Abstract
Peptide phosphorodiamidate morpholino oligomers (PPMOs) are synthetic DNA mimics that bind cRNA and inhibit bacterial gene expression. The PPMO (RFF)3RXB-AcpP (where R is arginine, F, phenylalanine, X is 6-aminohexanoic acid, B is β-alanine, and AcpP is acyl carrier protein) is complementary to 11 bases of the essential gene acpP (which encodes acyl carrier protein). The MIC of (RFF)3RXB-AcpP was 2.5 μM (14 μg/ml) in Escherichia coli W3110. The rate of spontaneous resistance of E. coli to (RFF)3RXB-AcpP was 4 × 10−7 mutations/cell division. A spontaneous (RFF)3RXB-AcpP-resistant mutant (PR200.1) was isolated. The MIC of (RFF)3RXB-AcpP was 40 μM (224 μg/ml) for PR200.1. The MICs of standard antibiotics for PR200.1 and W3110 were identical. The sequence of acpP was identical in PR200.1 and W3110. PR200.1 was also resistant to other PPMOs conjugated to (RFF)3RXB or peptides with a similar composition or pattern of cationic and nonpolar residues. Genomic sequencing of PR200.1 identified a mutation in sbmA, which encodes an active transport protein. In separate experiments, a (RFF)3RXB-AcpP-resistant isolate (RR3) was selected from a transposome library, and the insertion was mapped to sbmA. Genetic complementation of PR200.1 or RR3 with sbmA restored susceptibility to (RFF)3RXB-AcpP. Deletion of sbmA caused resistance to (RFF)3RXB-AcpP. We conclude that resistance to (RFF)3RXB-AcpP was linked to the peptide and not the phosphorodiamidate morpholino oligomer, dependent on the composition or repeating pattern of amino acids, and caused by mutations in sbmA. The data further suggest that (RFF)3R-XB PPMOs may be transported across the plasma membrane by SbmA.
INTRODUCTION
Antibiotic resistance in bacteria continues to be a serious problem. The number of antibiotic-resistant pathogens is increasing, the level of resistance to standard antibiotics is increasing, and the percentage of isolates with resistance to multiple antibiotics has risen dramatically in recent years (3, 37). At the same time, the number of antibiotics that are being developed has decreased significantly, particularly those targeting Gram-negative bacteria. Most of the new antibiotics that have been approved for use in the United States in the past 40 years are not new classes of antibiotics but are simply chemical derivatives of the same antibiotic classes that were discovered in the mid-20th century (8). There is an urgent need for new antibiotics, particularly those with novel or innovative strategies of targeting bacterial pathogens that cause serious diseases (3, 22).
Genomics has created an attractive potential for developing innovative strategies that address the problem of antibiotic resistance. Synthetic antisense oligomers, such as peptide nucleic acids (14), phosphorothioates (16), and phosphorodiamidate morpholino oligomers (PMOs) (11, 15), silence expression of bacterial genes. Gene-silencing oligomers decrease expression of reporter genes such as luciferase, activate endogenous genes such as β-galactosidase, and inhibit growth and kill bacteria by targeting essential genes (10). Antisense oligomers targeted to specific, essential bacterial genes reduce infections and increase survival in mouse models of infection (12, 15, 40).
Antisense oligomers require assistance to cross the outer membrane of Gram-negative bacteria because of their molecular weight and polar characteristics. Short amphipathic peptides have been attached to antisense oligomers, and this has greatly improved their entry into Gram-negative bacteria and increased their potency (11, 13, 27).
Membrane-penetrating peptides have diverse sequences, but many are cationic and amphipathic. Previous investigations suggest that a repeated peptide motif with one cationic residue followed by either one or two hydrophobic residues may be an important feature for efficient membrane penetration (39). More recently, we have compared a variety of membrane-penetrating peptides for their abilities to enhance the efficacy of peptide phosphorodiamidate morpholino oligomers (PPMOs) and found differences among peptides that vary in their pattern of alternating cationic and nonpolar residues and their amino acid compositions (27).
Despite the progress on improving the efficacy and potency of antisense oligomers that has been made, little is known about bacterial resistance to these compounds. Some naturally occurring antimicrobial peptides, which have some characteristics similar to those of the synthetic peptides used to make peptide oligomers, do not appear to cause resistance in bacteria (38). One report of resistance to an antisense morpholino oligomer found a mutation in the region of a virus genome targeted by the oligomer (28). Resistance to any antibiotic is always an important characteristic to be determined during drug development. The frequency of antibiotic resistance will ultimately manifest itself in the clinic and will play a role in its use for any particular indication.
In this report, we characterize spontaneous resistance to a PPMO and compare cross-resistance to other antibiotics, PPMOs with different peptides but the same PMO, and PPMOs with the same peptide but targeted to different genes. Furthermore, the same gene that causes PPMO resistance is identified in isolates from two independent strategies of selection.
MATERIALS AND METHODS
Bacterial strains.
Wild-type Escherichia coli K-12 strain W3110 was used for selecting spontaneous mutants that are resistant to the PPMO (RFF)3RXB-AcpP (where R is arginine, F, phenylalanine, X is 6-aminohexanoic acid, B is β-alanine, and AcpP is acyl carrier protein). Spontaneous mutants that are resistant to (RFF)3RXB-AcpP were selected by growth in Mueller-Hinton II broth supplemented with 8× MIC of (RFF)3RXB-AcpP. Liquid cultures were grown in either Mueller-Hinton II or LB broth. LB agar was used for growth on solid medium. Transformants with pSE380myc-luc (11) were grown in LB medium supplemented with 50 μg/ml ampicillin (LBA; Sigma-Aldrich, St. Louis, MO).
