Abstract
Squamate reptiles (lizards and snakes) are one of the most diverse groups of terrestrial vertebrates. Recent molecular analyses have suggested a very different squamate phylogeny relative to morphological hypotheses, but many aspects remain uncertain from molecular data. Here, we analyse higher-level squamate phylogeny with a molecular dataset of unprecedented size, including 161 squamate species for up to 44 nuclear genes each (33 717 base pairs), using both concatenated and species-tree methods for the first time. Our results strongly resolve most squamate relationships and reveal some surprising results. In contrast to most other recent studies, we find that dibamids and gekkotans are together the sister group to all other squamates. Remarkably, we find that the distinctive scolecophidians (blind snakes) are paraphyletic with respect to other snakes, suggesting that snakes were primitively burrowers and subsequently re-invaded surface habitats. Finally, we find that some clades remain poorly supported, despite our extensive data. Our analyses show that weakly supported clades are associated with relatively short branches for which individual genes often show conflicting relationships. These latter results have important implications for all studies that attempt to resolve phylogenies with large-scale phylogenomic datasets.
Keywords: phylogeny, phylogenomics, reptiles
1. Introduction
Squamate reptiles are one of the most diverse and well-known vertebrate groups, with approximately 9000 species among 61 families [1]. Squamates offer outstanding model systems in ecology and evolution, especially for studying origins of asexuality, viviparity, body form and venom [1]. Some squamates are also an important cause of human mortality, with tens of thousands of snakebite deaths every year [2].
Understanding these diverse aspects of squamate biology requires a well-resolved phylogeny. Recent molecular analyses have suggested a phylogeny that differs dramatically from morphological hypotheses, especially in placing iguanians with snakes and anguimorphans [3–5]. Although these molecular studies are generally concordant, several issues remain unclear [3–9], including: (i) the sister group to other squamates, (ii) the sister group to snakes, (iii) interrelationships of major snake clades and (iv) relationships of iguanian families. Many of these questions have resisted resolution even with datasets of 20 genes or more [5,7,9].
Here, we analyse squamate phylogeny using extensive sampling of taxa (161) and characters (44 loci), the largest dataset yet assembled. We also present the first analysis of higher squamate relationships using species-tree methods [10,11]. We generate a strongly supported hypothesis and reveal some surprising results. However, some branches remain weakly supported, and our analyses shed light on this unexpected pattern.
2. Material and methods
We sampled 161 squamate species and 10 outgroup taxa, including mammals (Homo, Mus, Tachyglossus), crocodilians (Alligator, Crocodylus), birds (Dromaius, Gallus), turtles (Chelydra, Podocnemis) and a rhyncocephalian (Sphenodon). We included all extant squamate families (excepting a few recently recognized groups, such as Cadeidae, Blanidae, Phyllodactylidae and Xenophiidae [1]), with two or more representatives from most families. We sequenced portions of 44 nuclear genes (exons approx. 500–1500 base pairs in length) carefully selected based on comparisons of vertebrate genomes [12], targeting single-copy genes evolving at appropriate rates. Standard methods of DNA extraction, amplification and sequencing were used. Nucleotide sequences were translated to amino acids to aid alignment, and alignment was straightforward. The total alignment consisted of 33 717 base pairs (available through the Dryad data depository (doi:10.5061/dryad.g1gd8)). Voucher and GenBank numbers and the names, lengths and sampling of genes are provided in the electronic supplementary material, appendices S1–S3).
On average, each gene had data for 143 species (84% complete). Simulations and empirical analyses suggest that missing data need not be problematic for concatenated phylogenetic analyses, particularly when sampling many characters [13].
Two general approaches to data analysis were used (concatenated and species tree). First, we performed a concatenated analysis of all taxa using likelihood (RAxML, v. 7.2.0; [14]), using 1000 bootstrap replicates integrated with 200 searches for the optimal tree. We used the GTR + Γ model and partitioned the data by genes and codon positions (see the electronic supplementary material, appendix S4). We also performed a Bayesian concatenated analysis using MrBayes v. 3.1.2 [15] (see the electronic supplementary material, appendix S4), with branch support based on posterior probabilities (Pp).
