Abstract
Measures of biodiversity encompass variation along several dimensions such as species richness (SR), phylogenetic diversity (PD) and functional/trait diversity (TD). At the global scale, it is widely recognized that SR and PD are strongly correlated, but the extent to which either tends to capture variation in TD is unclear. Here, we assess relationships among PD, SR and TD for a number of traits both across clades and regional assemblages of mammals. We also contrast results using two different measures of TD, trait variance and a new measure we refer to as trait bin filling (the number of orders of magnitude of variation that contain at least one species). When TD is defined as trait variance, PD is a much stronger correlate of TD than SR across clades, consistent with hypotheses about the conservation value of PD. However, when TD is defined as bin filling, PD and SR show similar correlations with TD across clades and space. We also investigate potential losses of SR, PD and TD if species that are currently threatened were to go extinct, and find that threatened PD is often a similar predictor of threatened TD as SR.
Keywords: biodiversity, species richness, phylogenetic diversity, trait diversity, mammals
1. Introduction
Quantifying biodiversity is a central issue in ecology, biogeography and conservation biology [1–3]. Considered broadly, biodiversity encompasses variation in the composition and characteristics of species assemblages (e.g. clades and communities). The diversity of assemblages can vary along several dimensions such as species richness (SR, taxonomic diversity sensu [4]), genetic diversity and trait diversity (TD) or functional diversity [1,4–6]. Of these, TD has been the least studied even though it is potentially one of the most important aspects of biodiversity from a conservation perspective. For example, communities with high TD and trait redundancy among species might be more resilient to perturbation [6,7]. Conversely, when species with distinctive ecological characteristics are lost from a local community, their extinction can have a disproportionate effect on the species that remain, and alter or reduce ecosystem function [8–11]. Unfortunately, despite the availability of many sophisticated mathematical algorithms that have been developed for calculating TD of species (reviewed in Pavoine & Bonsall [3]), in most real-world cases, it is not feasible to quantify TD for regional assemblages owing to our limited knowledge of species' distributions and traits. Even when a complete species list is available in many cases, the traits of all but a few well-studied species are poorly known. For example, in mammals, perhaps the best-studied class of vertebrates, estimates of adult body mass are available in the literature for only approximately 60 per cent of species [12]. Other characteristics such as litter size and population density have been quantified for even fewer species [12].
Recently, there has been much interest in quantifying the phylogenetic diversity (PD) of species assemblages [13,14], defined as the summed branch lengths of a phylogeny of the species in an assemblage [15]. PD reflects both the number and the evolutionary distinctiveness of species in an assemblage, and thus can potentially act as a ‘silver bullet’ ensconcing several dimensions of biodiversity [13,16–18]. We know from previous studies that PD is strongly correlated with SR in many systems [2,4,19–21], due in large part to the way PD is calculated (i.e. when you add more species to a tree, the summed branch lengths of the tree are bound to increase). When PD is calculated from a tree with branch lengths that reflect sequence divergence, it can be presumed to also capture genetic diversity above the species level [15,22]. Whether PD generally captures variation in TD remains an open question. Several studies have investigated the relationship between PD and trait variance in experimental plant systems [23–25], but with mixed results. A global study of mammals considered the relationships among SR, PD and a composite measure of functional diversity [5], and showed all three variables to be strongly correlated; however, this analysis included only four traits, and the study did not assess variation in the strength of correlations for individual traits.
If PD is, in general, correlated with TD for a wide range of traits, then it could serve as an important proxy for a dimension of biodiversity that is difficult to quantify otherwise. Knowledge of the phylogenetic structure of a community could also provide useful information such as which species might have unique ecological roles based on their phylogenetic distinctiveness. However, SR is easier to quantify than PD, the latter of which requires not only knowledge of which species occur in an assemblage but also their evolutionary relationships. If PD is not a stronger correlate of TD than SR, then the extra effort required to quantify PD (i.e. constructing a phylogeny) would not be warranted when the primary goal is to estimate expected TD. To date, this issue has rarely been investigated empirically. Fritz & Purvis [26] found that if threatened mammal species were to go extinct, the geographical patterns of PD losses across the world's ecoregions [27] would not be consistent with the losses of TD measured as adult body mass variance. However, the spatial scale they used (i.e. entire ecoregions) is arguably coarse for many conservation considerations, and they did not assess the relationship between total TD and total PD, examine traits other than mass, or consider measures of TD other than variance [26].
