Abstract
Redox chemistry and redox regulation are central to the operation of photosynthesis and respiration. However, the roles of different oxidants and antioxidants in the regulation of photosynthetic or respiratory gene expression remain poorly understood. Leaf transcriptome profiles of a range of Arabidopsis thaliana genotypes that are deficient in either hydrogen peroxide processing enzymes or in low molecular weight antioxidant were therefore compared to determine how different antioxidant systems that process hydrogen peroxide influence transcripts encoding proteins targeted to the chloroplasts or mitochondria. Less than 10 per cent overlap was observed in the transcriptome patterns of leaves that are deficient in either photorespiratory (catalase (cat)2) or chloroplastic (thylakoid ascorbate peroxidase (tapx)) hydrogen peroxide processing. Transcripts encoding photosystem II (PSII) repair cycle components were lower in glutathione-deficient leaves, as were the thylakoid NAD(P)H (nicotinamide adenine dinucleotide (phosphate)) dehydrogenases (NDH) mRNAs. Some thylakoid NDH mRNAs were also less abundant in tAPX-deficient and ascorbate-deficient leaves. Transcripts encoding the external and internal respiratory NDHs were increased by low glutathione and low ascorbate. Regulation of transcripts encoding specific components of the photosynthetic and respiratory electron transport chains by hydrogen peroxide, ascorbate and glutathione may serve to balance non-cyclic and cyclic electron flow pathways in relation to oxidant production and reductant availability.
Keywords: photosystem II repair cycle, cyclic electron flow, NAD(P)H dehydrogenase (NDH) complex, redox regulation, gene expression, respiration
1. Introduction
Photosynthesis is regulated by metabolic and environment controls that balance energy supply and demand in order to optimize photosynthetic efficiency while minimizing back reactions [1] that favour the production of reactive oxygen species (ROS). The regulation of the photosynthetic electron transport chain incorporates a range of mechanisms designed to prevent the over-reduction of electron carrier that inevitably leads to ROS production [2]. Flexibility in the production of adenosine triphosphate (ATP) and reductant by the photosynthetic electron transport system is required to match the changing demands of metabolism while accommodating variations in light and CO2 availability and avoiding over-reduction and uncontrolled oxidation [2,3]. This is achieved through rapid, short-term modifications in post-translational controls and longer term regulation through regulation of gene expression. Photosynthetic control of gene expression is observed in response to state transitions. It also serves to accommodate varying metabolic demands for ATP and reductant that occur when the rate of photorespiration is varied relative to photosynthesis [2]. In addition, the production and accumulation of different forms of ROS (superoxide (O2−), hydrogen peroxide (H2O2) and singlet oxygen (1O2)) by photosynthesis and respiration are important regulators of photosynthetic and respiratory gene expression, as are the reductants (reduced glutathione (GSH), ascorbate, thioredoxin (Trx) and peroxiredoxins) that are generated or regenerated by the reduced ferredoxin and NAD(P)H (nicotinamide adenine dinucleotide (phosphate)) produced by photosynthesis [1,2].
Superoxide and hydrogen peroxide are produced by photosynthesis, photorespiration and respiration, as well as other metabolic pathways [4,5]. Genetic evidence has demonstrated unequivocally that ROS are signalling molecules with important and specific roles in the control of gene expression [4,6–10]. Uncontrolled oxidation is prevented by the complex cellular antioxidant network, which limits not only the lifetime of ROS but also facilitates the propagation of oxidative and reductive signals [11].
The majority of proteins with functions in photosynthesis are encoded by the nuclear genome [12]. The coordination of the expression of photosynthetic genes in the nuclear and chloroplast genomes involves a complex network of signalling pathways. Of these, retrograde signals that convey information from the chloroplasts to the nucleus are considered to be essential in tailoring gene expression to match the varying requirements of electron transport and metabolism for different components under fluctuating environmental conditions [12,13]. The photosynthetic electron transport chain is an important source of retrograde signals such as singlet oxygen, which is generated by photosytem II (PSII) and superoxide, which is produced abundantly on the reducing side of photosystem I (PSI) [7,9,12,13]. If the hydrogen peroxide generated by photorespiration is not efficiently removed by catalase, salicylic acid (SA)-dependent cell death pathways are activated [8]. Similarly, singlet oxygen can trigger cell death via the activation of a signal transduction sequence involving the two plastid proteins called EXECUTER1 and EXECUTER2 [7,9]. The flexible control of oxidant production and removal is therefore a crucial determinant of cell fate. ROS signals participate in the chloroplast-to-nucleus signalling network, functioning alongside other factors such as the redox state of the plastoquinone pool, intermediates of chlorophyll biosynthesis, genomes uncoupled (GUN) proteins, abscisic acid (ABA) and sugar signalling [14].
2. Antioxidants and redox signalling
The metabolic and signalling action of ROS is ‘hormetic’ in function because controlled ROS production stimulates growth and stress tolerance. Hormesis describes the biological phenomenon whereby beneficial effects such as improved stress tolerance, growth or longevity are derived from the exposure to low levels (or higher amounts over short periods) of compounds that are toxic at high doses or by exposures to low doses over long periods. Thus, depending on the relative rates of production and removal, singlet oxygen and hydrogen peroxide can either activate the expression of defence genes such as glutathione S-transferases (GST) or trigger genetically programmed cell suicide pathways [7,9,11,13].
Perhaps because of the immense complexity of cellular oxidant/antioxidant interactions, the respective roles of each component in signal transduction pathways remain largely unexplored. However, accumulating evidence suggests that low molecular weight antioxidants such as ascorbate and GSH participate in redox signalling [15–17]. For example, the abundance of ascorbate might convey information concerning the redox buffering capacity of the chloroplast and mitochondria and other cellular compartments. Ascorbate regulates plant growth and defence through the activation of SA- and ABA-dependent pathways that also influence jasmonate signalling [15,17]. In contrast, the GSH pool may contain information concerning the production of γ-glutamyl cysteine and GSH by the chloroplasts, as well as abundance of thiols and the redox state of the cell [16]. GSH controls the cell cycle and is required for the correct establishment of the root meristem [18].
The abundance of ascorbate and GSH in leaves is regulated by the amount and quality of light [19–21]. While the last step of ascorbate synthesis occurs in the mitochondria as indicated in figure 1, the response of ascorbate synthesis to high light is considered to be controlled largely by the regulated expression of VTC (VITAMIN C)2/5, which encodes guanosine diphosphate-l-galactose phosphorylase, a cytosolic enzyme that catalyses the first dedicated biosynthetic step of the ascorbate synthesis pathway [21]. The leaves of the Arabidopsis vtc2 mutants accumulate less (75–85% lower) ascorbate than the wild-type [15,17]. Similarly, Arabidopsis vtc1 mutants, which carry a point mutation in a gene encoding an enzyme that catalyses an earlier step in the pathway, also accumulate considerably less (about 70% lower) ascorbate than the wild-type [15,17].
