Abstract
We have determined that the mutational inactivation of the SmeDEF efflux pump and the SmQnr quinolone resistance protein widens the mutant selection windows for ofloxacin and ciprofloxacin of Stenotrophomonas maltophilia by reducing their MICs. Resistant mutants arising from a strain lacking SmeDEF and SmQnr presented levels of susceptibility similar to those of the wild-type strain. This indicates that inactivation of intrinsic resistance determinants might increase the chances for selecting resistant mutants at low antibiotic concentrations.
TEXT
The mutant selection window has been defined as the antimicrobial concentration range extending from the MIC to the mutant preventive concentration (MPC), at which selective enrichment of antibiotic-resistant mutants occurs (4, 7, 17). This selective window is thus dependent on the MIC of a given antibiotic; different strains, presenting different antibiotic susceptibilities, might present different selective windows (11). This possibility has been discussed in the case of stepwise mutations providing high-level resistance to antibiotics. Once a mutation is acquired, MIC and MPC increase, and the mutant selection window shifts up (10), making it more difficult to prevent the emergence of new mutants (8). However, the converse situation, in other words the effect that the inactivation of elements involved in intrinsic resistance to antibiotics may have on the mutant selection window, has not been explored in detail. The predicted outcomes are diverse: (i) such inactivation should keep intact the size of the selection window but shift the window to lower values, both for MIC and MPC; this is the situation if the effects of inactivating the elements involved in intrinsic resistance are additive with the resistant alleles responsible for MPC; (ii) such inactivation could enlarge the selection window, by lowering the MIC but maintaining the MPC at least for some particular mutants; this is the situation if the effects of inactivating the elements involved in intrinsic resistance are independent of the resistant alleles responsible for MPC; (iii) such inactivation might reduce the size of the selection window, if the rise of mutants is dependent from epistatic effects with the inactivated genes, or if the population size required for the emergence of mutants is severely reduced by very low antibiotic concentrations. In the present work, we analyze these possibilities using as a model the opportunistic pathogen Stenotrophomonas maltophilia (19). A characteristic of this organism is that it presents high-level intrinsic resistance to many antibiotic classes (13).
The intrinsic resistome has been defined as the ensemble of determinants that contribute to the characteristic phenotype of susceptibility to antibiotics of a given bacterial species (9). It has been proposed that the inhibition of these intrinsic resistance elements will make bacteria more susceptible to antibiotics currently in use. As a consequence, the development of drugs to be used as adjuvants of antibiotics for improving their efficacy is a field of interest for obtaining novel antimicrobial agents (14). The expectation is that these putative inhibitors not only will improve the efficacy of antibiotics but also should most likely reduce their MPCs, although the effect they may have on the mutant selection window has not yet been analyzed. To get information on this topic, we have studied the role of two determinants that contribute to the intrinsic resistance to quinolones of S. maltophilia on the mutant selection window for quinolones in this bacterial species, namely, the SmQnr protein (20, 21) and the SmeDEF multidrug efflux pump (1). The strains used in our work are Stenotrophomonas maltophilia D457 (wild type) (12) and the mutants derived from this isolate, including MBS82 (ΔSmqnr) (21), MBS411 (ΔsmeE) (M. B. Sánchez, unpublished data), GGL199 (ΔsmeΔSmqnr GGL) (Sánchez, unpublished), and D457R (smeDEF overproducer) (3). The deletion mutants have been constructed by homologous recombination as described in reference 21. In all cases, MICs and MPCs were determined in solid Mueller-Hinton medium by using a double-dilution assay. For MICs, 105 cells were inoculated for each determination, whereas for MPCs the inoculum was 1010. The results are shown in Fig. 1. As could be predicted, the deletion of determinants involved in intrinsic resistance to quinolones of S. maltophilia reduced the MICs for quinolones. However, the effect on MPCs was smaller. As shown in Fig. 1, the same trend can be observed for the smeDEF-overproducing mutant.
