Abstract
Metallo-β-lactamases are important determinants of antibacterial resistance. In this study, we investigate the sequence-activity relationship between the closely related enzymes IMP-1, IMP-6, and IMP-25. While IMP-1 is the more efficient enzyme across the overall spectrum of tested β-lactam antibacterial agents, IMP-6 and IMP-25 seem to have evolved to specifically inactivate the newer carbapenem meropenem. Molecular modeling indicates that the G235S mutation distinguishing IMP-25 from IMP-1 and IMP-6 may affect enzyme activity via Asn233.
TEXT
Antibacterial resistance in combination with few new antibacterial agents being developed raises concerns for the future treatment of bacterial infections (13). β-Lactamases (EC 3.5.2.6.), enzymes that hydrolyze and inactivate β-lactam antibacterials, and in particular metallo-β-lactamases (MBLs), present a serious challenge to public health (4). The IMP-type enzymes are among the clinically most important MBLs (7, 14). These enzymes hydrolyze penicillins, cephalosporins, and carbapenems, and they are not inhibited by traditional serine β-lactamase inhibitors, such as clavulanic acid and sulbactam (25). They utilize two Zn(II) ions as metal cofactors to bind antibiotics and activate a water molecule for nucleophilic attack. Although they cannot hydrolyze aztreonam, they are often coproduced in bacteria that produce aztreonam-hydrolyzing extended-spectrum β-lactamases (3).
The IMP-1 enzyme was first isolated in 1991 in Japan from Serratia marcescens (19) and has since been found in several countries worldwide, as well as in multiple microorganisms, including Pseudomonas aeruginosa, members of the family Enterobacteriaceae, and Acinetobacter spp. (7, 14). IMP-6, which differs from IMP-1 by an S262G mutation, was first isolated in 1996 in Japan, also from Serratia marcescens (27), and has also been identified in P. aeruginosa and Enterobacteriaceae in Japan (28) and South Korea (20, 21, 29). The biochemical properties of IMP-1 and IMP-6 have been compared extensively (10, 16, 18, 27). Interestingly, IMP-1 confers higher resistance levels to the carbapenem imipenem and several other antibiotics (10, 16), whereas IMP-6 confers higher resistance levels to the newer carbapenems meropenem (27) and doripenem (this study).
IMP-25, which differs from IMP-6 by a G235S mutation and from IMP-1 by two mutations, S262G and G235S (Fig. 1A), was recently isolated from P. aeruginosa in South Korea (GenBank accession no. EU541448). In this study, we investigate the sequence-activity relationship between these closely related enzymes by determining and comparing their biochemical characteristics and abilities to confer β-lactam resistance to Escherichia coli cells. Furthermore, we use molecular modeling to examine the role of residue 235.
Fig 1.
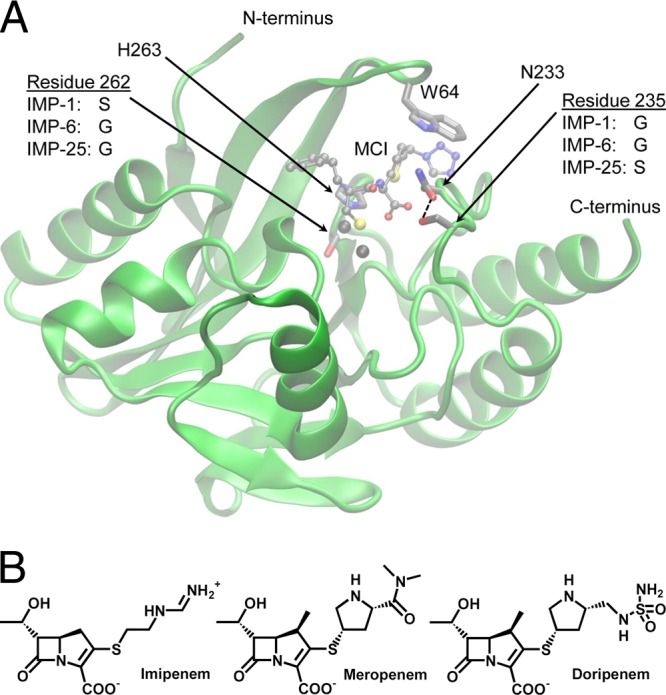
(A) Graphical representation of the MBLs studied generated with VMD (9). The backbone coordinates of IMP-1 (chain A of Protein Data Bank [PDB] accession no. 1DD6 [6]) in complex with a mercaptocarboxylate inhibitor (MCI) (shown as balls and sticks) are shown in green. The side chains of residues mutated in this study, residues 262 and 235, are shown as sticks. A few other key residues, the Zn(II) ligand His263, Asn233, which likely interacts with residue 235, and Trp64, which covers the active site, are shown as sticks. The color code of atoms is as follows: gray, C; red, O; blue, N; yellow, S. The Zn(II) ions are shown as black spheres. The enzyme shown is a hypothetical enzyme with both Ser262 and Ser235. IMP-1 would be obtained by removing the Ser235 side chain, IMP-25 by removing the Ser262 side chain, and IMP-6 by removing both. The close proximity of the Ser235 and Asn233 side chains (2.7 Å) allowing for hydrogen bonding is indicated by a dashed line. (B) Chemical structures of the three carbapenems studied. The pKa values of the R2 groups attached to the right of the carbapenem core are 10.4 (imipenem), 7.4 (meropenem), and 7.9 (doripenem). The kinetic and MIC data are consistent with the R2 groups of meropenem and doripenem being neutral in the enzyme active site.