Here, we investigate the relationship between TD, PD and SR for a variety of traits in terrestrial mammals for two types of assemblage: across phylogenetic clades and across geographical regions. We first investigate the relationship between PD and TD across clades, because if no correlation occurs at the clade level it seems unlikely that such a relationship would hold between the PD and TD of communities (which can be thought of as somewhat random subsamples of full mammalian clades). We quantify TD in both individual-level traits, such as litter size, as well as in population and species-level traits such as population density. We perform analyses to address three questions: (i) Is SR and PD correlated with TD across clades? (ii) Is PD generally a stronger correlate of TD than SR? Finally, (iii) does sample size or variation in phylogenetic signal explain differences in the strength of correlations between TD and PD or SR? Intuitively, the strength of phylogenetic signal in a trait could affect the degree to which PD is a better predictor than SR, because traits with lower signal exhibit variation that is more independent of phylogeny. Additionally, we expect that sample size for an individual trait might affect the observed patterns of TD. Because the amount of data available for mammals varies dramatically for different traits [12], low proportions of mammal species in a clade or a community that a trait has been sampled for could add ‘noise’ to estimates of correlations between TD and PD (or SR). We further conduct global spatial analyses using two traits (body mass and geographical range size) that have particularly large sample sizes to see (iv) whether PD (or SR) can be a good representative of TD across geographical regions; (v) whether the areas of highest PD and TD show geographical correspondence (i.e. are ‘hotspots’ [28,29] of PD and TD in mammals congruent) and (vi) from a conservation perspective, whether the number of threatened species and amount of PD loss in a region predict the amount of TD that stands to be lost.
2. Methods
(a). Defining trait diversity
We here use the term ‘trait diversity’ in preference to ‘functional diversity’ in acknowledgement of the fact that the biological traits we considered may or may not directly reflect the ecosystem function of a species (e.g. mice and shrews have very different ecological roles, though they are both small, have high reproductive rates, and can reach high population densities). Among existing measures of TD (reviewed in [3,30]), trait variance [26,31] and composite measures of functional diversity [4,32,33] are perhaps the most widely used in recent broad-scale studies. A previous global study of mammals [5] used a composite measure of trait variation based on dendrograms constructed using the unweighted pair group method with arithmetic averages (i.e. UPGMA). However, the summed branch lengths of a UPGMA tree will inevitably be correlated with the summed branch lengths of a phylogeny for the same number of species because both are sensitive to SR, even when UPGMA trees are generated using completely random trait data (see the electronic supplementary material, figure S1). Thus, their method is not well suited for testing for a general relationship between TD and PD. Other ways to examine composite TD include principal components and a multidimensional measure of functional richness such as FRV or FRIm (sensu [30]). However, the number of species that we could include using these methods would diminish rapidly as we included more traits because they all require complete case analysis (see the electronic supplementary material, figure S2). Both to maximize sample sizes and to avoid using a measure of TD that is auto-correlated with PD and SR, we focused on the variance of individual traits in this study.
While trait variance is a fairly standard measure of TD, it does not necessarily capture TD in the sense of niche filling. A measure of niche filling that has been used in previous studies is functional range, the range of species trait values in a local community compared with the range of species trait values in a region or globally [34]. While this does capture variation in the breadth of niche space occupied communities, it does not fully reflect ‘niche filling’ in that a community with two species (e.g. a community consisting of a mouse species and an elephant species) could potentially be considered as diverse as a community containing many species with a greater variety of trait values. We therefore propose a new measure of TD that we refer to as trait bin filling, which more closely reflects TD in the classical community ecological sense of the filling of semi-discrete niches (e.g. MacArthur's ‘broken stick’ model of niche partitioning; [35]) than either trait variance or other measures considering functional range [34]. Bin filling refers to the number of orders of magnitude of a given trait occupied by at least one species in a clade or community. For example, body mass spans roughly eight orders of magnitude in mammals, and so an assemblage of mammal species can potentially fill up to eight mass bins. If a community consisted of one large mammal of body mass 1.4 × 106 g and 10 small mammals of between 1.0 × 102 and 9.9 × 102 g, it would have two mass bins filled. In some cases where it was desirable to have larger numbers of bins, such as analyses of traits that span few orders of magnitude of variation in mammals and when producing global maps of bin filling, we used natural log bins instead of log10 bins.
We acknowledge that mammals similar in certain traits can fulfil very different ecological roles in a community (e.g. rodents versus insectivores). However, it at least stands to reason that species that differ by an order of magnitude in a trait such as mass or litter size have important ecological differences. Furthermore, the bin filling definition of TD is potentially easier to measure accurately than variance or many other TD measures. Most species traits exhibit some degree of allometric scaling [36]. In general, medium-to-large-bodied species that occur in communities are known, whereas most unknown species will tend to be small-bodied. Owing to the greater overall number of small-bodied species in most communities and the potential for such species to reach extremely high abundance, at least a few small-bodied species will generally have been recorded for any given community [37]. By definition, as long as one species is reported falling into a given trait bin, the bin is considered filled. Thus, estimates of bin filling are potentially more robust to incomplete species sampling than other TD estimates.
