Abstract
Human activities have reorganized the earth's biota resulting in spatially disparate locales becoming more or less similar in species composition over time through the processes of biotic homogenization and biotic differentiation, respectively. Despite mounting evidence suggesting that this process may be widespread in both aquatic and terrestrial systems, past studies have predominantly focused on single taxonomic groups at a single spatial scale. Furthermore, change in pairwise similarity is itself dependent on two distinct processes, spatial turnover in species composition and changes in gradients of species richness. Most past research has failed to disentangle the effect of these two mechanisms on homogenization patterns. Here, we use recent statistical advances and collate a global database of homogenization studies (20 studies, 50 datasets) to provide the first global investigation of the homogenization process across major faunal and floral groups and elucidate the relative role of changes in species richness and turnover. We found evidence of homogenization (change in similarity ranging from −0.02 to 0.09) across nearly all taxonomic groups, spatial extent and grain sizes. Partitioning of change in pairwise similarity shows that overall change in community similarity is driven by changes in species richness. Our results show that biotic homogenization is truly a global phenomenon and put into question many of the ecological mechanisms invoked in previous studies to explain patterns of homogenization.
Keywords: beta diversity, biotic homogenization, spatial turnover, species richness, taxonomic homogenization
1. Introduction
Invasions and extinctions have resulted in spatially disparate locales becoming more or less similar in species composition over time through the processes of biotic homogenization and differentiation, respectively [1–3]. Despite the growth in numbers of publications examining homogenization patterns in both aquatic and terrestrial systems, these efforts predominantly focused on single taxonomic groups at a single spatial scale [3]. Not surprisingly, we are left with a hodge-podge of results that highlight potential patterns of interest but that are never capable of documenting the universality of those patterns. For example, from previous studies, we know that patterns of homogenization and differentiation can vary depending on the taxonomic group examined [4], spatial extent and grain of the study [5–7], and the evolutionary history of the taxa included [8]. This fragmentation of evidence leaves the homogenization process both debated and untested globally.
A significant challenge to achieving a global understanding of homogenization patterns is that previous studies almost exclusively relied on metrics that measured changes in pairwise similarity (e.g. Jaccard's or Sorenson's index), never settling on one obviously suitable metric. Change in pairwise similarity is itself dependent on two distinct processes, spatial species turnover and changes in species richness [9]. Disentangling the effect of these two mechanisms is essential for interpreting homogenization patterns [10,11]. Spatial turnover involves the loss of species that are unique to each locale or the establishment of common invaders in the case of homogenization and the loss of species common to both locales or the establishment of different invaders in the case of differentiation. This process was invoked in McKinney & Lockwood's [1] seminal paper on biotic homogenization where it is defined as ‘the replacement of local biotas with non-indigenous species, usually introduced by humans’ (p. 450). Species losses and gains that reduce differences in richness between two locales can also increase similarity, whereas increased discrepancy in species richness between locales will decrease similarity.
The dynamics of spatial turnover and changes in species richness result in very different perceived mechanisms driving biotic homogenization with likely disparate ecological and evolutionary implications [2]. The richness component of similarity quantifies how richness gradients created by historical ecological and evolutionary processes (e.g. latitudinal, ecosystem size, island distance from mainland) are destroyed or accentuated by anthropogenic environmental change; but tells us nothing about change in compositional (and by extension functional or genetic) similarity [2,12]. Whereas the spatial turnover component tracks change in species similarity, shedding light on how the uniqueness of local biotas is altered by human activities driving anthropogenic change. As species are replaced through turnover, functional traits that provide a key link between diversity and ecosystem functioning [13] are also reshuffled; potentially resulting in a negative feedback loop further altering ecological systems [14]. So far, the role of richness and turnover in generating homogenization patterns has only been explored in one study [10], and cannot be distinguished by the majority of similarity measures thus far used to quantify homogenization.
Here, we use recent statistical advances to provide the first global investigation of the homogenization process across major faunal and floral groups and elucidate the relative role of changes in species richness and turnover for patterns of biotic homogenization and differentiation. By reanalysing a global database of previously published studies, we quantify how species invasions and extinctions have altered species composition across multiple spatial scales, thus providing new insights into the biogeography of the Homogecene era.