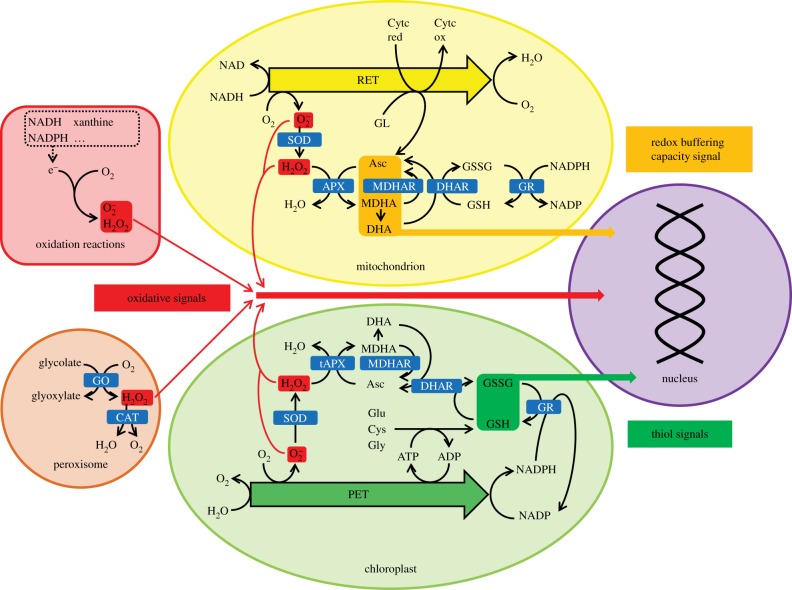
Figure 1.
Schematic of possible signalling pathways between organelles and the nucleus triggered by low glutathione synthesis transmitting thiol signals, low ascorbate transmitting signals concerning cellular redox buffering capacity and decreased hydrogen peroxide scavenging transmitting an oxidant signal. APX, ascorbate peroxidase; Asc, ascorbate; CAT, catalase; Cytc red, reduced cytochrome c, ox, oxidized; DHA, dehydroascorbate; DHAR, DHA reductase; GO, glycolate oxidase; GR, glutathione reductase; MDHA, monodehydroascorbate; MDHAR, MDHA reductase; PET, photosynthetic electron transfer chain; RET, respiratory electron transfer chain; SOD, superoxide dismutase; tAPX, thylakoid APX.
Catalases and ascorbate peroxidases (APXs) play important roles in the removal of hydrogen peroxide from the cellular environment (figure 1). While catalases are enzymes that catalyse the disproportionation of hydrogen peroxide to water and oxygen (figure 1), APXs reduce hydrogen peroxide to water, producing monodehydroascorbate and thereafter dehydroascorbate. Ascorbate is rapidly regenerated using photosynthetic reductants as illustrated in figure 1. Arabidopsis contains a small family of APX genes that are targeted to the cytosol, peroxisomes, mitochondria and chloroplasts, which has stromal (sAPX) and thylakoid-bound (tAPX) forms. Deficiencies in the sAPX and tAPX forms resulted in increased sensitivity to high light. Moreover, the expression of ROS-responsive genes under high light conditions was repressed in these mutants [22]. Hence, H2O2-derived signals produced by the chloroplasts may exert a negative influence on the expression of ROS-responsive genes, particularly under high light [22].
GSH is synthesized in plants from component amino acids by two ATP-dependent steps, the first of which is catalysed by γ-glutamyl cysteine synthetase (γ-ECS) and occurs exclusively in plastids in Arabidopsis. The second step is catalysed by glutathione synthetase and is located in the cytosol and chloroplasts [16,23]. Feedback control of GSH synthesis in the chloroplast by GSH [16] can be overcome by high γ-ECS activity [23], which increases the abundance of amino acids that are synthesized in chloroplasts particularly valine, leucine, tyrosine and isoleucine [24]. The rootmeristemless (rml)1 genotype has a point mutation in the GSH1 gene that encodes γ-ECS and results in impaired GSH synthesis such that the leaves accumulate less than 5 per cent of the GSH levels observed in the wild-type [18]. GSH is a powerful redox buffer that can prevent the oxidation of surface-exposed cysteines on proteins, which can be oxidized to form sulfenic acid, impairing protein function. Stabilization of the protein sulfenic acid residues can occur by disulfide linkage to another thiol group, or they can be hyper-oxidized to sulfinic and sulfonic acid forms. Along with disulfide reductases, glutaredoxins and Trxs, GSH participates in thiol–disulfide exchange reactions. Thiol–disulfide exchange regulation of proteins can have a profound effect on folding and activity. Crucially, disulfides formed during oxidation function as redox switches. For example, redox modification of transcription factors can alter their DNA-binding properties and activates signal transduction pathways. In addition, there are several examples of transcription factors that are induced by oxidative signalling pathways in plants. For example, the NAC (NAM, ATAF1,2, CUC2) transcription factor JUNGBRUNNEN (JUB)1 is induced by hydrogen peroxide; JUB1 expression serves to delay senescence and extends leaf longevity by promoting enhanced defence and tolerance to abiotic stresses [25].
The ratio of GSH to glutathione disulfide (GSSG) can also modulate protein activity through glutathionylation, which is a reversible post-translational modification consisting of the formation of a mixed disulfide between the free protein thiol and GSH. Glutathionylation of redox-sensitive protein cysteines can be achieved through various reactions, such as thiol–disulfide exchange between protein cysteines and GSSG, by ROS-catalysed production of protein thiyl radicals, or through the nitric oxide (NO)-mediated formation of S-nitrosothiols [16]. The removal of bound GSH from proteins is catalysed by glutaredoxins and Trxs. Glutathionylation may provide a mechanism for transmitting oxidative signals under stress conditions but the regulation of protein glutathionylation in plants remains largely uncharacterized and poorly understood. The formation of S-nitrosothiols particularly S-nitrosoglutathione (GSNO) provides a further mechanism by which GSH make a further contribution to post-translational modifications regulating cell signalling. GSNO acts as a reservoir of NO and it is also important in the transport, storage and delivery of NO to sites of action. GSH-dependent formaldehyde dehydrogenases (GSNO reductases) are class III alcohol dehydrogenases that catalyse the NADH-dependent reduction of GSNO.
3. Comparative analysis of redox transcriptome profiles
To provide insights into how gene expression patterns are changed in response to oxidants and antioxidants, we have compared the leaf transcriptome profiles of A. thaliana genotypes that are either deficient in either photorespiratory hydrogen peroxide processing (catalase 2 (cat2) [26]), chloroplastic hydrogen peroxide processing (thylakoid ascorbate peroxidase (tapx) [27]), ascorbate synthesis (vtc1 and vtc2 [17]) or GSH synthesis (rml1 [18]). This comparison might allow for the identification of overlapping patterns or unique transcript changes resulting from variations in hydrogen peroxide processing capacity in different cellular compartments, as well as low GSH (thiol) and low ascorbate availability. Figure 1 provides a very simple schematic model of the transmission of different oxidative and reductive signals from various cellular locations to the nucleus.
The data used in the following analysis of transcript patterns in leaves that are deficient in either ascorbate [17], GSH [18] or catalase [26] were collected by ourselves. The cat2 mutants and controls (Col0) were grown on soil in controlled environment cabinets, with an 8 h photoperiod (200 μmol m−2 s−1), and a 20°C day and 18°C night regime. Plants were grown for five weeks in a CO2-enriched atmosphere (3000 μl l−1) to suppress photorespiration [26]. Then, two batches of plants were transferred to air (400 μl l−1 CO2; photorespiratory conditions). One batch was grown under an 8 h photoperiod, whereas the second batch was grown under a 16 h photoperiod. Leaf samples were harvested in triplicate, 2 and 4 days after transfer to air. The vtc1 and vtc2 mutants and controls (Col0) were grown on soil in controlled environment cabinets, with a 12 h photoperiod (250 μmol m−2 s−1), at a constant temperature of 20°C. Whole rosettes were harvested in triplicate per genotype.
