Abstract
Cationic antimicrobial peptides pass across the outer membrane by interacting with negatively charged lipopolysaccharide (LPS), leading to outer membrane permeabilization in a process termed self-promoted uptake. Resistance can be mediated by the addition of positively charged arabinosamine through the action of the arnBCADTEF operon. We recently described a series of two-component regulators that lead to the activation of the arn operon after recognizing environmental signals, including low-Mg2+ (PhoPQ, PmrAB) or cationic (ParRS) peptides. However, some peptides did not activate the arn operon through ParRS. Here, we report the identification of a new two-component system, CprRS, which, upon exposure to a wide range of antimicrobial peptides, triggered the expression of the LPS modification operon. Thus, mutations in the cprRS operon blocked the induction of the arn operon in response to several antimicrobial peptides independently of ParRS but did not affect the response to low Mg2+. Distinct patterns of arn induction were identified. Thus, the responses to polymyxins were abrogated by either parR or cprR mutations, while responses to other peptides, including indolicidin, showed differential dependency on the CprRS and ParRS systems in a concentration-dependent manner. It was further demonstrated that, following exposure to inducing antimicrobial peptides, cprRS mutants did not become adaptively resistant to polymyxins as was observed for wild-type cells. Our microarray studies demonstrated that the CprRS system controlled a quite modest regulon, indicating that it was quite specific to adaptive peptide resistance. These findings provide greater insight into the complex regulation of LPS modification in Pseudomonas aeruginosa, which involves the participation of at least 4 two-component systems.
INTRODUCTION
Analysis of the Pseudomonas aeruginosa genome has revealed one of the main causes of the extraordinary versatility of this Gram-negative bacterium. With nearly 10% of all open reading frames (ORFs) encoding proteins with a predicted regulatory function, Pseudomonas can control the expression of different metabolic pathways and functional attributes with great precision, adapting to many circumstances imposed by the surrounding environment (37). This explains how P. aeruginosa can thrive in such diverse niches as soil, water, plants, and animals and how it can cause serious acute and chronic infections in humans when the immune system is compromised (22, 33). Indeed, it is responsible for a substantial proportion of hospital-acquired infections and is known to cause chronic infections associated with the deterioration of lung function in cystic fibrosis patients (2, 34).
Infections by P. aeruginosa are difficult to eradicate due to its multiple mechanisms of antibiotic resistance (3). Pseudomonas is intrinsically quite resistant to a wide range of antimicrobials due to the low permeability of its outer membrane and the cooperative action of efflux pumps and periplasmic enzymes, like β-lactamase. It can also acquire resistance from the horizontal transfer of genetic elements or mutational events. Furthermore, this pathogen displays adaptive resistance to antibiotics under certain environmental conditions. The effects of adaptive resistance are transient rather than stable, reverting upon removal of the triggering environmental factor, and can involve multiple resistance mechanisms (9); this makes this form of resistance more difficult to study in depth. Nevertheless, there is substantial evidence that adaptive resistance represents an important factor in determining the success of treatment with various antibiotics (9); indeed, adaptive resistance can explain the lack of consistency between the antimicrobial susceptibility determined in vitro and the clinical success or failure of an antimicrobial therapy program.
Adaptive resistance to antibiotics, in particular to aminoglycosides, is well established in P. aeruginosa (3, 9). Similarly, adaptive resistance to cationic antimicrobials, including the lipopeptide polymyxins, was first described decades ago (14, 15, 16, 29), although the specific mechanisms involved are only now being elucidated. For example, it is known that certain environmental cues lead to increased resistance to cationic peptides in P. aeruginosa, including low Mg2+ concentrations (23, 25) and phosphate deprivation (21). Under limiting Mg2+ growth conditions, there is independent activation of the two-component regulatory systems PhoPQ and PmrAB, comprising the sensor kinases PhoQ and PmrB, which recognize Mg2+-deficient conditions and either phosphorylate or dephosphorylate transcription factors termed response regulators PhoP and PmrA, respectively. Upon activation, these induce the transcription of the arnBCADTEF operon, which encodes genes that modify lipopolysaccharide (LPS) by the addition of 4-aminoarabinose to the lipid A moiety (25, 26, 27), thus reducing the negative charge on LPS and its ability to mediate the self-promoted uptake across the outer membrane of polycationic lipopeptide polymyxins and antimicrobial peptides. However, Mg2+ deficiency does not occur in vivo, and the likely trigger for the induction of this adaptive resistance mechanism is the cationic peptides themselves. McPhee et al. (25) demonstrated that, even in high Mg2+ concentrations, the expression levels of the pmrAB and arn operons were significantly induced by subinhibitory concentrations of the cationic peptide CP11CN, a derivative of the bovine host defense peptide indolicidin. Interestingly, this upregulation occurred independently of the existence of the functional PhoPQ or PmrAB two-component systems. Recently, we identified one of the regulatory systems involved in peptide-mediated induction of LPS modification, the two-component system ParRS (10). The sensor ParS can be activated by indolicidin and polymyxins, leading to an upregulation of the arn LPS modification operon. In turn, this results in increased resistance to various cationic antimicrobial peptides, including the polymyxins. Nevertheless, the induction of the arnB promoter by certain peptides, like CP28, was not compromised by mutations in the parRS operon and remained independent of the presence of phoPQ or pmrAB. Therefore, it appeared that at least one more two-component system had to be involved in peptide-induced adaptive resistance.
In this study, we demonstrate that the novel two-component regulator CprRS (PA3077/PA3078; cationic peptide resistance [cpr]) is involved in the complex regulatory network mediating adaptive resistance to cationic peptides in P. aeruginosa. Our results show that both CprRS and ParRS are necessary for sensing particular cationic peptides, although they appeared to be activated independently.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and antimicrobials.
Table 1 lists the bacterial strains and plasmids used in this study. Bacteria were routinely grown in Luria-Bertani (LB) broth or agar at 37°C. The defined medium BM2-glucose [62 mM potassium phosphate buffer (pH 7), 7 mM (NH4)2SO4, 10 μM FeSO4, 0.4% (wt/vol) glucose], containing 2 mM (high) or 20 μM (low) MgSO4, was used for most experiments. All antibiotics were purchased from Sigma. The concentrations used for selection were 50 μg/ml tetracycline, 500 μg/ml carbenicillin, and 30 μg/ml gentamicin for P. aeruginosa and 100 μg/ml ampicillin and 50 μg/ml kanamycin for Escherichia coli. The cationic peptides were synthesized by 9-fluorenylmethoxy carbonyl (Fmoc) methods at either the Brain Research Center (University of British Columbia, Vancouver, Canada) or GenScript (Piscataway, NJ) and were 95% pure by high-performance liquid chromatography (HPLC) and mass spectrometry (MS).
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or characteristicsb | Reference |
|---|---|---|
| Strains | ||
| P. aeruginosa | ||
| WT | Wild-type P. aeruginosa PAO1 strain H103 | 37 |
| UW-PA3077a | PA3077::ISlacZ/hah-Tcr; insertion at bp 502 (672) in PA3077; derived from UW-WT | 18 |
| UW-PA3078 | PA3078::ISpho/hah-Tcr; insertion at bp 739 (1296) in PA3078; derived from UW-WT | 18 |
| cprR | PA3077::ISlacZ/hah-Tcr, H103 background; Tcr | This study |
| cprS | PA3078::ISpho/hah-Tcr, H103 background; Tcr | This study |
| cprR+ | cprR mutant with Tn7-cprRS+ integrated; Tcr Gmr | This study |
| cprS+ | cprS mutant with Tn7-cprRS+ integrated; Tcr Gmr | This study |
| parR | PA1799::ISlacZ/hah-Tcr, H103 background; Tcr | 10 |
| E. coli TOP10 | F− mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 recA1 araΔ139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| Plasmids | ||
| pCR-Blunt II-TOPO | PCR cloning vector; Kanr | Invitrogen |
| pCR-cprRS+ | pCR-Blunt II-TOPO harboring 2.5-kb cprRS amplicon | This study |
| pUC18-mini-Tn7T-Gm | Suicide plasmid; Gmr Ampr | 5 |
| pUC-Tn7-cprRS+ | pUC18-mini-Tn7T-Gm with 2.5-kb cprRS fragment from pCR-cprRS | This study |
| pTNS2 | Transposition helper plasmid; Ampr | 5 |
| pUCPlux-PPA3552 | pUCP23 containing the entire intergenic region between PA3551 and PA3552 fused to luxCDABE | 25 |
UW, University of Washington Genome Science Center.