Oligopeptide transport mutants PA0183 (opp), PA0333 (opp dpp), PA0410 (opp tpp), PA0643 (opp dpp tpp), and PA0610 (opp dpp tpp), which were derived from parent strain Morse 2034 [trpE9851 leu 277 F−IN(rrnD-rrnE)], have been described previously (36) and were gifts from J. W. Payne (University of Wales, Bangor, United Kingdom).
In-frame, nonpolar knockout strains E. coli JW3496 (dctA knockout mutant), JW5730 (eptA knockout mutant), and JW0368 (sbmA knockout mutant) and their isogenic parent strain, BW25113 (2), were provided from the Keio collection by the National BioResource Project (NIG; Japan). The knockout strains were grown in LB broth with 50 μg/ml kanamycin (Sigma-Aldrich). The IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible sbmA expression plasmid (which we call pSbmA, from strain b0377) and empty vector control (pNTR-SD) (34) were also provided by the National BioResource Project (NIG; Japan) and grown in LB broth with 20 μg/ml ampicillin.
PPMOs.
PPMOs were synthesized at AVI BioPharma, Inc. (Corvallis, OR), as described previously (39). The base sequence of all PPMOs targeted to acpP (AcpP) is 5′-CTTCGATAGTG-3′, that of all PPMOs targeted to ftsZ (FtsZ) is 5′-TCCATTGGTTC-3′, and that of all PPMOs targeted to luc (Luc) is 5′-AACGTTGAGIG. Inosine in place of a guanine in the Luc PPMO was necessary to make the oligomer soluble in aqueous solutions by avoiding the guanine quartet structure. The scrambled-base-sequence (Scr) control is 5′-TCTCAGATGGT-3′.
Antibiotics.
All antibiotics except bleomycin were purchased from Sigma-Aldrich. Bleomycin was purchased from Enzo Life Science (Farmingdale, NY).
MICs.
MICs were determined by the microdilution method (5) in Mueller-Hinton II broth. For determination of MICs for strains XL1-Blue MRF′ and RR3, Mueller-Hinton II broth was supplemented with 1% tryptone.
Luciferase expression.
Spontaneous (RFF)3RXB-AcpP-resistant mutants were made chemically competent and transformed as described previously (29) with pSE380myc-Luc (11). Overnight cultures were grown aerobically at 37°C in LBA and then diluted 2 × 10−2 into LBA with or without various concentrations (8, 20, and 50 μM) of (RFF)3RXB-Luc or (RFF)3RXB-Scr and grown aerobically at 37°C for 7 h. Samples were analyzed for luciferase expression by luminometry as described previously (11).
Rate of spontaneous resistance.
The rate of spontaneous resistance to peptide PMO was measured by the method of Luria and Delbruck (23) as described previously (33). An overnight culture was diluted to 1 × 104 CFU/ml in LB medium and divided into 20 1-ml aliquots. Each aliquot was grown overnight at 37°C with aeration, and then 1 μl or 50 μl of each was spread on 20 agar plates (60 mm by 15 mm) of LB plus 20 μM peptide PMO. The cultures were grown overnight at 37°C with aeration, and colonies were enumerated.
Screening transposome mutants.
Transposome EZ-Tn5<R6Kγori/Kan-2>Tnp (Epicentre, Madison, WI) was electroporated into E. coli XL1-Blue MRF′, and 3 × 103 transductants were selected on LB-kanamycin (15 μg/ml) plates. The transductants were pooled and stored in phosphate-buffered saline with 15% glycerol at −75°C. The pooled transductants were thawed, and 1 × 104 CFU was spread on an LB plate that included 20 μM (RFF)3RXB-AcpP. Insertion mutations from PPMO-resistant mutants were sequenced by rescue cloning as described by the manufacturer (Epicentre). Insertions in sbmA were confirmed using PCR as described previously (18). Briefly, PCR mixtures contained chromosomal DNA extracted from bacteria using a commercial kit (DNeasy; Qiagen, Valencia, CA) and primers (IDT Technologies, Coralville, IA) that flank the insertion site: 5′-GATTGCCGTTATCTTCTGGC and 5′-GCTCAAGGTATGGGTTACTTCC. Thirty PCR cycles of denaturation at 95°C for 0 s, annealing at 45°C for 0 s, and extension at 72°C for 1 min were carried out. PCRs were run on a 1605 air thermocycler (Idaho Technology, Idaho Falls, ID).
Sequencing acpP.
The acpP allele from each strain analyzed was amplified by PCR (18), using as the template a single bacterial colony picked from a growth plate, Promega Taq polymerase (Madison, WI), and the following primers (Invitrogen, Carlsbad, CA): 5′-AACGTAAAATCGTGGTAAGACC-3′ and 5′-TAACGCCTGGTGGCCGTTGATG-3′. The PCR products were gel purified using a Qiagen MinElute PCR purification kit (Valencia, CA) and sequenced using the same primers shown above at the core laboratory of the Center for Genome Research, Oregon State University.
Genomic sequencing.
Genomic DNA from the W3110 wild type and PR200.1 was generated by standard procedures (1). DNA was sheared by sonication and processed for Illumina high-throughput sequencing as previously described (31, 32). Data analyses to find individual point mutations were carried out as described previously (32).
RESULTS
Spontaneous mutants resistant to peptide PMOs.
Spontaneous resistance was apparent from growth that occasionally occurred in some cultures that included (RFF)3RXB-AcpP (X is 6-aminohexanoic acid and B is β-alanine) at concentrations above the MIC. Growth above the MIC was never observed in cultures that included other AcpP PPMOs with different peptides attached to the same PMO, such as (RX)6B-AcpP or (RXR)4XB-AcpP. The rate of spontaneous resistance to (RFF)3RXB-AcpP was measured and found to be 4 × 10−7 mutations/cell generation.