2. Methods
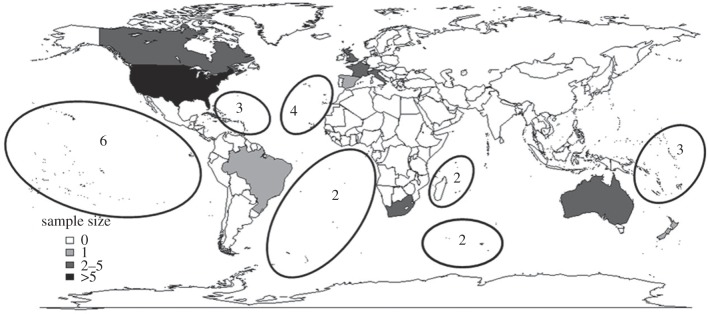
We collated 50 datasets compiled from 20 published studies that included major taxonomic groups across the world (figure 1; see the electronic supplementary material, appendix A and B). The spatial extent of each dataset was categorized as global, continental or provincial (e.g. country, state, province). Each dataset consisted of a group of spatially distinct subregions (hereafter termed locales) that ranged in grain size from small (<1 km2), moderate (1–100 km2), large (100–1000 km2), to very large (>1000 km2). Each dataset consisted of species presence–absence-by-locale matrices for a specific taxonomic group for two time periods (historical and contemporary; [15,16]) or across an anthropogenic gradient (e.g. natural to urban; [17]). For temporal studies, historical and contemporary data were compiled either by sampling at two time periods or considering native species only as the historical time period, and native + non-native species as the contemporary time period [18].
Figure 1.
Map showing the countries where homogenization datasets used in this analysis were collected. Circles denote oceanic regions where archipelagos or single islands were part of a dataset. Studies with a global spatial extent (n = 2) are excluded.
Pairwise (dis)similarity indices used to quantify β-diversity fall into two classes. The first is ‘broad-sense’ similarity that accounts for both spatial turnover and species richness gradients (e.g. Jaccard, Sorensen, Bray–Curtis). The second is ‘narrow-sense’ similarity that largely depends on spatial turnover alone (e.g. βsim, β3; [19]). Studies of biotic homogenization and differentiation predominantly use broad-sense measures ([20], but see [21–23]), leaving the influence of changes in species richness and turnover on similarity metrics indistinguishable.
Recently, methods have been developed for partitioning pairwise dissimilarity into its turnover and richness components [24–28]. The general mathematical partition:
shows that overall (broad-sense) dissimilarity is equal to the sum of dissimilarity owing to turnover and richness [24,27]. Here, we use the methods of Carvalho et al. [27] where broad-sense dissimilarity is quantified as the complement of Jaccard's similarity index, βcc [29]
where a is the number of species shared by two locales and, b and c are the number of species unique to each local, respectively. The narrow-sense or turnover component, β3 [30,31], is quantified as:
and the richness component, βrich [27], is quantified as:
Partitioning pairwise dissimilarity with βcc, β3, and βrich provides unbiased estimates of dissimilarity owing to richness and turnover, because all three components are scaled in the same manner (i.e. by total species richness of the two locales, a + b + c) and thus change proportionally to the replacement and gain/loss of species across locales [26–28].
While the mathematical partition requires the use of dissimilarity indices [27], we couched changes in these indices in terms of similarity through matrix subtraction. We subtracted the dissimilarity matrix representing the contemporary assemblages (i.e. the matrix from the more recent time period for temporal studies or the urban matrix for spatial studies) from the historical matrix (i.e. the matrix from the initial time period for temporal studies or the ‘natural’ matrix in spatial studies). Matrix subtraction was performed for all three dissimilarity measures, thereby creating three change matrices, Δβcc, Δβ3 and Δβrich, for each dataset. Positive values within the matrices Δβcc, Δβ3 and Δβrich indicate an increase in pairwise similarity between two locales in that region (i.e. homogenization), and negative values indicate a decrease in similarity between two locales (i.e. differentiation).