Antibiotic resistance phenotypes: Ampr, ampicillin for E. coli and carbenicillin for P. aeruginosa; Gmr, gentamicin; Kanr, kanamycin; Tcr, tetracycline; Strr, streptomycin.
Transfer of the UW-PA3077 and UW-PA3078 transposon mutations into a new PAO1 background.
The correct insertion of the transposon into the mutant strains UW-PA3077 and UW-PA3078 (Table 1) was first confirmed by colony PCR following the protocol recommended by the University of Washington Genome Science Center (18). Subsequent transfer of the mutations to the sequenced P. aeruginosa PAO1 strain H103, our laboratory wild type, was carried out by transformation with 1 μg of genomic DNA from each UW mutant into electrocompetent cells of H103, as described previously (6). Following transformation, the cells were recovered for 1 h at 37°C and then plated onto LB agar supplemented with 50 μg/ml tetracycline. The tetracycline-resistant colonies observed after 24 h were analyzed for the insertion of the transposon as described above.
Genetic complementation.
Complementation of the cprR and cprS mutations was carried out by inserting a single wild-type copy of the operon cprRS into the chromosome of the mutant strains by using vector pUC18-mini-Tn7T-Gm (5). First, the genes comprising cprRS were amplified from the wild-type strain H103 together with 336 bp of upstream DNA containing the putative native promoter and 184 bp of downstream DNA using Phusion DNA polymerase (Finnzymes). The primers used for this PCR were designed with program primer3 (35), and the sequences were PA3077-F (5′-CGCAGTATCCGAAGGAAGAA-3′) and PA3078-R (5′-TTCATGCTGCTCTGGAACAT-3′). The 2.5-kb PCR product was then ligated into pCR-Blunt II-TOPO (Invitrogen) and transformed into One Shot TOP10 cells (Invitrogen), generating pCR-cprRS+. The insert was then subcloned into the suicide vector pUC18-mini-Tn7T-Gm. The resulting construct, pUC-Tn7-cprRS+, was coelectroporated with the transposition helper plasmid pTNS2 into competent cells of the P. aeruginosa cprR and cprS mutants prepared as described previously (5, 6). The transformants were selected on LB plates containing gentamicin. The correct integration of the mini-Tn7T-Gm transposon into the chromosome was verified by PCR as recommended by Choi et al. (5).
Luminescence gene expression assays.
Transcription from the arn promoter was tested by using plasmid pUCPlux-PPA3552, which harbors a transcriptional fusion between this promoter and the luxCDABE cassette (25). This construct was transformed into different P. aeruginosa strains, which were then grown in LB broth supplemented with carbenicillin to maintain the plasmid. Overnight cultures of these strains were washed with BM2-glucose medium with a high (2 mM) or low (20 μM) concentration of Mg2+ and then diluted 1:20. Different antimicrobial peptides were added to high-Mg2+ medium when required. At specific time points, luminescence and growth, determined as the optical density at 620 nm, were quantified with a SpectraFluor Plus luminometer (Tecan, San Jose, CA).
Computer analysis of the induction of the arn operon in response to peptide exposure in the cprR and parR mutants.
To enable the structural modeling of our peptides in the absence of precise 3-dimensional structures, a set of 295 physicochemical descriptors was calculated for each of the peptides described in Table 2 using Molecular Operating Environment (MOE) software (Chemical Computing Group Inc., Montreal, Canada). All peptides were optimized on the basis of a linear starting structure followed by potential energy minimization using an all-atom force field parameterized algorithm for proteins (OPLS) and Born solvation (simulating a liquid environment). The biological effect that was modeled was the induction of the arn operon in the presence of different peptides at sub-MICs, defined as the percentage of luminescence production compared to the wild type, for both the parR and cprR mutants.
Table 2.
Antimicrobial peptides used in this study
| Peptide | Sequence | FCharge (pH 7)c | Hydrophilic vol (Å)d | Referencee |
|---|---|---|---|---|
| HH18 | IWVIWRR-NH2a | 3 | 965.1 | Unpublished |
| CP10A | ILAWKWAWWAWRR-NH2 | 4 | 1,255.1 | 11 |
| Indolicidin | ILPWKWPWWPWRR-NH2 | 4 | 1,027.3 | 36 |
| HH2 | VQLRIRVAVIRA-NH2 | 4 | 1,317.6 | Unpublished |
| HH17 | KIWVRWK-NH2 | 4 | 980.4 | Unpublished |
| Pleurocidin | GWGSFFKKAAHVGKHVGKAALTHYL | 4 | 1,938.0 | 8a |
| Bac2A | RLARIVVIRVAR-NH2 | 5 | 1,169.9 | 42 |
| Polymyxin B | fa-BTB(BBFDLBBT)b | 5 | 305.9 | 41 |
| Colistin | fa-BTB(BBLDLBBT) | 5 | 1,144.8 | 41 |
| IDR-1002 | VQRWLIVWRIRK-NH2 | 5 | 1,269.8 | 30 |
| IDR-1012 | IFWRRIVIVKKF-NH2 | 5 | 1,147.9 | Unpublished |
| IDR-1018 | VRLIVAVRIWRR-NH2 | 5 | 1,072.1 | 38 |
| HHC36 | KRWWKWWRR-NH2 | 6 | 1,426.4 | 4 |
| IDR-1010 | IRWRIRVWVRRI-NH2 | 6 | 1,431.1 | Unpublished |
| IDR-1020 | VRLRIRWWVLRK-NH2 | 6 | 1,398.4 | Unpublished |
| CP29 | KWKSFIKKLTTAVKKVLTTGLPALIS | 6 | 1,329.9 | 12 |
| CRAMP | GLLRKGGEKIGEKLKKIGQKIKNFFQKLVPQPEQ | 6 | 2,908.1 | 13 |
| CP26 | KWKSFIKKLTSAAKKVVTTAKPLISS | 7 | 1,362.1 | 12 |
| CP28 | KWKLFKKIGIGAVLKVLTTGLPALKLTK | 7 | 1,145.4 | 32 |
Sequence in the one-letter amino acid code; NH2 indicates the amidation of the carboxyl terminus.
Sequence in the one-letter amino acid code; B indicates positively charged α,γ-diaminobutyrate; a superscript D indicates that amino acid is the d-enantiomer; fa indicates a 6-methyloctanoyl or 6-methylheptanoyl fatty acid chain.
Sum of formal charges, descriptor 83 (FCharge) calculated using MOE software.
Hydrophilic volume, descriptor 247 (vsurf_W3).
Unpublished, R. E. W. Hancock and K. Hilpert, unpublished data.
To compare structure (estimated through the use of descriptors) to function (measured as operon induction), both principal component analysis (PCA) and the partial least-squares (PLS) regression were carried out using the Simca-P 10.0 package (Umetrics, Umeå, Sweden). PCA is a multivariate projection method designed to extract and display systematic variation in a data matrix by transforming correlated variables into a smaller number of defined variables or principal components. PLS regression, on the other hand, is a linear regression model where the predicted variables are projected onto the observed variables in an n-dimensional space. All descriptor values and biological readouts used for PCA and PLS modeling were scaled to unit variance to ensure equal importance.