Second, we performed Bayesian species-tree analyses (using *BEAST v. 1.6.2 [11]). We used 31 species (29 squamates, two outgroups, Sphenodon and Gallus) for computational feasibility. The selected species represented all major squamate clades, and had relatively complete sampling of genes (mean = 41.5 genes), given that the impact of missing data on species-tree methods remains poorly known. Details of methods are provided in the electronic supplementary material, appendix S4.
To address why some clades are strongly versus weakly supported, we used 49 interfamilial clades from the concatenated-data likelihood tree and analysed relationships between bootstrap support (bs), branch lengths and congruence among genes [7]. Branch lengths from the concatenated-data tree were used, and these lengths were strongly correlated with mean lengths (for comparable clades) from separately analysed genes (Rho = 0.840; p = 0.0001; see the electronic supplementary material, appendix S5). We evaluated the proportion of separately analysed genes supporting each node in the concatenated tree, and the bootstrap support for supporting and conflicting clades. Relationships were tested using non-parametric Spearman's rank correlation (data in the electronic supplementary material, appendix S5).
3. Results
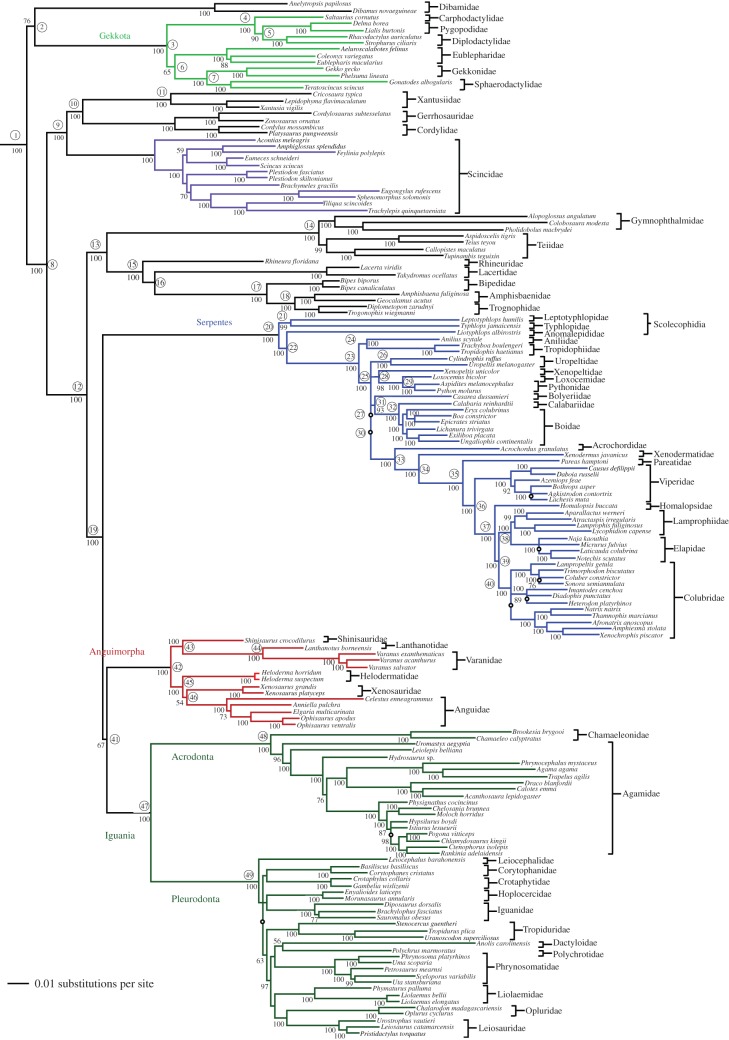
Analyses of the concatenated data using likelihood (figure 1) and Bayesian (see the electronic supplementary material, figure S1) methods yield similar phylogenies and support values and provide strong support for most higher-level squamate relationships. Many aspects of the tree are consistent with other recent molecular analyses [e.g. 3–5] such as the clade of snakes, anguimorphs and iguanians (Toxicofera).