Susceptibility to antibiotics and growth rate.
Colonies were isolated from a single liquid culture of W3110 grown with 8× MIC (20 μM, or 112 μg/ml) of (RFF)3RXB-AcpP. One colony (PR200.1) was picked at random and further characterized. PR200.1 was equally as susceptible as parent strain W3110 to each antibiotic tested (MICs, 4 μg/ml, 1.25 μg/ml, 1.25 μg/ml, 0.125 μg/ml, and 10 μg/ml for ampicillin, tetracycline, kanamycin, polymyxin B, and rifampin, respectively). These results indicate that this particular PPMO-resistant isolate was not resistant to antibiotics in general.
The doubling times of strains PR200.1 and W3110 were identical, and no difference in growth rate was observed in liquid or solid medium.
Sequences of acpP alleles.
The target of the PMO, acpP, was sequenced in PR200.1 and W3110, and the sequences were found to be identical (data not shown).
MICs of AcpP PPMOs attached to various peptides.
MICs for different AcpP PPMOs were measured using strains PR200.1 and W3110 as indicators (Table 1). All of the AcpP PPMOs tested had the same base sequence but had different peptides attached. The attached peptides differed not only in their amino acid compositions but also in the pattern of repeating sequences of cationic and nonpolar residues. Repeating patterns of amino acids, often including cationic and nonpolar residues, are important features of membrane-penetrating peptides (17, 41, 42). Four of the AcpP PPMOs, including (RFF)3RXB-AcpP, had peptides with a repeating amino acid motif of cationic-nonpolar-nonpolar (C-N-N), and one of these was composed of d-amino acids instead of the usual l-amino acids. One AcpP PPMO was conjugated to (RX)6B, which has a repeating motif of cationic-nonpolar (C-N). Two other AcpP PPMOs were conjugated to peptides with a repeating motif of cationic-nonpolar-cationic (C-N-C): (RXR)4XB and (RFR)4XB. Another AcpP PMO was conjugated to RTRTRFLRRTXB, which has a repeat pattern that does not conform to any of the other repeat patterns. All of these PPMOs with various peptides attached to the same AcpP PMO have been previously characterized and found to be effective in inhibiting growth of E. coli (27).
Table 1.
MIC of AcpP PPMOs in pure cultures of E. coli
| Motif, PPMO no.a | Conjugated peptideb | MIC (μM [μg/ml]) |
|
|---|---|---|---|
| W3110 | PR200.1 | ||
| Motif 1 (C-N-N) | |||
| NG-05-0200 | RFFRFFRFFRXB | 2.5 (14) | 40 (222) |
| NG-05-0653 | DRDFDFDRDFDFDRDFDFDRXB | 2.5 (14) | 40 (222) |
| NG-23-248 | RXXRXXRXXRXB | 20 (102) | 80 (204) |
| NG-06-0199 | KFFKFFKFFKXB | 10 (54) | 80 (435) |
| Motif 2 (C-N), NG-06-0073 | RXRXRXRXRXRXB | 1.25 (7) | 1.25 (7) |
| Motif 3 (C-N-C) | |||
| NG-06-0076 | RXRRXRRXRRXRXB | 1.25 (7) | 1.25 (7) |
| NG-07-0795 | RFRRFRRFRRFRXB | 1 (6) | 16 (94) |
| No motif, NG-05-0246 | RTRTRFLRRTXB | 20 (111) | 40 (111) |
Motif 1 is cationic-nonpolar-nonpolar (C-N-N). Motif 2 is cationic-nonpolar (C-N). Motif 3 is cationic-nonpolar-cationic (C-N-C).
X is 6-aminohexanoic acid, B is β-alanine, O is ornithine, and D indicates the isomeric form or the residue that follows.
The results show that PR200.1 was resistant to every AcpP PMO with the C-N-N peptide motif tested but was fully susceptible to the (RX)6B-AcpP PMO and (RXR)4XB-AcpP PMO (Table 1). However, PR200.1 was resistant to (RFR)4XB-AcpP, which shares the C-N-C motif with (RXR)4XB-AcpP but, like (RFF)3RXB-AcpP, contains phenylalanine instead of 6-aminohexaonoic acid. Compared to the susceptible parent strain W3110, resistance to (RFF)3RXB-AcpP in PR200.1 increased the MIC 16-fold. PR200.1 was also resistant to the d-isomeric form of (RFF)3RXB. The MIC of RTRTRFLRRTXB-AcpP, which lacks a repeating amino acid motif but includes one phenylalanine, increased only 2-fold when PR200.1 was used as the indicator compared to that obtained when W3110 was used as the indicator. The MICs of scrambled-base-sequence PPMOs composed with each of the same peptides used for the AcpP PPMOs were undetectable (>80 μM) in every case.
(RFF)3RXB-PPMOs targeted to various genes.
PR200.1 was tested for susceptibility to two PPMOs, each with the (RFF)3 peptide motif but different base sequences. One PPMO is complementary to ftsZ, which is an essential gene involved in cell division. The other PPMO is targeted to a luciferase reporter gene (luc).