Killing curves.
Overnight cultures of P. aeruginosa strains were diluted 1:50 into fresh BM2-glucose medium with or without 6 μg/ml CP28 and then grown to an OD600 of approximately 0.5. One milliliter of culture was harvested and washed with 1× BM2 salts and subsequently diluted 1:10 into 1× BM2 salts. The cells were then challenged with 2.0 μg/ml polymyxin B sulfate (Sigma). Killing was carried out at room temperature under constant shaking with a benchtop Nutator (model no. 1105; BD Clay Adams). Fifty-microliter aliquots were taken at specific time points and were then serially diluted and plated on LB agar plates in order to assay for survivors.
MIC determination.
MICs in BM2-glucose medium were determined using the standard broth microdilution method (7) according to CLSI guidelines (M7-A6 [7]) and M100-S15 [8]) as described previously (39). The MIC represented the minimal concentration at which no growth could be observed with the naked eye after 24 h of growth at 37°C. To prevent artificially high MICs due to binding of the peptides to polystyrene, polypropylene microtiter plates were used as described previously (39).
Microarray analysis.
The transcriptomes of the cprR mutant and the wild type upon exposure to CP28 were compared by microarray analysis. Three independent overnight cultures from each strain were diluted 1:50 in fresh BM2-glucose medium and then grown to an OD600 of approximately 0.5, which corresponds to the mid-log phase of growth. Subsequently, 1-ml aliquots from each culture were withdrawn and incubated for 30 min in the presence of 12 μg/ml CP28 under constant shaking using a benchtop Nutator rotator platform (model no. 1105; BD Clay Adams). RNA preparation as well as cDNA synthesis, hybridization, and analysis was carried out as described previously (10, 26). The microarray slides were provided by the Institute for Genomic Research (TIGR) Pathogenic Functional Genomics Resource Center (http://pfgrc.jcvi.org/). Assessments of the slide quality, normalization, detection of differential gene expression, and statistical analysis were carried out using ArrayPipe v1.7, and the genome annotation is available at www.pseudomonas.com. ImaGene 6.0 software standard edition (BioDiscovery, Inc., El Segundo, CA) was used to quantify the slide images. Overall, changes for the cprR mutant compared to the wild type were calculated by averaging the results obtained for the three replicates. Only changes that were ≥2-fold with a P value of ≤0.05 in a two-sided one-sample Student's t test were considered significant.
Reverse transcriptase quantitative PCR.
Three independent cultures of P. aeruginosa wild type and the cprR mutant were grown to an OD600 of approximately 0.5 in BM2-glucose minimal medium containing 2 mM Mg2+ with or without indolicidin. Afterward, an aliquot from each culture was exposed for 30 min to 12 μg/ml peptide CP28. RNA from these cultures was prepared as described above. cDNA synthesis and quantitative PCR (qPCR) were performed as described previously (26). The template for the reaction was 2.5 μl of a 1:100 dilution of cDNA, to which SYBR green PCR master mix (Applied Biosystems, Foster, CA) was added. The reaction was carried out in an ABI Prism 7000 instrument (Applied Biosystems). Primer design was carried out using Primer Express (Applied Biosystems). Three independent cultures, each repeated in duplicate, were tested for each strain and condition. Calculations of the fold-changes were done according to the comparative CT method using PA1544 as a housekeeping gene.
Microarray data accession number.
The ArrayExpress accession number corresponding to this work is E-MTAB-1276.
RESULTS
Identification of the CprRS two-component system.
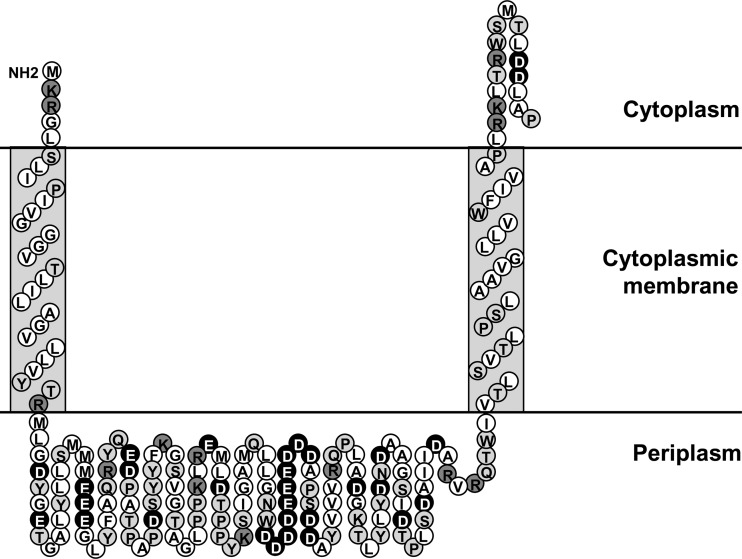
In a previous study, we demonstrated the involvement of the two-component system ParRS in the induction of the arn operon by certain antimicrobial peptides, in particular indolicidin and polymyxins (10). However, this regulatory system does not play a significant role in induction by certain other peptides, for example, the α-helical cecropin A-melittin hybrid peptide CP28 (10), also known as CEMA (32). Here, we set out to identify the two-component system that mediates this induction response. Preliminary data indicated that the well-characterized PmrAB and PhoPQ regulatory systems were not essential for the upregulation of the LPS modification operon by CP28 (data not shown); as a result, our efforts were concentrated on uncharacterized two-component systems. It is known that Salmonella PhoQ, which is responsible for sensing the presence of antimicrobial peptides in this microorganism, is unlike Pseudomonas PhoQ in that it has a periplasmic loop with an overall high negative charge (1). Therefore, we analyzed the predicted amino acid sequence of all Pseudomonas sensor kinases with the program Sosui 1.11 to determine the amino acids forming this loop according to their predicted membrane topology (17). The net charge at pH 7 of the periplasmic loop from different sensor kinases in the P. aeruginosa genome was then calculated using the program Protein Calculator v3.3 (http://www.scripps.edu/∼cdputnam/protcalc.html). The sensor kinase with the greatest net negative charge, −18, in its periplasmic loop was encoded by gene PA3078 and was part of an operon together with gene PA3077, encoding the cognate response regulator (24). This value is substantially more negative than that for ParS, PmrB, or PhoQ with respective periplasmic loop charges of −6.6, −8.6, and −7.6. Indeed, observation of the residues present in the loop of CprS revealed a very high proportion of negatively charged amino acids; in particular, there was a negative patch of 10 consecutive acidic Asp and Glu residues (Fig. 1).
Fig 1.
Topology prediction of the N-terminal portion (amino acids 1 to 187) of the sensor kinase CprS, showing the two transmembrane helices and the periplasmic sensor domain. Hydrophobic residues are represented by the white circles, polar residues are represented by the light-gray circles, and positively and negatively charged residues are represented by the dark-gray and black circles, respectively. This figure was developed based on the output of the program Sosui 1.11 (17).
Mutants in PA3077 and PA3078 (Table 1) were then taken from the University of Washington Genome Science Center (UW) transposon library for further analysis (18). To determine the effects of these mutants on peptide-induced expression of the arn genes, we utilized the plasmid pUCPlux-PPA3552 (25), which harbors a transcriptional fusion between the promoter of arnB (PA3552) and the promoterless luxCDABE operon. Transcription from the arn promoter was then quantitated by measuring the production of luminescence. The strains H103 (wild type), UW-PA3077, and UW-PA3078 (mutants of PA3077 and PA3078 were from the University of Washington transposon mutant library) containing plasmid pUCPlux-PPA3552 were assayed for the production of luminescence in BM2-glucose with or without 6 μg/ml CP28. The two mutants displayed a defective induction of the arn promoter in response to CP28 (Fig. 2A). The genes PA3077 and PA3078 were named cprR (cationic peptide resistance regulator) and cprS (cationic peptide resistance sensor), respectively.