Fig 1.
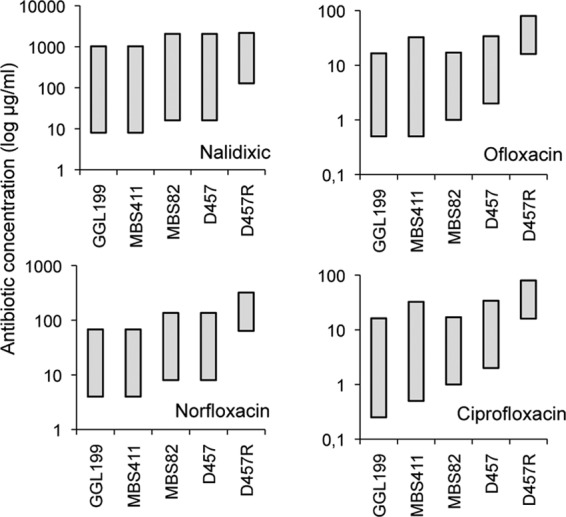
Mutant selection window for quinolones of S. maltophilia strains expressing different amounts of the intrinsic resistance elements SmQnr and SmeDEF. The mutant selection windows for ofloxacin, nalidixic acid, norfloxacin, and ciprofloxacin were determined for the S. maltophilia strains D457 (wild type), MBS82 (SmQnr defective), MBS411 (SmeDEF defective), GGL199 (SmQnr and SmeDEF defective), and D457R (SmeDEF overproducer). Mutant selection windows are represented with gray boxes in which the upper part of each box stands for the MPC and the lower part of each box stands for the MIC. As shown, the size of the mutant selection window increases with the inactivation of intrinsic resistance determinants and decreases upon overexpression.
The results of our study indicate that the effect of deleting or overproducing intrinsic resistance determinants on the mutant selection window is asymmetric, being higher on MICs than on MPCs. Since MPC is an estimation of the MIC of the least susceptible single-step resistant mutant, this indicates that the effect on antibiotic susceptibility of deleting (or overproducing) intrinsic resistance determinants cannot be extrapolated to the susceptibility of the resistant mutants that can arise from each strain. On the other hand, the accumulation of resistance narrows the mutant selection window. Conversely, deleting (or inhibiting) resistance determinants can make this window wider, hence increasing the chances for selection of resistance at low concentrations of antibiotics. We are conscious that the effect of the deletion of intrinsic resistance might be better documented for MICs, and eventually equivalent reductions in MPCs could be overlooked. But the effect of lowering MICs might have more clinical relevance than lowering (equivalently) MPCs, as the length of time in which the antibiotics are in low concentrations exceeds by far the time in which they reach the MPC concentrations (4). This extended selective time caused by inhibition of the intrinsic resistome may thus increase the chances of selection of low-level resistance mutants in spatial and temporal compartments, where high concentrations of antibiotics are unavailable (4, 5), or under conditions such as stationary growth phase or on biofilms, in which antibiotics are less effective (15, 16). By reducing MICs, there is also an increased opportunity to convert in selectable traits a number of mutations of small effect that will remain cryptic when covered by intrinsic resistance. All together, our results indicate that the inhibition of intrinsic resistance may increase the chances for the selection of antibiotic-resistant mutants at low concentrations of antibiotics.