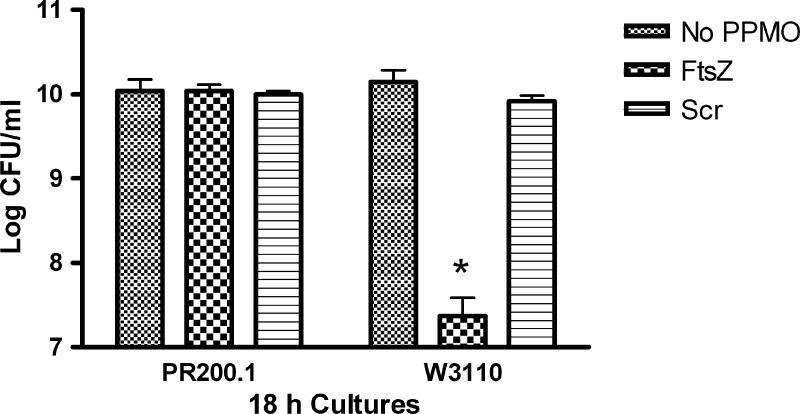
Exponential cultures were grown for 18 h with (RFF)3RXB-FtsZ, which is targeted to ftsZ, or a scrambled-base-sequence (Scr) control. Samples of each culture were then plated, and the viable cells were counted. PR200.1 grew to normal cell density, whereas the viable cell count of parent strain W3110 was reduced by over 2 orders of magnitude in the presence of (RFF)3RXB-FtsZ (Fig. 1). The scrambled-base-sequence control had no effect on the growth of either W3110 or PR200.1.
Fig 1.
Viable cell count of 18-h cultures. Stationary cultures of W3110 or PR200.1 were diluted to 5 × 105 CFU/ml in Mueller-Hinton broth and divided in three. (RFF)3RXB-FtsZ (FtsZ) or scrambled-base-sequence (Scr) control PPMO (160 μM) or no PPMO was added. Cultures were grown aerobically at 37°C for 18 h, and then samples of each were diluted and plated to determine viable cells. Error bars indicate standard deviations. *, highly significant (P < 0.01) difference compared to either the no-PPMO or scrambled-base-sequence (Scr) control-treated culture.
In other experiments, exponential cultures of W3110 and PR200.1 were grown for 7 h with various concentrations of (RFF)3RXB-Luc, which is targeted to a luciferase reporter gene, or the scrambled-base-sequence control, (RFF)3RXB-Scr. A plasmid that expresses luciferase had been transferred into PR200.1 prior to the experiment. Samples of each culture were then analyzed by luminometry for luciferase activity. The results show that (RFF)3RXB-Luc did not inhibit luciferase in PR200.1 at any of the 3 concentrations tested (Fig. 2). In comparison, W3110 showed inhibition of luciferase that was proportional to the concentration of PPMO added. The scrambled-base-sequence control did not inhibit luciferase in either strain. There were no differences in the growth (optical density) of any of the cultures (data not shown).
Fig 2.
Luciferase activity of cultures treated with PPMOs. Growing cultures were treated for 7 h without PPMO (no PPMO) or with 3 concentrations of a PPMO [(RFF)3RXB-Luc] targeted to a luciferase reporter gene or a scrambled-base-sequence control [(RFF)3RXB-Scr]. After 7 h, samples of each culture were measured for luciferase activity by luminometry. The experiment was repeated 3 times, and the error bars indicate standard deviations. *, a highly significant (P < 0.01) difference compared to cultures of PR200.1 with the same concentrations of (RFF)3RXB-Luc, the cultures of W3110 with the same concentrations of (RFF)3RXB-Scr, or the culture without PPMO.
Peptide transport mutants.
The results presented above indicate that resistance to PPMOs is linked to the peptide moiety. We hypothesized that PPMO resistance could be caused by a mutation in one of three known oligopeptide transporters. To test this, the MIC was measured using various strains with mutations in one, two, or all three oligopeptide transporters (Table 2). The results show that (RFF)3RXB-AcpP had the same MIC for PR200.1 as for the parent (nonmutant) strain. The scrambled-base-sequence control (RFF)3RXB-Scr showed no detectable MIC (>160 μM).
Table 2.
MIC of (RFF)3R-AcpP (NG-05-0200) using oligopeptide transport mutants
| E. coli strain | Mutation/phenotype | MIC |
|
|---|---|---|---|
| μM | μg/ml | ||
| Morse 2034 | Wild-type oligopeptide transport | 5 | 28 |
| PA0183 | opp knockout, oligopeptide permease deletion | 5 | 28 |
| PA0333 | dpp and opp knockout, PA0183 plus dipeptide permease deletion | 5 | 28 |
| PA0643 | tpp, dpp, and opp knockout, PA0333 plus tripeptide permease mutant | 5 | 28 |
Genomic sequencing.
The genomes of PR200.1 and its parent strain, W3110, were sequence and compared. The results indicated that a total of 3 genes had mutations in PR200.1 compared to the W3110 sequence: dctA, eptA, and sbmA. In dctA, there were 2 transition mutations at bases 3958154 (T → A) and 395153 (A → G), both of which are in codon 396, that caused a missense from Ile to Ala. In eptA, there was one transversion mutation at base 4339795 (T → A) that affected codon 259 and caused a missense from Ser to Thr. In sbmA, there was one transversion mutation at base 396121 (T → G) that changed codon 87 (Ser to Ala). No deletions or insertions were detected in any gene.
Characterization of deletion mutants.
Mutants with in-frame knockout mutations of dctA, eptA, and sbmA were tested for susceptibility to (RFF)3RXB-AcpP, and susceptibilities were compared to those of the parent strain (BW25113). The MIC of (RFF)3RXB-AcpP was the same (2 μM, or 11 μg/ml) using either the dctA or eptA knockout strain or the parent strain. The MIC obtained using the sbmA knockout strain was 32 μM (179 μg/ml).
The MIC of (RXR)4XB-AcpP was measured using the sbmA-knockout strain as an indicator and found to be 2 μM (11 μg/ml), the same as that for its isogenic parent strain.
Complementation with pSbmA.
PR200.1 was genetically complemented with an IPTG-inducible expression plasmid that encodes sbmA (pSbma) or its empty control. The complemented strain was grown with IPTG and used to measure the MIC of (RFF)3RXB-AcpP. The MICs were 1 μM and 32 μM for the induced, sbmA complemented strain and the empty vector control strain, respectively.