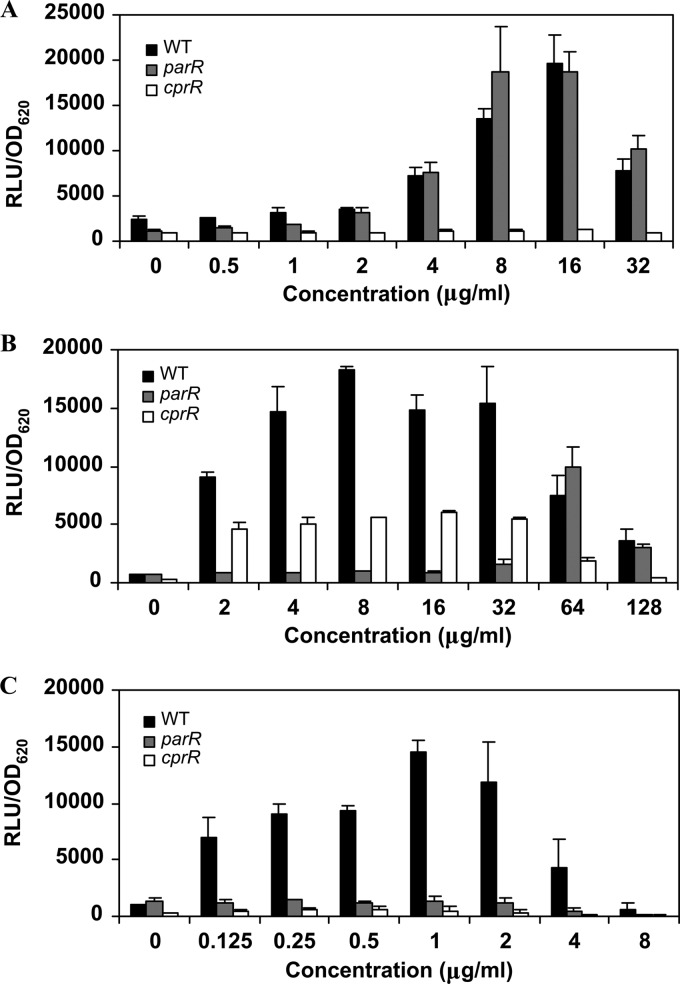
Fig 2.
Fold induction of the PPA3552::lux fusion in BM2-glucose minimal medium supplemented with 2 mM Mg2+ in the presence of 6 μg/ml CP28 (A) or without peptides (B) or grown in a low (20 μM) concentration of Mg2+ (C). The results represent the averages and standard deviations of three independent assays carried out with the WT (H103), parR, UW-PA1798, and UW-PA1799 (A) strains or with the WT strain (H103), the cprR and cprS mutants, and their respective cprR+ and cprS+ complemented mutants carrying a single copy of the cprRS operon in their chromosome (B and C).
To analyze further the mutations in this novel two-component system, the transposon insertions were transferred to a clean genetic background in P. aeruginosa PAO1 strain H103, the sequenced wild-type strain. These mutants, named the cprR and cprS mutants, were used for all subsequent experiments described in this study. The plasmid pUCPlux-PPA3552 was transformed into the cprR and cprS mutants as well as into their respective complemented strains, the cprR+ and cprS+ strains. The induction of the arn operon by 6 μg/ml CP28 was assessed by the increase in luminescence. Both mutants were defective in the upregulation of the arn operon by the peptide CP28 (P < 0.05 by Student's t test), and this defect could be restored to near-wild-type levels by complementation with a single copy of the cprRS operon into the chromosome of the mutants (Fig. 2B) (P > 0.05 for the differences between the complemented strains and the wild type). However, mutations in the cprRS operon did not lead to a defective induction of the arn promoter by a low (20 μM) concentration of Mg2+ (Fig. 2C) (P > 0.05).
Effects of parR and cprR on the induction of the arn operon by different peptides.
To provide clues as to which characteristics of the peptides were responsible for the induction of adaptive resistance in P. aeruginosa, we examined which of the 2 known peptide-sensing two-component systems, ParRS or CprRS, was required for the induction of the arn operon by different peptides. The strain H103, a parR mutant, and a cprR mutant, each carrying the pUCPlux-PPA3552 plasmid, were incubated in the presence of increasing concentrations of a battery of peptides, and induction of the arn operon was determined by the relative level of luminescence. The results obtained varied depending on the specific peptide used. For instance, when using peptide CP28 as an inducer, only the cprR mutant showed a lack of induction, whereas the parR mutant showed luminescence levels similar to those of the wild type at all concentrations tested (Fig. 3A). In contrast, exposure to indolicidin revealed a very interesting pattern of induction depending on the concentration used. Thus, the cprR mutant showed a 2- to 3-fold lower production of luminescence at all indolicidin concentrations tested (Fig. 3B); this was relatively more pronounced at higher concentrations. However, at subinhibitory concentrations of indolicidin, the ParRS two-component system proved to be more critical for the induction of the arn promoter. Interestingly, at doses around the MIC, the expression of the LPS modification operon in the parR mutant was similar to that of the wild type (Fig. 3B). We can speculate that this effect is related to the possibility that these two regulators are jointly required for maximum activation and that the differing affinities of the sensor kinases ParS and CprS for each peptide explain the variable requirement for activation at subinhibitory and near-inhibitory concentrations. The pattern exhibited by cells treated with the lipopeptide polymyxin B was different from both of the patterns discussed above in that the mutation of either cprR or parR abolished induction, indicating that both were obligately involved in the induction of the LPS modification operon at all concentrations tested (Fig. 3C).
Fig 3.
Production of luminescence in the presence of increasing concentrations of CP28 (A), indolicidin, (B) and polymyxin B (C) in the WT, parR, and cprR mutant strains. RLU, relative light units.
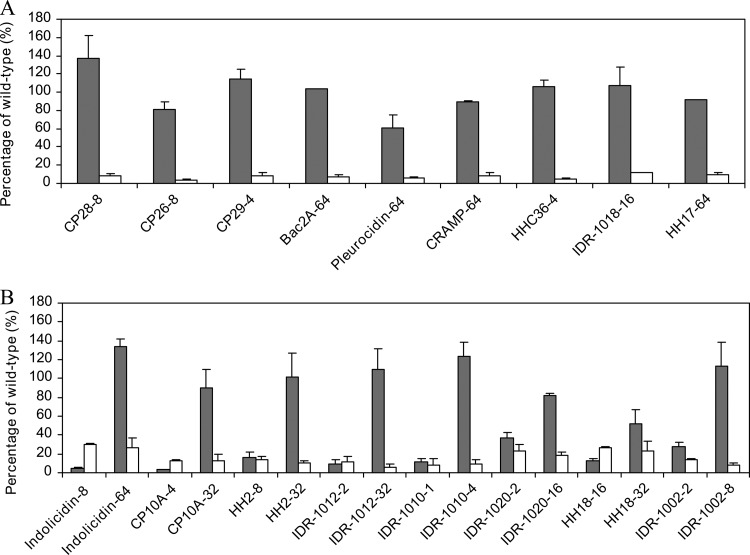
The remaining peptides that were analyzed showed a pattern similar to one of three models: CP28, indolicidin, or polymyxin B. Thus, the following peptides showed a response similar to that of CP28, in which only the CprRS system was involved in arn induction: CP26, CP29, Bac2A, pleurocidin, cathelin-related antimicrobial peptide (CRAMP), HHC36, IDR-1018, and HH17 (Fig. 4A). Other peptides, however, displayed a pattern of induction like that of indolicidin, where both mutants, the parR and cprR mutants, influenced the induction of the LPS modification operon, but parRS was important only at lower concentrations of the peptide (Fig. 4B). These peptides included CP10A, HH2, IDR-1012, IDR-1010, IDR-1020, HH18, and IDR-1002. Finally, colistin induced adaptive resistance in a way similar to polymyxin B (data not shown).