Plasmids pET30a-blaIMP-6 for heterologous overexpression and pBC SK(+)-blaIMP-6 for MIC assays were created by PCR-based site-directed mutagenesis using pET30a-blaIMP-1 and pBC SK(+)-blaIMP-1 [obtained by subcloning blaIMP-1 with leader sequence from pET26b into pBC SK(+)], respectively, as the templates. Plasmids pET30a-blaIMP-25 and pBC SK(+)-blaIMP-25 were created in an analogous way with pET30a-blaIMP-6 and pBC SK(+)-blaIMP-6 as the templates.
IMP-1, IMP-6, and IMP-25 were overexpressed, purified, and biophysically characterized as described previously (16), except that E. coli OverExpress C43 (DE3) cells (Lucigen, Middleton, WI) were used. In short, the expression levels of the three enzymes were similar, enzymes were purified to >95% homogeneity, circular dichroism scans were superimposable, the molecular masses determined by electrospray ionization mass spectrometry were as calculated, and all enzymes bound approximately two Zn(II) ions.
Kinetic constants were determined as described previously (16) and using published wavelengths and extinction coefficients for cefoxitin, meropenem (12), and doripenem (23). Data reported in Table 1 are averages ± standard deviations, and any changes by a factor of 2 or more are considered significant. All three enzymes were able to efficiently hydrolyze all tested β-lactams except aztreonam (Table 1). The kinetic parameters of IMP-1 and IMP-6 are in good agreement with our previous study (16) and trends are in agreement with a study from another laboratory (10). The three substrates cefoxitin, meropenem, and doripenem added in this study behaved like “type I substrates” (kcat/Km values are similar for IMP-1 and IMP-6 [16]). In this respect, the newer carbapenems meropenem and doripenem differ from imipenem, which behaves like a “type II substrate” (higher kcat/Km for IMP-1 than IMP-6 [10, 16, 27]). The difference between IMP-1 and IMP-6 in their kcat/Km ratios toward imipenem, which has a positively charged 2-(aminomethylideneamino)ethylsulfanyl R2 group (pKa = 10.4 [24]) (Fig. 1B), can be explained by a domino effect, where Ser262 supports the His263 Zn(II) ligand in the presence of substrates with bulky or positively charged R2 groups (18). The pKas of the corresponding 5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl and 5-[(sulfamoylamino)methyl]pyrrolidin-3-yl]sulfanyl R2 groups in meropenem (AstraZeneca, Mississauga, Ontario, Canada) and doripenem (Janssen-Cilag Pty Ltd., North Ryde, NSW, Australia) are 7.4 and 7.9, respectively (Fig. 1B). These moieties may be neutral within the enzyme active site, consistent with their type I substrate-like behavior. IMP-25's kcat/Km values are comparable to those of IMP-6 (cefoxitin, penicillins, and carbapenems), slightly lower (cephalothin and cefotaxime), or slightly higher (ceftazidime).
Table 1.