(b). Clade-level analyses
Twelve traits extracted from PanTHERIA [12] and IUCN (see below) were used for clade level analyses. Included traits span a wide variety of sample sizes from 535 to 4642 species (table 1; electronic supplementary material, figure S2) and a wide range of phylogenetic dependence (Blomberg's K [38,39]), from K = 0.079 to 1.855 (table 1). Despite the wide variation in signal observed, all traits showed statistically significant signal (sensu [38]) when compared to randomized data (p < 0.01 in every case). Analyses were conducted for both trait variance and trait bins using a super-tree of all extant mammals [10]. We analysed correlations using Spearman's rank-order test, and compared Spearman's ρ-values to see which of SR and PD was more tightly correlated with TD (see the electronic supplementary material for more details). All the clade analyses were conducted in R v. 2.12.2 [40], phylogenetic analyses were performed using the package APE v. 2.7 [41].
Table 1.
Traits included in clade analyses.
| trait | sample size | Blomberg's K |
|---|---|---|
| geographical range area (km2) | 4762 | 0.079 |
| adult body mass (g) | 3468 | 2.146 |
| litter size (no. of offspring) | 2478 | 0.504 |
| adult head-body length (mm) | 1910 | 1.554 |
| gestation length (day) | 1359 | 3.172 |
| population density (number km−2) | 950 | 0.286 |
| adult forearm lengh (mm) | 892 | 1.671 |
| litters per year (no. of litters) | 889 | 0.358 |
| home range (km2) | 703 | 0.419 |
| teat number (no. of teats) | 628 | 0.454 |
| basal metabolic rate (ml O2 h−1) | 571 | 0.834 |
| mass-specific metabolic rate (ml O2 h−1/g) | 571 | 0.888 |
(c). Geographical analyses
Geographical analyses focused on two traits for which we had particularly good species-level sampling, adult body mass and geographic range area (table 1; electronic supplementary material, figure S3). Both traits are considered important in ecology, evolution, and conservation biology [8,26,37,42]. Geographical distribution data and threat status data of all terrestrial mammals were obtained from the IUCN database (www.iucnredlist.org) and were matched to the taxonomy [43] used in the phylogenetic and trait data. SR, PD and the two measures of TD using two traits, and the hypothetical reduction of them given extinction of threatened species were calculated for assemblages occurring in each 100 × 100 km2 grid cell of an equal-area projection of the world's land surface. We analysed spatial correlations using Spearman's rank (see the electronic supplementary material for more details). All the geographical data and results were processed in ArcGIS and all the spatial analyses were conducted in R [40].
3. Results
(a). Clade-level analyses
Variances of eight of 12 traits showed significant correlations with PD, whereas variances of only three showed significant correlations with SR (see the electronic supplementary material, table S1). The observed correlations (Spearman's ρ) were generally higher between PD and TD compared with those between SR and TD (seven of eight traits that showed significant correlations). However, both PD and SR were similar predictors of bin filling. Five of 12 traits showed significant correlations for both PD and SR, whereas one additional trait showed a correlation with SR alone and one with PD alone. Whether bin filling shows a stronger correlation with PD or SR varied among traits. Blomberg's K did not show a significant correlation with the strength of correlations observed across traits (n = 12; trait variance versus PD: ρ = 0.164, p = 0.609; trait variance versus SR: ρ = 0.217, p = 0.499; trait bins versus PD: ρ = −0.245, p = 0.444; trait bins versus SR: ρ = −0.168, p = 0.604) or the degree to which correlations with PD and SR differed (n = 12; trait variance: ρ = 0.147, p = 0.651; trait bins: ρ = 0.111, p = 0.733). There was no correlation between sample size and the strength of correlations observed for trait variance (n = 12; PD: r2 = 0.021, p = 0.591; SR: r2 = 0.193, p = 0.088) or trait bins (n = 12; PD: r2 = 0.046, p = 0.427; SR: r2 = 0.017, p = 0.589). Simulations showed that trait variance shows no autocorrelation with PD or SR (see the electronic supplementary material, figure S4). However, bin filling did show significant autocorrelation with both PD and SR, with either variable explaining roughly 45 per cent of the variation in the number of ‘bins’ filled with draws from a random lognormal distribution (see the electronic supplementary material, figure S5).