Fig 4.
Induction of the arn operon by different peptides. Percentage of induction displayed by the parR and the cprR mutants compared to the wild type in the presence of different peptides that triggered responses similar to those of CP28 (A) or indolicidin (B). The gray and white bars represent the parR and the cprR mutants, respectively. The labels on the x axis show the name and concentration of each peptide in μg/ml used to obtain the values shown. The values correspond to the averages and standard deviations of three independent experiments.
Computer modeling of the induction of the arn operon in response to peptide exposure in the cprR and parR mutants.
The luminescence experiments described above showed that different peptides had differing abilities to induce the LPS modification arn operon through the ParRS and/or CprRS two-component systems. Therefore, the influence on this effect of the charge and hydrophilicity of the peptides was analyzed in an attempt to establish a pattern. However, no particular relationship could be easily established between these properties and the induction of the arn operon in the parR and cprR mutant strains. For this reason, the expression of the arn operon, as determined in the luminescence assay upon exposure of the parR and cprR mutants to different peptides, was analyzed using computer-based modeling using physicochemical descriptors as a surrogate for structure determination, as described previously for the analysis of antimicrobial peptides (19, 20).
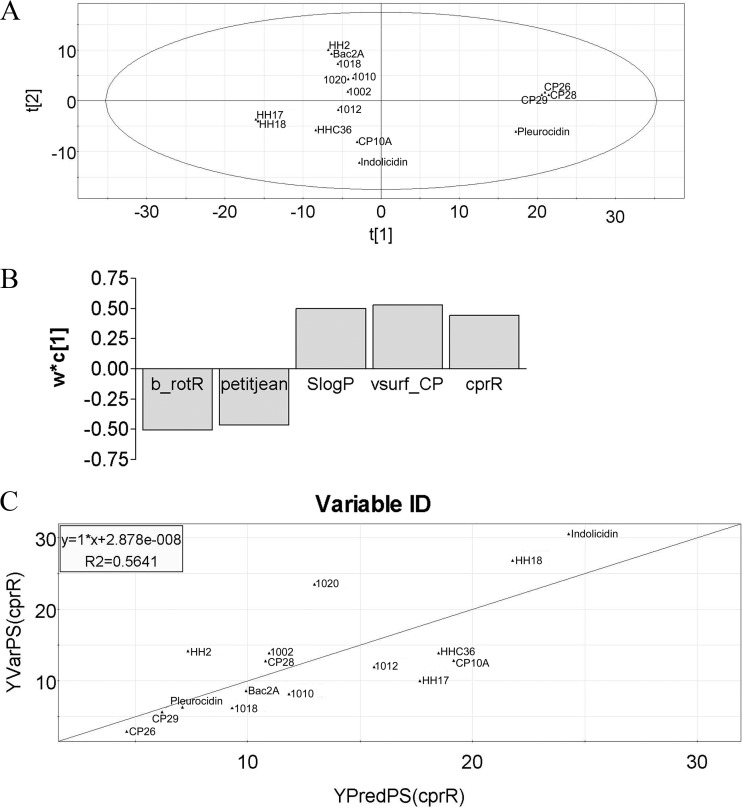
Principal component analysis (PCA) was first used to enable the identification of subgroups or clusters of peptides as well as outliers. PCA analysis of the data set enabled the identification of polymyxin B and CRAMP as outliers (data not shown), possibly due to their inherent structural difference (polymyxin) and considerably greater length (CRAMP), respectively. Thus, we excluded these peptides and also colistin from subsequent analyses. PCA analyses on the remaining members of the peptide library tested at sub-MICs for their potential to induce arn expression in the parR and cprR mutants resulted in seven significant components explaining 94% of the variation in the data with a cumulative cross-validation of 73%, with all peptides nicely spread within the 95% confidence limit (Fig. 5A). To identify any possible correlations between any of the independent variables, i.e., descriptors (X matrix), and the dependent variables, i.e., measured activation of the arn operon in the parR and cprR mutants (Y matrix), a partial least squares (PLS) regression was performed. Only the data corresponding to the inducibility by peptides of the cprR mutant could be modeled. This resulted in a model with one significant component explaining 54% of the variation in X and 32% of the variation in Y, with a weak cross-validation of 12%; part of the Y matrix was very poorly explained. By stepwise refinement of the PLS analysis and evaluation of variable importance, it became apparent that the 10 most influential descriptors could be grouped into four classes: adjacency and distance matrix descriptors, atom counts and bond counts, physical properties, and surface area, volume, and shape descriptors. Hence, selecting the most important descriptor from any one of these classes (Table 3) resulted in a PLS model with one significant component explaining 73% and 56% of the variation in X and Y, respectively, with a significant increase in the cross-validation to 44%. The loadings plot for this model (Fig. 5B) illustrated how the X variables were positively/negatively correlated with the Y variables, and the scatter plot indicated a good correlation between the predicted and observed levels of induction in the cprR mutant (Fig. 5C). Although a precise chemical understanding of the descriptors in Table 3 is somewhat difficult, these data indicate that the ability of a peptide to induce the arn operon through the activation of CprS involves particular properties of the peptide as defined by the correlative descriptors, and numerical values for these descriptors can be easily calculated for any given peptide sequence.
Fig 5.
PCA and PLS modeling of arn induction in the cprR mutant. (A) First two components from the principal component analysis of the optimized peptide set. t[1] and t[2] stand for the scores for principal components 1 and 2, respectively. (B) Loadings of the first components from the PLS analysis of the optimized peptide set. b_rotR and petitjean are negatively correlated, while SlogP and vsurf_CP are positively correlated with the levels of arn induction in the cprR mutant. w*c[1] stands for weight of variables. (C) Predicted versus observed peptide induction of the arn operon in the cprR mutant using the 4 selected descriptors.
Table 3.
Four most influential physicochemical descriptors used for modeling
| Descriptor | Description | Family |
|---|---|---|
| B_rotR | Fraction of rotatable bonds: b_rotN divided by b_heavy | Atom counts and bond counts |
| Petitjean | Value of (diam − radius)/diam (31) | Adjacency and distance matrix descriptors |
| SlogP | Log of the octanol/water (o/w) partition coefficient. This property is an atomic contribution model (40) that calculates logP from the given structure; i.e., the correct protonation state. Results may vary from the logP(o/w) descriptor. The training set for SlogP was ∼7,000 structures | Physical properties |
| Vsurf_CP | Critical packing parameter | Surface area, vol, and shape descriptors |
Reduced induction of polymyxin B adaptive resistance in the cprR and cprS mutants.
The polymyxin B MICs of the cprR and cprS mutants were the same as those of the wild type in both high- and low-Mg2+ conditions (Table 4). However, the results obtained in the luminescence assay suggested that these strains would have a substantially reduced level of adaptive resistance. Since CP28 and polymyxin B exhibited a high degree of synergy, which resulted in total killing of the bacterial cells at all concentrations of polymyxin B, we tested the acquisition of adaptive resistance to polymyxin B by means of killing curves. The results obtained were in good agreement with the values of fold induction observed in the luminescence assay in that both the cprR and cprS mutants were killed more rapidly by polymyxin B than the wild-type strain following growth in the presence of peptide CP28, with a two-log difference in the percentage of survivors after 10 min of killing (Fig. 6A). The reduced adaptive resistance to peptides seen here could be complemented by the chromosomal integration of the Tn7 transposon carrying the cprRS operon into the chromosome of the mutants (Fig. 6A). Conversely, a parR mutant did not show any significant difference in CP28-induced adaptive resistance to polymyxin B compared to the wild type (Fig. 6B). In the case of indolicidin, adaptive resistance was evaluated by means of MIC determination. The results showed increases in susceptibility for both mutants, which could be fully or partially complemented in the parS and parR mutants, respectively (Table 4). The partial complementation could be due to an imbalance between the levels of the sensor kinase and the response regulator.
