Abstract
Five VanN-type vancomycin-resistant Enterococcus faecium strains were isolated from a sample of domestic chicken meat in Japan. All isolates showed low-level resistance to vancomycin (MIC, 12 mg/liter) and had the same pulsed-field gel electrophoresis profile. The vancomycin resistance was encoded on a large plasmid (160 kbp) and was expressed constitutively. The VanN-type resistance operon was identical to the first resistance operon to be reported, with the exception of a 1-bp deletion in vanTN and a 1-bp substitution in vanSN.
TEXT
Since the first reports of vancomycin resistance in Enterococcus faecium in 1988 (12, 20), the glycopeptide-resistant enterococci (GRE) have become increasingly widespread throughout the world and are found as multiresistant opportunistic pathogens in hospitals and also in the environment (food animals). To date, nine types of operon structure conferring resistance to glycopeptides have been reported (5, 11). They are designated according to the characteristics of a key ligase gene that encodes either a d-alanyl–d-lactate or a d-alanyl–d-serine ligase (2). The d-alanyl–d-lactate ligase group includes the vanA, vanB, vanD, and vanM genes. The d-alanyl–d-serine group includes the vanC1, vanC2, vanC3, vanE, vanG, vanL, and vanN genes. Except for vanC-type resistance, which is intrinsic to Enterococcus gallinarum and Enterococcus casseliflavus, all resistance types are acquired externally. The d-Ala–d-Lac-type operons may be located on either plasmids or the chromosome. While the d-Ala–d-Ser-type vanG, vanE, and vanL operons have been detected only in the chromosome of Enterococcus faecalis, the location of VanN in E. faecium has not been clearly identified to date. VanG and VanN are transferable d-Ala–d-Ser resistance-type operons. The recently reported VanN type was identified in Enterococcus faecium isolated from a blood culture, showed a low level of vancomycin resistance (MIC, 16 mg/liter), and was susceptible to teicoplanin (0.5 mg/liter) (11). The vanN resistance operon was reported to be found on a transferable element. VanN-type resistance was detected in E. faecium strains isolated from a patient in France in 2008, which is the only report of VanN-type GRE to date. So far, there have not been any epidemiological data reported for VanN-type GRE strains obtained anywhere in the world and the significance of the acquisition of VanN-type resistance by enterococci is not clear.
In Japan, there is a lower incidence of GRE in humans and animals than in other countries (16). However, there are several reports showing the possible transmission of GRE or glycopeptide resistance between humans and food animals through food products such as chicken meat (8, 16). More than 10 years ago, we isolated GRE strains from both imported and domestic meats, including chicken meat (9). Since then, we have examined both imported and domestic meat samples as part of a surveillance program looking at GRE contamination (16, 19). While VanA- and VanB-type GRE strains, which show high-level resistance to glycopeptides, are occasionally detected in the samples, most of the GRE isolates from the meat samples had VanC-type resistance, which is carried naturally by enterococci that show low-level resistance to glycopeptides. We recently identified VanN-type GRE strains isolated from a sample of domestic chicken meat. Here we present the results of the analysis of those strains.