For cat2, samples were hybridized to Affymetrix GeneChip Arabidopsis ATH1 Genome Arrays (https://www.affymetrix.com) at the VIB Microarray Facility (Leuven, Belgium). Raw data processing and normalization were performed with the statistical algorithm robust multiarray average (gcRMA) and the affy package of R/Bioconductor (http://www.bioconductor.org). The whole microarray data are available at: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE27985. In the following analysis, only data from plants transferred to the 8 h photoperiod have been used. Statistical analyses were performed by a t-test comparison using Limma algorithm (http://www.ncbi.nlm.nih.gov/geo/geo2r); only genes showing false discovery rate (FDR)-corrected p-values below 0.05 and a minimum 2-fold change in expression, 2 or 4 days after transfer from high CO2 growth conditions to air at 8 h photoperiod were used for the analysis.
For vtc1 and vtc2, data were analysed using two methods. Firstly, raw intensity values from the scanned Affymetrix GeneChip Arabidopsis ATH1 Genome Arrays (https://www.affymetrix.com) were analysed using the robust multichip average method using RMAExpress (http://rmaexpress.bmbolstad.com). A significance test was undertaken by calculating a null hypothesis through a permuted modified t-statistic across the replicates for each array comparison. The log2 means of the normalized intensity values of the biological replicates were compared as ratios, where a log2 ratio of ±1.0 and with a p-value less than 0.01 was selected as worthy of further study. p-values were further used to apply significance where the probe log2 ratios were more than or equal to 1. Gene expression was also analysed using GeneSpring GX 11.00 and p-values were calculated by asymptotic unpaired t-test and subjected to multiple testing corrections (Benjamini Hochberg FDR). A cut-off with a p-value less than 0.05 and log2 expression ratio ±1 was adopted. The probe targets were as defined by Affymetrix (https://www.affymetrix.com/analysis/netaffx/index.affx) and these targets were used to guide annotation of the genes through the use of Blast against the Tair (http://www.arabidopsis.org) database. All the datasets reported here are deposited at: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE23331.
GSH is required for cell proliferation at the root tip after the root primordia have fully formed. The rml1 seeds germinate and the shoot meristem begins to develop as in the wild-type. However, the growth of the embryonic root meristem arrests after approximately 17 longitudinal cell divisions. The rml1 mutants are therefore deficient in postembryonic root (but not shoot) development. However, the arrested mutant root cells can carry out normal root functions, such as the production of lateral roots from the pericycle. The deficiency in postembryonic root (but not shoot) development means that plants have to be grown on plates. Mutants were therefore grown in controlled environment cabinets, together with controls (Col0) on vertical plates containing 1 per cent agar in PANG 2 medium for 7 days under a photoperiod of 16 h at a constant temperature of 20°C. The gene expression data used in the present analysis was obtained from shoot samples that were harvested in triplicate. Samples were hybridized to Agronomics1 tiling arrays [28] at the VIB core service (www.nucleomics.be). Quality control and normalization of the data were performed using Affymetix package and statistical analysis was performed by t-test using Limma package from the Bioconductor software (http://www.bioconductor.org). Transcripts were identified based on a minimum 2 fold-change in expression between Col0 and rml1 and a FDR corrected p-value threshold below 0.05 (see http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE36893 for full dataset).
The data for the Arabidopsis plants in which the tAPX was silenced were taken from Maruta et al. [27]. In this study an oestrogen inducible tAPX-silencing line (RNAi, IS-tAPX-19–23) was grown together with control genotype (IS-GUS-2-17) on Murashige and Skoog medium containing 3 per cent sucrose for 7 days under continuous light (100 μmol m−2 s−1) and at 25°C. Seedlings were then transferred on soil and grown for a further 10 days under the same conditions. The 17-day-old transgenic plants were then treated with oestrogen for 2 days to induce silencing of the tAPX. Microarray analysis was then performed on two leaf replicates by using Agilent Arabidopsis 3 Oligo Microarrays. The data were normalized and analysed using GeneSpring and are available at: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE35526. Genes showing a minimum twofold change in expression ratio in the tAPX-silenced plants were selected for further analysis [27].
4. Hydrogen peroxide- and antioxidant-triggered transcriptome patterns
(a). Transcript responses to low ascorbate, low GSH and enhanced oxidation
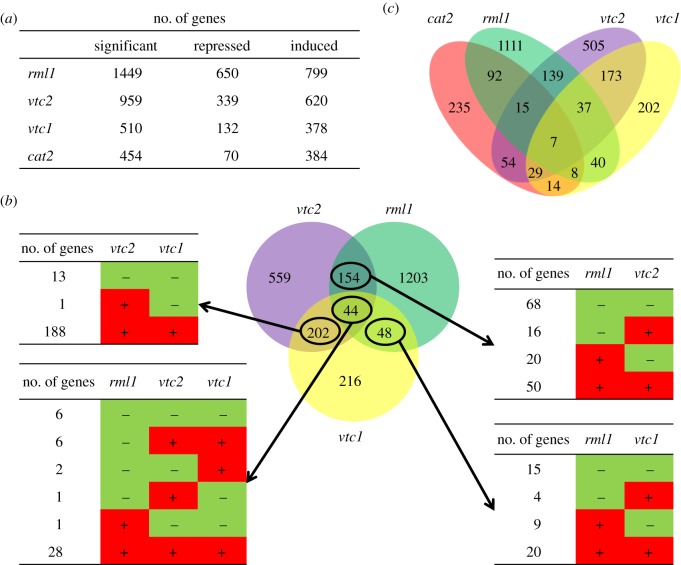
The leaves of the rml1, vtc1 and vtc2 mutants showed significant transcriptome reprogramming relative to the wild-type leaves, with 1449 genes regulated by low GSH and between 510 (vtc1) and 959 (vtc2) genes regulated in response to low ascorbate (figure 2a). Of the ascorbate regulated genes only 44 showed GSH-dependent expression changes (figure 2b,c), suggesting that low GSH and low ascorbate have differential effects on gene expression.
Figure 2.
Comparisons of the leaf transcriptome profiles of the rml1 [18], vtc2, vtc1 [17] and cat2 [26] mutants. (a) Number of significantly repressed and induced genes in the four genotypes. (b) Venn diagram showing the gene overlaps between the rml1, vtc1 and vtc2 transcriptomes. Numbers indicate the quantities of repressed and induced genes. (c) Venn diagram showing the numbers of overlapping genes in the four genotypes.
A degree of overlap has been reported in genes regulated by H2O2 and by low GSH in plants and animals [13,29]. In human promyelocytic leukemia cells (HL)-60 for example, 2016 genes were regulated by H2O2, of which 215 also showed GSH-dependent expression changes [29]. Of the 454 genes that were regulated by H2O2 in the A. thaliana cat2 mutant, 122 showed GSH-dependent expression changes (figure 2c). Of the genes whose expression was similarly regulated by H2O2 and by low GSH, only six transcripts were decreased and 95 were increased in abundance (see the electronic supplementary material, table S1). The genes that were similarly modified in expression in the cat2 and rml1 genotypes included several transcription factors (ATAF1, NAC domain transcription factors ANAC13, ANAC032, ANAC052 and ANAC102, the zinc finger transcription factors ZINC TRANSPORTER (ZAT)6 and ZAT12, and only one WRKY domain transcription factor, WRKY 40), several UDP-glycosyltransferase genes such as IAGLU, UGT73B2, UGT73B4, UGT73B5, UGT73C1, UGT75B1, UGT84A2 and UGT84A3, and GSTs (GSTU1, GSTU4, GSTU7, GSTU8, GSTU24 and GSTU25).