(b). Geographical analyses
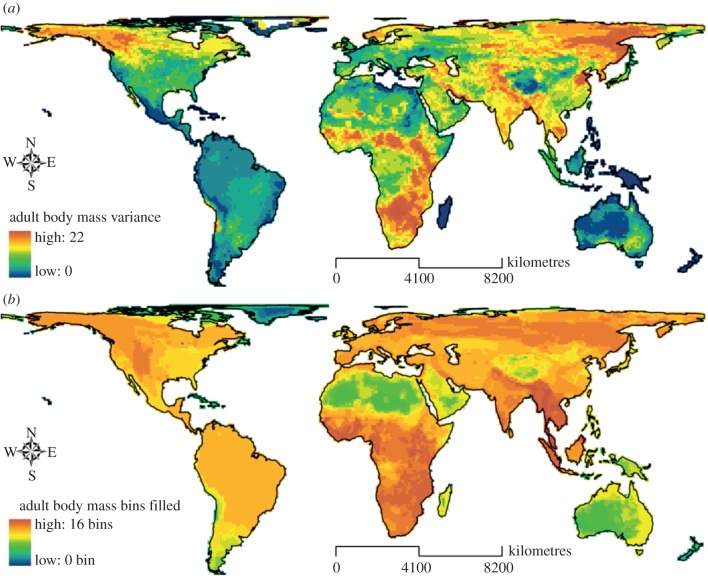
All three dimensions of biodiversity showed complex patterns of variation across the globe (figure 1; electronic supplementary material, figures S6 and S7). When trait variance was used as our measure of TD, extremely weak but (generally) statistically significant global correlations between the TD and the PD or SR of regional assemblages were observed (see the electronic supplementary material, table S2). By contrast, strong correlations were observed at a continental scale (see the electronic supplementary material, table S3). PD loss appeared to be a good predictor of TD loss in terms of trait variance, especially for body mass, at both global and continental scales; the number of threatened species (i.e. threatened SR) was also correlated with TD loss though to a lower degree for body mass (see the electronic supplementary material, table S2 and S3 and for additional discussion of these results). We further found that TD was significantly correlated with PD and SR in cells where residual PD was either very high (top five percentile), such as those in the eastern to southern Africa and India, or very low (bottom five percentile), such as those in western north America and the Amazon area (see the electronic supplementary material, table S4 and figure S6). However, in these areas, PD did not appear to be a much stronger predictor of TD than SR.
Figure 1.
Global variations of mammalian trait diversity in adult body mass measured using (a) variance and (b) bin filling.
When the number of trait bins filled by species in a grid cell was used as our measure of TD, PD and SR showed strong positive correlations with TD (see the electronic supplementary material, table S2 and S3). On the basis of comparison of the correlations, PD had a stronger predictive power than SR for the TD of geographical range area but not adult body mass. SR and PD losses also showed significant correlations with the number of bins that would be ‘emptied out’ if all threatened species were to go extinct, though the correlations observed were much stronger for mass than range area (see the electronic supplementary material, table S2 and S3 and for additional discussion of these results). At a global scale, there was relatively little overlap between the areas of highest diversity for PD and the two measures of TD. Areas of high PD and trait variance were generally outside CI hotspots. However, trait bin ‘hotspots’ were generally well captured by the CI hotspots (see the electronic supplementary material, figure S8).
4. Discussion
Whether or not PD is a ‘silver bullet’ that captures TD in addition to SR depends on the definition of TD used and the scale of analysis. At the clade level, when TD is defined as trait variance, models based on PD are generally a better fit than models based on SR (see the electronic supplementary material, table S1). However, when TD was defined as bin filling, which of the two was a better fit varied from one trait to the next and both showed similar overall performance (i.e. when SR showed a significant correlation PD generally also did and vice versa). In general, PD is at worst a similar fit as SR to models of TD for clades, and it is often a better fit. When both PD and SR are available for mammal clades PD is generally to be preferred as a surrogate for expected TD, though the difference between the two measures is sometimes modest (i.e. Δ ρ < 0.2 in some cases). Surprisingly neither phylogenetic signal nor sample size seemed to explain the strength of correlations observed between PD, SR and TD across traits. Phylogenetic signal also did not explain the degree to which PD was a superior predictor of TD than SR. The strength of observed correlations varied in a highly idiosyncratic way across traits in clade-level analyses, and further study is needed to investigate the factors that cause the diversity of a trait to show strong or weak correlations with SR and PD. In particular, factors such as the model of trait evolution that best describes variation in different traits or the degree to which various traits seem to be under stabilizing selection may be informative.