During the period from February to May 2011, a total of 322 meat and swab samples from meat destined for consumption in Japan were collected and investigated. The samples were obtained from two major national quarantine stations (Yokohama and Kobe) and from three meat inspection offices (Gunma, Kagoshima, and Miyazaki) in Japan. They included samples from 90 domestic chickens, 45 domestic pork meat samples, 85 imported chickens, and 102 imported pork meat samples. The country or region of origin of each sample is listed in Table 1. A hundred grams of each meat sample (mincemeat) was smashed and homogenized using an EXNIZER 400 (Organo, Japan) in 150 ml of buffered peptone water (Nissui, Japan). Eight milliliters of the supernatant from the homogenized meat sample was mixed with 32 ml of esculin saline buffer containing vancomycin at a concentration of 6 mg/liter and preincubated for 48 h at 37°C. After preincubation, 0.1 ml of each culture broth was spread on bile esculin azide agar (BEAA) plates containing vancomycin at a concentration of 6 mg/liter. The meat swab samples were preincubated for 48 h in 40 ml of brain heart infusion broth (Difco, Detroit, MI) containing vancomycin at a concentration of 6 mg/liter, and then 0.1 ml of each culture broth was spread on BEAA plates containing vancomycin at a concentration of 6 mg/liter. In the present study, enterococci showing a vancomycin MIC of >12 mg/liter were isolated and further analyzed as GRE, because the VanA-type GRE strain occasionally shows low-level resistance to vancomycin (18). Two colonies were picked at random from each GRE-positive sample giving multiple colonies on the selective plate. A total of 128 samples (40%) were positive for GRE, and 371 GRE isolates were obtained in total. To genotype the vancomycin resistance of the GRE isolates, multiplex PCR of the key ligase genes was performed (6). Three hundred forty-nine VanC1-type GRE strains, 17 VanC2-type GRE strains, and 5 unknown-type GRE strains were isolated (Table 1). It was considered that the VanC1-type GRE isolates would be E. gallinarum strains and the VanC2-type GRE isolates would be E. casseliflavus strains, as both species naturally exhibit low-level resistance to glycopeptides (5). At first, two unknown-type GRE strains were isolated from one domestic chicken that had been bred in Miyazaki Prefecture, Kyusyu Island, Japan. During this study, an additional three isolates of the unknown-type GRE strain were picked up from the master plate which produced the original two unknown-type GRE strains to give five isolates in total. These five isolates were then used to examine whether they arose from a single clone or from multiple clones. The five GRE isolates, named GU121-1, GU121-2, GU121-3, GU121-4, and GU121-5, were all identified as E. faecium strains on the basis of their ddl gene sequences (15). They showed low-level resistance to vancomycin (MIC, 12 mg/liter) and were susceptible to teicoplanin (MIC, 1.5 mg/liter). The five isolates were susceptible or showed intermediate resistance to other antibiotics (MICs [mg/liter]: ampicillin, ≤4; chloramphenicol, ≤4, ciprofloxacin, 1; erythromycin, 2; fosfomycin, 32; gentamicin, ≤125; kanamycin, ≤125; linezolid, ≤1; rifampin, 2; tetracycline, ≤2). Pulsed-field gel electrophoresis (PFGE) analysis confirmed that they were indistinguishable and were from a single GRE clone (see Fig. S1 in the supplemental material) (16). Of the five isolates, E. faecium GU121-1 was picked for further analysis as the representative strain in this study.
Table 1.
GRE isolates from meat samplesa
| Country (prefecture) of origin | No. of chicken, pig samples | Corresponding no. of GRE strains/samplesb |
||
|---|---|---|---|---|
| vanC1 | vanC2 | vanN | ||
| Japan (Gunma) | 30, 15 | 62/21, 6/3 | 3/1, 3/1 | 0, 0 |
| Japan (Miyazaki) | 30, 15 | 77/27, 0 | 0, 0 | 5/1, 0 |
| Japan (Kagoshima) | 30, 15 | 15/5, 0 | 3/1, 0 | 0, 0 |
| United States | 3, 44 | 3/1, 0 | 0, 0 | 0, 0 |
| Brazil | 71, 0 | 173/59, 0 | 0, 0 | 0, 0 |
| France | 6, 0 | 0, 0 | 0, 0 | 0, 0 |
| Philippines | 5, 0 | 13/5, 0 | 0, 0 | 0, 0 |
| Canada | 0, 25 | 0, 0 | 0, 3/1 | 0, 0 |
| Denmark | 0, 19 | 0, 0 | 0, 2/1 | 0, 0 |
| Mexico | 0, 7 | 0, 0 | 3/1, 0 | 0, 0 |
| Chile | 0, 3 | 0, 0 | 0, 0, | 0, 0 |
| Spain | 0, 2 | 0, 0 | 0, 0 | 0, 0 |
| Hungary | 0, 1 | 0, 0 | 0, 0 | 0, 0 |
| Netherlands | 0, 1 | 0, 0 | 0, 0 | 0, 0 |
| Chicken, pig totals | 175, 147 | 343/118, 6/3 | 9/3, 8/3 | 5/1, 0 |
The vancomycin MICs of GRE isolates were more than 12 mg/liter.