Of the 454 genes that were regulated by H2O2 in the A. thaliana cat2 mutant, 47 showed similar ascorbate-dependent expression changes in the vtc2 mutants (see the electronic supplementary material, table S1). The expression of 58 genes including WRKY38, HEAT SHOCK PROTEIN (HSP)70, SMALL UBIQUITIN-LIKE MODIFIER (SUMO)3, UGT74D1 and UGT76E11 was reversed in the cat2 relative to the vtc2 mutant (see the electronic supplementary material, table S1). Only 26 genes were similarly regulated by H2O2 and by low ascorbate in both the vtc1 and vtc2 mutants (see the electronic supplementary material, table S1). It is worth noting that relatively few genes such as WRKY40, PINOID-BINDING PROTEIN (PBP)1, CYTOCHROME P450 (CYP)81D8 and the TRANSLOCON AT THE INNER ENVELOPE MEMBRANE OF CHLOROPLASTS (TIC)20-IV were expressed in a similar manner in response to H2O2, low GSH and low ascorbate, whereas EARLY RESPONSE TO DEHYDRATION (ERD)9, CARBONIC ANHYDRASE1 and a LATE EMBRYOGENESIS ABUNDANT (LEA)3 family protein were induced in response to H2O2 and repressed by low GSH and low ascorbate (see the electronic supplementary material, table S1).
(b). Regulation of photosynthetic gene expression by low ascorbate and low GSH
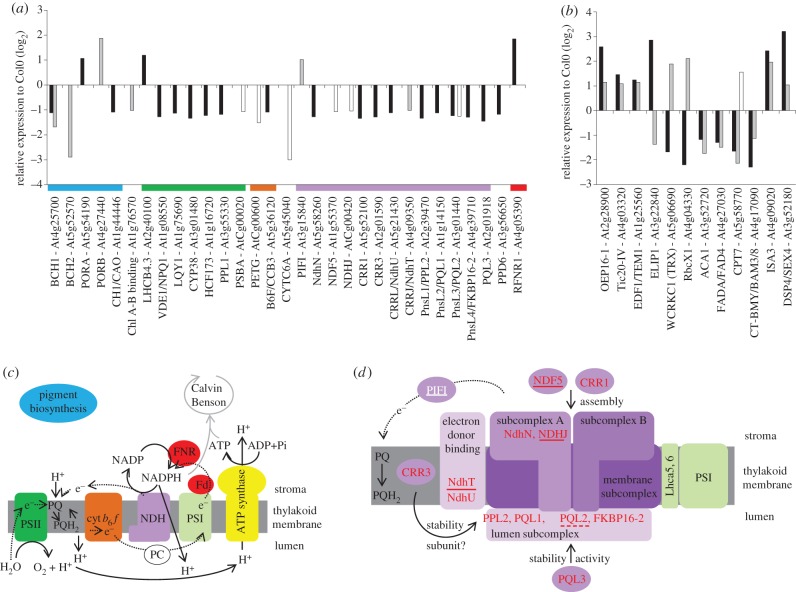
Very few genes encoding chloroplast-localized proteins were regulated in response to low ascorbate or low GSH (figure 3). However, low glutathione increased the abundance of transcripts encoding NADPH: protochlorophyllide oxidoreductase (POR)A, while low ascorbate increased the expression of PORB (figure 3a). Ethylene signalling converges on the ETHYLENE-INSENSITIVE3 (EIN3)/EIN3-Like (EIL) transcription factors that bind directly to the specific elements present in the PORA and PORB promoters. In co-operation with the PHYTOCHROME-INTERACTING FACTORS such as PIF1, which also shows ascorbate-dependent expression changes, the EIN3 prevents photo-oxidative damage by repression of the accumulation of protochlorophyllide, a phototoxic intermediate of chlorophyll synthesis [30]. Although the expression of EIN3 was not induced in the different genotypes studied here, EIN3-BINDING F BOX PROTEIN (EBF)2 was induced by low GSH. These findings suggest that GSH-dependent expression of genes that regulate protochorophyllide biosynthesis is important in ensuring survival in conditions of low thiol availability.
Figure 3.
The effects of ascorbate and glutathione on the abundance of transcripts encoding chloroplast proteins. (a) Transcripts encoding components of the photosynthetic electron transfer chain: the colour of the bar below each group indicates the location of the encoded protein as illustrated in diagram (c) or (d). (b) Transcripts encoding chloroplast-associated proteins in the vtc2 and rml1 mutants, respectively; for (a) and (b): black bars, low glutathione in rml1; grey bars, low ascorbate in vtc2; white bars, low ascorbate in vtc1. (c) Schematic of the photosynthetic electron transfer chain. (d) Schematic of the NDH complex indicating the localization of proteins encoded by genes identified in (a). The NDH subcomplexes and related proteins are highlighted in purple, the association with PSI is shown in green (white, induced genes; red, repressed genes; not underlined indicates responsive to low GSH; solid underline indicates responsive to low ascorbate; dashed underline indicates responsive to low GSH and low ascorbate).
Low ascorbate decreased the expression of β-CAROTENE HYDROXYLASE (BCH)1 and BCH2 (figure 3a). The activities of these β-carotene hydroxylases are required for carotenoid synthesis. Overexpression of BCH1 in Arabidopsis leads to an increase in the xanthophyll cycle pool (violaxanthin and zeaxanthin) and enhances stress tolerance [31]. Transcripts encoding violoxanthin de-epoxidase, an enzyme that requires ascorbate as a cofactor in zeaxanthin synthesis by the xanthophyll cycle, were decreased by low GSH (figure 3a) but not by low ascorbate. Zeaxanthin is a key component of non-photochemical quenching (NPQ) and thermal energy dissipation in photosynthesis. While photosynthesis in the vtc1 and vtc2 mutants is more sensitive to high light stress, the low levels of ascorbate in these mutants have little effect on NPQ even at high light intensities [32,33]. The vtc2 mutants had lower PSII quantum efficiencies than the wild plants when grown under very high light [33]. Double mutants (such as vtc2npq4 and vtc2npq1) that are defective in NPQ as well as ascorbate synthesis were able to grow under the high-light conditions [33]. However, the PSII quantum efficiencies of the vtc2npq4 and vtc2npq1 leaves were similar to those determined in vtc2. Taken together, these data suggest that depletion of leaf ascorbate to 20–30% wild-type levels does not severely impair the protective mechanisms that underpin thermal energy dissipation in photosynthesis.
Other genes that were repressed by low ascorbate or low GSH encode components that are not on the main route of non-cyclic electron flow, particularly those associated with the chloroplastic NAD(P)H dehydrogenase (NDH) complex (figure 3a), as well as other genes such as CHLORORESPIRATORY REDUCTION (CRR)1 and CRR3 that are classified as co-expressing with NDH core complex genes [34,35]. In addition, a small number of other genes associated with photosynthesis are differentially expressed in response to low ascorbate and low GSH (figure 3b). One of these, WCRKC1, the expression of which is repressed by low GSH but is activated by low ascorbate, encodes a member of the thioredoxin (Trx) superfamily, which fulfil diverse roles in plants ranging from protein disulfide reductases to disulfide isomerases or to disulfide transferases. WCRKC1 encodes a Trx that has an atypical redox active site and a less reducing redox midpoint potential than that of the classic chloroplast Trxs [36]. This Trx reduces the chloroplast 2-Cys peroxiredoxin (Prx) using electrons derived from non-cyclic photosynthetic electron flow.
