Abstract
A biochemical test (Carba NP test II) was developed to identify carbapenemase production in Enterobacteriaceae and Pseudomonas spp. and to discriminate between the different types of carbapenemases (classes A, B, and D). It is based on the detection of the acidification resulting from imipenem hydrolysis, coupled with tazobactam and EDTA as inhibitors. This is an easy and reliable technique (100% sensitivity and specificity) for detection of not only carbapenemase activity but also carbapenemase types in Enterobacteriaceae and Pseudomonas aeruginosa.
TEXT
Multidrug resistance is now emerging worldwide and at an alarming rate among a variety of bacterial species, causing both community-acquired and nosocomial infections (13). The last line of therapy, carbapenems, is now frequently needed to treat nosocomial infections, and increasing resistance to this class of ß-lactams leaves us with almost no effective drugs for infection due to Gram-negative bacteria (13). However, carbapenem-resistant Enterobacteriaceae and Pseudomonas spp. have been increasingly reported (5, 8). Thus, detection of carbapenemase producers in clinical laboratories is now of utmost importance for the determination of appropriate therapeutic schemes and the implementation of infection control measures (7, 9). Recently, the Carba NP test has been developed for an early identification of carbapenemases in Enterobacteriaceae (10). This technique, based on the detection of hydrolysis of the β-lactam ring of a carbapenem, imipenem, is rapid, sensitive, and specific (10). A variety of carbapenemases in Enterobacteriaceae (8) and Pseudomonas spp. (3, 5) have been reported. They are grouped into three classes according to their amino acid identity. They belong to Ambler class A (mostly KPC), class B (metallo-ß-lactamases [MBLs] of VIM, IMP, and NDM types), and class D (mostly OXA-48). Ambler class A ß-lactamases are, at least partially, inhibited by ß-lactamase inhibitors such as clavulanic acid and tazobactam, whereas MBLs are inhibited by divalent cation chelators such as EDTA (2, 6, 7, 9). Usually, in vitro complementary tests based on the inhibition of carbapenemase activity require an additional 24- to 48-h culture step (2, 6) and do not possess high sensitivity and specificity, especially when additional mechanisms of resistance do occur or when isolates exhibit only low-level carbapenem resistance (11). In addition, detection of OXA-48 producers is challenging since there is no available chemical inhibitor(s) for those carbapenemases (4). Therefore, we have developed here the Carba NP test version II to rapidly identify the types of carbapenemase produced in Enterobacteriaceae and P. aeruginosa.
A panel of carbapenemase-producing Enterobacteriaceae (n = 97) and P. aeruginosa (n = 21) strains were included in the study. The carbapenemase types were as follows: KPC (n = 20), SME (n = 2), VIM (n = 20), IMP (n = 20), NDM (n = 20), IMI (n = 1), SPM (n = 1), AIM (n = 3), GIM (n = 6), OXA-48 (n = 20), and OXA-181 (n = 2) (OXA-181 is a derivative of OXA-48). In addition, one Citrobacter freundii isolate coproducing the carbapenemases NDM-1, VIM-4, and OXA-181 (12) was included. Strains of non-carbapenemase-producing Enterobacteriaceae (n = 40, 6 being resistant to carbapenems) and P. aeruginosa (n = 50, half being resistant to carbapenems) were used as controls (1, 10). All strains had previously been characterized for their ß-lactamase content and carbapenemase resistance mechanisms at the molecular level.