Neither VanA-type nor VanB-type GRE was detected in this study. VanC1-type and VanC2-type resistances are carried naturally by E. gallinarum and E. casseliflavus.
Multilocus sequence typing (MLST) analysis showed that one of seven genes, purK, was a new allele (number 58), and E. faecium isolate GU121-1 was categorized as a new sequence type (ST) designated ST669 (gene, allele number: AtpA, 9; Ddl, 8; Gdh, 14; PurK, 58; Gyd, 6; PstS, 27; Adk, 6). This new type showed the greatest similarity to previously reported ST329 (gene, allele number: AtpA, 9; Ddl, 8; Gdh, 14; PurK, 8; Gyd, 10; PstS, 27; Adk, 6) on the basis of information in the database (MLST database [http://efaecium.mlst.net/]). ST669 was a double-locus variant of ST329 (we found two nucleotide substitutions in purK and one in gyd). The VanA-type GRE strain categorized as ST329 was isolated from a blood sample from a patient hospitalized in the Netherlands in 1999. Both ST669 and ST329 do not belong to well-characterized clonal complex 17, which is found in the hospital-adapted and epidemic E. faecium strain cluster. ST669 was grouped as a satellite sequence type on the basis of the MLST analysis (10).
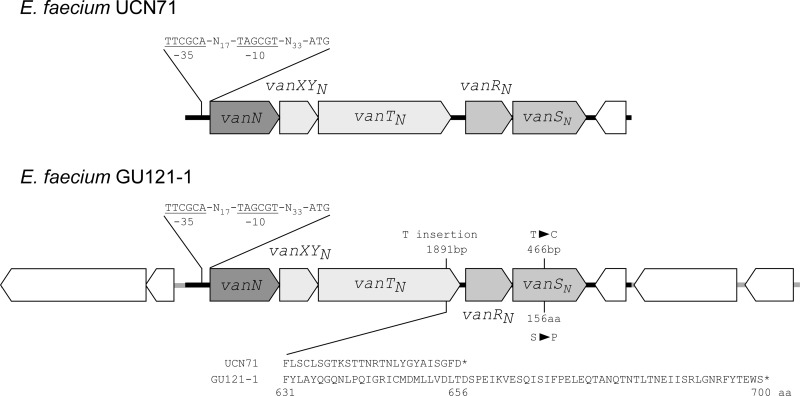
The specific PCR primer sets used to amplify the internal region of the reported resistance ligase genes (vanA, vanB, vanC1, vanC2, and vanC3) did not work, and no PCR product was obtained (6). Previously reported primers oligo V1 and oligo V2, which were designed on the basis of the ligase amino acid sequences, were then used for PCR amplification of the unknown ligase gene (7). The PCR product successfully amplified using this primer set was analyzed by direct DNA sequencing. The DNA sequence obtained for the ligase gene was homologous to the vanL ligase (around 70% identity at the base pair level) (4). On the basis of the DNA sequence obtained for the ligase gene, a pair of primers for inverse PCR was designed in order to examine the entire resistance gene cluster (operon) around the ligase gene. Several repeated inverse PCR amplifications with restriction enzymes HindIII, BamHI, and SalI were performed to determine the entire DNA sequence of the vanL-like resistance operon structure. A DNA region covering 12,344 bp that was located between a SalI site and an EcoRI site and included the predicted vancomycin resistance operon (including the vanL-like ligase gene) was determined in this study (Fig. 1). Analyses of the DNA sequence data showed that the unknown resistance operon structure containing the vanL-like gene was almost identical to the newly reported d-Ala–d-Ser VanN-type ligase and was composed of five genes designated vanN, vanXYN, vanTN, vanRN, and vanSN (11). Compared to the previously reported VanN-type operon found in the E. faecium UCN71 strain, there was a 1-bp insertion and a 1-bp substitution in the VanN-type operon of E. faecium GU121-1. A thymidine residue was inserted at bp 1891 in the vanTN gene. The insertion resulted in a frameshift in the C-terminal region of vanTN and the production of an elongated VanTN peptide compared with the prototype protein (a length increase from 656 to 700 amino acids) (Fig. 1). A 1-bp substitution was located in vanSN gene at 466 bp. This caused an amino acid change of Ser to Pro at residue 156 of the VanSN protein.