The abundance of WCRKC1 transcripts is specifically decreased by low GSH, together with marked GSH-dependent decreases in the expression of LOW QUANTUM YIELD OF PHOTOSYSTEM II (LQY)1, CYCLOPHILIN (CYP)38 and HIGH CHLOROPHYLL FLUORESCENCE (HCF)173 (figure 3a,c). These genes encode thylakoid accessory proteins that are required for PSII core protein repair and assembly. HCF173 is involved in the initiation of translation of the psbA mRNA and is essential for D1 synthesis [37]. LQY1 is a Zn-finger protein with disulfide isomerase activity that interacts with the PSII core complex [38]. Although LQY1 is not absolutely essential for PSII core repair, it participates in the repair and reassembly of photo-damaged PSII proteins. CYP38, which has a peptidyl–prolyl cis–trans isomerase protein-folding activity, plays an important role in the assembly and stabilization of PSII particularly under high light stress [39]. The half-life of the D1 and D2 proteins in mutants lacking CYP38 or LQY1 is shorter under high light than the wild-type [38,39]. Another chloroplast cyclophilin (CYP20) has foldase activity, which is enhanced by reduced thioredoxin [40]. In this case, the CYP20-3 protein interacts with serine acetyltransferase (SAT1), which catalyses the rate-limiting enzyme of cysteine biosynthesis (the precursor of GSH biosynthesis). Although CYP20-3 activity links photosynthetic electron transport processes to the redox-dependent folding of SAT1, no such activity has been described for CYP38 to date. Nevertheless, γ-glutamyl cysteine and GSH biosynthesis in chloroplasts requires ATP produced by photosynthesis. Moreover, the rate of GSH synthesis is dependent on that of cysteine biosynthesis, which is directly regulated by the photosynthetic electron transport system.
Transcripts encoding other extrinsic subunits of PSII that show GSH-dependent regulation include PPD6, which encodes a PsbP-domain protein, PSBP-LIKE (PPL)1 and PPL2 (figure 3a,c). While the PsbP protein optimizes the water-splitting reaction, the PsbP-domain-like protein PPL1 is expressed under stress conditions, and PPL2 together with subunits of chloroplast NDH complex. Mutants lacking PPL2 show decreased NDH complex activity while ppl1 mutants are more sensitive to high-intensity light and show slow recovery from photoinhibition of PSII activity [41]. Of the NDH transcripts that are less abundant in rml1 leaves, PPL2, PSBQ-LIKE (PQL)1, PQL2 and CCR3 are induced after transfer from enriched CO2 atmosphere to air [2]. PQL1 and PQL2 are necessary for NDH activity and localize together with PPL2 to the lumenal side of the NDH complex (figure 3d) [42,43]. While CRR3 stabilizes the NDH complex and localizes to the membrane fraction of chloroplast, the identity of CRR3 as an NDH subunit remains to be established (figure 3d) [43,44].
(c). Regulation of respiratory gene expression by low ascorbate and low GSH
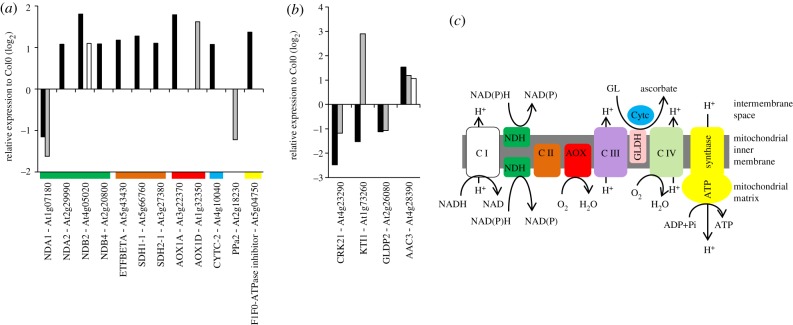
Ascorbic acid is synthesized from galactono-γ-lactone (GL) in leaves through the activity of galactono-γ-lactone dehydrogenase (GLDH), a mitochondrial enzyme, which uses reduced cytochrome c as a cofactor [45]. However, low ascorbate had little effect on the expression of genes encoding mitochondrial proteins (figure 4a,b). Transcripts encoding the ALTERNATIVE OXIDASE (AOX)1D and the internal NAD(P)H DEHYDROGENASES (ND)A1 were regulated by low ascorbate in the vtc2 mutant (figure 4a). While NDA1 showed a similar GSH-dependent expression pattern, AOX1A was induced by low GSH compared with AOX1D in the low ascorbate mutants (figure 4a). AOX1A and AOX1D are among the most stress responsive of all the known genes encoding mitochondrial proteins.
Figure 4.
Acorbate- and glutathione-regulated expression of mitochondrial genes in the rml1, vtc1 and vtc2 mutants. (a) Expression changes in genes encoding components of the respiratory electron transfer chain. The colour bar below each group indicates the location of the encoded protein as illustrated in diagram (c). (b) Ascorbate- and glutathione-regulated changes in the expression of genes encoding other mitochondrial proteins. (c) Schematic of the respiratory electron transfer chain. (a) and (b): black bars, rml1; grey bars, vtc2; white bars, vtc1.
The AOX is part of the inducible non-phosphorylating alternative pathway of the plant respiratory electron transport chain that also consist of type II NAD(P)H dehydrogenases on both sides of the inner membrane linked through the ubiquinone pool to the AOX. NDA1, which is considered to play a key role in integrating metabolic activities of chloroplasts and mitochondria, was decreased in response to low ascorbate and low GSH. However, transcripts (NDB2 and NDB4) encoding external NAD(P)H dehydrogenases forms were increased in abundance in response to low GSH, as were transcripts encoding the flavoprotein subunit (SUCCINATE DEHYDROGENASE (SDH)1 and SDH2) of complex II (figure 4a,c). Since a mild reduction in mitochondrial SDH activity had a positive impact on photosynthetic CO2 assimilation rates [46], the GSH-dependent increase in SDH1 and SDH2 might suggest a trend towards a slightly lower photosynthetic performance in plants deficient in GSH.
Complex I (NDH) and Complex III (ubiquinol–cytochrome bc1 reductase) are the major sites for ROS production in the mitochondrial electron transport chain. Mitochondria with reduced respiration are considered to generate a permanent stress condition by producing ROS as a constitutive stress signal for activation of cellular defense responses. In Arabidopsis, this typical stress response is controlled by transcriptional upregulation of AOX1A and AOX1D. AOX activity bypasses a block in proton pumping Complex III and so maintains electron flux through the respiratory electron transport chain, thus minimizing the possibility of ROS production. By enabling the tricarboxylic acid cycle to continue functioning when the electron transport in the cytochrome c pathway is blocked, the induction of the AOX gives essential flexibility to plant respiration as well as contributing to the re-programming of metabolism in response to changing environmental conditions. Taken together, these data suggest that low ascorbate and low GSH induce the expression of key components of the non-phosphorylating alternative pathway of respiration to decrease the possibility of ROS generation (figure 4).