We performed univariate Mantel tests [32] where we designated the response matrix as the broad-sense change in compositional similarity, Δβcc, and the predictor matrix as either the change in similarity owing to turnover (Δβ3) or as the matrix showing change in similarity owing to species richness, Δβrich. We did not consider these two predictor variables within the same statistical model using a partial Mantel test because Δβrich and Δβ3 are additive components of the response Δβcc, so conditioning on either predictor in a partial Mantel test would have been nonsensical. To test for differences in the amount of variability in Δβcc explained by Δβrich and Δβ3 across all 50 datasets, we conducted a one-way analysis of variance (ANOVA) with type II sums of squares in which the response was the squared value of the Mantel correlation coefficient (r) output from the univariate Mantel tests, and the predictor variable was the type of β metric (i.e. βrich or β3).
It is possible that unique aspects of each dataset will impact the degree to which changes in richness or turnover will influence homogenization patterns. To gauge this effect, we tested whether the amount of variability in Δβcc explained by Δβrich and Δβ3 varied between major taxonomic groups (e.g. birds, plants, fish, reptiles, amphibians, mammals, specifically, ungulates), across the spatial extent of study (e.g. region, continent and globe), and across the sampling resolution or grain size of the study (e.g. small, moderate, large, very large). We performed three separate two-way ANOVAs with type II sums of squares in which the response variables were the squared value of the Mantel correlation coefficient (r) and the predictors were the type β metric (e.g. βrich or β3) and one of the three grouping variables (i.e. taxa, extent or grain). Our main purpose of running the two-way ANOVAs was to test the interaction term between the grouping variable and the type of β component to see whether the relationship between the Mantel R2 value and βrich and β3 varied by taxa, extent or grain. Prior to running the ANOVAs, we confirmed that the data met assumptions of normality of the distributions of residuals and homoscedasticity of variances. We excluded four taxonomic groups (amphibians, algae, reptiles and ungulates) in the taxa analysis, because there was two or less datasets for each of those taxonomic groups and excluded the group ‘global’ from the spatial extent analysis owing to only two studies that examined homogenization at this spatial scale. We performed all analyses, using R statistical software (v. 2.13.2) and the Ecodist, Car and Vegan packages.
3. Results
Our results provide evidence for the global homogenization of biotas. The mean similarity across all datasets increased by 8 per cent for βcc (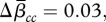 s.d. = 0.08), 2 per cent for βrich (
s.d. = 0.08), 2 per cent for βrich (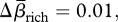 s.d. = 0.07) and 0.4 per cent for β3 (
s.d. = 0.07) and 0.4 per cent for β3 (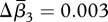 s.d. = 0.01) indicating homogenization (positive values) for each metric. Birds and plants homogenized according to all three metrics (table 1), whereas fish differentiated for Δβcc and Δβrich and homogenized according to β3 (table 1). The mean change in similarity indicated homogenization in all three metrics for both the continental and regional spatial extent. All three metrics indicated homogenization for studies using small, high and very high grain size, whereas studies sampling at a moderate grain size showed differentiation for all three metrics (table 1).
s.d. = 0.01) indicating homogenization (positive values) for each metric. Birds and plants homogenized according to all three metrics (table 1), whereas fish differentiated for Δβcc and Δβrich and homogenized according to β3 (table 1). The mean change in similarity indicated homogenization in all three metrics for both the continental and regional spatial extent. All three metrics indicated homogenization for studies using small, high and very high grain size, whereas studies sampling at a moderate grain size showed differentiation for all three metrics (table 1).
Table 1.
Mean and standard deviation for broad-sense (Δβcc) and narrow-sense (spatial turnover; Δβ3) change in similarity and change in similarity owing to differences in species richness (Δβrich). Algae, ungulates, reptiles and amphibians for taxonomic groups and global for spatial extent were not included owing to a sample size ≤2.