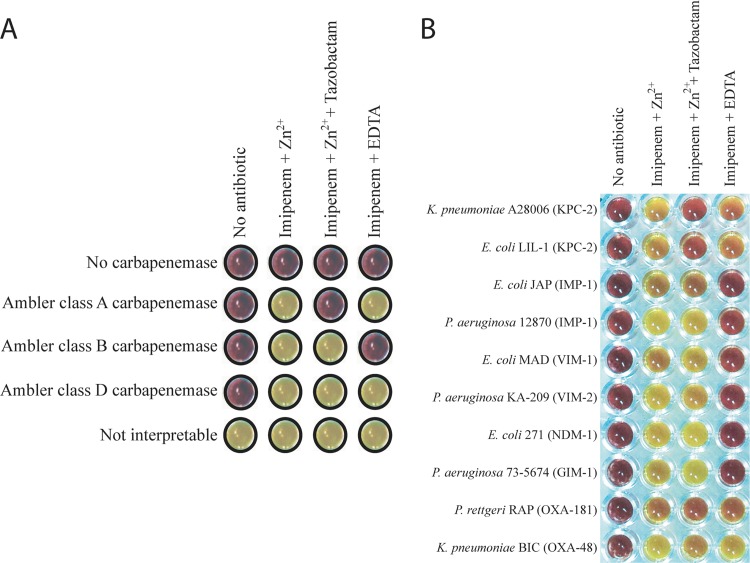
The Carba NP test II was as follows. Two calibrated inoculated loops (10 μl) of the tested strain directly recovered from a Mueller-Hinton agar (Becton-Dickinson, Le-Pont-de-Chaix, France) were resuspended in 200 μl Tris-HCl lysis buffer (B-PER II bacterial protein extraction reagent; Thermo Scientific [Pierce], Rockford, IL) (20 mM), subjected to a vortex procedure for 1 min, and further incubated at room temperature for 30 min. This bacterial suspension was centrifuged at 10,000 × g at room temperature for 5 min. Thirty microliters of the supernatant, corresponding to the enzymatic bacterial suspension, was mixed in a microwell with 100 μl of (i) a diluted red phenol solution containing 0.1 mM ZnSO4 (Merck Millipore, Guyancourt, France), (ii) a diluted red phenol solution containing 0.1 mM ZnSO4 and 3 mg/ml of imipenem monohydrate (Sigma-Aldrich, Saint-Quentin Fallavier, France), (iii) a diluted red phenol solution containing 0.1 mM ZnSO4, 3 mg/ml of imipenem monohydrate, and 4 mg/ml of tazobactam sodium salt (Sigma-Aldrich), and (iv) a diluted red phenol solution containing 3 mg/ml of imipenem monohydrate and 0.003 M EDTA. The diluted red phenol solution used was prepared by taking 2 ml of a phenol red (Merck Millipore) solution (0.5% [wt/vol]) to which 16.6 ml of distilled water was added. The pH value was then adjusted to 7.8 by adding drops of 1 N NaOH. Mixtures of the revealing solutions and the enzymatic suspension being tested were incubated at 37°C for a maximum of 2 h. In the presence of any carbapenemase, imipenem was hydrolyzed and transformed into its carboxylic form, thus leading to a pH decrease which was detected by a color change of phenol red solution (red to yellow-orange). The Carba NP test has been validated on colonies grown on Mueller-Hinton agar (Becton, Dickinson). Other culture media are being tested to check whether they support similar specificity and sensitivity of the test (10). The Carba NP test II was interpreted as follows (Fig. 1A): (i) if the color of the wells containing imipenem plus ZnSO4 and imipenem plus EDTA turned from red to yellow-orange but the well containing imipenem plus ZnSO4 plus tazobactam remained red, the strain produced an Ambler class A carbapenemase; (ii) if the color of the wells containing imipenem plus ZnSO4 and imipenem plus ZnSO4 plus tazobactam turned from red to yellow-orange but the well containing imipenem plus EDTA remained red, the strain was an MBL producer; (iii) if the color of the wells containing imipenem plus ZnSO4, imipenem plus ZnSO4 plus tazobactam, and imipenem plus EDTA turned from red to yellow-orange, the strain produced a carbapenemase belonging to neither Ambler class A nor class B that was likely a class D carbapenemase; (iv) if the color of all wells remained red, the strain was not a carbapenemase producer; and (v) if all well colors turned from red to yellow-orange, the test was considered not interpretable.
Fig 1.
Representative results of the Carba NP test II. (A) Carba NP test II interpretation scheme. (B) Results for carbapenemase-producing Enterobacteriaceae and Pseudomonas spp.