Transposome mutants.
E. coli XL1-Blue MRF′ was mutagenized with the transposome EZ-Tn5, and a library of 1 × 104 mutants was spread on selection plates that included 20 μM (RFF)3R-AcpP. Two colonies grew on the selection plate, and the mutated gene in each was sequenced. The sequences of both isolates indicated that the transposome had inserted into the exact same position in sbmA in each isolate, suggesting that the two colonies were clones. The isolates were named RR3.
RR3 was characterized by measuring the MICs of various standard antibiotics (Table 3). All standard antibiotics tested had the same MIC using either XL1-Blue MRF′ or RR3 as the indicator, including two peptide antibiotics, colistin and polymyxin B. However, RR3 was about 4-fold resistant to each of the peptide antibiotics bleomycin and phleomycin. PR200.1 was also 4-fold resistant to bleomycin (MIC = 5.6 μM [8 μg/ml]) and phleomycin (MIC = 5.2 μM [8 μg/ml]) than W3110 (bleomycin MIC = 1.4 μM [2 μg/ml]; phleomycin MIC = 1.3 μM [2 μg/ml]).
Table 3.
MICs of standard antibiotics and PPMOs using transposome mutant RR3, isogenic parent strain XL1-Blue MRF′, and their sbmA complemented strains
| PPMO no. | Antibiotic or PPMO | MIC (μM [μg/ml]) |
|||
|---|---|---|---|---|---|
| XL1-Blue MRF′ | RR3 | XL1-Blue MRF′ (pSbmA) | RR3(pSbmA) | ||
| Polymyxin B | 0.8 (1) | 0.8 (1) | 0.8 (1) | 0.8 (1) | |
| Colistin | 0.9 (1) | 0.9 (1) | 0.9 (1) | 0.9 (1) | |
| Erythromycin | 34 (25) | 34 (25) | 34 (25) | 34 (25) | |
| Rifampin | 6 (5) | 6 (5) | 6 (5) | 6 (5) | |
| Bleomycin | 0.2 (0.25) | 0.7 (1) | 0.01 (0.016) | 0.01 (0.016) | |
| Phleomycin | 0.3 (0.5) | 1.3 (2) | 0.02 (0.03) | 0.02 (0.03) | |
| NG-05-0200 | (RFF)3RXB-AcpP | 2 (11) | 64 (355) | 2 (11) | 2 (11) |
| NG-05-0653 | [d-(RFF)3R]XB-AcpP | 2 (11) | 64 (355) | 2 (11) | 2 (11) |
| NG-06-0076 | (RXR)4XB-AcpP | 1 (6) | 8 (48) | 0.1 (0.6) | 0.1 (0.6) |
| NG-05-0655 | (RFF)3RXB-Scr | >128 (>714) | >128 (>714) | >128 (>714) | >128 (>714) |
| NG-06-0078 | (RXR)4XB-Scr | >128 (>714) | >128 (>714) | >128 (>714) | >128 (>714) |
The MICs of (RFF)3R-AcpP and (RXR)4XB-AcpP were measured using RR3 or XL1-Blue MRF′ as indicator strains. The results show that RR3 was 32-fold more resistant to (RFF)3RXB-AcpP and 8-fold more resistant to (RXR)4XB-AcpP (Table 3). RR3 was also resistant to the PPMO made with d-amino acids ([d-(RFF)3R]XB-AcpP). Scrambled-base-sequence control PPMOs did not inhibit the growth of either RR3 or XL1-Blue MRF′.
RR3 and XL1-Blue MRF′ were genetically complemented with pSbmA and used to measure the MIC of (RFF)3RXB-AcpP. pSbma fully restored the susceptibility of RR3 to the PPMO, when induced with IPTG (Table 3). Interestingly, the MIC was significantly less when the complemented strains were used as indicators than when the strains without pSbmA were used as indicators. Complementation with pSbmA also restored susceptibility to bleomycin and phleomycin.
DISCUSSION
This is the first study that we are aware of to characterize bacterial resistance to an antisense antibacterial compound. Initially, growth was occasionally and unexpectedly observed during routine MIC assays with (RFF)3XB-AcpP present in cultures at levels 4- to 8-fold above the MIC. Similar growth was never observed during MIC assays with (RXR)4XB-AcpP or (RX)6B-AcpP. We speculate that the greater number of X (6-aminohexanoic acid) residues or the lack of F in the latter two PPMOs may be responsible for the apparent lack of spontaneous resistance to these PPMOs under the conditions used for the MIC assay. Alternatively, there could be more genetic loci involved in resistance to (RFF)3RXB PPMOs than in any (putative) resistance to PPMOs conjugated to other peptides, such as (RXR)4XB or (RX)6B. However, we have not yet rigorously pursued resistance to the latter two PPMOs, and it is certainly possible that spontaneous resistance may occur under appropriate conditions.
The rate of spontaneous resistance to (RFF)3RXB-AcpP was similar to the rate of spontaneous mutation for individual genes in E. coli, which is typically between 10−6 and 10−7 mutations/gene/generation (6, 24). This suggests that there are few genes which, when mutated, can give rise to the PPMO-resistant phenotype. However, the rate of mutation can vary widely, and any measurement of the rate of mutation is a function of many variables (25), including the concentration of antibiotic used for selection and the number of genes or loci capable of causing a resistance phenotype. Ultimately, the rate of resistance to PPMOs under in vivo conditions for specific infections will be the most meaningful measure of their usefulness in the clinic.