| group | metric | n | Δβcc mean (s.d.) | Δβrich mean (s.d.) | Δβ3 mean (s.d.) |
|---|---|---|---|---|---|
| all | 50 | 0.03 (0.08) | 0.01 (0.07) | 0.003 (0.01) | |
| taxa | birds | 26 | 0.04 (0.08) | 0.01 (0.06) | 0.02 (0.04) |
| fish | 12 | −0.002 (0.1) | −0.01 (0.09) | 0.002 (0.03) | |
| plants | 7 | 0.04 (0.06) | 0.03 (0.05) | 0.005 (0.01) | |
| spatial extent | continent | 10 | 0.05 (0.08) | 0.02 (0.07) | 0.03 (0.04) |
| region | 38 | 0.01 (0.09) | 0.004 (0.07) | 0.009 (0.04) | |
| grain size | small | 11 | 0.09 (0.12) | 0.04 (0.1) | 0.05 (0.05) |
| moderate | 15 | −0.02 (0.07) | −0.02 (0.07) | −0.003 (0.03) | |
| high | 12 | 0.02 (0.06) | 0.006 (0.05) | 0.01 (0.02) | |
| very high | 12 | 0.02 (0.04) | 0.01 (0.02) | 0.006 (0.02) |
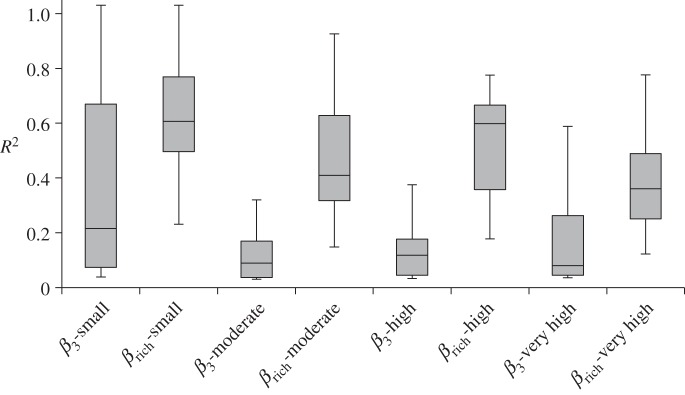
Across all studies, change in broad-sense similarity, Δβcc, was predominantly explained by changes in species richness Δβrich (Mantel correlation coefficient R2: mean = 0.47, s.d. = 0.23) rather than by spatial turnover β3 (Mantel correlation coefficient R2: mean = 0.17, s.d. = 0.25; figure 2). The amount of variation in βcc explained by βrich was significantly greater than variation attributable to β3 (ANOVA: F1,98 = 39.57, p ≤ 0.001). βrich explained more variation in βcc, regardless of taxonomic group (ANOVA: F1,84 = 34.14, p ≤ 0.001; figure 2), spatial extent (ANOVA: F1,92 = 39.67, p ≤ 0.001; figure 3) or spatial grain (ANOVA: F1,92 = 43.60, p ≤ 0.001; figure 4). Testing the interaction term between β metric and taxonomic group (ANOVA: F2,84 = 2.77, p = 0.07; figure 2), spatial extent (ANOVA: F2,92 = 0.47, p = 0.5; figure 3) or spatial grain (ANOVA: F2,92 = 1.19, p = 0.32; figure 4) in the two-way ANOVAs yielded no significant differences across these three categories.
Figure 2.
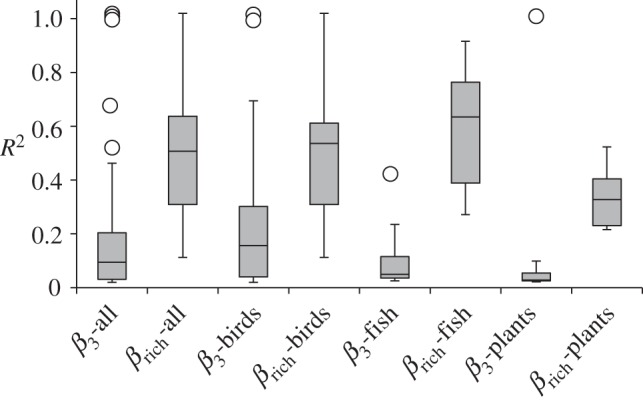
Box plot comparing the mean mantel R2 of Δβcc versus Δβ3 and Δβcc versus Δβrich for all taxonomic groups (all) and across taxonomic groups (birds, fish and plants).
Figure 3.
