1111/j.1432-1033.1992.tb16640.x) [DOI] [PubMed] [Google Scholar]