Table 4.
MICs to polymyxins of the wild-type H103, the cprR and cprS mutants, and their respective cprR+ and cprS+ complemented strains under different conditionsa
| Antibiotic | MIC (μg/ml) |
||||
|---|---|---|---|---|---|
| WT | cprR | cprS | cprR+ | cprS+ | |
| In minimal medium with high (2 mM) Mg2+ | |||||
| Polymyxin B | 1 | 1 | 1 | 1 | 1 |
| Colistin | 1 | 1 | 1 | 1 | 1 |
| In minimal medium with high (2 mM) Mg2+ supplemented with 4 μg/ml indolicidinb | |||||
| Polymyxin Bb | 8 | 0.5 | 0.5 | 1–2 | 8 |
| Colistinb | >8 | 0.5 | 0.5 | 2–4 | >8 |
| In minimal medium with low (20 μM) Mg2+ | |||||
| Polymyxin B | 16 | 8 | 8 | 8 | 8 |
| Colistin | 32 | 32 | 16 | 32 | 32 |
MICs were determined by serial 2-fold dilutions in BM2-glucose minimal medium with 2 mM Mg2+, unless indicated otherwise. The MIC represents the concentration at which no growth was observed after 24 h of incubation at 37°C. The values shown are the modes of 4 to 8 independent experiments.
The cultures used to inoculate the MIC plates were also grown in the presence of indolicidin.
Fig 6.
Killing curves. Different P. aeruginosa strains, including the WT (⧫), cprR (○), cprS (□), cprR+ (●), and cprS+ (■) strains (A) or the WT (⧫), cprR (○), and parR (▲) strains (B), were grown to mid-log phase in the presence of 6 μg/ml CP28 and then challenged with 2 μg/ml of polymyxin B. Survivors were counted at specific time points. Each graphic shows data from one representative experiment of three with the same trends.
Transcriptional analysis.
After observing the different responses of the wild-type strain and the cprR mutant upon exposure to the peptide CP28, we sought to compare the transcriptional responses of the two strains under these conditions. Surprisingly, microarray analyses revealed that the differences in gene expression caused by the cprR mutation were very small, with only 15 genes showing differences of 2-fold or greater (Table 5). As expected, among these genes was the LPS modification (arn) operon, which showed a considerably lower expression in the mutant. Also, the PA4773-PA4775-pmrAB operon (where PA4773-PA4775 represents PA4773 to PA4775) and the PmrA-regulated genes PA1559-PA1560 were downregulated in the mutant. The greatest dysregulation was displayed by the predicted operon PA1559-PA1560, currently of unknown function, which we previously showed not to participate in adaptive peptide resistance (10). Thus, the genes affected by the CprRS two-component system seem to be very specific to peptide adaptive resistance. The downregulation of the genes observed in the microarray in the cprR mutant was confirmed by reverse transcriptase (RT)-qPCR analysis. In addition to validating the microarray, RT-qPCR allowed us to analyze in more detail the transcriptional levels of specific genes under different conditions, namely, growth in BM2-glucose, growth in BM2-glucose supplemented with indolicidin, and treatment with CP28 after growth in BM2-glucose (Table 6). The results obtained demonstrated that the cprRS system is more important for the induction of the arn operon by CP28 than by indolicidin. Thus, upon mutation of cprR, the induction of the gene arnB by CP28 and indolicidin was reduced by 58- and 5-fold, respectively, compared to the wild type. Also significant was the relative participation of CprRS in the upregulation of the genes encoding the PmrAB two-component system. Although the pmrAB genes were induced by both peptides, CP28 and indolicidin, the cprR mutation only affected the induction by CP28, while normal induction levels were observed when cells were grown in subinhibitory indolicidin concentrations. In addition, these results showed that there was no significant dysregulation of the cprRS or parRS operons caused by peptide exposure, indicating that these operons are not autoregulatory.
Table 5.
Microarray analysis of genes differentially expressed in the cprR mutant compared to the WT after exposure to 12 μg/ml CP28 for 30 mina
| Gene ID | Name | Fold change | P value | Description |
|---|---|---|---|---|
| PA1559 | −15.20 | 1.27e−08 | Hypothetical protein | |
| PA1560 | −13.88 | 1.89e−05 | Hypothetical protein | |
| PA3552 | arnB | −8.26 | 1.53e−07 | Hypothetical protein |
| PA3553 | arnC | −6.70 | 1.67e−07 | Probable glycosyl transferase |
| PA3554 | arnA | −8.06 | 1.55e−07 | Hypothetical protein |
| PA3555 | arnD | −8.58 | 9.81e−08 | Hypothetical protein |
| PA3556 | arnT | −7.87 | 2.38e−06 | Inner membrane l-Ara4N transferase ArnT |
| PA3557 | arnE | −5.65 | 6.29e−07 | Hypothetical protein |
| PA3558 | arnF | −7.92 | 1.53e−07 | Hypothetical protein |
| PA3559 | −7.06 | 1.53e−07 | Probable nucleotide sugar dehydrogenase | |
| PA4773 | −3.58 | 1.68e−05 | Putative S-adenosylmethionine decarboxylase proenzyme | |
| PA4774 | −5.16 | 4.84e−07 | Putative spermidine synthase | |
| PA4775 | −4.10 | 0.0077 | Hypothetical protein | |
| PA4776 | pmrA | −4.00 | 2.74e−06 | Two-component system response regulator |
| PA4777 | pmrB | −3.73 | 9.29e−06 | Two-component system signal sensor kinase |
A negative value signifies downregulation.
Table 6.
RT-qPCR analysis of the WT as well as the cprR and parR mutants under different conditions
| Strains and conditionsa | Fold change in expression of the indicated gene |
|||
|---|---|---|---|---|
| arnB | pmrB | parR | cprR | |
| WT plus CP28/WT | 335.6 ± 30.4 | 21.6 ± 1.2 | 1.9 ± 0.1 | 0.8 ± 0.1 |
| WT plus CP28/cprR mutant plus CP28 | 58.1 ± 30.8 | 24.4 ± 1.2 | NTb | NT |
| WT plus CP28/parR mutant plus CP28 | 1.1 ± 0.3 | 0.8 ± 0.1 | NT | NT |
| WT plus indolicidin/WT | 150.0 ± 50.1 | 15.19 ± 1.9 | 1.9 ± 0.2 | 1.6 ± 0.1 |
| WT plus indolicidin/cprR mutant plus indolicidin | 4.9 ± 2.6 | 1.34 ± 0.1 | NT | NT |
Grown in BM2 medium with or without indolicidin and grown in BM2 and then exposed to 12 μg/ml CP28 for 30 min.
NT, not tested.
DISCUSSION
The phenomenon of adaptive resistance is gaining increasing attention due to its potential involvement in therapeutic failure and in the global increase of resistance to antimicrobial agents. Understanding this phenomenon may be crucial in attempts to reduce the rate of increase of antibiotic resistance in human pathogens. P. aeruginosa is a good example of a highly resistant microorganism (3), and therapeutic options are extremely limited in the case of multidrug-resistant strains. In many such cases, polymyxins, peptides of bacterial origin, have been used as a drug of last resort despite early toxicity concerns. Consequently, the development of resistance to this type of antimicrobial could leave some patients without therapeutic alternatives. In a previous study (10), we described the participation of the two-component system ParRS in the acquisition of adaptive resistance to peptides and aminoglycosides after exposure of Pseudomonas cells to subinhibitory levels of indolicidin or polymyxins. More recently, Muller et al. (28) demonstrated that point mutations resulting in constitutive activation of the ParRS system lead to resistance to polymyxins, aminoglycosides, fluoroquinolones, and β-lactams. Furthermore, such mutations have been observed in multiresistant clinical strains (28). These findings highlight the importance of characterizing in detail the regulatory network involved in the inducible resistance of P. aeruginosa to antimicrobial peptides.