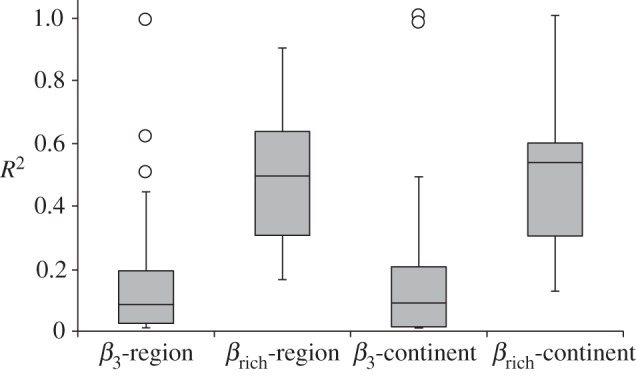
Box plot comparing the mean mantel R2 of Δβcc versus Δβ3 and Δβcc versus Δβrich across spatial extent (continental and regional). Global spatial extent was not included because only two of the 50 datasets studied homogenization at this scale.
Figure 4.
Box plot comparing the mean mantel R2 of Δβcc versus Δβ3 and Δβcc versus Δβrich across spatial grain (small <1 km2, moderate 1–100 km2, large 100–1000 km2, to very large >1000 km2).
The Mantel correlation coefficient (r) was positive for both βrich and β3 for 40 of the datasets. However, for seven datasets, the Mantel correlation coefficient (r) for β3 was negative, whereas βrich was positive and, for three datasets, the opposite was true. This result implies that there are scenarios where a pair of locales can become more similar according to one metric (e.g. species richness) and less similar based on another (e.g. species composition).
4. Discussion
We show that biotic homogenization is truly a global phenomenon, where species invasions and extinctions dramatically reorganize a number of major faunal and floral groups. We provide the first multi-taxa investigation of homogenization using the same metrics for all datasets, which, for the first time, allows a cross-taxa comparison of homogenization patterns across spatial extent and grain. We show a tendency towards homogenization, regardless of the metric used across nearly all taxonomic groups, spatial extent and grain sizes. The exceptions to this trend are fish, which showed differentiation for Δβcc and Δβrich, and studies that sampled at a moderate grain size (1–1000 km2), which showed differentiation for all three metrics (table 1). These results suggest that most taxonomic groups evaluated at most spatial scales are becoming more similar overall (Δβcc) owing to both spatial turnover (Δβ3) and changes in species richness (Δβrich). However, the standard deviations are large for all measures (table 1) suggesting that all groups assessed here include some locales that have differentiated.
Overall (broad-sense) change in similarity is driven by changes in species richness as opposed to spatial turnover. Thus, what we perceive as biotic homogenization or differentiation is largely dependent on how invasions and extinctions either diminish species richness gradients or accentuate them. Spatial turnover consistently plays a lesser role in driving patterns of community similarity despite this mechanism being suggested as the basis of homogenization [1]. This result puts into question many of the ecological mechanisms invoked in previous studies to explain patterns of homogenization across taxonomic groups.
While our results show that overall change in pairwise similarity is largely driven by changes in species richness, there is evidence that either metric can be negatively correlated with overall change in similarity. This result opens the door for misinterpretations of homogenization patterns when only using a broad-sense metric, as most homogenization studies do [18]. In the case that Δβrich and Δβ3 have opposite signs (i.e. one shows homogenization and one differentiation), the resulting broad-sense similarity measure can show little or no change [10]. One would then conclude, based on their broad-sense metric, that homogenization or differentiation is not occurring when, in fact, there are large changes in species richness and spatial turnover. On the other hand, using only a narrow-sense metric could fail to identify a large invasion or extinction event if species losses or gains do not affect spatial turnover.
A further benefit of partitioning change in community similarity into its species richness and turnover components is the opportunity to identify mechanisms and test hypotheses about how invasions and extinctions drive biotic homogenization and differentiation [9]. Change in species richness—measured as βrich—results in homogenization when species richness gradients get smaller (i.e. the rich get poorer and/or the poor get richer) and differentiation when species richness gradients get larger (i.e. the rich get richer and/or the poor get poorer). If invasion dynamics follow the same pattern as the initial assembly dynamics that created the original species richness gradient, then differences in species richness will increase resulting in differentiation according to βrich. An example comes from the theory of island biogeography [33] where larger islands, or islands close to the mainland accumulate more species during assembly than smaller and more distant islands. If the theory of island biogeography applies to invasive species [34], then species rich islands will accumulate more invaders than species poor islands, thereby accentuating the species richness gradient. Here, we present a framework to test the hypothesis that pairwise Δβrich will be negatively correlated with pairwise difference in island size.