(d). Hydrogen peroxide-derived signals arising from chloroplasts and peroxisomes
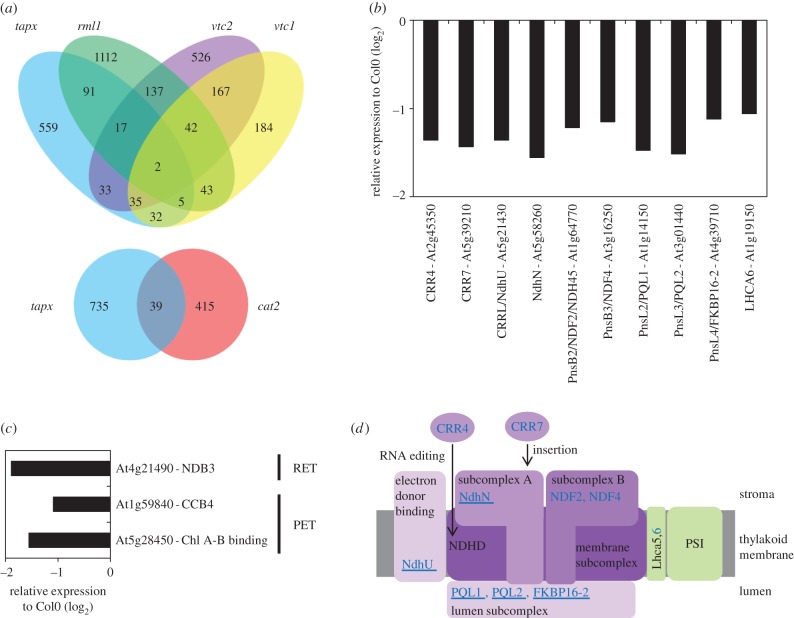
Of the transcripts that were regulated by H2O2 in the oestrogen inducible tAPX-silencing line, 115 were also changed in a similar manner in rml1 leaves (figure 5a). Similarly, 87 transcripts were commonly altered in abundance to tapx and vtc2 leaves and 74 transcripts were common to tapx and vtc1 leaves (figure 5a). The transcript profile of the tapx leaves was distinct from that of the cat2 mutants, with only 39 transcripts changed in a similar manner in both genotypes (figure 5a). The absence of a common transcript profile may suggest that hydrogen peroxide-derived signals arising from chloroplasts and peroxisomes have differential effects on gene expression. Of the transcripts that were changed in abundance in the tapx leaves, very few encoded proteins that were targeted at the chloroplasts and were associated with photosynthesis (figure 5b,c). Similarly, very few transcripts encoding components of the mitochondrial electron transfer chains were altered in abundance in the tapx leaves (figure 5c). However, 10 transcripts encoding proteins associated with the NDH complex were decreased in abundance in the tapx leaves (figure 5b). The location of the encoded proteins within the NDH subcomplexes is shown in figure 5d, which also indicates the location of proteins encoded by transcripts that are decreased in abundance both in tapx and rml1 leaves.
Figure 5.
Comparisons of the leaf transcriptome profiles of the tapx [27], rml1 [18], vtc2 and vtc1 [17] mutants and expression changes in genes encoding electron transfer chain components in tapx. (a) Venn diagram showing the gene overlaps between the tapx, rml1, vtc1 and vtc2 transcriptomes, and between tapx and cat2. (b) Expression changes in genes encoding components of NDH in tapx. (c) Expression changes in genes encoding other components of the photosynthetic (PET) and respiratory (RET) electron transfer chains in tapx. (d) Schematic of the NDH complex indicating the localization of proteins encoded by genes identified in (b). The NDH subcomplexes and related proteins are highlighted in purple, the association with PSI is shown in green (blue, repressed genes in tapx; underline indicates repression in rml1).
5. Summary and conclusions
Although the conclusions derived from the global transcriptome comparisons reported here must be regarded with a degree of caution as the growth conditions were not the same for all of the mutants, the findings of this study also provide some intriguing new insights into the relationships between different components of the cellular reduction–oxidation (redox) network and photosynthetic gene expression. A common action on leaf transcriptome patterns is demonstrated by the approximate 30 per cent overlap in the transcript changes in cat2, rml1, vtc1 and vtc2 mutants and in tapx, rml1, vtc1 and vtc2 mutants. The common responses in these gene expression patterns must reflect the general role of antioxidants in processing ROS signals. However, less than 10 per cent overlap was observed in the transcriptome patterns of cat2 and tapx even though both enzymes are involved in hydrogen peroxide processing. Although there were marked differences in the growth conditions used for the studies of gene expression in the mutants, it is nevertheless tempting to suggest that at least some of the differences arise from the location of hydrogen peroxide-derived signals, photorespiratory signals in the case of cat2 and chloroplast signals in the case of tapx. The differential transcriptome changes described here suggest that the site of enhanced oxidation (chloroplast or peroxisome) and the nature of the antioxidant exert specific effects on transcript abundance.
The suites of transcripts that encode proteins that can be classified as ROS-regulated are similar to those described previously [4] and they have therefore not been discussed in detail here. However, it is worthy of note that very few photosynthetic or respiratory pathway genes showed hydrogen peroxide-dependent gene expression. The most striking effect of depletion of several antioxidants, particularly in the rml1 mutants with low GSH and the tapx mutants, was on the abundance of transcripts encoding core and accessory components of the thylakoid NDH complex. The decrease in transcripts encoding NDH complex components by low antioxidant capacity is interesting particularly because of the low abundance of this complex in the thylakoid membranes and its importance in cyclic electron flow and chlororespiration. It is generally considered that the NDH complex associates with PSI to form NDH–PSI supercomplexes that re-direct electron flow into a cyclic pathway in which electrons are transported from PSI to the plastoquinone (PQ) pool via NADPH and the NDH complex in order to prevent over-reduction of the stroma. Since the stroma might be perceived as more oxidized when antioxidant capacity is diminished, it is perhaps surprising that low GSH, and to a lower extent low ascorbate and low tAPX activity, favours a decreased abundance of transcripts encoding NDH complex components that are required to support cyclic electron transport.
Transcripts encoding auxiliary proteins involved in the PSII repair cycle were also specifically decreased in plants with low GSH, as were transcripts encoding proteins that repress protochlorophyllide accumulation. The PSII repair cycle, which involves the replacement of damaged D1 subunits, occurs much more frequently than does de novo PSII biogenesis in mature chloroplasts [47]. This interaction may provide a further link between PSII activity and the redox state of the stroma, in addition to that involving TRX- and cyclophilin-dependent regulation of cysteine synthesis in the chloroplast [40]. Cysteine levels are tightly regulated in plant cells, particularly in relation to GSH synthesis, which is a major sink for cysteine [48]. The reduced demand for cysteine in the chloroplast caused by impaired γ-glutamyl cysteine synthesis in the rml1 mutant may also have a feedback effect on SAT1 activity [48].
Taken together, these findings indicate that manipulation of the abundance of individual components of the antioxidant network results in specific patterns of transcript changes. Regulated changes in gene expression patterns may hence be tailored to address the perceived imbalances in antioxidant strength and composition relative to oxidant production.
Acknowledgements
The authors acknowledge funding from the EU Marie-Curie ITN ‘COSI’ project (Orsay, Leeds). Guillaume Queval is funded by a Marie Curie Individual Fellowship: PIEF-GA-2009-252927: ROXNP.
