Abstract
Although carotenoids serve important biological functions, animals are generally unable to synthesize these pigments and instead obtain them from food. However, many animals, such as sap-feeding insects, may have limited access to carotenoids in their diet, and it was recently shown that aphids have acquired the ability to produce carotenoids by lateral transfer of fungal genes. Whiteflies also contain carotenoids but show no evidence of the fungus-derived genes found in aphids. Because many sap-feeding insects harbour intracellular bacteria, it has long been hypothesized that these endosymbionts could serve as an alternative source of carotenoid biosynthesis. We sequenced the genome of the obligate bacterial endosymbiont Portiera from the whitefly Bemisia tabaci. The genome exhibits typical signatures of obligate endosymbionts in sap-feeding insects, including extensive size reduction (358.2 kb) and enrichment for genes involved in essential amino acid biosynthesis. Unlike other sequenced insect endosymbionts, however, Portiera has bacterial homologues of the fungal carotenoid biosynthesis genes in aphids. Therefore, related lineages of sap-feeding insects appear to have convergently acquired the same functional trait by distinct evolutionary mechanisms—bacterial endosymbiosis versus fungal lateral gene transfer.
Keywords: Bemisia tabaci, carotenoids, endosymbionts, genome reduction, Candidatus Portiera aleyrodidarum
1. Introduction
Carotenoids are a widespread class of pigments that play a variety of important functional roles based on their ability to absorb light and prevent oxidation [1,2]. They are synthesized by diverse lineages of eubacteria, archaea, protists, fungi and plants, but animals generally lack the ability to produce carotenoids and must sequester them from their diet. However, at least two lineages of arthropods have independently acquired a set of carotenoid biosynthesis genes by lateral transfer from fungi [3–5]. This was first shown in aphids, which are phloem sap-feeding insects within the suborder Sternorrhyncha (Hemiptera). Interestingly, other sap-feeding insects within the Sternorrhyncha, including the whitefly Bemisia tabaci, have also been found to contain carotenoids, raising the possibility that the transfer event occurred prior to their divergence from aphids [5]. But PCR-based screens did not find evidence of carotenoid biosynthetic genes with fungal ancestry in the B. tabaci genome [5]. Thus, the source of carotenoids in whiteflies remains unknown.
Bemisia tabaci is a species complex of major agricultural pests that affect a wide range of crop species [6]. Like many sap-feeding insects, whiteflies have evolved ancient relationships with intracellular bacteria that reside within a brightly pigmented abdominal organ known as the bacteriome [7]. Such bacteria commonly provision their hosts with essential amino acids, but they may play other important roles as well. For example, it has been hypothesized that endosymbiotic bacteria could serve as a source of carotenoids for their insect hosts [8,9]. However, direct evidence for this hypothesis is lacking and it has been undermined by the absence of carotenoid genes in sequenced endosymbiont genomes [10,11] and the emergence of lateral gene transfer to host nuclear genomes as an alternative explanation [3].
Here, we report the complete genome sequence of the obligate or ‘primary’ whitefly endosymbiont, Candidatus Portiera aleyrodidarum (hereafter referred to as Portiera). The genome contains genes coding for enzymes that catalyse the key steps in carotenoid biosynthesis, supporting the hypothesis that endosymbiotic bacteria can provide a source of carotenoids and indicating that related lineages of sap-feeding insects have acquired the ability to synthesize carotenoids by independent mechanisms.
2. Material and methods
Bemisia tabaci (B biotype) specimens from a laboratory colony founded with insects collected on cotton plants in Maricopa County, AZ, USA in October 2009 were kindly provided by Dr Martha Hunter. Total insect DNA was extracted from multiple pooled individuals and used for paired-end Illumina sequencing and endosymbiont genome assembly with a combination of Velvet v. 1.1.06 [12], SOAP v. 2.21 [13] and MIRA v. 3.4.0 [14] as described previously [15]. Initial analysis with Velvet assembled the Portiera genome into two scaffolds, consisting of 20 contigs, which were distinguished from sequences corresponding to the host and other symbionts on the basis of their depth of coverage, homology to published sequences and paired-end connections that could be linked into a closed circular assembly. Following gap closing, the genome was annotated with the JGI IMG-ER pipeline [16] and submitted to GenBank (CP003708).