Conversely, if non-native species establishment is driven by a factor unrelated to initial community assembly and thus these species decrease the natural difference in species richness, the locales will homogenize. Once again using the theory of island biogeography, if human settlement and thus introduction of non-native species occurred on smaller or more distant islands, then these islands would accumulate relatively more species and eventually close the gap in species richness with their neighbouring species rich unsettled islands. The hypothesis to test in this case is that pairwise Δβrich is positively correlated with pairwise difference in colonization date, human population size or number of ports. By using our approach here, hypotheses can be developed for invasion and extinction patterns for both Δβrich and Δβ3 based on knowledge of the study system.
The anthropogenic reshuffling of the earth's biota has resulted in taxonomic homogenization, irrespective of taxonomic group and spatial scale. Species extinctions and invasions will undoubtedly continue, and understanding the role of changes in turnover and richness gradients can help predict future patterns of homogenization and ecosystem change. Although enhancing our knowledge of the homogenization process and resultant patterns can help inform conservation efforts at biogeographic scales [35], the selection of quantitative metrics is crucial to accurately understanding the underlying ecological mechanisms.
Acknowledgements
We graciously thank the following people for sharing their data: T. M. Blackburn, R. B. Blair, P. Cassey, M. Clavero, O. Filippi-Codaccioni, C. W. Hoagstrom, S. A. Keith, D. Lobo, M. P. Marchetti, L. Piazzi, J. Pino, J. Plue, M. W. Schwartz, S. M. Smart, K. G. Smith, D. Spear, E. B. Taylor, B. J. van Rensburg and all of their co-authors. Without their excellent work and willingness to share, this paper would not have been possible. Datasets were contributed for use in this publication only and as a result, we could not make them publically available. Please contact the authors directly for inquiries regarding these datasets. We also thank Aaron Ellison for comments on this manuscript.
References
- 1.McKinney M. L., Lockwood J. L. 1999. Biotic homogenization: a few winners replacing many losers in the next mass extinction. Trends Ecol. Evol. 14, 450–453 10.1016/S0169-5347(99)01679-1 (doi:10.1016/S0169-5347(99)01679-1) [DOI] [PubMed] [Google Scholar]
- 2.Olden J. D., Poff N. L., Douglas M. R., Douglas M. E., Fausch K. D. 2004. Ecological and evolutionary consequences of biotic homogenization. Trends Ecol. Evol. 19, 18–24 10.1016/j.tree.2003.09.010 (doi:10.1016/j.tree.2003.09.010) [DOI] [PubMed] [Google Scholar]
- 3.Olden J. D. 2006. Biotic homogenization: a new research agenda for conservation biogeography. J. Biogeogr. 33, 2027–2039 10.1111/j.1365-2699.2006.01572.x (doi:10.1111/j.1365-2699.2006.01572.x) [DOI] [Google Scholar]
- 4.Olden J. D., Poff N. L., McKinney M. L. 2006. Forecasting faunal and floral homogenization associated with human population geography in North America. Biol. Conserv. 127, 261–271 10.1016/j.biocon.2005.04.027 (doi:10.1016/j.biocon.2005.04.027) [DOI] [Google Scholar]
- 5.Cassey P., Blackburn T. M., Lockwood J. L., Sax D. F. 2006. A stochastic model for integrating changes in species richness and community similarity across spatial scales. Oikos 115, 207–218 10.1111/j.2006.0030-1299.15223.x (doi:10.1111/j.2006.0030-1299.15223.x) [DOI] [Google Scholar]
- 6.Marchetti M. P., Lockwood J. L., Light T. 2006. Effects of urbanization on California's fish diversity: differentiation, homogenization and the influence of spatial scale. Biol. Conserv. 127, 310–318 10.1016/j.biocon.2005.04.025 (doi:10.1016/j.biocon.2005.04.025) [DOI] [Google Scholar]