Kinetic parameters of the three MBLs toward 10 tested β-lactams
| β-Lactam | Kinetic parameter of the MBL to β-lactama |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| IMP-1 |
IMP-6 |
IMP-25 |
|||||||
| kcat (s−1) | Km (μM) | kcat/Km (μM−1 · s−1) | kcat (s−1) | Km (μM) | kcat/Km (μM−1 · s−1) | kcat (s−1) | Km (μM) | kcat/Km (μM−1 · s−1) | |
| Cephalothin | 50 ± 1 | 4.5 ± 0.2 | 11.1 ± 0.2 | 131 ± 4 | 15 ± 2 | 9 ± 1 | 66 ± 3 | 17 ± 1 | 3.9 ± 0.1 |
| Cefotaxime | 16.4 ± 0.4 | 5.1 ± 0.8 | 3.2 ± 0.5 | 26 ± 1 | 6 ± 1 | 4.3 ± 0.4 | 19 ± 2 | 17 ± 1 | 1.1 ± 0.1 |
| Ceftazidime | 14 ± 1 | 55 ± 7 | 0.25 ± 0.02 | 3.6 ± 0.4 | 80 ± 10 | 0.050 ± 0.002 | 5.2 ± 0.5 | 48 ± 6 | 0.11 ± 0.01 |
| Cefoxitin | 16.2 ± 0.1 | 4.0 ± 0.1 | 4.0 ± 0.1 | 70 ± 10 | 17 ± 4 | 4.3 ± 0.4 | 30 ± 10 | 25 ± 7 | 3.6 ± 0.4 |
| Benzylpenicillin | 2,000 ± 100 | 530 ± 50 | 3.8 ± 0.1 | 220 ± 30 | 900 ± 100 | 0.25 ± 0.01 | 60 ± 10 | 230 ± 90 | 0.26 ± 0.05 |
| Ampicillin | 260 ± 30 | 210 ± 30 | 1.2 ± 0.1 | 24 ± 1 | 290 ± 30 | 0.09 ± 0.01 | 67 ± 4 | 720 ± 60 | 0.065 ± 0.005 |
| Imipenem | 105 ± 5 | 35 ± 4 | 3.0 ± 0.2 | 60 ± 10 | 70 ± 8 | 0.9 ± 0.1 | 100 ± 30 | 140 ± 60 | 0.7 ± 0.1 |
| Meropenem | 22 ± 1 | 2.9 ± 0.2 | 7.7 ± 0.4 | 60 ± 10 | 8 ± 2 | 7.6 ± 0.6 | 100 ± 10 | 19 ± 4 | 5.6 ± 0.5 |
| Doripenem | 37 ± 8 | 35 ± 9 | 1.1 ± 0.1 | 260 ± 40 | 80 ± 20 | 3.2 ± 0.2 | 160 ± 40 | 70 ± 20 | 2.3 ± 0.1 |
| Aztreonam | ND | ND | ND | ND | ND | ND | ND | ND | ND |
Values are means ± standard deviations. ND, not detectable.
MIC assays were performed 6-fold in a microbroth dilution format following CLSI guidelines (5) using E. coli DH10B cells, and medians are reported. Changes by one or more serial dilutions are considered significant. IMP-1 confers higher resistance levels than IMP-6 toward the type II substrates ceftazidime (2-fold), penicillins (4- to 16-fold), and imipenem (4-fold), while IMP-6 confers higher or equal resistance levels toward the type I substrates cephalothin (2-fold), meropenem and doripenem (4-fold), and cefotaxime and cefoxitin (equal) (Table 2). The increase in MIC of meropenem for IMP-6 was not as dramatic as the 128-fold difference reported by Yano et al. (27), which could be due to the use of different plasmids in their study. This is supported by the fact that they obtained equal MICs, while we obtained an increased MIC of imipenem with IMP-1 versus IMP-6, in agreement with Iyobe et al. (10) and consistent with a 3-fold-higher kcat/Km for IMP-1 (Table 1). The MICs of IMP-25 are generally in the range of the values of IMP-1 and IMP-6, except for cefotaxime and meropenem (higher) and cefoxitin (lower). Interestingly, the MICs of meropenem (16 μg/ml for IMP-1, 64 μg/ml for IMP-6, and 128 μg/ml for IMP-25) do not correlate with the kcat/Km values, as those of imipenem and doripenem do, but rather correlate with kcat (22 ± 1 s−1 for IMP-1, 60 ± 10 s−1 for IMP-6, and 100 ± 10 s−1 for IMP-25). This observation is consistent with the smaller Km values for meropenem (1 order of magnitude smaller) relative to imipenem and doripenem, suggesting that the MBLs may be saturated with meropenem under the MIC assay conditions.