Figure 1.
Phylogeny of squamate reptiles from concatenated likelihood analysis of 44 nuclear genes (see the electronic supplementary material, figure S1 for Bayesian tree). Uncircled numbers at nodes indicate bootstrap values >50% (branches too short to depict here have clades indicated with an open circle); circled numbers correspond to clades in electronic supplementary material, appendix S5. Branch lengths are estimated by likelihood (length for root arbitrarily shortened to facilitate showing ingroup branch lengths).
However, we also find some surprising relationships (figure 1; electronic supplementary material, figure S1). First, we find that dibamids and gekkotans are together the sister group to other squamates (bs = 76%; Pp = 1.00), whereas previous studies have generally placed either dibamids or gekkotans as sister to other squamates [4,6,16], but not both (but see [5]). Second, we find strong support for non-monophyly of Scolecophidia, the blind snakes (Anomalepididae, Leptotyphlopidae and Typhlopidae). Specifically, leptotyphlopids + typhlopids are strongly supported as sister to all other snakes, whereas anomalepidids are sister to all non-scolecophidians (bs = 100%; Pp = 1.00). We also find a strongly supported clade (bs = 97%; Pp = 1.00) within pleurodont iguanians that is inconsistent with relationships from 29 nuclear loci [9]. This clade includes oplurids, leiosaurids, polychrotids, dactyloids, liolaemids and phrynosomatids, whereas the earlier study found phrynosomatids as sister to other pleurodonts (bs = 88%; Pp = 1.00).
Unexpectedly, some aspects of squamate phylogeny still remain weakly supported. These include placement of uropeltids among snakes and relationships among many pleurodont iguanian families (figure 1; see the electronic supplementary material, figure S1). We find a strong relationship between bootstrap support and branch lengths, and between congruence and branch lengths, such that shorter branches tend to be weakly supported and have greater incongruence among genes (table 1).
Table 1.
Relationships between likelihood branch lengths, support and congruence for 49 clades of squamate reptiles.
| variables | Rho | p |
|---|---|---|
| branch length versus bootstrap support | 0.667 | 0.0001 |
| branch length versus proportion genes supporting | 0.921 | 0.0001 |
| branch length versus proportion genes strongly supporting (bs ≥70%) | 0.931 | 0.0001 |
| branch length versus proportion genes strongly supporting (bs ≥95%) | 0.897 | 0.0001 |
| branch length versus proportion genes strongly rejecting (bs ≥70%) | −0.491 | 0.0003 |
| branch length versus proportion genes strongly rejecting (bs ≥95%) | −0.266 | 0.0646 |
The phylogenetic estimate from the species-tree analysis (see the electronic supplementary material, figure S2) is generally similar to the concatenated-data trees (but with some differences), and strongly supports some of the more surprising relationships. Specifically, there is very strong support for placing dibamids and gekkotans as sister taxa, and for non-monophyly of scolecophidians (but with a different arrangement of taxa). Interestingly, this analysis strongly supports iguanians and anguimorphs as sister taxa, a particularly controversial aspect of squamate phylogeny. This clade is also supported by the concatenated analyses (figure 1; see the electronic supplementary material, figure S1), as sister to snakes.
4. Discussion
We present the most extensive analysis of higher squamate phylogeny to date (in terms of characters and taxa), and the first to apply species-tree methods to these relationships. Our results support some aspects of previous hypotheses, but also show some surprising findings. First, both approaches support dibamids and gekkotans as the sister group to all other squamates, in contrast to most previous studies. Second, we find strong support for paraphyly of scolecophidian snakes. This result has appeared in some previous studies [e.g. 7], and we are unaware of molecular studies that have both included all three families and strongly supported scolecophidian monophyly.