In the current study, we set out to identify additional two-component systems that participate in sensing the presence of cationic peptides and in promoting the addition of aminoarabinose to lipid A. To provide lead candidates, computational analyses were performed on the amino-terminal sequences of all sensor kinases in the P. aeruginosa genome, with the aim of identifying those with a highly negative charge in the periplasmic loop that might participate in cationic peptide binding. These analyses led to the identification of a sensor kinase, subsequently named CprS, with a predicted periplasmic loop having a charge of −18, the most negative of all sensor kinases in P. aeruginosa. Mutants with mutations in the cprRS operon showed a defective induction of the LPS modification operon after exposure to the peptide CP28, which was not recognized by the previously identified ParRS operon (10). However, as for ParRS, the presence of a functional CprRS system was not necessary for the induction of LPS modification upon growth in low-Mg2+ conditions. This emphasizes that P. aeruginosa senses cationic peptides and the levels of divalent cations through different systems, which contrasts considerably with the situation in other pathogens, e.g., the common binding site in Salmonella PhoQ for cations and antimicrobial peptides (1). Thus, under noninducing conditions, divalent cations are bound to an acidic surface in the amino-terminal region of PhoQ (1). The displacement of these cations by peptides or by a decrease in the concentration of cations will result in a conformational change that results in the activation of the sensor kinase. This appears to be different from Pseudomonas, where PhoQ and PmrB are able to detect low magnesium concentrations but not peptides, whereas ParS and CprS can sense the presence of particular cationic antimicrobial peptides. Thus, the existence of at least two regulatory systems to sense and respond to antimicrobial peptides highlights the complexity of this response in P. aeruginosa.
Interestingly, the relative participation of ParRS and CprRS in peptide adaptive resistance varied depending on the specific peptide and the concentration of exposure. For instance, the aforementioned CP28, as well as CP26, CP29, Bac2A, pleurocidin, CRAMP, HHC36, IDR-1018, and HH17, required the presence of an intact CprRS system for the upregulation of the LPS modification operon, while mutations in ParRS had no effect on this response. Conversely, exposure to other peptides, like indolicidin, triggered a complex response involving both two-component systems that varied according to the concentration of peptide. The regulation at low peptide concentrations was decreased significantly by the lack of either two-component system, although ParRS appeared to play a more critical role. In contrast, at concentrations closer to the MIC of the peptide, the influence of ParRS was insignificant, and only mutations in the cprRS operon reduced the induction of the arn operon. Other peptides that triggered a similar response include CP10A, HH2, IDR-1012, IDR-1010, IDR-1020, HH18, and IDR-1002. Interestingly, the polymyxins showed a third pattern of induction, in which both two-component systems participated at all concentrations, with a greater involvement of ParRS. Thus, these data indicate that functional ParRS and CprRS systems are necessary for a full adaptive response to the antimicrobial peptides in this pathogen. Our results indicate the possibility that the ParS and CprS sensor proteins recognize different properties of the peptides or different effects of the peptide on the bacterial cell at specific concentrations. Attempts to determine the roles of charge and hydrophobicity in the activation of ParRS or CprRS showed no clear trend. Computational modeling using descriptors as a surrogate for 3-dimensional peptide structures enabled the establishment of a model for the activation of the arn operon in the cprR mutant, but not in the parR mutant, corresponding to a role in activation through the CprRS and ParRS systems, respectively. Although the degree of correlation obtained for this model was not very strong, in part due to the limited number of peptides tested, it should facilitate the design of weaker inducers of arn via CprRS, which would consequently be weaker in triggering adaptive resistance.
The abrogation of peptide-mediated upregulation of the arn operon resulted in a lower ability of the cprR and cprS mutants to acquire adaptive resistance. The behaviors of these mutants, however, differed from those of the parR and parS mutant strains. Thus, transposon insertions within the cprRS operon had a lesser impact on indolicidin- or polymyxin-induced adaptive resistance than did parRS mutations. In contrast, a functional CprR, but not ParR, was essential for the induction of resistance to polymyxin B by the α-helical peptide CP28.
The transcriptional response mediated by CprRS upon CP28 challenge appears to be very specific, as it involves only the dysregulation of 15 genes, which includes the arn operon together with the PmrA-regulated PA4773-PA4775-pmrAB and PA1559-PA1560 operons. This is the smallest regulon observed by us for any of the two-component regulators studied to date. Thus, we feel it is extremely likely that CprRS is a dedicated peptide-sensing and response system. In the case of indolicidin, CprRS seems to play a lesser role than ParRS. Thus, whereas parR mutations practically abolished indolicidin-mediated induction of arnB (10), the lack of a functional CprR only slightly reduced the induction of the LPS modification operon. We can conclude that both ParRS and CprRS are decisive for the ability of P. aeruginosa to sense cationic antimicrobial peptides and that they respond by inducing LPS modification and, consequently, adaptive resistance. Furthermore, despite the importance of ParRS and CprRS for peptide-mediated adaptive resistance, neither of the two-component systems themselves were upregulated in the presence of peptides. However, the main difference between these two regulatory systems seems to be the specific signal that activates each one of them, which seems to be dependent on the characteristics of the peptide as well as the concentration of exposure. Further research should focus on identifying the nature of these signals, as well as the possible involvement of these two-component regulatory systems in the pathogenesis of P. aeruginosa. Another significant difference is the specificity of the response mediated by CprRS in contrast to the wider transcriptional response triggered by ParRS.
Overall, this study highlights the complexity of the regulatory cascades involved in adaptive resistance to peptides in P. aeruginosa. The identification of CprRS brings us closer to attaining a good comprehension of how this network works, although further research is necessary to characterize the molecular mechanisms involved in more detail. Knowledge of these systems will be very helpful in designing novel antimicrobial peptides with low capacities to induce LPS modification and determining the best administration regimes for polymyxins and other peptides that might potentially be used in the clinic. Furthermore, it seems possible that gain-of-function mutations in CprRS could be relevant to clinical resistance to polymyxins, as was demonstrated recently for ParRS (28).
ACKNOWLEDGMENTS
This work was funded by a grant from Cystic Fibrosis Canada (CFC). W.J.G. was supported by a CFC studentship, and L.F. received a postdoctoral fellowship from the Fundación Alfonso Martín Escudero (Spain). I.W. was supported by the Juergen Manchot Foundation and the Mukoviszidose e.V., Bonn, Germany (German Cystic Fibrosis Association). R.E.W.H. holds a Canada Research Chair in Microbiology.
We thank Herbert Schweizer (Colorado State University) for kindly providing us with the Tn7 plasmids.