Table 2.
In vitro susceptibility assay of E. coli DH10B cells with pBC SK(+) containing different bla genesa
| β-Lactam | MIC (μg/ml) of β-lactam for E. coli carrying the following plasmid: |
||||
|---|---|---|---|---|---|
| pBC SK(+)-blaIMP-1 | pBC SK(+)-blaIMP-6 | pBC SK(+)-blaIMP-25 | pBC SK(+) | No plasmid | |
| Cephalothin | 512 | 1,024 | 1,024 | 16 | 16 |
| Cefotaxime | 256 | 256 | 512 | 0.125 | 0.25 |
| Ceftazidime | 256 | 128 | 256 | 0.5 | 0.5 |
| Cefoxitin | 1,024 | 1,024 | 256 | 8 | 16 |
| Benzylpenicillin | 256 | 64 | 64 | 32 | 32 |
| Ampicillin | 256 | 16 | 64 | 4 | 8 |
| Imipenem | 4 | 1 | 2 | 0.25 | 0.25 |
| Meropenem | 16 | 64 | 128 | <0.0625 | 0.0625 |
| Doripenem | 16 | 64 | 64 | 0.0625 | 0.125 |
| Aztreonam | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 |
E. coli DH10B cells with pBC SK(+) containing blaIMP-1, blaIMP-6, blaIMP-25, or no bla gene and control cells with no plasmid.
IMP-1, IMP-6, and IMP-25 differ minimally in amino acid sequence: IMP-6 from IMP-1 by an S262G mutation in the active site, and IMP-25 from IMP-6 by an additional G235S mutation on the protein surface (Fig. 1A). Because of the comparable expression levels, biophysical characteristics, and overall similar MICs conferred to E. coli, it is expected that these mutations do not alter the level of functional enzyme in vivo significantly. In the absence of crystal structures of IMP-6 and IMP-25, molecular modeling can provide some insights into the role of mutations, as shown previously for S262G distinguishing IMP-6 from IMP-1 (18). Position 235 is located on the opposite side of the substrate binding site relative to position 262 (Fig. 1A) and has not been studied previously. However, residue 233 in the same loop, a conserved asparagine in all but two (IMP-14 and IMP-35) IMP-type enzymes, has been studied extensively (2). The results indicate that substitutions at position 233 significantly alter the kinetic parameters but that most substituted enzymes still provide high levels of resistance (2). In two crystal structures of IMP-1 with a mercaptocarboxylate inhibitor (6) and a succinic acid inhibitor (22), Asn233 is aligned parallel to the inhibitor. In molecular dynamics (MD) simulations, we did not find the side chain of Asn233 to form a stable oxyanion hole with a cephalothin intermediate (17), and both the side and main chains of Asn233 interacted with different parts of the substrate, including the R2 group (18). When virtually mutating Gly235 in both crystal structures to serine using the “mutate” tool in Swiss-PDB Viewer (http://spdbv.vital-it.ch/), which selects the most favorable rotamer, the serine hydroxyl group is placed within hydrogen bonding distance (≤3.2 Å between heavy atoms) of the Asn233 side chain (Fig. 1A). Asn233 is located between residue 235 and Trp64, which covers the active site. Thus, it is reasonable to assume that Ser235 hydrogen bonds to Asn233 and alters or rigidifies the substrate binding site, causing the altered kinetic constants and MIC values. It is noteworthy that IMP-25 is the only IMP-type enzyme with serine at position 235, but several other enzymes (IMP-11, IMP-12, IMP-16, IMP-21, IMP-22, and IMP-29) contain aspartate at this position (26). Virtual mutation of Gly235 to aspartate also positions the Asp235 side chain within hydrogen bonding distance of the Asn233 side chain. Further studies are needed to elucidate the exact molecular basis of the substrate-specific effects of mutations at position 235.