Paraphyly of scolecophidians is surprising in that these families share many morphological and ecological traits, including highly reduced eyes [17]. All scolecophidians are specialized burrowers. Thus, paraphyly of scolecophidians at the base of snake phylogeny suggests that snakes may have been burrowers ancestrally, and that most snake species evolved from an ancestor that subsequently returned to surface dwelling. This hypothesis is also supported by the morphology of snakes relative to other limb-reduced lizards: for example, snakes have short tails and elongate trunks (as do burrowing snake-like lizards), whereas surface-dwelling snake-like lizards have elongate tails relative to the trunk [16]. However, some scolecophidian features may have evolved convergently in the two clades, rather than being ancestral for snakes.
Our results also show that some aspects of squamate phylogeny remain weakly supported, especially relationships among some snake and iguanian families. Many of these relationships were also weakly supported in analyses of 20–29 loci [5,7,9]. Thus, roughly doubling the number of loci fails to lead to strong support. In contrast, many relationships supported here were also found in analyses of only one or two nuclear loci [3].
Our analyses of branch lengths, support and congruence suggest that these patterns are related to branch lengths [7]. Specifically, we find weaker support and greater incongruence among genes (and more strongly supported conflicts) on shorter branches, suggesting that these nodes may continue to be problematic as more loci are added. This incongruence seems to arise from incomplete lineage sorting on short branches [7] and may plague many other phylogenomic studies [18]. Importantly, many shorter branches are strongly resolved by the coalescent-based species-tree approach, which incorporates incomplete lineage sorting [10,11]. Our results highlight the potential value of this general approach for resolving short branches with phylogenomic data, although further advances may be needed to make these methods practical for large datasets with incomplete sampling of genes.
Acknowledgements
This project was supported by a US National Science Foundation-AToL grant, with awards to T.W.R. (EF 0334967), J.W.S. (EF 0334966) and J.J.W. (EF 0334923). We thank the many institutions and individuals who provided tissue samples (see the electronic supplementary material, appendix S1), Jyoti Aggrawal and Arianna Klein for assistance with congruence analyses, and two anonymous reviewers for helpful comments on the manuscript.
References
- 1.Sites J. W., Jr, Reeder T. W., Wiens J. J. 2011. Phylogenetic insights on evolutionary novelties in lizards and snakes: sex, birth, bodies, niches, and venom. Annu. Rev. Ecol. Evol. Syst. 42, 227–244 10.1146/annurev-ecolsys-102710-145051 (doi:10.1146/annurev-ecolsys-102710-145051) [DOI] [Google Scholar]
- 2.Kasturiratne A., Wickremasinghe A. R., de Silva N., Gunawardena N. K., Pathmeswaran A., Premaratna R., Savioli L., Lalloo D. G., de Silva J. 2008. Estimation of the global burden of snakebite. PLoS Med. 5, e218. 10.1371/journal.pmed.0050218 (doi:10.1371/journal.pmed.0050218) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Townsend T., Larson A., Louis E. J., Macey J. R. 2004. Molecular phylogenetics of Squamata: the position of snakes, amphisbaenians, and dibamids, and the root of the squamate tree. Syst. Biol. 53, 735–757 10.1080/10635150490522340 (doi:10.1080/10635150490522340) [DOI] [PubMed] [Google Scholar]
- 4.Vidal N., Hedges S. B. 2005. The phylogeny of squamate reptiles (lizards, snakes, and amphisbaenians) inferred from nine nuclear protein-coding genes. C. R. Biol. 328, 1000–1008 10.1016/j.crvi.2005.10.001 (doi:10.1016/j.crvi.2005.10.001) [DOI] [PubMed] [Google Scholar]