Geographical analyses showed that neither SR nor PD is correlated with adult body mass variance of regional assemblages. By contrast, both SR and PD were significantly correlated with geographical range variance (see the electronic supplementary material, table S2). In addition to differences in their biological and ecological significance, differences in data availability and the range of variation between range area and mass may also have affected their correlations with SR and PD. Estimates of range area were available for almost all species of mammals, whereas estimates of adult body mass were available for only approximately 60 per cent of all species. In addition, range area spans 11 orders of magnitude in terrestrial mammals, whereas body mass spans only seven. We also found that within continental regions, correlations of PD and trait variance do frequently occur, but the relationship varies among different continents (see the electronic supplementary material, table S3). We suspect that these patterns are shaped at least partly by the evolutionary history of mammal assemblages that occur in each region. When we focused our analyses on areas where PD departs from what would be expected from SR, both PD and SR showed statistically significant correlations with TD (see the electronic supplementary material, table S4). In high-residual PD areas, mainly located in central Africa, PD is positively correlated with mass variance as expected from the clade-level analyses. Within these high-residual PD areas, species diverged from each other in the relatively distant past and so in many cases are more morphologically distinctive from each other than species occurring in other regions of the world [13,44]. Thus, as more species are sampled trait variance is likely to increase. By contrast, in areas such as western north America, Amazon basin and south Asian islands where PD is much lower than expected from SR, there is a negative correlation between PD and TD. This is probably because these areas contain clades that arose from recent speciation events, resulting in high redundancy of traits among species and leading to a reduction in trait variance as more (relatively similar) species are sampled [13,44].
Our analyses of geographical range variance showed very different results from those of body mass variance. Not only were consistent correlations among PD, SR and range area variance observed, but smaller scale analyses also showed that correlations between PD and geographical range variance were negative in areas of both high- and low-residual PD. This is likely related to the fact that areas of high- and low-residual PD are often also areas of high SR [13], and within these regions most species tend to have small geographical ranges (see the electronic supplementary material, figure S5, see also Gaston [45]). Thus, as more species are sampled species with similarly small geographical ranges will tend to be sampled, deflating variance. More generally, because geographical range area in mammals has very low phylogenetic signal (table 1), the relationship between range variance and PD is likely mostly shaped by biogeographic mechanisms not directly considered by our study.
When we used a definition of TD that is more closely related to niche filling (i.e. trait bins) than trait variance, we found a strong relationship among PD, SR and TD in regional assemblages. For body mass, the correlation between trait bins and PD (and SR) in grid cells with high- and low-residual PD is similar to what we found for trait variance. However, for range area, grid cells with high-residual PD actually showed a strong positive correlation between range area bins and PD (see the electronic supplementary material, table S4), and areas of low-residual PD did not show any significant correlations. The latter pattern likely occurs due to the fact that areas with low-residual PD are also areas where there are many species that are the product of recent speciation events [13]. Such species will tend to have (similarly) small geographical ranges, and will tend to be ecologically similar (i.e. exhibit similar body mass).
Our results support bin filling as a useful measure of TD that captures different information about variation in TD among communities than trait variance. As bin filling is closer to the idea of niche filling than simple variance, it conceptually resembles how community ecologists and conservation biologists tend to think of TD more than trait variance. For example, if two of 10 order of magnitude body mass bins have been emptied in a community owing to local extinction, it is safe to conclude that the ecological structure of the community has been profoundly altered; some ecological niches have been entirely vacated. By contrast, noting that the body mass variance in a community has been reduced by 20 per cent owing to local extinction conveys relatively little information on impacts to the overall structure of the community. Mass variance can even increase if species with intermediate traits go extinct from a community. Bin filling also has an analytical advantage over trait variance in that it can be much easier to quantify. In order to accurately quantify the trait variance of an assemblage, a complete list of species and their trait values are needed. In order to accurately assess bin filling, it is only necessary to know of at least one species that falls into each bin interval. An important caveat is that bin filling does show some degree of autocorrelation with SR, and thus with PD (see the electronic supplementary material, figure S5). While the autocorrelation is not nearly as extreme as that shown for functional diversity (sensu [5]; electronic supplementary material, figure S1), in cases where a measure of TD that is completely independent of SR is desired bin filling should not be used. We also note that an aspect of TD not directly quantified by either trait variance or bin filling is the range of species trait values (e.g. functional range sensu [34]). An ideal measure of niche filling would combine information on both the number and range of niches that are filled.
Finally, preliminary analyses showed that there is relatively little overlap between the geographical regions that show the highest levels of TD and PD, and relatively little correspondence between the two measures of TD (see the electronic supplementary material, figure S8). Current identified biodiversity hotspots also encompassed relatively few areas of high PD and trait variance. Somewhat surprisingly, however, they encompassed many areas where TD in terms of bin filling is highest, particularly in Asia. We speculate that such areas represent relatively intact assemblages where species with very small geographical ranges and with large body mass still occur. This represents an exciting area for future research.