IMP-25 was isolated from Pseudomonas aeruginosa in a South Korean hospital (GenBank accession no. EU541448), just like IMP-6 (20, 21, 29), suggesting that it might be a derivative of IMP-6, which in turn could be a derivative of IMP-1 (19). When going from IMP-1 over IMP-6 to IMP-25, the resistance levels to meropenem steadily increase, consistent with the idea that they present an evolutionary pathway for the adaptation to meropenem exposure. This implication is based on the assumption that trends in resistance levels conferred to E. coli cells used in this study reflect trends in resistance levels conferred to Pseudomonas aeruginosa. Because of its broad spectrum of antibacterial activity, good tolerability, low toxicity, and cost-effectiveness compared to imipenem/cilastatin, meropenem is widely used as an initial empirical therapy in severe nosocomial infections (1). However, the spread of meropenem and doripenem resistance factors, such as IMP-6 and IMP-25, could severely harm patient outcomes. Infection control and sentinel programs are needed to impede the further spread of such resistance factors and to monitor resistance patterns, so that suitable antibacterial agents can be selected “right the first time” in order to improve survival rates and diminish the evolution of antibacterial resistance (11). The results presented here indicate that positively charged R2 groups in carbapenems (imipenem) pose a bigger challenge to be overcome by evolving IMP enzymes, consistent with previous observations (8, 15), than neutral R2 groups (meropenem and doripenem).
ACKNOWLEDGMENTS
This research was supported by an award from the Research Corporation for Science Advancement (award 10493 to P.O.). The Center for Macromolecular Modeling and Materials Design at California State Polytechnic University, Pomona, where circular dichroism experiments were conducted, is supported by a grant from the W. M. Keck Foundation.
We thank James Spencer (University of Bristol, United Kingdom) for the pET26b expression vector containing the IMP-1 gene with its native leader sequence and Merck, Sharp & Dohme Corp. for the pET30a expression vector containing the IMP-1 gene without leader sequence and imipenem. We thank H. Howard Xu and Jennifer Fleischer (California State University, Los Angeles) for assistance with the MIC assays and Antony Huang (California State Polytechnic University, Pomona) for assistance with the circular dichroism experiments.
Footnotes
Published ahead of print 24 September 2012
REFERENCES
- 1. Baldwin CM, Lyseng-Williamson KA, Keam SJ. 2008. Meropenem: a review of its use in the treatment of serious bacterial infections. Drugs 68:803–838 [DOI] [PubMed] [Google Scholar]
- 2. Brown NG, Horton LB, Huang W, Vongpunsawad S, Palzkill T. 2011. Analysis of the functional contributions of Asn233 in metallo-β-lactamase IMP-1. Antimicrob. Agents Chemother. 55:5696–5702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bush K. 2010. Alarming β-lactamase-mediated resistance in multidrug-resistant Enterobacteriaceae. Curr. Opin. Microbiol. 13:558–564 [DOI] [PubMed] [Google Scholar]
- 4. Bush K, Fisher JF. 2011. Epidemiological expansion, structural studies, and clinical challenges of new β-lactamases from gram-negative bacteria. Annu. Rev. Microbiol. 65:455–478 [DOI] [PubMed] [Google Scholar]
- 5. Clinical and Laboratory Standards Institute 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard—ninth edition. M07-A9 Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 6. Concha NO, et al. 2000. Crystal structure of the IMP-1 metallo-β-lactamase from Pseudomonas aeruginosa and its complex with a mercaptocarboxylate inhibitor: binding determinants of a potent, broad-spectrum inhibitor. Biochemistry 39:4288–4298 [DOI] [PubMed] [Google Scholar]
- 7. Cornaglia G, Giamarellou H, Rossolini GM. 2011. Metallo-β-lactamases: a last frontier for β-lactams? Lancet Infect. Dis. 11:381–393 [DOI] [PubMed] [Google Scholar]
- 8. Hall BG. 2004. In vitro evolution predicts that the IMP-1 metallo-β-lactamase does not have the potential to evolve increased activity against imipenem. Antimicrob. Agents Chemother. 48:1032–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Humphrey W, Dalke A, Schulten K. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 14:33–38 [DOI] [PubMed] [Google Scholar]