Fig 1.
The VanN-type resistance operon structure and the predicted ORFs found around the resistance region of E. faecium GU121-1. The upper panel shows the operon structure located on the conjugative plasmid found in the first reported VanN-type vancomycin-resistant E. faecium UCN71 clinical isolate in France (GenBank accession number: JF802084). The lower panel shows the operon structure and the predicted ORFs located on the chromosomal DNA of E. faecium GU121-1 (AB701345). The horizontal open arrows indicate ORFs and their direction of transcription. The gray-colored ORFs show the VanN-type resistance gene clusters. The thick horizontal lines behind the ORFs show the DNA sequence regions determined. The thick black lines indicate regions corresponding to the published DNA sequence data of UCN71 (6,750 bp). The thick gray lines indicate the plasmid DNA sequence regions determined in this study (12,344 bp). The promoter regions (−35 and −10 sequences) for vanN genes are shown in detail.
Both the upstream and downstream regions of the vanN operon structure of E. faecium GU121-1 were examined in the present study. A DNA sequence of about 3,400 bp located upstream of the vanN ligase gene and a 3,400-bp DNA sequence located downstream of vanSN were determined (Fig. 1). Five open reading frames (ORFs) were identified in these regions; two ORFs were located upstream and three ORFs were downstream of the van resistance operon. These ORFs were transcribed in the opposite direction from the resistance genes. Homology analysis showed that most of the ORFs encoded unknown hypothetical proteins. It was obvious that there is no predicted gene related to mobile elements, including insertion sequence-related structures such as transposase and inverted repeat sequences. The ORF located downstream of the vanN operon was identical to that found in the reported DNA sequence, though its function is unknown (Fig. 1).
The first reported d-Ala–d-Ser VanN-type resistance was transferable between E. faecium strains at a low frequency (on the order of 10−10) by filter mating experiments. We examined whether the VanN-type resistance could be transferred by conjugation in vitro. Vancomycin-resistant transconjugants could not be obtained from the five VanN-type GRE isolates by solid-surface mating experiments with E. faecium BM4105RF and E. faecalis FA2-2 as the recipient strains. Although no transconjugant was obtained in this study, we cannot dismiss the possibility of transferability of the VanN-type resistance of the GU121 isolates. If the resistances were transferable, the transfer frequencies were less than 4 × 10−8 per donor cell, which was the limit of detection of transconjugants in the present study. Further experimental studies and DNA sequence analysis are needed to clarify this point.
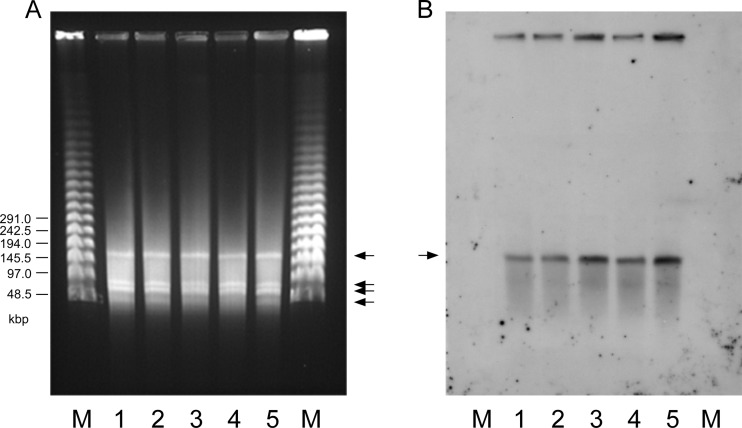
The plasmid DNAs of VanN-type strains were isolated by the alkaline-lysis method and analyzed (17). All five VanN-type E. faecium isolates showed the same plasmid profiles in agarose gel electrophoresis analysis (data not shown). The result of PFGE analyses using the S1 nuclease (Promega) showed that VanN-type strains harbor at least four plasmids (Fig. 2) (3). The conditions for electrophoresis were as follows: 19.5 h at 6 V/cm, 5.3 to 66.0 s nonlinear 21% (for 50 to 1,000 kbp), 0.5× Tris-borate-EDTA buffer, and 1% agarose gel. Four plasmids of approximately 160, 70, 60, and 40 kbp were identified. Standard PFGE analysis with the I-Ceu I endonuclease enzyme and Southern blotting hybridization using the specific probe for 23S rRNA confirmed that the four bands did not correspond to chromosomal DNA but were plasmid DNA (see Fig. S2 in the supplemental material) (13). The location of the VanN-type operon of the strains was also determined by Southern blotting hybridization using the vanN probe (Fig. 2; see Fig. S2 in the supplemental material). The 1,315-bp vanN-specific probe was constructed by PCR amplification using forward primer 5′-AGGAACATCACACTTCGAGG-3′ and reverse primer 5′-CGCATAGGTCGCTTGAACAA-3′. Hybridization analyses of PFGE showed that the vanN probe hybridized to the largest plasmid DNA band. The results clearly indicated that the vanN-type operon was located on the 160-kbp plasmid.