In summary, our results support the idea that PD is often a reliable measure of the overall biodiversity of an assemblage, including TD. However, two caveats apply. First, PD will generally only be a useful surrogate measure of spatial patterns of trait variance in areas with higher PD than would be expected from their SR, and the difference in the variation explained by the PD versus SR alone is generally modest (e.g. PD only generally outperforms SR by Δρ = 0.15–0.35 in clade-level analyses). Second, PD and SR show similar correlations with bin filling. In cases where researchers are primarily interested in bin filling or similar measures of TD (e.g. niche filling), the additional effort needed to quantify PD compared with SR (i.e. estimating the phylogenetic relationships of species) may not be warranted. Nevertheless, PD remains the only measure of biodiversity that can simultaneously capture variation in SR, TD and genetic diversity above the species level.
Acknowledgements
We thank Nate Nibbelink, Jonathan Davies, Susanne Fritz and the Altizer laboratory group and Ezenwa lab group at Odum School of Ecology for comments. This work was partially supported by a NSF grant (no. 0642619) and the Odum School of Ecology.
References
- 1.Purvis A., Hector A. 2000. Getting the measure of biodiversity. Nature 405, 212–219 10.1038/35012221 (doi:10.1038/35012221) [DOI] [PubMed] [Google Scholar]
- 2.Brooks T. M., Mittermeier R. A., da Fonseca G. A. B., Gerlach J., Hoffmann M., Lamoreux J. F., Mittermeier C. G., Pilgrim J. D., Rodrigues A. S. L. 2006. Global biodiversity conservation priorities. Science 313, 58–61 10.1126/science.1127609 (doi:10.1126/science.1127609) [DOI] [PubMed] [Google Scholar]
- 3.Pavoine S., Bonsall M. B. 2011. Measuring biodiversity to explain community assembly: a unified approach. Biol. Rev. 86, 792–812 10.1111/j.1469-185X.2010.00171.x (doi:10.1111/j.1469-185X.2010.00171.x) [DOI] [PubMed] [Google Scholar]
- 4.Devictor V., Mouillot D., Meynard C., Jiguet F., Thuiller W., Mouquet N. 2010. Spatial mismatch and congruence between taxonomic, phylogenetic and functional diversity: the need for integrative conservation strategies in a changing world. Ecol. Lett. 13, 1030–1040 10.1111/j.1461-0248.2010.01493.x (doi:10.1111/j.1461-0248.2010.01493.x) [DOI] [PubMed] [Google Scholar]
- 5.Safi K., Cianciaruso M. V., Loyola R. D., Brito D., Armour-Marshall K., Diniz-Filho J. A. F. 2011. Understanding global patterns of mammalian functional and phylogenetic diversity. Phil. Trans. R. Soc. B 366, 2536–2544 10.1098/rstb.2011.0024 (doi:10.1098/rstb.2011.0024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crozier R. H. 1997. Preserving the information content of species:genetic diversity, phylogeny, and conservation worth. Annu. Rev. Ecol. Syst. 28, 243–268 10.1146/annurev.ecolsys.28.1.243 (doi:10.1146/annurev.ecolsys.28.1.243) [DOI] [Google Scholar]
- 7.McNeely J. A., Miller K. R., Reid W. V., Mittermeier R. A., Werner T. B. 1990. Conserving the world's biological diversity. Washington, DC: International Union for Conservation of Nature and Natural Resources [Google Scholar]
- 8.Cardillo M., Mace G. M., Gittleman J. L., Jones K. E., Bielby J., Purvis A. 2008. The predictability of extinction: biological and external correlates of decline in mammals. Proc. R. Soc. B 275, 1441–1448 10.1098/rspb.2008.0179 (doi:10.1098/rspb.2008.0179) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cardillo M., Mace G. M., Jones K. E., Bielby J., Bininda-Emonds O. R. P., Sechrest W., Orme C. D. L., Purvis A. 2005. Multiple causes of high extinction risk in large mammal species. Science 309, 1239–1241 10.1126/science.1116030 (doi:10.1126/science.1116030) [DOI] [PubMed] [Google Scholar]
- 10.Fritz S. A., Bininda-Emonds O. R. P., Purvis A. 2009. Geographical variation in predictors of mammalian extinction risk: big is bad, but only in the tropics. Ecol. Lett. 12, 538–549 10.1111/j.1461-0248.2009.01307.x (doi:10.1111/j.1461-0248.2009.01307.x) [DOI] [PubMed] [Google Scholar]