- 10. Iyobe S, et al. 2000. Amino acid substitutions in a variant of IMP-1 metallo-β-lactamase. Antimicrob. Agents Chemother. 44:2023–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kollef MH. 2003. Appropriate empirical antibacterial therapy for nosocomial infections: getting it right the first time. Drugs 63:2157–2168 [DOI] [PubMed] [Google Scholar]
- 12. Laraki N, et al. 1999. Biochemical characterization of the Pseudomonas aeruginosa 101/1477 metallo-β-lactamase IMP-1 produced by Escherichia coli. Antimicrob. Agents Chemother. 43:902–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Livermore DM. 2009. Has the era of untreatable infections arrived? J. Antimicrob. Chemother. 64(Suppl 1):i29–i36 [DOI] [PubMed] [Google Scholar]
- 14. Oelschlaeger P, Ai N, Duprez KT, Welsh WJ, Toney JH. 2010. Evolving carbapenemases: can medicinal chemists advance one step ahead of the coming storm? J. Med. Chem. 53:3013–3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Oelschlaeger P, Mayo SL. 2005. Hydroxyl groups in the ββ sandwich of metallo-β-lactamases favor enzyme activity: a computational protein design study. J. Mol. Biol. 350:395–401 [DOI] [PubMed] [Google Scholar]
- 16. Oelschlaeger P, Mayo SL, Pleiss J. 2005. Impact of remote mutations on metallo-β-lactamase substrate specificity: implications for the evolution of antibiotic resistance. Protein Sci. 14:765–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Oelschlaeger P, Schmid RD, Pleiss J. 2003. Insight into the mechanism of the IMP-1 metallo-β-lactamase by molecular dynamics simulations. Protein Eng. 16:341–350 [DOI] [PubMed] [Google Scholar]
- 18. Oelschlaeger P, Schmid RD, Pleiss J. 2003. Modeling domino effects in enzymes: molecular basis of the substrate specificity of the bacterial metallo-β-lactamases IMP-1 and IMP-6. Biochemistry 42:8945–8956 [DOI] [PubMed] [Google Scholar]
- 19. Osano E, et al. 1994. Molecular characterization of an enterobacterial metallo β-lactamase found in a clinical isolate of Serratia marcescens that shows imipenem resistance. Antimicrob. Agents Chemother. 38:71–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ryoo NH, et al. 2009. Outbreak by meropenem-resistant Pseudomonas aeruginosa producing IMP-6 metallo-β-lactamase in a Korean hospital. Diagn. Microbiol. Infect. Dis. 63:115–117 [DOI] [PubMed] [Google Scholar]
- 21. Seok Y, et al. 2011. Dissemination of IMP-6 metallo-β-lactamase-producing Pseudomonas aeruginosa sequence type 235 in Korea. J. Antimicrob. Chemother. 66:2791–2796 [DOI] [PubMed] [Google Scholar]
- 22. Toney JH, et al. 2001. Succinic acids as potent inhibitors of plasmid-borne IMP-1 metallo-β-lactamase. J. Biol. Chem. 276:31913–31918 [DOI] [PubMed] [Google Scholar]
- 23. Tremblay LW, Fan F, Blanchard JS. 2010. Biochemical and structural characterization of Mycobacterium tuberculosis β-lactamase with the carbapenems ertapenem and doripenem. Biochemistry 49:3766–3773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Viaene E, Chanteux H, Servais H, Mingeot-Leclercq MP, Tulkens PM. 2002. Comparative stability studies of antipseudomonal β-lactams for potential administration through portable elastomeric pumps (home therapy for cystic fibrosis patients) and motor-operated syringes (intensive care units). Antimicrob. Agents Chemother. 46:2327–2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Walsh TR, Toleman MA, Poirel L, Nordmann P. 2005. Metallo-β-lactamases: the quiet before the storm? Clin. Microbiol. Rev. 18:306–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Widmann M, Pleiss J, Oelschlaeger P. 2012. Systematic analysis of metallo-β-lactamases using an automated database. Antimicrob. Agents Chemother. 56:3481–3491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yano H, et al. 2001. Plasmid-encoded metallo-β-lactamase (IMP-6) conferring resistance to carbapenems, especially meropenem. Antimicrob. Agents Chemother. 45:1343–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yano H, et al. 2012. High frequency of IMP-6 among clinical isolates of metallo-β-lactamase-producing Escherichia coli in Japan. Antimicrob. Agents Chemother. 56:4554–4555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yoo JS, et al. 2012. Dissemination of genetically related IMP-6-producing multidrug-resistant Pseudomonas aeruginosa ST235 in South Korea. Int. J. Antimicrob. Agents 39:300–304 [DOI] [PubMed] [Google Scholar]