It has been shown that S. maltophilia quinolone-resistant mutants do not present mutations in the quinolone determining resistance regions (QRDRs) of the topoisomerase genes (18, 23). This situation has been suggested to be the consequence of the presence in the chromosome of this bacterial species of a large number of antibiotic resistance genes, expression of which can confer clinically relevant resistance to this family of drugs (6, 19). Indeed, a correlation between quinolone resistance and overproduction of the SmeDEF efflux pump has been found in clinical isolates of this bacterial species (2, 22). It has been suggested that low antibiotic concentrations usually select low-level resistance mutants (5). However, we note that the MPCs for all the strains analyzed in our study are always equal or above the MICs for the D457R strain, which overproduces SmeDEF (Fig. 1). These results indicate that mutants presenting clinically relevant resistance can be selected, at low concentrations, even in strains in which intrinsic resistance determinants have been eliminated. To further support this statement, the susceptibilities to quinolones of mutants from the strains D457 (wt) and GGL199 (defective in both SmeDEF and SmQnr) selected at different concentrations of ofloxacin (2, 4, 8 μg/ml) were measured. For all selective concentrations and for all the tested quinolones, MICs of the mutants presenting the highest level of resistance were the same, regardless of whether the original strain was the wild-type D457 or the hypersusceptible GGL199 (Table 1). The only difference that could be observed was that the range of MICs of the different selected mutants was larger for D457 than for GGL199, most likely because the latter already lacks two elements, overexpression of which may confer resistance to quinolones. Together with the aforementioned effect in widening the mutant selection window, this indicates that besides the benefits for improving antibiotic efficacy, the use of inhibitors of intrinsic resistance determinants might increase the risk for selecting clinically relevant antibiotic-resistant mutants at low concentrations of antibiotics.
Table 1.
Susceptibility to quinolones of mutants obtained at different concentrations of ofloxacin
| Strain | Concn (μg/ml) of ofloxacin for selection (no. of mutants tested) | Highest MIC (MIC range) (μg/ml)a |
|||
|---|---|---|---|---|---|
| Norfloxacin | Ofloxacin | Nalidixic | Ciprofloxacin | ||
| GGL199 | 2 (12) | 128 (64–128) | 16 (8–16) | >128 (128–>128) | 16 (4–16) |
| 4 (12) | 128 (64–128) | 16 (4–16) | >128 (128–>128) | 16 (4–16) | |
| 8 (9) | 128 (A) | 16 (A) | >128 (A) | 16 (A) | |
| D457 | 2 (10) | 64 (16–64) | 16 (2–16) | 64 (16–64) | 16 (2–16) |
| 4 (12) | 64 (16–64) | 16 (4–16) | 128 (16–128) | 16 (4–16) | |
| 8 (16) | 64 (A) | 16 (8–16) | >128 (64–>128) | 16 (A) | |
Among the several tested mutants in each situation, the table shows the MIC of the mutant presenting the highest MIC and the range of MICs for all of them. A, all the tested mutants presented the same MIC.
ACKNOWLEDGMENTS
Work in our laboratory is supported by grants BIO2011-25255 from the Spanish Ministry of Science and Innovation, PROMPT from CAM, and HEALTH-F3-2011-282004 (EVOTAR) and HEALTH-F3-2010-241476 (PAR) from the European Union. G.G.-L. is the recipient of an FPI fellowship.