- 11.Huang S., Davies T. J., Gittleman J. L. 2012. How global extinctions impact regional biodiversity in mammals. Biol. Lett. (doi:10.1098/rsbl.2011.0752) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones K. E., et al. 2009. PanTHERIA: a species-level database of life history, ecology, and geography of extant and recently extinct mammals. Ecology 90, 2648. 10.1890/08-1494.1 (doi:10.1890/08-1494.1) [DOI] [Google Scholar]
- 13.Davies T. J., et al. 2008. Phylogenetic trees and the future of mammalian biodiversity. Proc. Natl Acad. Sci. USA 105(Suppl. 1), 11 556–11 563 10.1073/pnas.0801917105 (doi:10.1073/pnas.0801917105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodrigues A. S. L., et al. 2011. Complete, accurate, mammalian phylogenies aid conservation planning, but not much. Phil. Trans. R. Soc. B 366, 2652–2660 10.1098/rstb.2011.0104 (doi:10.1098/rstb.2011.0104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faith D. P. 1992. Conservation evaluation and phylogenetic diversity. Biol. Conserv. 61, 1–10 10.1016/0006-3207(92)91201-3 (doi:10.1016/0006-3207(92)91201-3) [DOI] [Google Scholar]
- 16.Faith D. P. 2002. Quantifying biodiversity: a phylogenetic perspective. Conserv. Biol. 16, 248–252 10.1046/j.1523-1739.2002.00503.x (doi:10.1046/j.1523-1739.2002.00503.x) [DOI] [PubMed] [Google Scholar]
- 17.Mace G. M., Purvis A. 2008. Evolutionary biology and practical conservation: bridging a widening gap. Mol. Ecol. 17, 9–19 10.1111/j.1365-294X.2007.03455.x (doi:10.1111/j.1365-294X.2007.03455.x) [DOI] [PubMed] [Google Scholar]
- 18.Soutullo A., Dodsworth S., Heard S. B., Mooers A. 2005. Distribution and correlates of carnivore phylogenetic diversity across the Americas. Anim. Conserv. 8, 249–258 10.1017/S136794300500226X (doi:10.1017/S136794300500226X) [DOI] [Google Scholar]
- 19.Schipper J., et al. 2008. The status of the world's land and marine mammals: diversity, threat, and knowledge. Science 322, 225–230 10.1126/science.1165115 (doi:10.1126/science.1165115) [DOI] [PubMed] [Google Scholar]
- 20.Morlon H., Schwilk D. W., Bryant J. A., Marquet P. A., Rebelo A. G., Tauss C., Bohannan B. J. M., Green J. L. 2011. Spatial patterns of phylogenetic diversity. Ecol. Lett. 14, 141–149 10.1111/j.1461-0248.2010.01563.x (doi:10.1111/j.1461-0248.2010.01563.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Polasky S., Csuti B., Vossler C. A., Meyers S. M. 2001. A comparison of taxonomic distinctness versus richness as criteria for setting conservation priorities for North American birds. Biol. Conserv. 97, 99–105 10.1016/S0006-3207(00)00103-8 (doi:10.1016/S0006-3207(00)00103-8) [DOI] [Google Scholar]
- 22.Bininda-Emonds O. R. P., et al. 2007. The delayed rise of present-day mammals. Nature 446, 507–512 10.1038/nature05634 (doi:10.1038/nature05634) [DOI] [PubMed] [Google Scholar]
- 23.Flynn D. F. B., Mirotchnick N., Jain M., Palmer M. I., Naeem S. 2011. Functional and phylogenetic diversity as predictors of biodiversity, ecosystem–function relationships. Ecology 92, 1573–1581 10.1890/10-1245.1 (doi:10.1890/10-1245.1) [DOI] [PubMed] [Google Scholar]
- 24.Cadotte M. W., Cardinale B. J., Oakley T. H. 2008. Evolutionary history and the effect of biodiversity on plant productivity. Proc. Natl Acad. Sci. USA 105, 17 012–17 017 10.1073/pnas.0805962105 (doi:10.1073/pnas.0805962105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maherali H., Klironomos J. N. 2007. Influence of phylogeny on fungal community assembly and ecosystem functioning. Science 316, 1746–1748 10.1126/science.1143082 (doi:10.1126/science.1143082) [DOI] [PubMed] [Google Scholar]
- 26.Fritz S. A., Purvis A. 2010. Phylogenetic diversity does not capture body size variation at risk in the world's mammals. Proc. R. Soc. B 277, 2435–2441 10.1098/rspb.2010.0030 (doi:10.1098/rspb.2010.0030) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olson D. M., et al. 2001. Terrestrial ecoregions of the world: a new map of life on earth. BioScience 51, 933–938 10.1641/0006-3568(2001)051[0933:TEOTWA]2.0.CO;2 (doi:10.1641/0006-3568(2001)051[0933:TEOTWA]2.0.CO;2) [DOI] [Google Scholar]