The spontaneous mutant PR200.1 was susceptible to all small-molecule antibiotics tested. This shows that resistance to the PPMO is not caused by a change in physiology that might result in resistance to antibiotics in general. Such general changes are known to occur and include a reduction of the net negative charge of the lipopolysaccharide of Gram-negative bacteria (7, 30), changes in capsule polysaccharide (4), changes in expression of outer membrane porins (30), alterations in outer membrane lipid composition that result in decreased membrane permeability (30), and activation or overexpression of multidrug efflux pumps (21). We tested a variety of antibiotics, some of which are hydrophilic (ampicillin, kanamycin), hydrophobic (rifampin, tetracycline), or amphiphilic (polymyxin B) and some of which enter Gram-negative bacteria through the outer membrane porins (ampicillin, tetracycline) or through the outer membrane lipid bilayer (rifampin, polymyxin B). The results suggested that the mutation in PR200.1 is specific for (RFF)3RXB-AcpP or PPMOs with similar peptide moieties. Later, following the identification of the mutation in sbmA, PR200.1 was found to be mildly resistant (4-fold) to the peptide antibiotics bleomycin and phleomycin.
We hypothesized that resistance was caused by a mutation in the sequence of acpP targeted by the PPMO. We have previously shown that a one-base mismatch near the 3′end of a PPMO targeted to acpP in the Burkholderia cepacia complex raised the MIC by a factor of at least 8- to 32-fold (15). However, the present study found no mutation in the target region of acpP in this one resistant mutant. Therefore, the hypothesis in this case was disproven. However, this is not to say that target-site mutations cannot or do not occur on other as of yet uncharacterized mutants. Nevertheless, target-site mutations would be statistically improbable considering that there are only 4 wobble bases in the target region of acpP that might possibly lead to a decrease in efficacy without changing the amino acid sequence of the targeted protein.
Another hypothesis was that resistance in PR200.1 was caused by a mutation in an oligopeptide transporter. However, oligopeptide transport mutants were just as susceptible to (RFF)3RXB-AcpP as the isogenic parent strain. This showed that resistance in PR200.1 was not caused by a mutation in the known oligopeptide transporters that were tested.
In another effort to identify the mutation in PR200.1 that is responsible for resistance to (RFF)3RXB-AcpP, the genome of PR200.1 was sequenced. The results showed missense mutations in only 3 genes compared to the sequence of the PPMO-susceptible strain: dctA, eptA, and sbmA. In-frame, nonpolar deletion mutations of each gene showed that of the three, only the strain with the sbmA deletion was resistant to (RFF)3RXB-AcpP. Furthermore, complementation of PR200.1 with sbmA restored susceptibility to the PPMO. These results show that mutations in sbmA cause resistance to (RFF)3RXB-AcpP.
The MIC of (RFF)3RXB-AcpP was slightly lower when the strain with the sbmA deletion than when PR200.1 was used as the indicator. However, the strains originated from different parent strains, and this probably accounts for the difference in susceptibility. The parent strain of the strain with the sbmA deletion was also slightly more susceptible to the PPMO than the parent strain of PR200.1.
The transposome mutant RR3 was resistant to both (RFF)3RXB-AcpP and (RXR)4XB-AcpP. This differs from the result for the spontaneous mutant PR200.1 and the strain with the sbmA deletion, which were resistant to (RFF)3RXB-AcpP but not (RXR)4XB-AcpP. This suggests that in RR3 a polar effect on the gene downstream from sbmA (yaiW) may be responsible for resistance to (RXR)4XB-AcpP. yaiW is a predicted DNA-binding transcriptional regulator. A mutation in sbmA is apparently sufficient to cause resistance to (RFF)3RXB-AcpP, but not (RXR)4-AcpP.
PR200.1 was susceptible to (RXR)4XB-AcpP but resistant to (RFR)4XB-AcpP, although these two PPMOs share the same C-N-C repeat motif. This indicates that the amino acid composition of the PPMOs may be more important than the repeating pattern of amino acids in determining resistance in PR200.1. The similarity in the resistance of PR200.1 to either (RFF)3RXB-AcpP or (RFR)4XB-AcpP but complete susceptibility to (RXR)4XB-AcpP may suggest that X (6-aminohexanoic acid) accounts for the difference. This is supported by the result (Table 1) that the level of resistance to (RXX)3RXB-AcpP is 4-fold less than the level of resistance to (RFF)3RXB-AcpP, even though they have the same repeating pattern of cationic and nonpolar amino acids but the former contains more X. Perhaps the unusual 6-carbon backbone of X causes a conformational change that disallows interaction with SbmA. Alternatively, sbmA mutants seem to be more resistant to peptides with F (phenylalanine). There is a positive trend between the number of F residues in the peptide and resistance. This is supported by the results that show higher resistance to PPMOs with more F [such as (RFF)3RXB-AcpP and (RFR)4XB-AcpP], less resistance to PPMOs with fewer F residues (such as RTRTRFLRRTXB-AcpP), and no resistance to PPMOs with no F [such as (RXR)4XB-AcpP and (RXR)6XB-AcpP], although (RXX)3RXB-AcpP is an exception to this trend.
sbmA encodes an active transporter for bleomycin and other peptide antibiotics (19, 26, 35, 43). Our results are consistent with SbmA acting as the active transporter for (RFF)3RXB-AcpP. sbmA homologs are widely conserved among bacteria (9). The homolog of sbmA in Rhizobium meliloti, bacA, is required for symbiosis with alfalfa (9). The homolog in Brucella abortus is a virulence factor important for intracellular survival in macrophages (20). It has been proposed that the physiological substrates of SbmA are organic signaling molecules (43). An assay to measure uptake of PPMOs is currently not available but could be used to define further the role of SbmA in resistance to PPMO.