The color of the well containing imipenem and ZnSO4 turned from red to orange or yellow for all tested strains producing carbapenemases (Table 1), whereas it remained red for bacterial extracts corresponding to non-carbapenemase producers (data not shown) (10). The Carba NP test II discriminates, with a specificity of 100%, the three main types of carbapenemases (Ambler classes A, B, and D) (Table 1, Fig. 1B). The inhibition properties of tazobactam and EDTA perfectly identified class A carbapenemases and MBL producers (Table 1, Fig. 1B), whereas class D carbapenemase production was deduced from a lack of inhibition by both tazobactam and EDTA (Table 1, Fig. 1B). As expected, results obtained with the NDM-1/VIM-4/OXA-181-producing strain did not allow differentiation between those different types of carbapenemases, since positive signals were obtained in all wells. Further development of the test may include inhibition of class A and D carbapenemases by the novel inhibitor avibactam (NXL-104) (3).
Table 1.
Results of the Carba NP test II on carbapenemase-producing strains
| Ambler class | Strain | na | Carbapenemase(s) | Carba NP test IIb |
|||
|---|---|---|---|---|---|---|---|
| No antibiotic | IMP + Zn2+ | IMP + Zn2+ + TZB | IMP + EDTA | ||||
| A | E. coli | 4 | KPC-2 | − | + | − | + |
| K. pneumoniae | 4 | KPC-2 | − | + | − | + | |
| 1 | KPC-3 | − | + | − | + | ||
| E. cloacae | 5 | KPC-2 | − | + | − | + | |
| C. freundii | 1 | KPC-2 | − | + | − | + | |
| S. marcescens | 2 | KPC-2 | − | + | − | + | |
| P. aeruginosa | 3 | KPC-2 | − | + | − | + | |
| S. marscescens | 1 | Sme-1 | − | + | − | + | |
| 1 | Sme-2 | − | + | − | + | ||
| B | E. coli | 1 | VIM-1 | − | + | + | − |
| 2 | VIM-4 | − | + | + | − | ||
| 1 | VIM-19 | − | + | + | − | ||
| K. pneumoniae | 3 | VIM-1 | − | + | + | − | |
| 3 | VIM-4 | − | + | + | − | ||
| 1 | VIM-19 | − | + | + | − | ||
| E. cloacae | 1 | VIM-1 | − | + | + | − | |
| 1 | VIM-4 | − | + | + | − | ||
| C. freundii | 2 | VIM-2 | − | + | + | − | |
| P. aeruginosa | 3 | VIM-2 | − | + | + | − | |
| 2 | VIM-4 | − | + | + | − | ||
| E. coli | 2 | IMP-1 | − | + | + | − | |
| 1 | IMP-8 | − | + | + | − | ||
| K. pneumoniae | 4 | IMP-1 | − | + | + | − | |
| 2 | IMP-8 | − | + | + | − | ||
| E. cloacae | 3 | IMP-1 | − | + | + | − | |
| 2 | IMP-8 | − | + | + | − | ||
| S. marcescens | 1 | IMP-1 | − | + | + | − | |
| 1 | IMP-11 | − | + | + | − | ||
| P. aeruginosa | 2 | IMP-1 | − | + | + | − | |
| 1 | IMP-2 | − | + | + | − | ||
| 1 | IMP-13 | − | + | + | − | ||
| E. coli | 5 | NDM-1 | − | + | + | − | |
| 2 | NDM-4 | − | + | + | − | ||
| K. pneumoniae | 8 | NDM-1 | − | + | + | − | |
| P. stuartii | 1 | NDM-1 | − | + | + | − | |
| P. rettgeri | 1 | NDM-1 | − | + | + | − | |
| Salmonella spp. | 1 | NDM-1 | − | + | + | − | |
| C. freundii | 1 | NDM-1 + VIM-4 + OXA-181 | − | + | + | + | |
| P. aeruginosa | 2 | NDM-1 | − | + | + | − | |
| E. asburiae | 1 | IMI-2 | − | + | + | − | |
| P. aeruginosa | 1 | SPM-1 | − | + | + | − | |
| P. aeruginosa | 3 | AIM-1 | − | + | + | − | |
| E. cloacae | 2 | GIM-1 | − | + | + | − | |
| P. aeruginosa | 4 | GIM-1 | − | + | + | − | |
| D | E. coli | 6 | OXA-48 | − | + | + | + |
| K. pneumoniae | 10 | OXA-48 | − | + | + | + | |
| E. cloacae | 3 | OXA-48 | − | + | + | + | |
| C. koseri | 1 | OXA-48 | − | + | + | + | |
| K. pneumoniae | 1 | OXA-181 | − | + | + | + | |
| P. rettgeri | 1 | OXA-181 | − | + | + | + | |
n, number of isolates tested.