References
- 1.Rutherford A. W., Osyczka A., Rappaport F. 2012. Back-reactions, short-circuits, leaks and other energy wasteful reactions in biological electron transfer: redox tuning to survive life in O2. FEBS Lett. 586, 603–616 10.1016/j.febslet.2011.12.039 (doi:10.1016/j.febslet.2011.12.039) [DOI] [PubMed] [Google Scholar]
- 2.Foyer C. H., Neukermans J., Queval G., Noctor G., Harbinson J. 2012. Photosynthetic control of electron transport and the regulation of gene expression. J. Exp. Bot. 63, 1637–1661 10.1093/jxb/ers013 (doi:10.1093/jxb/ers013) [DOI] [PubMed] [Google Scholar]
- 3.Foyer C. H., Noctor G. 2009. Redox regulation in photosynthetic organisms: signaling, acclimation and practical implications. Antioxid. Redox Signal. 11, 862–905 10.1089/ars.2008.2177 (doi:10.1089/ars.2008.2177) [DOI] [PubMed] [Google Scholar]
- 4.Miller G., Schlauch K., Tam R., Cortes D., Torres M. A., Shulaev V., Dangl J. L., Mittler R. 2009. The plant NADPH oxidase RBOHD mediates rapid systemic signaling in response to diverse stimuli. Sci. Signal. 2, ra45. 10.1126/scisignal.2000448 (doi:10.1126/scisignal.2000448) [DOI] [PubMed] [Google Scholar]
- 5.Foyer C. H., Bloom A., Queval G., Noctor G. 2009. Photorespiratory metabolism: genes, mutants, energetics, and redox signaling. Annu. Rev. Plant Biol. 60, 455–484 10.1146/annurev.arplant.043008.091948 (doi:10.1146/annurev.arplant.043008.091948) [DOI] [PubMed] [Google Scholar]
- 6.Queval G., Issakidis-Bourguet E., Hoeberichts F. A., Vandorpe M., Gakière B., Vanacker H., Miginiac-Maslow M., Van Breusegem F., Noctor G. 2007. Conditional oxidative stress responses in the Arabidopsis photorespiratory mutant cat2 demonstrate that redox state is a key modulator of daylength-dependent gene expression and define photoperiod as a crucial factor in the regulation of H2O2-induced cell death. Plant J. 52, 640–657 10.1111/j.1365-313X.2007.03263.x (doi:10.1111/j.1365-313X.2007.03263.x) [DOI] [PubMed] [Google Scholar]
- 7.Wagner D., et al. 2004. The genetic basis of singlet oxygen-induced stress responses of Arabidopsis thaliana. Science 306, 1183–1185 10.1126/science.1103178 (doi:10.1126/science.1103178) [DOI] [PubMed] [Google Scholar]
- 8.Chaouch S., Queval G., Vanderauwera S., Mhamdi A., Vandorpe M., Langlois-Meurinne M., Van Breusegem F., Saindrenan P., Noctor G. 2010. Peroxisomal hydrogen peroxide is coupled to biotic defense responses by ISOCHORISMATE SYNTHASE1 in a daylength-related manner. Plant Physiol. 153, 1692–1705 10.1104/pp.110.153957 (doi:10.1104/pp.110.153957) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee K. P., Kim C., Landgraf F., Apel K. 2007. EXECUTER1- and EXECUTER2-dependent transfer of stress-related signals from the plastid to the nucleus of Arabidopsis thaliana. Proc. Natl Acad. Sci. USA. 104, 10 270–10 275 10.1073/pnas.0702061104 (doi:10.1073/pnas.0702061104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foyer C. H., Noctor G. 2005. Redox homeostasis and antioxidant signalling: a metabolic interface between stress perception and physiological responses. Plant Cell 17, 1866–1875 10.1105/tpc.105.033589 (doi:10.1105/tpc.105.033589) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foyer C. H., Shigeoka S. 2011. Understanding oxidative stress and antioxidant functions to enhance photosynthesis. Plant Physiol. 155, 93–100 10.1104/pp.110.166181 (doi:10.1104/pp.110.166181) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Surpin M., Larkin R. M., Chory J. 2002. Signal transduction between the chloroplast and the nucleus. Plant Cell 14(Suppl.), S327–S338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galvez-Valdivieso G., Mullineaux P. M. 2010. The role of reactive oxygen species in signalling from chloroplasts to nucleus. Physiol. Plant. 138, 430–439 10.1111/j.1399-3054.2009.01331.x (doi:10.1111/j.1399-3054.2009.01331.x) [DOI] [PubMed] [Google Scholar]
- 14.Cottage C., Gray J. C. 2011. Timing the switch to phototrophic growth. Plant Signal. Behav. 6, 578–582 10.4161/psb.6.4.14900 (doi:10.4161/psb.6.4.14900) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pastori G. M., Kiddle G., Antoniw J., Bernard S., Veljovic-Jovanovic S., Verrier P. J., Noctor G., Foyer C. H. 2003. Leaf vitamin C contents modulate plant defense transcripts and regulate genes controlling development through hormone signaling. Plant Cell 15, 939–951 10.1105/tpc.010538 (doi:10.1105/tpc.010538) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noctor G., Mhamdi A., Chaouch S., Han Y., Neukermans J., Marquez-Garcia B., Queval G., Foyer C. H. 2012. Glutathione functions in plants: an integrated overview. Plant Cell Environ. 35, 454–484 10.1111/j.1365-3040.2011.02400.x (doi:10.1111/j.1365-3040.2011.02400.x) [DOI] [PubMed] [Google Scholar]
- 17.Kerchev P. I., et al. 2011. The transcription factor ABI-4 is required for the ascorbic acid-dependent regulation of growth and regulation of jasmonate-dependent defense signaling pathways in Arabidopsis. Plant Cell 23, 3319–3334 10.1105/tpc.111.090100 (doi:10.1105/tpc.111.090100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vernoux T., et al. 2000. The ROOT MERISTEMLESS1/CADMIUM SENSITIVE2 gene defines a glutathione-dependent pathway involved in initiation and maintenance of cell division during postembryonic root development. Plant Cell 12, 97–110 10.1105/tpc.12.1.97 (doi:10.1105/tpc.12.1.97) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartoli C. G., Gómez F., Fernández L., Yu J., McIntosh L., Foyer C. H. 2006. Inter-relationships between light and respiration in the control of ascorbic acid synthesis and accumulation in Arabidopsis thaliana leaves. J. Exp. Bot. 57, 1621–1631 10.1093/jxb/erl005 (doi:10.1093/jxb/erl005) [DOI] [PubMed] [Google Scholar]
- 20.Bartoli C. G., Tambussi E. A., Fanello D., Foyer C. H. 2009. Control of ascorbic acid synthesis and accumulation and glutathione by the incident light red/far red ratio in Phaseolus vulgaris leaves. FEBS Lett. 583, 118–122 10.1016/j.febslet.2008.11.034 (doi:10.1016/j.febslet.2008.11.034) [DOI] [PubMed] [Google Scholar]
- 21.Dowdle J., Ishikawa T., Gatzek S., Rolinski S., Smirnoff N. 2007. Two genes in Arabidopsis thaliana encoding GDP-l-galactose phosphorylase are required for ascorbate biosynthesis and seedling viability. Plant J. 52, 673–689 10.1111/j.1365-313X.2007.03266.x (doi:10.1111/j.1365-313X.2007.03266.x) [DOI] [PubMed] [Google Scholar]
- 22.Maruta T., Tanouchi A., Tamoi M., Yabuta Y., Yoshimura K., Ishikawa T., Shigeoka S. 2010. Arabidopsis chloroplastic ascorbate peroxidise isoenzymes play a dual role in photoprotection and gene regulation under photooxidative stress. Plant Cell Physiol. 51, 190–200 10.1093/pcp/pcp177 (doi:10.1093/pcp/pcp177) [DOI] [PubMed] [Google Scholar]
- 23.Wachter A., Wolf S., Steiniger H., Bogs J., Rausch T. 2005. Differential targeting of GSH1 and GSH2 is achieved by multiple transcription initiation: implications for the compartmentation of glutathione biosynthesis in the Brassicaceae. Plant J. 41, 15–30 10.1111/j.1365-313X.2004.02269.x (doi:10.1111/j.1365-313X.2004.02269.x) [DOI] [PubMed] [Google Scholar]