Footnotes
Published ahead of print 24 September 2012
REFERENCES
- 1. Bader MW, et al. 2005. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell 122:461–472 [DOI] [PubMed] [Google Scholar]
- 2. Bonomo RA, Szabo D. 2006. Mechanisms of multidrug resistance in Acinetobacter species and Pseudomonas aeruginosa. Clin. Infect. Dis. 43:SS49–SS56 [DOI] [PubMed] [Google Scholar]
- 3. Breidenstein EBM, de la Fuente-Núñez C, Hancock REW. 2011. Pseudomonas aeruginosa: all roads lead to resistance. Trends Microbiol. 19:419–426 [DOI] [PubMed] [Google Scholar]
- 4. Cherkasov A, et al. 2009. Use of artificial intelligence in the design of small peptide antibiotics effective against a broad spectrum of highly antibiotic-resistant superbugs. ACS Chem. Biol. 4:65–74 [DOI] [PubMed] [Google Scholar]
- 5. Choi KH, et al. 2005. A Tn7-based broad-range bacterial cloning and expression system. Nat. Methods 2:443–448 [DOI] [PubMed] [Google Scholar]
- 6. Choi KH, Kumar A, Schweizer HP. 2006. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods 64:391–397 [DOI] [PubMed] [Google Scholar]
- 7. Clinical and Laboratory Standards Institute 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 7th ed Approved standard M7-A7. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 8. Clinical and Laboratory Standards Institute 2007. Performance standards for antimicrobial susceptibility testing. CLSI approved standard M100-S17. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 8a. Cole AM, Weis P, Diamond G. 1997. Isolation and characterization of pleurocidin, an antimicrobial peptide in the skin secretions of winter flounder. J. Biol. Chem. 272:12008–12013 [DOI] [PubMed] [Google Scholar]
- 9. Fernández L, Breidenstein EBM, Hancock REW. 2011. Creeping baselines and adaptive resistance to antibiotics. Drug Resist. Updat. 14:1–21 [DOI] [PubMed] [Google Scholar]
- 10. Fernández L, et al. 2010. Adaptive resistance to the “last hope” antibiotics polymyxin B and colistin in Pseudomonas aeruginosa is mediated by the novel two-component regulatory system ParR-ParS. Antimicrob. Agents Chemother. 54:3372–3382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Friedrich CL, Moyles D, Beveridge TJ, Hancock REW. 2000. Antibacterial action of structurally diverse cationic peptides on Gram-positive bacteria. Antimicrob. Agents Chemother. 44:2086–2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Friedrich C, Scott MG, Karunaratne N, Yan H, Hancock REW. 1999. Salt-resistant alpha-helical cationic antimicrobial peptides. Antimicrob. Agents Chemother. 43:1542–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gallo RL, et al. 1997. Identification of CRAMP, a cathelin-related antimicrobial peptide expressed in the embryonic and adult mouse. J. Biol. Chem. 272:13088–13093 [DOI] [PubMed] [Google Scholar]
- 14. Gilleland HE, Jr, Conrad RS. 1982. Chemical alterations in cell envelopes of polymyxin-resistant mutants of Pseudomonas aeruginosa grown in the absence or presence of polymyxin. Antimicrob. Agents Chemother. 22:1012–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gilleland HE, Jr, Lyle RD. 1979. Chemical alterations in cell envelopes of polymyxin-resistant Pseudomonas aeruginosa isolates. J. Bacteriol. 138:839–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gilleland HE, Jr, Murray RG. 1976. Ultrastructural study of polymyxin-resistant isolates of Pseudomonas aeruginosa. J. Bacteriol. 125:267–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hirokawa T, Boon-Chieng S, Mitaku S. 1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14:378–379 [DOI] [PubMed] [Google Scholar]
- 18. Jacobs MA, et al. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 100:14339–14344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jenssen H, et al. 2007. Evaluating different descriptors for model design of antimicrobial 12-mer peptides with enhanced activity towards Pseudomonas aeruginosa. Chem. Biol. Drug Des. 70:134–142 [DOI] [PubMed] [Google Scholar]
- 20. Jenssen H, Fjell C, Cherkasov A, Hancock REW. 2008. QSAR modeling and computer-aided design of antimicrobial peptides. J. Pept. Sci. 14:110–114 [DOI] [PubMed] [Google Scholar]
- 21. Lewenza S, et al. 2011. The olsA gene mediates the synthesis of an ornithine lipid in Pseudomonas aeruginosa during growth under phosphate-limiting conditions, but is not involved in antimicrobial peptide susceptibility. FEMS Microbiol. Lett. 320:92–102 [DOI] [PubMed] [Google Scholar]
- 22. Lyczak JB, Cannon CL, Pier GB. 2000. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect. 2:1051–1060 [DOI] [PubMed] [Google Scholar]
- 23. Macfarlane ELA, Kwasnicka A, Ochs MM, Hancock REW. 1999. PhoP-PhoQ homologues in Pseudomonas aeruginosa regulate expression of the outer-membrane protein OprH and polymyxin B resistance. Mol. Microbiol. 34:305–316 [DOI] [PubMed] [Google Scholar]
- 24. Mao F, Dam P, Chou J, Olman V, Xu Y. 2009. DOOR: a database for prokaryotic operons. Nucleic Acids Res. 37:D459–D463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McPhee JB, Lewenza S, Hancock REW. 2003. Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol. Microbiol. 50:205–217 [DOI] [PubMed] [Google Scholar]
- 26. McPhee JB, et al. 2006. Contribution of the PhoP-PhoQ and PmrA-PmrB two-component regulatory systems to Mg2+-induced gene regulation in Pseudomonas aeruginosa. J. Bacteriol. 188:3995–4006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Moskowitz SM, Ernst RK, Miller SI. 2004. PmrAB, a two-component regulatory system of Pseudomonas aeruginosa that modulates resistance to cationic antimicrobial peptides and addition of aminoarabinose to lipid A. J. Bacteriol. 186:575–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Muller C, Plésiat P, Jeannot K. 2011. A two-component regulatory system interconnects resistance to polymyxins, aminoglycosides, fluoroquinolones, and β-lactams in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 55:1211–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nicas TI, Hancock REW. 1980. Outer membrane protein H1 of Pseudomonas aeruginosa: involvement in adaptive and mutational resistance to ethylenediaminetetraacetate, polymyxin B and gentamicin. J. Bacteriol. 143:872–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nijnik A, et al. 2010. Synthetic cationic peptide IDR-1002 provides protection against bacterial infections through chemokine induction and enhanced leukocyte recruitment. J. Immunol. 184:2539–2550 [DOI] [PubMed] [Google Scholar]
- 31. Petitjean M. 1992. Applications of the radius-diameter diagram to the classification of topological and geometrical shapes of chemical compounds. J. Chem. Inf. Comput. Sci. 32:331–337 [Google Scholar]
- 32. Piers KL, Brown MH, Hancock REW. 1994. Improvement of outer membrane-permeabilizing and lipopolysaccharide-binding activities of an antimicrobial cationic peptide by C-terminal modification. Antimicrob. Agents Chemother. 38:2311–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rahme LG, et al. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899–1902 [DOI] [PubMed] [Google Scholar]
- 34. Rowe SM, Miller S, Sorscher EJ. 2005. Cystic fibrosis. N. Engl. J. Med. 352:1992–2001 [DOI] [PubMed] [Google Scholar]
- 35. Rozen S, Skaletsky HJ. 2000. Primer3 on the WWW for general users and for biologist programmers, p 365–386 In Krawetz S, Misener S. (ed), Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, NJ: [DOI] [PubMed] [Google Scholar]
- 36. Selsted ME, et al. 1992. Indolicidin, a novel bactericidal tridecapeptide amide from neutrophils. J. Biol. Chem. 267:4292–4295 [PubMed] [Google Scholar]
- 37. Stover CK, et al. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959–964 [DOI] [PubMed] [Google Scholar]
- 38. Wieczorek M, et al. 2010. Structural studies of a peptide with immune modulating and direct antimicrobial activity. Chem. Biol. 17:970–980 [DOI] [PubMed] [Google Scholar]
- 39. Wiegand I, Hilpert K, Hancock REW. 2008. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 3:163–175 [DOI] [PubMed] [Google Scholar]
- 40. Wildman SA, Crippen GM. 1999. Prediction of physiochemical parameters by atomic contributions. J. Chem. Inf. Comput. Sci. 39:868–873 [Google Scholar]
- 41. Windholz M, Budavari S, Stroumtsos L, Fertig N. 1976. The Merck index. Merck, Rahway, NJ [Google Scholar]
- 42. Wu M, Hancock REW. 1999. Improved derivatives of bactenecin, a cyclic dodecameric antimicrobial cationic peptide. Antimicrob. Agents Chemother. 43:1274–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]