All reported phylogenetic analyses were conducted with RAxML v. 7.2.6 [17], using amino acid sequences aligned with MUSCLE v. 3.7 [18] and trimmed with Gblocks v. 0.91b [19]. Substitution models were chosen based on analysis with ProtTest v. 2.4 [20].
3. Results
Sequencing of total insect DNA from B. tabaci produced deep genomic coverage of its obligate endosymbiont and yielded a closed, circular genome. Of 16.4 million Illumina read pairs, 2.9 per cent mapped to the Portiera genome, yielding a median coverage of 255×. Lower coverage contigs corresponding to the known ‘secondary’ endosymbionts Rickettsia sp. and Hamiltonella defensa were also detected but not analysed further. The Portiera genome exhibits common signatures of obligate endosymbionts, including a dramatic reduction in size and low GC content (table 1). It contains two pairs of direct repeats (172 and 349 bp in size) that are associated with paired-end sequencing conflicts in the assembly (see the electronic supplementary material, figure S1), raising the possibility that recombination between repeat pairs generates alternative genome structures as observed in Tremblaya, an obligate endosymbiont of mealybugs [21]. However, because DNA was pooled from multiple individual insects from a potentially genetically heterogeneous laboratory colony, it is not clear how this variation might be structured within and among host individuals, and we cannot exclude the possibility that PCR-mediated recombination could be an artefactual source of read-pair conflicts. Mapping reads against the Portiera genome identified several SNPs (ca 0.2 kb–1 with a minor allele frequency of at least 10%), further supporting the existence of multiple Portiera variants within the pooled sample.
Table 1.
Portiera genome summary.
| gene count | total length (kb) | coverage (%) | GC content (%) | |
|---|---|---|---|---|
| genic | 248.5 | 69.4 | 27.2 | |
| protein coding | 256 | 241.6 | 67.4 | 26.7 |
| rRNA | 3 | 4.4 | 1.2 | 45.9 |
| tRNA | 33 | 2.5 | 0.7 | 49.8 |
| intergenic | 109.8 | 30.6 | 23.6 | |
| total | 292 | 358.2 | 100.0 | 26.2 |