The substrate specificity of SbmA has been investigated and found to be quite flexible. Initially, the specificity was proposed to be associated with a thiazole or oxazole structural motif (43). Later, proline-rich antimicrobial peptides were shown to be transported by SbmA (26). However, (RFF)3RXB-AcpP has none of these structural features. If SbmA is the transporter of (RFF)3RXB-AcpP, the specificity of SbmA is apparently not limited to thiazole- or oxazole-containing compounds or to proline-rich peptides. Our results suggest that the substrate specificity of SbmA is flexible enough to accommodate polypeptides without thiazole, oxazole, or proline. With the peptides that we used in our conjugates, the specificity appears to be linked to the spacing of cationic and nonpolar amino acid residues within the context of the peptide. It is also noteworthy that our all-d-enantiomer conjugate (NG-05-0653) had the same MIC values as the all-l-enantiomer conjugate (NG-05-0200) for parental and resistant strains. This is in contrast to results shown for an all-d-isomer of the proline-rich antimicrobial peptide Bac7(1-35), which was ineffective compared to the all-l-isomer form (26). It was suggested that the stereospecificity of Bac7(1-35) was attributable to its interaction with SbmA, although uptake of all-d-isomer Bac7(1-35) was not demonstrated. Perhaps the stereospecificity of Bac7(1-35) is caused by its interaction with its cytoplasmic target and not SbmA. Our results suggest that the specificity of SbmA is not necessarily limited to either the l- or d-enantiomeric form of a peptide and is broader than previously known.
If SbmA is the plasma membrane transporter for (RFF)3R-AcpP, we speculate that other mechanisms for PPMOs to cross the plasma membrane exist. Strains PR200.1 and RR3 and the strain with the sbmA deletion are still somewhat susceptible to (RFF)3RXB-AcpP, albeit at high concentrations. We speculate that PPMOs may be able to cross the plasma membrane by passing through the lipid bilayer in the same manner that they cross the outer membrane. There also may be additional active transporters with specificities for nucleic acid oligomers. The latter possibility is suggested by the ability of PMOs (not conjugated to a peptide) to inhibit gene expression in strains with porous outer membranes that allow passage of large oligomers (11, 12).
In summary, the results suggest that bacterial resistance to a PPMO can be determined by the peptide and not the PMO. The rate of occurrence of spontaneous resistance to (RFF)3RXB-AcpP is similar to that of spontaneous changes in other bacterial phenotypes. In PR200.1 and RR3, resistance is caused by mutations in sbmA. Our results, in combination with the known role of SbmA in peptide antibiotic uptake, suggest that SbmA acts as a transporter of (RFF)3RXB-AcpP from the periplasm to the cytoplasm.
ACKNOWLEDGMENTS
This work was supported by AVI BioPharma, Inc., and the Howard Hughes Medical Institute (through undergraduate student research fellowships to Susan E. Puckett and Valerie Mullen). Preparation of Illumina sequencing libraries and data analyses were supported by start-up funds from the OSU Computational and Genome Biology Initiative to Michael Freitag.
We thank Andrew Karplus for a critical discussion.
Bruce L. Geller was employed by both AVI BioPharma, Inc., and Oregon State University.
Footnotes
Published ahead of print 17 September 2012
REFERENCES
- 1. Ausubel FM, et al. 1998. Current protocols in molecular biology. John Wiley & Sons, Inc, New York, NY [Google Scholar]
- 2. Baba T, et al. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. doi:10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boucher HW, et al. 2009. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 48:1–12 [DOI] [PubMed] [Google Scholar]
- 4. Campos MA, et al. 2004. Capsule polysaccharide mediates bacterial resistance to antimicrobial peptides. Infect. Immun. 72:7107–7114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Clinical and Laboratory Standards Institute 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, p 10.2–10.3 Approved standard, 7th ed Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 6. Drake J, Charlesworth WB, Charlesworth D, Crow JF. 1998. Rates of spontaneous mutation. Genetics 148:1667–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ernst RK, Guina T, Miller SI. 2001. Salmonella typhimurium outer membrane remodeling: role in resistance to host immunity. Microbes Infect. 3:1327–1334 [DOI] [PubMed] [Google Scholar]
- 8. Fischback MA, Walsh CT. 2009. Antibiotics for emerging pathogens. Science 325:1089–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gazebrook J, Ichige A, Walker GC. 1993. A Rhizobium meliloti homolog of the Escherichia coli peptide antibiotic-transport protein SbmA is essential for bacteroid development. Genes Dev. 7:1485–1497 [DOI] [PubMed] [Google Scholar]
- 10. Geller BL. 2005. Antisense antibiotics. Curr. Opin. Mol. Ther. 7:109–113 [PubMed] [Google Scholar]
- 11. Geller BL, et al. 2003. Inhibition of gene expression in Escherichia coli by antisense phosphorodiamidate morpholino oligomers. Antimicrob. Agents Chemother. 47:3233–3239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Geller BL, Deere J, Tilley L, Iversen PL. 2005. Antisense phosphorodiamidate morpholino oligomer inhibits viability of Escherichia coli in pure culture and in mouse peritonitis. J. Antimicrob. Chemother. 55:983–988 [DOI] [PubMed] [Google Scholar]
- 13. Good L, Awasthi SK, Dryselius R, Larsson O, Nielsen PE. 2001. Bactericidal antisense effects of peptide-PNA conjugates. Nat. Biotechnol. 19:360–364 [DOI] [PubMed] [Google Scholar]
- 14. Good L, Nielsen PE. 1998. Antisense inhibition of gene expression in bacteria by PNA targeted to mRNA. Nat. Biotechnol. 16:355–358 [DOI] [PubMed] [Google Scholar]