Fig 2.
PFGE analysis of the plasmid DNAs of VanN-type GRE isolates using S1 nuclease enzyme and Southern hybridization with the vanN ligase gene probe. (A) PFGE analysis using S1 nuclease enzyme. At least four plasmid DNA bands were detected on the gel (arrows). (B) Southern hybridization of panel A blotted with the vanN ligase gene labeled using a nonisotopic digoxigenin system (Roche). Of the four plasmid bands, the largest, a 160-kbp plasmid DNA band, bound to the vanN-specific probe (arrow). Lanes M, lambda ladder PFGE molecular size marker (NEB); lanes 1, E. faecium GU121-1; lanes 2, GU121-2; lanes 3, GU121-3; lanes 4, GU121-4; lanes 5, GU121-5.
Gene expression in the VanN-type resistance operon was examined by the real-time PCR (RT-PCR) method to detect the transcription of the vanN ligase gene. An Applied Biosystems 7500 Fast Real-Time PCR System machine and SYBR premix EX Taq II (TaKaRa, Tokyo, Japan) were used for the RT-PCR experiment (1). The expression of rrsA was used as the endogenous control. The chromosomal d-Ala–d-Ala ligase gene of E. faecium BM4105S was also used as an internal control. The transcription level of vanN was measured in the presence of different concentrations of vancomycin in the culture (0, 1, 2, and 4 mg/liter). The transcription of vanN was detected without vancomycin in the medium, and the expression levels remained unchanged after the addition of vancomycin (see Fig. S3 in the supplemental material). These data indicated that the VanN-type vancomycin resistance gene was expressed constitutively and was not induced by the addition of glycopeptide to the culture medium. This observation is consistent with the report describing the VanN-type E. faecium UCN71 strain, in which the resistance operon is considered to be expressed constitutively and which has the same promoter sequence for the VanN-type operon (vanN ligase gene) (Fig. 1) (11).
In the present study, VanN-type vancomycin resistance encoded on a 160-kbp plasmid was identified in E. faecium isolates obtained from the Japanese environment (chicken meat). In our experiment, we were unable to demonstrate that the VanN-type vancomycin resistant plasmid is mobile; however, it might be acquired from another organism through mobilization by a self-transferable plasmid or by another mechanism (14). Our data indicate that VanN-type GRE strains have already spread within the environment.
Nucleotide sequence accession number.
The sequence of the vanL-like resistance operon structure has been submitted to the DNA databases and was assigned accession number AB701345.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the Japanese Ministry of Education, Culture, Sport, Science and Technology [Kiban (B), Kiban (C), Gunma University Operation Grants] and the Japanese Ministry of Health, Labor and Welfare (H24-Shinkou-Ippan-010).
We thank E. Kamei for helpful advice on the manuscript.