- 24.Noctor G., Arisi A. C., Jouanin L., Foyer C. H. 1998. Manipulation of glutathione and amino acid biosynthesis in the chloroplast. Plant Physiol. 118, 471–482 10.1104/pp.118.2.471 (doi:10.1104/pp.118.2.471) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu A., et al. 2012. JUNGBRUNNEN1, a reactive oxygen species responsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell 24, 482–506 10.1105/tpc.111.090894 (doi:10.1105/tpc.111.090894) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Queval G., Neukermans J., Vanderauwera S., Van Breusegem F., Noctor G. 2012. Day length is a key regulator of transcriptomic responses to both CO2 and H2O2 in Arabidopsis. Plant Cell Environ. 35, 374–387 10.1111/j.1365-3040.2011.02368.x (doi:10.1111/j.1365-3040.2011.02368.x) [DOI] [PubMed] [Google Scholar]
- 27.Maruta T., Noshi M., Tanouchi M., Tamoi M., Yabuta Y., Yoshimura K., Ishikawa T., Shigeoka S. 2012. H2O2-triggered retrograde signaling from chloroplasts to nucleus plays specific role in response to stress . J. Biol. Chem. 287, 11 717–11 729 10.1074/jbc.M111.292847 (doi:10.1074/jbc.M111.292847) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rehrauer H., et al. 2010. AGRONOMICS1: a new resource for Arabidopsis transcriptome profiling. Plant Physiol. 152, 487–499 10.1104/pp.109.150185 (doi:10.1104/pp.109.150185) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fratelli M., Goodwin L. O., Ørom U. A., Lombardi S., Tonelli R., Mengozzi M., Ghezzi P. 2005. Gene expression profiling reveals a signaling role of glutathione in redox regulation. Proc. Natl Acad. Sci. USA 102, 13 998–14 003 10.1073/pnas.0504398102 (doi:10.1073/pnas.0504398102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhong S., Zhao M., Shi T., Shi H., An F., Zhao Q., Guo H. 2009. EIN3/EIL1 cooperate with PIF1 to prevent photo-oxidation and to promote greening of Arabidopsis seedlings. Proc. Natl Acad. Sci. USA. 106, 21 431–21 436 10.1073/pnas.0907670106 (doi:10.1073/pnas.0907670106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davison P. A., Hunter C. N., Horton P. 2002. Overexpression of β-carotene hydroxylase enhances stress tolerance in Arabidopsis. Nature 418, 203–206 10.1038/nature00861 (doi:10.1038/nature00861) [DOI] [PubMed] [Google Scholar]
- 32.Veljovic-Jovanovic S., Pignocchi C., Noctor G., Foyer C. H. 2001. Low vitamin C in the vtc 1 mutant of Arabidopsis thaliana is associated with decreased growth and intracellular redistribution of the antioxidant system. Plant Physiol. 127, 426–435 10.1104/pp.010141 (doi:10.1104/pp.010141) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muller-Moulé P., Golan T., Niyogi K. K. 2004. Ascorbate-deficient mutants of Arabidopsis grow in high light despite chronic photooxidative stress. Plant Physiol. 134, 1163–1172 10.1104/pp.103.032375 (doi:10.1104/pp.103.032375) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takabayashi A., Ishikawa N., Obayashi T., Ishida S., Obokata T., Endo J., Sato S. 2009. Three novel subunits of Arabidopsis chloroplastic NAD(P)H dehydrogenase identified by bioinformatic and reverse genetic approaches. Plant J. 57, 207–219 10.1111/j.1365-313X.2008.03680.x (doi:10.1111/j.1365-313X.2008.03680.x) [DOI] [PubMed] [Google Scholar]
- 35.Shimizu H., Shikanai T. 2007. Dihydrodipicolinate reductase-like protein, CRR1, is essential for chloroplast NAD(P)H dehydrogenase in Arabidopsis. Plant J. 52, 539–547 10.1111/j.1365-313X.2007.03256.x (doi:10.1111/j.1365-313X.2007.03256.x) [DOI] [PubMed] [Google Scholar]
- 36.Dangoor I., Peled-Zehavi H., Levitan A., Pasand O., Danon A. 2009. A small family of chloroplast atypical thioredoxins. Plant Physiol. 149, 1240–1250 10.1104/pp.108.128314 (doi:10.1104/pp.108.128314) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schult K., Meierhoff K., Paradies S., Töller T., Wolff P., Westhoff P. 2007. The nuclear-encoded factor HCF173 is involved in the initiation of translation of the psbA mRNA in Arabidopsis thaliana. Plant Cell 19, 1329–1346 10.1105/tpc.106.042895 (doi:10.1105/tpc.106.042895) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu Y., Hall D. A., Last R. L. 2012. A small zinc finger thylakoid protein plays a role in maintenance of photosystem II in Arabidopsis thaliana. Plant Cell. 23, 1861–1875 10.1105/tpc.111.085456 (doi:10.1105/tpc.111.085456) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fu A., He Z., Cho H. S., Lima A., Buchanan B. B., Luan S. 2007. A chloroplast cyclophilin and maintenance of functions in the assembly and maintenance of photosystem II in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA 104, 15 947–15 952 10.1073/pnas.0707851104 (doi:10.1073/pnas.0707851104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dominguez-Solis J. R., He Z. Y., Lima A., Ting J., Buchanan B. B., Luan S. 2008. A cyclophilin links redox and light signals to cysteine biosynthesis and stress responses in chloroplasts. Proc. Natl Acad. Sci. USA 105, 16 386–16 391 10.1073/pnas.0808204105 (doi:10.1073/pnas.0808204105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ishihara S., Takabayashi A., Ido K., Endo T., Ifuku K., Sato F. 2007. Distinct functions for the two PsbP-like proteins PPL1 and PPL2 in the chloroplast thylakoid lumen of Arabidopsis. Plant Physiol. 145, 668–679 10.1104/pp.107.105866 (doi:10.1104/pp.107.105866) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yabuta S., Ifuku K., Takabayashi A., Ishihara S., Ido K., Ishikawa N., Endo T., Sato F. 2010. Three PsbQ-like proteins are required for the function of the chloroplast NAD(P)H dehydrogenase complex in Arabidopsis. Plant Cell Physiol. 51, 866–876 10.1093/pcp/pcq060 (doi:10.1093/pcp/pcq060) [DOI] [PubMed] [Google Scholar]
- 43.Ifuku K., Endo T., Shikanai T., Aro E. M. 2011. Structure of the chloroplast NADH dehydrogenase-like complex: nomenclature for nuclear-encoded subunits. Plant Cell Physiol. 52, 1560–1568 10.1093/pcp/pcr098 (doi:10.1093/pcp/pcr098) [DOI] [PubMed] [Google Scholar]
- 44.Muraoka R., Okuda K., Kobayashi Y., Shikanai T. 2006. A eukaryotic factor required for accumulation of the chloroplast NAD(P)H dehydrogenase complex in Arabidopsis. Plant Physiol. 142, 1683–1689 10.1104/pp.106.088682 (doi:10.1104/pp.106.088682) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bartoli C., Pastori G., Foyer C. H. 2000. Ascorbate biosynthesis in mitochondria is linked to the electron transport chain between complexes III and IV. Plant Physiol. 123, 335–343 10.1104/pp.123.1.335 (doi:10.1104/pp.123.1.335) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fuentes D., et al. 2011. A deficiency in the flavoprotein of Arabidopsis mitochondrial complex II results in elevated photosynthesis and better growth in nitrogen-limiting conditions. Plant Physiol. 157, 1114–1127 10.1104/pp.111.183939 (doi:10.1104/pp.111.183939) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nixon P. J., Michoux F., Yu J., Boehm M., Komenda J. 2010. Recent advances in understanding the assembly and repair of photosystem II. Ann. Bot. 106, 1–16 10.1093/aob/mcq059 (doi:10.1093/aob/mcq059) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Noctor G., Gomez L., Vanacker H., Foyer C. H. 2002. Glutathione homeostasis and signalling: the influence of biosynthesis, compartmentation and transport. J. Exp. Bot. 53, 1283–1304 10.1093/jexbot/53.372.1283 (doi:10.1093/jexbot/53.372.1283) [DOI] [PubMed] [Google Scholar]