With intergenic sequences representing 30.6 per cent of the genome, Portiera has an unusually low gene density compared with other obligate endosymbionts [22], but its overall gene content shows many parallels with bacteria from related sap-feeding insects. The extremely reduced genome is highly enriched for genes involved in translation and essential amino acid biosynthesis (see the electronic supplementary material, figures S2 and S3), supporting the expectation that Portiera plays a major role in supplying amino acids to its host.
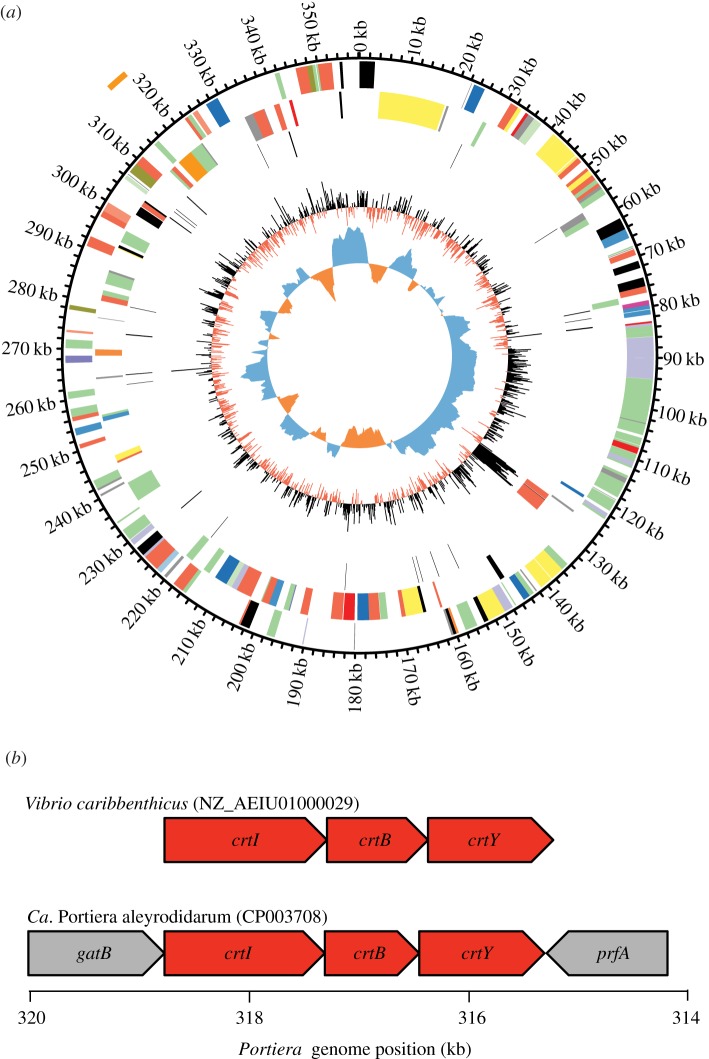
Notably, the genome contains three genes coding for proteins with clear homology to enzymes that carry out the core steps of carotenoid biosynthesis, including the synthesis of phytoene and subsequent desaturation and cyclization reactions (table 2). The same enzymes have been acquired by aphids and spider mites via lateral gene transfer from fungi [3,4], but the sequence and syntenic organization of the Portiera genes confirm that they are bacterial, not fungal, in origin (table 2; figures 1b and 2; see the electronic supplementary material, S4). BLAST searches against transcriptome sequences from another whitefly species, Trialeurodes vaporariorum, identified at least one read corresponding to orthologues of each of the three carotenoid biosynthesis genes, indicating that they were ancestrally present in Portiera across divergent host lineages [24].
Table 2.
Carotenoid biosynthesis genes in Portiera genome.
| gene | description | length (AA) | top BLAST hit | species | E-value | AA identity (%) |
|---|---|---|---|---|---|---|
| crtB | phytoene synthase | 287 | ABL97779 | uncultured marine bacterium HF10_29C11 | 6 × 10–38 | 34.2 |
| crtI | phytoene desaturase | 487 | ZP_07741622 | Vibrio caribbenthicus (Gammaproteobacteria) | 3 × 10–116 | 43.8 |
| crtY | lycopene cyclase | 379 | ZP_05052929 | Octadecabacter antarcticus (Alphaproteobacteria) | 2 × 10–32 | 28.4 |
Figure 1.
(a) Portiera chromosome map generated with Circos v. 0.56 [23]. From outside to inside, tracks depict: protein genes on (i) forward and (ii) reverse strands, (iii) RNA genes with rRNAs in red and tRNAs in black, (iv) GC content, and (v) GC skew. Protein-coding sequences are coloured by COG category. The orange line outside the circle notes the position of the three carotenoid biosynthesis genes. (b) Structural organization of carotenoid biosynthesis genes (red) and functionally unrelated flanking genes (grey) in Portiera and comparison with top BLAST hit, Vibrio caribbenthicus.
Figure 2.