- 7.Olden J. D., Kennard M. J., Pusey B. J. 2008. Species invasions and the changing biogeography of Australian freshwater fishes. Glob. Ecol. Biogeogr. 17, 25–37 10.1111/j.1466-8238.2007.00340.x (doi:10.1111/j.1466-8238.2007.00340.x) [DOI] [Google Scholar]
- 8.Cassey P., Lockwood J. L., Blackburn T. M., Olden J. D. 2007. Spatial scale and evolutionary history determine the degree of taxonomic homogenization across island bird assemblages. Diversity Distrib. 13, 458–466 10.1111/j.1472-4642.2007.00366.x (doi:10.1111/j.1472-4642.2007.00366.x) [DOI] [Google Scholar]
- 9.Olden J. D., Poff N. L. 2003. Toward a mechanistic understanding and prediction of biotic homogenization. Am. Nat. 162, 442–460 10.1086/378212 (doi:10.1086/378212) [DOI] [PubMed] [Google Scholar]
- 10.Baeten L., Vagansbeke P., Hermy M., Petrken G., Vanhuyse K., Verheyen K. 2012. Distinguishing between turnover and nestedness in the quantification of biotic homogenization. Biodivers. Conserv. 21, 1399–1409 10.1007/s10531-012-0251-0 (doi:10.1007/s10531-012-0251-0) [DOI] [Google Scholar]
- 11.Villéger S., Brosse S. 2012. Measuring changes in taxonomic dissimilarity following species introductions and extirpations. Ecol. Indicat. 18, 552–558 10.1016/j.ecolind.2012.01.009 (doi:10.1016/j.ecolind.2012.01.009) [DOI] [Google Scholar]
- 12.Su J. C., Debinski D. M., Jakubauskas M. E., Kindscher K. 2004. Beyond species richness: community similarity as a measure of cross-taxon congruence for coarse-filter conservation. Conserv. Biol. 18, 167–173 10.1111/j.1523-1739.2004.00337.x (doi:10.1111/j.1523-1739.2004.00337.x) [DOI] [Google Scholar]
- 13.Baiser B., Lockwood J. L. 2011. The relationship between functional and taxonomic homogenization. Glob. Ecol. Biogeogr. 20, 134–144 10.1111/j.1466-8238.2010.00583.x (doi:10.1111/j.1466-8238.2010.00583.x) [DOI] [Google Scholar]
- 14.Simberloff D., Von Holle B. 1999. Positive interactions of nonindigenous species: invasional meltdown? Biol. Invasions 1, 21–32 10.1023/A:1010086329619 (doi:10.1023/A:1010086329619) [DOI] [Google Scholar]
- 15.Smart S. M., Thompson K., Marrs R. H., Le Duc M., Lindsay G., Maskell C., Firbank L. G. 2006. Biotic homogenization and changes in species diversity across human-modified ecosystems. Proc. R. Soc. B 273, 2659–2665 10.1098/rspb.2006.3630 (doi:10.1098/rspb.2006.3630) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keith S. A., Newton A. C., Morecroft M. D., Bealey C. E., Bullock J. M. 2009. Taxonomic homogenization of woodland plant communities over 70 years. Proc. R. Soc. B 276, 3539–3544 10.1098/rspb.2009.0938 (doi:10.1098/rspb.2009.0938) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blair R. B., Johnson E. M. 2008. Suburban habitats and their role for birds in the urban-rural habitat network, points of local invasion and extinction?. Landscape Ecol. 23, 1157–1169 10.1007/s10980-008-9267-y (doi:10.1007/s10980-008-9267-y) [DOI] [Google Scholar]
- 18.Olden J. D., Rooney T. P. 2006. On defining and quantifying biotic homogenization. Glob. Ecol. Biogeogr. 15, 113–120 10.1111/j.1466-822X.2006.00214.x (doi:10.1111/j.1466-822X.2006.00214.x) [DOI] [Google Scholar]
- 19.Koleff P., Gaston K. J., Lennon J. 2003. Measuring beta diversity for presence–absence data. J. Anim. Ecol. 72, 367–382 10.1046/j.1365-2656.2003.00710.x (doi:10.1046/j.1365-2656.2003.00710.x) [DOI] [Google Scholar]
- 20.Olden J. D., Lockwood J. L., Parr C. L. 2011. Species invasions and the biotic homogenization of faunas and floras. In Conservation biogeography (eds Whittaker R. J., Ladle R. J.), pp. 224–243 Oxford, UK: Wiley-Blackwell [Google Scholar]