−, red (unchanged color); +, yellow-orange; IMP, imipenem; TZB, tazobactam.
This test combines excellent sensitivity (100%), specificity (100%), cost-effectiveness, and rapidity with the discrimination properties of tazobactam and EDTA as inhibitors of Ambler class A and class B ß-lactamases, respectively. Thus, this test could be performed for an early detection using bacterial isolates recovered directly from the antibiogram or from screening media, with a gain of time of at least 24 h compared to any other phenotypic method. One of the disadvantages of this technique may be the difficulty of identifying the carbapenemase types when two or more carbapenemase types are produced, which is a rare event. This technique is cheaper and faster than molecular techniques, which are useful mostly for epidemiological and research purposes. Accordingly, the results from the Carba NP test II may be used for selecting strains that are to be submitted to a targeted PCR analysis.
ACKNOWLEDGMENT
This work was funded by a grant from the INSERM (UMR914).
Footnotes
Published ahead of print 15 October 2012
REFERENCES
- 1. Dortet L, Poirel L, Nordmann P. 12 September 2012. Rapid detection of carbapenemase-producing Pseudomonas spp. J. Clin. Microbiol. doi:10.1128/JCM.01597-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Giske CG, et al. 2011. A sensitive and specific phenotypic assay for detection of metallo-β-lactamases and KPC in Klebsiella pneumoniae with the use of meropenem disks supplemented with aminophenylboronic acid, dipicolinic acid and cloxacillin. Clin. Microbiol. Infect. 17:552–556 [DOI] [PubMed] [Google Scholar]
- 3. Gupta V. 2008. Metallo-β-lactamases in Pseudomonas aeruginosa and Acinetobacter species. Expert Opin. Invest. Drugs 17:131–143 [DOI] [PubMed] [Google Scholar]
- 4. Livermore DM, et al. 2011. Activities of NXL104 combinations with ceftazidime and aztreonam against carbapenemase-producing Enterobacteriaceae. Antimicrob. Agents Chemother. 55:390–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mesaros N, et al. 2007. Pseudomonas aeruginosa: resistance and therapeutic options at the turn of the new millennium. Clin. Microbiol. Infect. 13:560–578 [DOI] [PubMed] [Google Scholar]
- 6. Migliavacca R, et al. 2002. Simple microdilution test for detection of metallo-β-lactamase production in Pseudomonas aeruginosa. J. Clin. Microbiol. 40:4388–4390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Miriagou V, et al. 2010. Acquired carbapenemases in Gram-negative bacterial pathogens: detection and surveillance issues. Clin. Microbiol. Infect. 16:112–122 [DOI] [PubMed] [Google Scholar]
- 8. Nordmann P, Dortet L, Poirel L. 2012. Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol. Med. 18:263–272 [DOI] [PubMed] [Google Scholar]
- 9. Nordmann P, et al. 2012. Identification and screening of carbapenemase-producing Enterobacteriaceae. Clin. Microbiol. Infect. 18:432–438 [DOI] [PubMed] [Google Scholar]
- 10. Nordmann P, Poirel L, Dortet L. 2012. Rapid detection of carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 18:1503–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Picão RC, et al. 2008. Metallo-β-lactamase detection: comparative evaluation of double-disk synergy versus combined disk tests for IMP-, GIM-, SIM-, SPM-, or VIM-producing isolates. J. Clin. Microbiol. 46:2028–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Poirel L, et al. 2011. Extremely drug-resistant Citrobacter freundii isolate producing NDM-1 and other carbapenemases identified in a patient returning from India. Antimicrob. Agents Chemother. 55:447–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Spellberg B, et al. 2011. Combating antimicrobial resistance: policy recommendations to save lives. Clin. Infect. Dis. 52(Suppl 5):S397–S428 [DOI] [PMC free article] [PubMed] [Google Scholar]