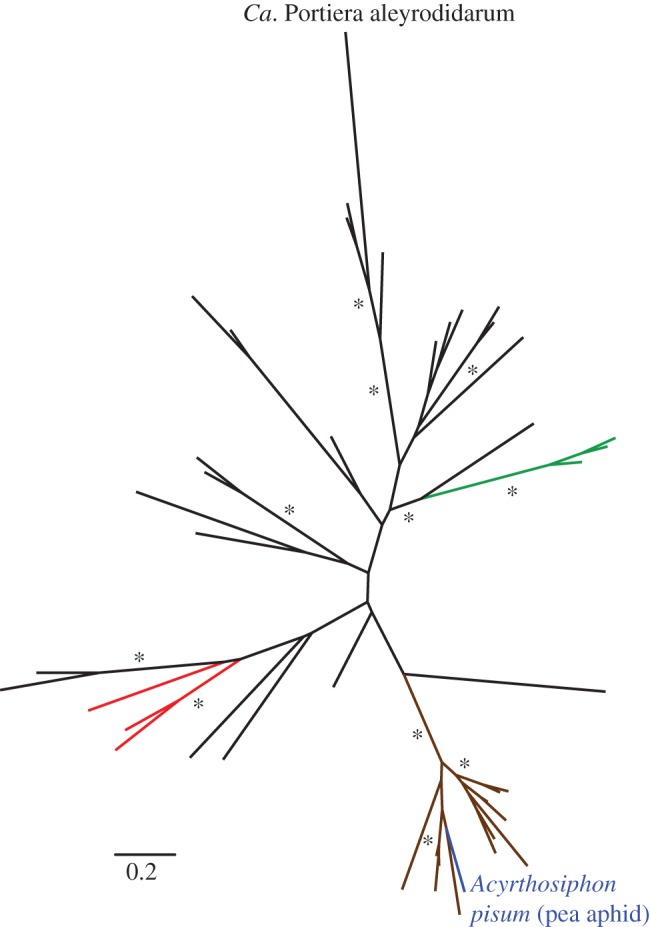
Unrooted phylogeny of carotenoid desaturase genes found in genomes of bacteria, archaea, plants, fungi and animals (black, red, green, brown and blue branches, respectively). Asterisks indicate bootstrap bipartition support of more than 90%. An expanded version with full taxon name, accession and bootstrap labelling is provided in electronic supplementary material, figure S4.
4. Discussion
The finding that carotenoid biosynthesis genes are retained in the highly reduced Portiera genome supports the hypothesis that, in at least some insects, endosymbiotic bacteria provide a source of carotenoids. This apparently novel mechanism for carotenoid biosynthesis in animals is clearly distinct from the cases of lateral gene transfer recently observed in other arthropods. The evidence for independent origins of carotenoid biosynthesis within the Sternorrhyncha indicates that sap-feeding insects may be under strong selection to acquire this ability. Although the specific functional roles of carotenoids in whiteflies are not clear, the distinctive yellow/orange pigmentation of the bacteriome raises the possibility that they may be concentrated in this organ. Given the intense metabolic activity within insect cells that house endosymbiotic bacteria [25], we hypothesize that bacterial-derived carotenoids may play an important role in preventing oxidative damage in this tissue. In addition, carotenoids have been found to protect against DNA damage and genomic instability [26], which may be of heightened importance in the context of an endosymbiont that has lost almost all genes involved in DNA repair. Speculatively, phloem-feeding insects, such as aphids and whiteflies, which consume high-sugar diets and are exposed to high levels of light, may experience a greater need for the protective effects of carotenoids.
Previous studies based on 16S rRNA sequences have grouped Portiera with members of the Halomonadaceae (Gammaproteobacteria), and suggested that it is the closest known relative of Carsonella, the obligate endosymbiont of psyllids, another group of sap-feeding insects within the Sternorrhyncha [27,28]. This relationship is also supported by protein-based analysis (see the electronic supplementary material, figure S5). Interestingly, the psyllid Pachypsylla venusta has also been found to contain carotenoids [5], but its Carsonella genome lacks genes required for their synthesis [11], suggesting that they are obtained from an alternative and as yet unidentified source. Whiteflies may also use additional sources of carotenoids. The B. tabaci carotenoid profile was found to contain beta-zeacarotene and the xanthophyll lutein [5]. The synthesis of xanthophylls, which are abundant in plants, requires a carotene hydroxylase [2], an enzyme commonly found in plants but not encoded in the Portiera genome. Therefore, whitefly carotenoid content potentially reflects a combination of dietary and de novo sources.
The acquisition of novel functions from bacterial endosymbionts is a defining theme in eukaryotic evolution. For example, the ubiquitous presence of carotenoids in plants reflects the ancient endosymbiotic origins of plastids from cyanobacteria [1]. In aphids, facultative endosymbionts have also been shown to affect host pigmentation through biochemical pathways unrelated to carotenoids [29]. Therefore, it is likely that endosymbiotic bacteria play a more widespread role in shaping host pigmentation than currently recognized.
Acknowledgements
We thank Molly Hunter and Kevin Vogel for providing B. tabaci specimens, and John Overton for advice on Illumina library construction. This work was supported by Yale University and a National Institutes of Health Post-doctoral Fellowship (1F32GM099334).