Footnotes
Published ahead of print 24 September 2012
REFERENCES
- 1. Alonso A, Martinez JL. 2000. Cloning and characterization of SmeDEF, a novel multidrug efflux pump from Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 44:3079–3086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alonso A, Martinez JL. 2001. Expression of multidrug efflux pump SmeDEF by clinical isolates of Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 45:1879–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alonso A, Martinez JL. 1997. Multiple antibiotic resistance in Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 41:1140–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baquero F, Negri MC. 1997. Selective compartments for resistant microorganisms in antibiotic gradients. Bioessays 19:731–736 [DOI] [PubMed] [Google Scholar]
- 5. Baquero F, Negri MC, Morosini MI, Blazquez J. 1998. Antibiotic-selective environments. Clin. Infect. Dis. 27(Suppl 1):S5–S11 [DOI] [PubMed] [Google Scholar]
- 6. Crossman LC, et al. 2008. The complete genome, comparative and functional analysis of Stenotrophomonas maltophilia reveals an organism heavily shielded by drug resistance determinants. Genome Biol. 9:R74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Drlica K. 2003. The mutant selection window and antimicrobial resistance. J. Antimicrob. Chemother. 52:11–17 [DOI] [PubMed] [Google Scholar]
- 8. Drlica K, Zhao X. 2007. Mutant selection window hypothesis updated. Clin. Infect. Dis. 44:681–688 [DOI] [PubMed] [Google Scholar]
- 9. Fajardo A, et al. 2008. The neglected intrinsic resistome of bacterial pathogens. PLoS One 3:e1619 doi:10.1371/journal.pone.0001619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li X, Mariano N, Rahal JJ, Urban CM, Drlica K. 2004. Quinolone-resistant Haemophilus influenzae: determination of mutant selection window for ciprofloxacin, garenoxacin, levofloxacin, and moxifloxacin. Antimicrob. Agents Chemother. 48:4460–4462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liang B, et al. 2011. Mutant prevention concentration-based pharmacokinetic/pharmacodynamic indices as dosing targets for suppressing the enrichment of levofloxacin-resistant subpopulations of Staphylococcus aureus. Antimicrob. Agents Chemother. 55:2409–2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lira F, et al. 2012. Whole-genome sequence of Stenotrophomonas maltophilia D457, a clinical isolate and a model strain. J. Bacteriol. 194:3563–3564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Looney WJ, Narita M, Muhlemann K. 2009. Stenotrophomonas maltophilia: an emerging opportunist human pathogen. Lancet Infect. Dis. 9:312–323 [DOI] [PubMed] [Google Scholar]
- 14. Martinez JL. 2012. The antibiotic resistome: challenge and opportunity for therapeutic intervention. Future Med. Chem. 4:347–359 [DOI] [PubMed] [Google Scholar]
- 15. Martinez JL, et al. 2009. A global view of antibiotic resistance. FEMS Microbiol. Rev. 33:44–65 [DOI] [PubMed] [Google Scholar]
- 16. Martinez JL, Rojo F. 2011. Metabolic regulation of antibiotic resistance. FEMS Microbiol. Rev. 35:768–789 [DOI] [PubMed] [Google Scholar]
- 17. Negri MC, Lipsitch M, Blazquez J, Levin BR, Baquero F. 2000. Concentration-dependent selection of small phenotypic differences in TEM beta-lactamase-mediated antibiotic resistance. Antimicrob. Agents Chemother. 44:2485–2491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ribera A, et al. 2002. Mutations in gyrA and parC QRDRs are not relevant for quinolone resistance in epidemiological unrelated Stenotrophomonas maltophilia clinical isolates. Microb. Drug Resist. 8:245–251 [DOI] [PubMed] [Google Scholar]
- 19. Sanchez MB, Hernandez A, Martinez JL. 2009. Stenotrophomonas maltophilia drug resistance. Future Microbiol. 4:655–660 [DOI] [PubMed] [Google Scholar]
- 20. Sanchez MB, Hernandez A, Rodriguez-Martinez JM, Martinez-Martinez L, Martinez JL. 2008. Predictive analysis of transmissible quinolone resistance indicates Stenotrophomonas maltophilia as a potential source of a novel family of Qnr determinants. BMC Microbiol. 8:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sanchez MB, Martinez JL. 2010. SmQnr contributes to intrinsic resistance to quinolones in Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 54:580–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sanchez P, Alonso A, Martinez JL. 2004. Regulatory regions of smeDEF in Stenotrophomonas maltophilia strains expressing different amounts of the multidrug efflux pump SmeDEF. Antimicrob. Agents Chemother. 48:2274–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Valdezate S, Vindel A, Saez-Nieto JA, Baquero F, Canton R. 2005. Preservation of topoisomerase genetic sequences during in vivo and in vitro development of high-level resistance to ciprofloxacin in isogenic Stenotrophomonas maltophilia strains. J. Antimicrob. Chemother. 56:220–223 [DOI] [PubMed] [Google Scholar]