- 5.Wiens J. J., Kuczynski C. A., Townsend T., Reeder T. W., Mulcahy D. G., Sites J. W., Jr 2010. Combining phylogenomics and fossils in higher-level squamate reptile phylogeny: molecular data change the placement of fossil taxa. Syst. Biol. 59, 674–688 10.1093/sysbio/syq048 (doi:10.1093/sysbio/syq048) [DOI] [PubMed] [Google Scholar]
- 6.Hugall A. F., Foster R., Lee M. S. Y. 2007. Calibration choice, rate smoothing, and the pattern of tetrapod diversification according to the long nuclear gene RAG-1. Syst. Biol. 56, 543–563 10.1080/10635150701477825 (doi:10.1080/10635150701477825) [DOI] [PubMed] [Google Scholar]
- 7.Wiens J. J., Kuczynski C. A., Smith S. A., Mulcahy D., Sites J. W., Jr, Townsend T. M., Reeder T. W. 2008. Branch lengths, support, and congruence: testing the phylogenomic approach with 20 nuclear loci in snakes. Syst. Biol. 57, 420–431 10.1080/10635150802166053 (doi:10.1080/10635150802166053) [DOI] [PubMed] [Google Scholar]
- 8.Lee M. S. Y. 2009. Hidden support from unpromising datasets strongly unites snakes and anguimorph lizards. J. Evol. Biol. 22, 1308–1316 10.1111/j.1420-9101.2009.01751.x (doi:10.1111/j.1420-9101.2009.01751.x) [DOI] [PubMed] [Google Scholar]
- 9.Townsend T. M., Mulcahy D. G., Sites J. W., Jr, Kuczynski C. A., Wiens J. J., Reeder T. W. 2011. Phylogeny of iguanian lizards inferred from 29 nuclear loci, and a comparison of concatenated and species-tree approaches for an ancient, rapid radiation. Mol. Phylogenet. Evol. 61, 363–380 10.1016/j.ympev.2011.07.008 (doi:10.1016/j.ympev.2011.07.008) [DOI] [PubMed] [Google Scholar]
- 10.Edwards S. V., Liu L., Pearl D. K. 2007. High-resolution species trees without concatenation. Proc. Natl Acad. Sci. USA 104, 5936–5941 10.1073/pnas.0607004104 (doi:10.1073/pnas.0607004104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heled J., Drummond A. J. 2010. Bayesian inference of species trees from multilocus data. Mol. Biol. Evol. 27, 570–580 10.1093/molbev/msp274 (doi:10.1093/molbev/msp274) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Townsend T. M., Alegre E. R., Kelley S. T., Wiens J. J., Reeder T. W. 2008. Rapid development of multiple nuclear loci for phylogenetic analysis using genomic resources: an example from squamate reptiles. Mol. Phylogenet. Evol. 47, 129–142 10.1016/j.ympev.2008.01.008 (doi:10.1016/j.ympev.2008.01.008) [DOI] [PubMed] [Google Scholar]
- 13.Wiens J. J., Morrill M. C. 2011. Missing data in phylogenetic analysis: reconciling results from simulations and empirical data. Syst. Biol. 60, 719–731 10.1093/sysbio/syr025 (doi:10.1093/sysbio/syr025) [DOI] [PubMed] [Google Scholar]
- 14.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 10.1093/bioinformatics/btl446 (doi:10.1093/bioinformatics/btl446) [DOI] [PubMed] [Google Scholar]
- 15.Huelsenbeck J. P., Ronquist F. 2001. MrBayes: Bayesian inference of phylogeny. Bioinformatics 17, 754–755 10.1093/bioinformatics/17.8.754 (doi:10.1093/bioinformatics/17.8.754) [DOI] [PubMed] [Google Scholar]
- 16.Wiens J. J., Brandley M. C., Reeder T. W. 2006. Why does a trait evolve multiple times within a clade? Repeated evolution of snake-like body form in squamate reptiles. Evolution 60, 123–141 10.1111/j.0014-3820.2006.tb01088.x (doi:10.1111/j.0014-3820.2006.tb01088.x) [DOI] [PubMed] [Google Scholar]
- 17.Vitt L. J., Caldwell J. P. 2009. Herpetology: an introductory biology of amphibians and reptiles, 3rd edn San Diego, CA: Academic Press [Google Scholar]
- 18.Rokas A., Carroll S. B. 2006. Bushes in the tree of life. PLoS Biol. 4, e352. 10.1371/journal.pbio.0040352 (doi:10.1371/journal.pbio.0040352) [DOI] [PMC free article] [PubMed] [Google Scholar]