References
- 1.Sandmann G. 2002. Molecular evolution of carotenoid biosynthesis from bacteria to plants . Physiol. Plantarum. 116, 431–440 10.1034/j.1399-3054.2002.1160401.x (doi:10.1034/j.1399-3054.2002.1160401.x) [DOI] [Google Scholar]
- 2.Fraser P. D., Bramley P. M. 2004. The biosynthesis and nutritional uses of carotenoids . Prog. Lipid Res. 43, 228–265 10.1016/j.plipres.2003.10.002 (doi:10.1016/j.plipres.2003.10.002) [DOI] [PubMed] [Google Scholar]
- 3.Moran N. A., Jarvik T. 2010. Lateral transfer of genes from fungi underlies carotenoid production in aphids . Science 328, 624–627 10.1126/science.1187113 (doi:10.1126/science.1187113) [DOI] [PubMed] [Google Scholar]
- 4.Altincicek B., Kovacs J. L., Gerardo N. M. 2012. Horizontally transferred fungal carotenoid genes in the two-spotted spider mite Tetranychus urticae . Biol. Lett. 8, 253–257 10.1098/rsbl.2011.0704 (doi:10.1098/rsbl.2011.0704) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nováková E., Moran N. A. 2012. Diversification of genes for carotenoid biosynthesis in aphids following an ancient transfer from a fungus . Mol. Biol. Evol. 29, 313–323 10.1093/molbev/msr206 (doi:10.1093/molbev/msr206) [DOI] [PubMed] [Google Scholar]
- 6.De Barro P. J., Liu S. S., Boykin L. M., Dinsdale A. B. 2011. Bemisia tabaci: a statement of species status . Annu. Rev. Entomol. 56, 1–19 10.1146/annurev-ento-112408-085504 (doi:10.1146/annurev-ento-112408-085504) [DOI] [PubMed] [Google Scholar]
- 7.Buchner P. 1965. Endosymbiosis of animals with plant microorganisms. New York, NY: John Wiley & Sons [Google Scholar]
- 8.Brown K. S. 1975. The chemistry of aphids and scale insects . Chem. Soc. Rev. 4, 263–288 10.1039/cs9750400263 (doi:10.1039/cs9750400263) [DOI] [Google Scholar]
- 9.Jenkins R. L., Loxdale H. D., Brookes C. P., Dixon A. F. G. 1999. The major carotenoid pigments of the grain aphid, Sitobion avenae (F.) (Hemiptera: Aphididae) . Physiol. Entomol. 24, 171–178 10.1046/j.1365-3032.1999.00128.x (doi:10.1046/j.1365-3032.1999.00128.x) [DOI] [Google Scholar]
- 10.Shigenobu S., Watanabe H., Hattori M., Sakaki Y., Ishikawa H. 2000. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 407, 81–86 10.1038/35024074 (doi:10.1038/35024074) [DOI] [PubMed] [Google Scholar]
- 11.Nakabachi A., Yamashita A., Toh H., Ishikawa H., Dunbar H. E., Moran N. A., Hattori M. 2006. The 160-kilobase genome of the bacterial endosymbiont Carsonella . Science 314, 267. 10.1126/science.1134196 (doi:10.1126/science.1134196) [DOI] [PubMed] [Google Scholar]
- 12.Zerbino D. R., Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs . Genome Res. 18, 821–829 10.1101/gr.074492.107 (doi:10.1101/gr.074492.107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li R., Yu C., Li Y., Lam T. W., Yiu S. M., Kristiansen K., Wang J. 2009. SOAP2: an improved ultrafast tool for short read alignment . Bioinformatics 25, 1966–1967 10.1093/bioinformatics/btp336 (doi:10.1093/bioinformatics/btp336) [DOI] [PubMed] [Google Scholar]
- 14.Chevreux B., Wetter T., Suhai S. 1999. Genome sequence assembly using trace signals and additional sequence information . Comp. Sci. Biol. Proc. German Conf. Bioinform. 99, 45–56 [Google Scholar]