- 15. Greenberg DE, et al. 2010. Antisense phosphorodiamidate morpholino oligomers targeted to an essential gene inhibit Burkholderia cepacia complex. J. Infect. Dis. 201:1822–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Harth G, Zamecnik PC, Tabatadze D, Pierson K, Horwitz MA. 2007. Hairpin extensions enhance the efficacy of mycolyl transferase-specific antisense oligonucleotides targeting Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 104:7199–7204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Henriques ST, Melo MN, Castanho MARB. 2006. Cell-penetrating peptides and antimicrobial peptides: how different are they? Biochem. J. 399:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kramer MF, Coen DM. 2001. Enzymatic amplification of DNA by PCR: standard procedures and optimization, unit 15.1. In Ausubel FM, et al. (ed), Current protocols in molecular biology. John Wiley & Sons, Inc, New York, NY: [DOI] [PubMed] [Google Scholar]
- 19. Lavina M, Pugsley AP, Moreno F. 1986. Identification, mapping, cloning and characterization of a gene (sbmA) required for mirocin B17 action on E. coli K12. J. Gen. Microbiol. 132:1685–1693 [DOI] [PubMed] [Google Scholar]
- 20. LeVier K, Phillips RW, Gripper VK, Roop RM, II, Walker GC. 2000. Similar requirements of a plant symbiont and a mammalian pathogen for prolonged intracellular survival. Science 287:2492–2493 [DOI] [PubMed] [Google Scholar]
- 21. Li XZ, Nikaido H. 2009. Efflux-mediated drug resistance in bacteria: an update. Drugs 69:1555–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Livermore DM. 2009. Has the era of untreatable infections arrived? J. Antimicrob. Chemother. 64(Suppl 1):i29–i36 [DOI] [PubMed] [Google Scholar]
- 23. Luria SE, Delbruck M. 1943. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28:491–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maloy S.Mutation rates. 2011. http://www.sci.sdsu.edu/∼smaloy/MicrobialGenetics/topics/mutations/fluctuation.html.
- 25. Martinez JL, Baquero F. 2000. Mutation frequencies and antibiotic resistance. Antimicrob. Agents Chemother. 44:1771–1777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mattimuzzo M, et al. 2007. Role of the Escherichia coli SbmA in the antimicrobial activity of proline-rich peptides. Mol. Microbiol. 66:151–163 [DOI] [PubMed] [Google Scholar]
- 27. Mellbye BL, Puckett SE, Tilley LD, Iversen PL, Geller BL. 2009. Variations in amino acid composition of antisense peptide-phosphorodiamidate morpholino oligomers affect potency against Escherichia coli in vitro and in vivo. Antimicrob. Agents Chemother. 53:525–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Neuman BW, et al. 2005. Inhibition, escape, and attenuated growth of severe acute respiratory syndrome coronavirus treated with antisense morpholino oligomers. J. Virol. 79:9665–9676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. New England Biolabs, Inc Rubidium chloride method. New England Biolabs, Inc, Ipswich, MA [Google Scholar]
- 30. Nikaido H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67:593–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pomraning KR, Smith KM, Freitag M. 2009. Genome-wide high throughput analysis of DNA methylation in eukaryotes. Methods 47:142–150 [DOI] [PubMed] [Google Scholar]
- 32. Pomraning KR, Smith KM, Freitag M. 2011. Bulk segregant analysis followed by high-throughput sequencing reveals the Neurospora cell cycle gene, ndc-1, to be allelic with the gene for ornithine decarboxylase, spe-1. Eukaryot. Cell 10:724–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rosche WA, Foster PL. 2000. Determining mutation rates in bacterial populations. Methods 20:4–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Saka K, et al. 2005. A complete set of Escherichia coli open reading frames in mobile plasmids facilitating genetic studies. DNA Res. 12:63–68 [DOI] [PubMed] [Google Scholar]
- 35. Salomon RA, Farias RN. 1995. The peptide antibiotic microcin 25 is important through the TonB pathway and the SbmA protein. J. Bacteriol. 177:3323–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Smith MW, Tyreman DR, Payne GM, Marshall NJ, Payne JW. 1999. Substrate specificity of the periplasmic dipeptide-binding protein from Escherichia coli: experimental basis for the design of peptide prodrugs. Microbiology 145:2891–2901 [DOI] [PubMed] [Google Scholar]
- 37. Spellberg B, et al. 2008. The epidemic of antibiotic-resistant infections: a call to action for the medical community from the Infectious Diseases Society of America. Clin. Infect. Dis. 46:155–164 [DOI] [PubMed] [Google Scholar]
- 38. Splith K, Neundorf I. 2011. Antimicrobial peptides with cell-penetrating peptide properties and vice versa. Eur. Biophys. J. 40:387–397 [DOI] [PubMed] [Google Scholar]
- 39. Tilley LD, et al. 2006. Gene-specific effects of antisense phosphorodiamidate morpholino oligomer-peptide conjugates on Escherichia coli and Salmonella enterica serovar Typhimurium in pure culture and in tissue culture. Antimicrob. Agents Chemother. 50:2789–2796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tilley LD, Mellbye BL, Puckett SE, Iversen PL, Geller BL. 2007. Antisense peptide-phosphorodiamidate morpholino oligomer conjugate: dose-response in mice infected with Escherichia coli. J. Antimicrob. Chemother. 59:66–73 [DOI] [PubMed] [Google Scholar]
- 41. Vaara M, Porro M. 1996. Group of peptides that act synergistically with hydrophobic antibiotics against gram-negative enteric bacteria. Antimicrob. Agents Chemother. 40:1801–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yeaman MR, Yount NY. 2003. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 55:27–55 [DOI] [PubMed] [Google Scholar]
- 43. Yorgey P, et al. 1994. Posttranslational modifications in microcin B17 define an additional class of DNA gyrase inhibitor. Proc. Natl. Acad. Sci. U. S. A. 91:4519–4523 [DOI] [PMC free article] [PubMed] [Google Scholar]