- 28.Myers N., Mittermeier R. A., Mittermeier C. G., da Fonseca G. A. B., Kent J. 2000. Biodiversity hotspots for conservation priorities. Nature 403, 853–858 10.1038/35002501 (doi:10.1038/35002501) [DOI] [PubMed] [Google Scholar]
- 29.Sechrest W., Brooks T. M., da Fonseca G. A. B., Konstant W. R., Mittermeier R. A., Purvis A., Rylands A. B., Gittleman J. L. 2002. Hotspots and the conservation of evolutionary history. Proc. Natl Acad. Sci. USA 99, 2067. 10.1073/pnas.251680798 (doi:10.1073/pnas.251680798) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schleuter D., Daufresne M., Massol F., Argillier C. 2010. A user's guide to functional diversity indices. Ecol. Monogr. 80, 469–484 10.1890/08-2225.1 (doi:10.1890/08-2225.1) [DOI] [Google Scholar]
- 31.Mason N. W. H., MacGillivray K., Steel J. B., Wilson J. B. 2003. An index of functional diversity. J. Vegetation Sci. 14, 571–578 10.1111/j.1654-1103.2003.tb02184.x (doi:10.1111/j.1654-1103.2003.tb02184.x) [DOI] [Google Scholar]
- 32.Cornwell W. K., Schwilk D. W., Ackerly D. D. 2006. A trait-based test for habitat filtering: convex hull volume. Ecology 87, 1465–1471 10.1890/0012-9658(2006)87[1465:ATTFHF]2.0.CO;2 (doi:10.1890/0012-9658(2006)87[1465:ATTFHF]2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- 33.Mouchet M., Guilhaumon F., Villéger S., Mason N. W. H., Tomasini J. A., Mouillot D. 2008. Towards a consensus for calculating dendrogram-based functional diversity indices. Oikos 117, 794–800 10.1111/j.0030-1299.2008.16594.x (doi:10.1111/j.0030-1299.2008.16594.x) [DOI] [Google Scholar]
- 34.Mason N. W. H., Mouillot D., Lee W. G., Wilson J. B. 2005. Functional richness, functional evenness and functional divergence: the primary components of functional diversity. Oikos 111, 112–118 10.1111/j.0030-1299.2005.13886.x (doi:10.1111/j.0030-1299.2005.13886.x) [DOI] [Google Scholar]
- 35.MacArthur R. H. 1957. On the relative abundance of bird species. Proc. Natl Acad. Sci. USA 43, 293. 10.1073/pnas.43.3.293 (doi:10.1073/pnas.43.3.293) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.West G. B., Brown J. H., Enquist B. J. 1997. A general model for the origin of allometric scaling laws in biology. Science 276, 122–126 10.1126/science.276.5309.122 (doi:10.1126/science.276.5309.122) [DOI] [PubMed] [Google Scholar]
- 37.Brown J. 1995. Macroecology. Chicago, IL: University of Chicago Press [Google Scholar]
- 38.Blomberg S., Garland T., Jr 2002. Tempo and mode in evolution: phylogenetic inertia, adaptation and comparative methods. J. Evol. Biol. 15, 899. 10.1046/j.1420-9101.2002.00472.x (doi:10.1046/j.1420-9101.2002.00472.x) [DOI] [Google Scholar]
- 39.Blomberg S. P., Garland T., Jr, Ives A. R. 2003. Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution 57, 717–745 10.1111/j.0014-3820.2003.tboo285.x (doi:10.1111/j.0014-3820.2003.tboo285.x) [DOI] [PubMed] [Google Scholar]
- 40.R Development Core Team 2011. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing [Google Scholar]
- 41.Paradis E., Claude J., Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290 10.1093/bioinformatics/btg412 (doi:10.1093/bioinformatics/btg412) [DOI] [PubMed] [Google Scholar]
- 42.Davies T. J., Buckley L. B., Grenyer R., Gittleman J. L. 2011. The influence of past and present climate on the biogeography of modern mammal diversity. Phil. Trans. R. Soc. B 366, 2526–2535 10.1098/rstb.2011.0018 (doi:10.1098/rstb.2011.0018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilson D., Reeder D. 2005. Mammal species of the world: a taxonomic and geographic reference. Baltimore, MD: Johns Hopkins University Press [Google Scholar]
- 44.Forest F., et al. 2007. Preserving the evolutionary potential of floras in biodiversity hotspots. Nature 445, 757–760 10.1038/nature05587 (doi:10.1038/nature05587) [DOI] [PubMed] [Google Scholar]
- 45.Gaston K. J. 1996. Species-range-size distributions: patterns, mechanisms and implications. Trends Ecol. Evol. 11, 197–201 10.1016/0169-5347(96)10027-6 (doi:10.1016/0169-5347(96)10027-6) [DOI] [PubMed] [Google Scholar]