Footnotes
Published ahead of print 24 September 2012
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1. Arthur M, Molinas C, Courvalin P. 1992. The VanS-VanR two-component regulatory system controls synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J. Bacteriol. 174:2582–2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arthur M, Reynolds P, Courvalin P. 1996. Glycopeptide resistance in enterococci. Trends Microbiol. 4:401–407 [DOI] [PubMed] [Google Scholar]
- 3. Barton BM, Harding GP, Zuccarelli AJ. 1995. A general method for detecting and sizing large plasmids. Anal. Biochem. 226:235–240 [DOI] [PubMed] [Google Scholar]
- 4. Boyd DA, Willey BM, Fawcett D, Gillani N, Mulvey MR. 2008. Molecular characterization of Enterococcus faecalis N06-0364 with low-level vancomycin resistance harboring a novel d-Ala–d-Ser gene cluster, vanL. Antimicrob. Agents Chemother. 52:2667–2672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Courvalin P. 2006. Vancomycin resistance in gram-positive cocci. Clin. Infect. Dis. 42(Suppl. 1):S25–S34 [DOI] [PubMed] [Google Scholar]
- 6. Dutka-Malen S, Evers S, Courvalin P. 1995. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J. Clin. Microbiol. 33:24–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dutka-Malen S, Molinas C, Arthur M, Courvalin P. 1992. Sequence of the vanC gene of Enterococcus gallinarum BM4147 encoding a d-alanine:d-alanine ligase-related protein necessary for vancomycin resistance. Gene 112:53–58 [DOI] [PubMed] [Google Scholar]
- 8. Hashimoto Y, Tanimoto K, Ozawa Y, Murata T, Ike Y. 2000. Amino acid substitutions in the VanS sensor of the VanA-type vancomycin-resistant Enterococcus strains result in high-level vancomycin resistance and low-level teicoplanin resistance. FEMS Microbiol. Lett. 185:247–254 [DOI] [PubMed] [Google Scholar]
- 9. Ike Y, et al. 1999. Vancomycin-resistant enterococci in imported chickens in Japan. Lancet 353:1854. [DOI] [PubMed] [Google Scholar]
- 10. Leavis HL, Bonten MJJ, Willems RJL. 2006. Identification of high-risk enterococcal clonal complexes: global dispersion and antibiotic resistance. Curr. Opin. Microbiol. 9:454–460 [DOI] [PubMed] [Google Scholar]
- 11. Lebreton F, et al. 2011. d-Ala–d-Ser VanN-type transferable vancomycin resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 55:4606–4612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Leclercq R, Derlot E, Duval J, Courvalin P. 1988. Plasmid-mediated resistance to vancomycin and teicoplanin in Enterococcus faecium. N. Engl. J. Med. 319:157–161 [DOI] [PubMed] [Google Scholar]
- 13. Liu SL, Hessel A, Sanderson KE. 1993. Genomic mapping with I-Ceu I, an intron-encoded endonuclease specific for genes for ribosomal RNA, in Salmonella spp., Escherichia coli, and other bacteria. Proc. Natl. Acad. Sci. U. S. A. 90:6874–6878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Manson JM, Hancock LE, Gilmore MS. 2010. Mechanisms of chromosomal transfer of Enterococcus faecalis pathogenicity island, capsule, antimicrobial resistance and other traits. Proc. Natl. Acad. Sci. U. S. A. 107:12269–12274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ozawa Y, Courvalin Gaiimand M. 2000. Identification of enterococci at the species level by sequencing of the genes for d-alanine:d-alanine ligases. Syst. Appl. Microbiol. 23:230–237 [DOI] [PubMed] [Google Scholar]
- 16. Ozawa Y, et al. 2002. Vancomycin-resistant enterococci in humans and imported chickens in Japan. Appl. Environ. Microbiol. 68:6457–6461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 18. Shin E, et al. 2006. VanB-vanA incongruent VRE isolated from animals and humans in 1999. J. Microbiol. 44:453–456 [PubMed] [Google Scholar]
- 19. Tanimoto K, Nomura T, Hamatani H, Xiao YH, Ike Y. 2005. A vancomycin-dependent VanA-type Enterococcus faecalis strain isolated in Japan from chicken imported from China. Lett. Appl. Microbiol. 41:157–162 [DOI] [PubMed] [Google Scholar]
- 20. Uttley AH, Collins CH, Naidoo J, George RC. 1988. Vancomycin-resistant enterococci. Lancet i:57–58 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.