- 15.Sloan D. B., Moran N. A. 2012. Genome reduction and co-evolution between the primary and secondary bacterial symbionts of psyllids. Mol. Biol. Evol. (doi:10.1093/molbev/mss180) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Markowitz V. M., Mavromatis K., Ivanova N. N., Chen I. M., Chu K., Kyrpides N. C. 2009. IMG ER: a system for microbial genome annotation expert review and curation . Bioinformatics 25, 2271–2278 10.1093/bioinformatics/btp393 (doi:10.1093/bioinformatics/btp393) [DOI] [PubMed] [Google Scholar]
- 17.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models . Bioinformatics 22, 2688–2690 10.1093/bioinformatics/btl446 (doi:10.1093/bioinformatics/btl446) [DOI] [PubMed] [Google Scholar]
- 18.Edgar R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput . Nucleic Acids Res. 32, 1792–1797 10.1093/nar/gkh340 (doi:10.1093/nar/gkh340) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis . Mol. Biol. Evol. 17, 540–552 10.1093/oxfordjournals.molbev.a026334 (doi:10.1093/oxfordjournals.molbev.a026334) [DOI] [PubMed] [Google Scholar]
- 20.Abascal F., Zardoya R., Posada D. 2005. ProtTest: selection of best-fit models of protein evolution . Bioinformatics 21, 2104–2105 10.1093/bioinformatics/bti263 (doi:10.1093/bioinformatics/bti263) [DOI] [PubMed] [Google Scholar]
- 21.McCutcheon J. P., von Dohlen C. D. 2011. An interdependent metabolic patchwork in the nested symbiosis of mealybugs . Curr. Biol. 21, 1366–1372 10.1016/j.cub.2011.06.051 (doi:10.1016/j.cub.2011.06.051) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baumann P. 2005. Biology of bacteriocyte-associated endosymbionts of plant sap-sucking insects . Annu. Rev. Microbiol. 59, 155–189 10.1146/annurev.micro.59.030804.121041 (doi:10.1146/annurev.micro.59.030804.121041) [DOI] [PubMed] [Google Scholar]
- 23.Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S. J., Marra M. A. 2009. Circos: an information aesthetic for comparative genomics . Genome Res. 19, 1639–1645 10.1101/gr.092759.109 (doi:10.1101/gr.092759.109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karatolos N., et al. 2011. Pyrosequencing the transcriptome of the greenhouse whitefly, Trialeurodes vaporariorum reveals multiple transcripts encoding insecticide targets and detoxifying enzymes . BMC Genomics 12, 56. 10.1186/1471-2164-12-56 (doi:10.1186/1471-2164-12-56) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hansen A. K., Moran N. A. 2011. Aphid genome expression reveals host–symbiont cooperation in the production of amino acids . Proc. Natl Acad. Sci. USA 108, 2849–2854 10.1073/pnas.1013465108 (doi:10.1073/pnas.1013465108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Collins A. R. 2001. Carotenoids and genomic stability . Mutat. Res. 475, 21–28 10.1016/S0027-5107(01)00071-9 (doi:10.1016/S0027-5107(01)00071-9) [DOI] [PubMed] [Google Scholar]
- 27.Spaulding A. W., von Dohlen C. D. 1998. Phylogenetic characterization and molecular evolution of bacterial endosymbionts in psyllids (Hemiptera: Sternorrhyncha). Mol. Biol. Evol. 15, 1506–1513 10.1093/oxfordjournals.molbev.a025878 (doi:10.1093/oxfordjournals.molbev.a025878) [DOI] [PubMed] [Google Scholar]
- 28.Thao M. L., Baumann P. 2004. Evolutionary relationships of primary prokaryotic endosymbionts of whiteflies and their hosts . Appl. Environ. Microbiol. 70, 3401–3406 10.1128/AEM.70.6.3401-3406.2004 (doi:10.1128/AEM.70.6.3401-3406.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsuchida T., Koga R., Horikawa M., Tsunoda T., Maoka T., Matsumoto S., Simon J. C., Fukatsu T. 2010. Symbiotic bacterium modifies aphid body color . Science 330, 1102–1104 10.1126/science.1195463 (doi:10.1126/science.1195463) [DOI] [PubMed] [Google Scholar]