- 21.La Sorte F. A., Boecklen W. J. 2005. Temporal turnover of common species in avian assemblages in North America. J. Biogeogr. 32, 1151–1160 10.1111/j.1365-2699.2005.01271.x (doi:10.1111/j.1365-2699.2005.01271.x) [DOI] [Google Scholar]
- 22.Leprieur F., Olden J. D., Lek S., Brosse S. 2009. Contrasting patterns and mechanisms of spatial turnover for native and exotic freshwater fishes in Europe. J. Biogeogr. 36, 1899–1912 10.1111/j.1365-2699.2009.02107.x (doi:10.1111/j.1365-2699.2009.02107.x) [DOI] [Google Scholar]
- 23.Pool T. K., Olden J. D. 2012. Taxonomic and functional homogenization of a globally endemic desert fish fauna. Diversity Distrib. 18, 366–376 10.1111/j.1472-4642.2011.00836.x (doi:10.1111/j.1472-4642.2011.00836.x) [DOI] [Google Scholar]
- 24.Baselga A. 2010. Partitioning the turnover and nestedness components of beta diversity. Glob. Ecol. Biogeogr. 19, 134–143 10.1111/j.1466-8238.2009.00490.x (doi:10.1111/j.1466-8238.2009.00490.x) [DOI] [Google Scholar]
- 25.Baselga A. 2012. The relationship between species replacement, dissimilarity derived from nestedness, and nestedness. Glob. Ecol. Biogeogr. 10.1111/j.1466-8238.2011.00756.x (doi:10.1111/j.1466-8238.2011.00756.x) [DOI] [Google Scholar]
- 26.Podani J., Schmera D. 2011. A new conceptual and methodological framework for exploring and explaining pattern in presence–absence data. Oikos 120, 1625–1638 10.1111/j.1600-0706.2011.19451.x (doi:10.1111/j.1600-0706.2011.19451.x) [DOI] [Google Scholar]
- 27.Carvalho J. C., Cardoso P., Gomes P. 2012. Determining the relative roles of species replacement and species richness differences in generating beta-diversity patterns. Glob. Ecol. Biogeogr. 21, 760–771 10.1111/j.1466-8238.2011.00694.x (doi:10.1111/j.1466-8238.2011.00694.x) [DOI] [Google Scholar]
- 28.Carvalho J. C., Cardoso P., Borges P. A. V., Schmera D., Podani J. In press Measuring fractions of beta diversity and their relationships to nestedness: a theoretical and empirical comparison of novel approaches. Oikos (doi:10.1111/j.1600-0706.2012.20980.x) [Google Scholar]
- 29.Colwell R. K., Coddington J. A. 1994. Estimating terrestrial biodiversity through extrapolation. Phil. Trans. R. Soc. Lond. B 345, 101–118 10.1098/rstb.1994.0091 (doi:10.1098/rstb.1994.0091) [DOI] [PubMed] [Google Scholar]
- 30.Williams P. H. 1996. Mapping variations in the strength and breadth of biogeographic transition zones using species turnover. Proc. R. Soc. Lond. B 263, 579–588 10.1098/rspb.1996.0087 (doi:10.1098/rspb.1996.0087) [DOI] [Google Scholar]
- 31.Cardoso P., Borges P. A. V., Veech J. A. 2009. Testing the performance of beta diversity measures based on incidence data: the robustness to undersampling. Diversity Distrib. 15, 1081–1090 10.1111/j.1472-4642.2009.00607.x (doi:10.1111/j.1472-4642.2009.00607.x) [DOI] [Google Scholar]
- 32.Mantel N. 1967. The detection of disease clustering and a generalized regression approach. Cancer Res. 27, 209–220 [PubMed] [Google Scholar]
- 33.MacArthur R. H., Wilson E. O. 1967. The theory of island biogeography. Princeton, NJ: Princeton University Press [Google Scholar]
- 34.Blackburn T. M., Cassey P., Lockwood J. L. 2008. The island biogeography of exotic bird species. Glob. Ecol. Biogeogr. 17, 246–251 10.1111/j.1466-8238.2007.00361.x (doi:10.1111/j.1466-8238.2007.00361.x) [DOI] [Google Scholar]
- 35.Rooney T. P., Olden J. D., Leach M. K., Rogers D. A. 2007. Biotic homogenization and conservation prioritization. Biol. Conserv. 134, 447–450 10.1016/j.biocon.2006.07.008 (doi:10.1016/j.biocon.2006.07.008) [DOI] [Google Scholar]