Abstract
Aggressive behaviour associated with territorial defence is widespread and has fitness consequences. However, excess aggression can interfere with other important biological functions such as immunity and energy homeostasis. How the expression of complex behaviours such as aggression is regulated in the brain has long intrigued ethologists, but has only recently become amenable for molecular dissection in non-model organisms. We investigated the transcriptomic response to territorial intrusion in four brain regions in breeding male threespined sticklebacks using expression microarrays and quantitative polymerase chain reaction (qPCR). Each region of the brain had a distinct genomic response to a territorial challenge. We identified a set of genes that were upregulated in the diencephalon and downregulated in the cerebellum and the brain stem. Cis-regulatory network analysis suggested transcription factors that regulated or co-regulated genes that were consistently regulated in all brain regions and others that regulated gene expression in opposing directions across brain regions. Our results support the hypothesis that territorial animals respond to social challenges via transcriptional regulation of genes in different brain regions. Finally, we found a remarkably close association between gene expression and aggressive behaviour at the individual level. This study sheds light on the molecular mechanisms in the brain that underlie the response to social challenges.
Keywords: microarray, gene expression regulation, Gasterosteus aculeatus, stickleback, sociogenomics, aggression
1. Introduction
Many animals restrict all or part of their activities to a territory that they defend vigorously for resources important to their survival and reproductive success such as food, mates, shelter or offspring. Consequently, territorial defence involves frequent aggressive confrontations with intruders, including predators and both conspecific and heterospecific competitors. However, the overt expression of aggression can be costly [1,2]. For example, highly aggressive animals often experience higher levels of stress that can impair important biological functions, such as immunity and energy homeostasis [1]. Moreover, aggression can be maladaptive when it is misdirected towards offspring, mates or potentially deadly predators [3]. Thus, aggression should be carefully regulated [4]. Understanding how complex behaviours such as aggression are modulated at the molecular level necessitates careful dissection of the brain's response to social interactions.
However, a challenge for studies of the molecular bases of complex behaviours such as aggression associated with territorial defence is that hundreds of genes influence aggression [5–11], and aggression involves many brain regions, each of which is specialized for different processes [12,13]. Therefore, a promising strategy for tackling the molecular bases of complex behaviours is to measure the transcriptional response of thousands of genes simultaneously in several brain areas rather than focusing on single genes or single brain areas in isolation.
Recent studies suggest that the brain responds to social stimuli via the modulation of transcription regulatory networks [14,15], a complex biological process that involves interactions between proteins (transcription factors, TFs) and DNA (cis-regulatory sequences in gene promoters and enhancers) to govern the rate at which genes in the network are transcribed into mRNA [16]. However, we are only beginning to understand the interactions between elements in a biological system (systems biology) of social behaviour, and we know little about how transcription regulatory networks operate within and across brain regions [17]. A study on brain gene expression in aggressive dogs revealed that genes that were upregulated in one brain region were downregulated in other brain regions [18], pointing to differences in the transcriptional regulatory activities invoked in different brain regions. An interesting question raised by such an observation is: are the opposing directions of regulation in different brain regions the result of completely different regulatory effects, or could the same TF be partnering with different region-specific TFs to give opposing results? The latter mode of regulation, where the region-specificity arises from combinatorial regulation by multiple TFs, was suggested to play an important role in the transcriptional response of honeybee brains during behavioural maturation [15].
The threespined stickleback, Gasterosteus aculeatus, is a teleost fish that, for many years, has been a model system in animal behaviour [19]. Despite the solid role that sticklebacks have played in studies of animal behaviour, we are only beginning to understand the molecular mechanisms underlying their rich behavioural repertoire [20–23]. The reproductive behaviour of this fish is well characterized both in the laboratory and in the field [24]. During the breeding season, male sticklebacks defend nesting territories, and they are especially aggressive toward other male sticklebacks that intrude into their territory. Besides these highly energetically demanding defensive activities, territorial male sticklebacks also actively court females and provide all of the parental care to the developing offspring [24]. Therefore, breeding male sticklebacks engage in a variety of activities, all of which are important to reproductive success. The sequencing of the stickleback genome and access to sophisticated bioinformatic tools now allow us to exploit the well-characterized territorial behaviour of this fish to examine how complex behaviours are regulated within and across brain regions.
Here, we used microarray gene expression profiling in four brain regions to understand how the expression of genes in several brain regions is regulated in response to a social challenge in wild-caught male sticklebacks. We focused on four macroscopically dissected regions of the brain (homologies to nodes in the vertebrate social behavioural network [25,26] are in parentheses): the telencephalon (medial amygdala, bed nucleus of the stria terminalis), the diencephalon (ventromedial and anterior hypothalamus, preoptic area), the cerebellum (including the anterior midbrain) and the brain stem (midbrain). The rich literature on the neuroendocrine regulation of aggressive behaviour generates predictions about some of the key biological processes and brain areas that are likely to be involved in the neural regulation of aggression in sticklebacks. For example, processes involving brain monoamines, neuropeptides and, in particular, sex steroids (from whole genome surveys, see [9,10,27]) have all been implicated for the response to social challenges in vertebrates (reviewed in [28]). Recent studies suggest that gene products known to modulate social behaviour such as androgen receptors are expressed in fish brain regions that correspond to the nodes of the vertebrate social behaviour network [29,30]. Therefore, we predicted that territorial intrusion would induce the transcription of genes involved in the regulation of sex steroids in the telencephalon and diencephalon, the two brain regions that we considered which include many homologous nodes in the social behaviour network.
In addition, we used a cis-regulatory network analysis to test the hypothesis that shifts in neurogenomic states [17] in response to territorial intrusion were modulated by specific cis-regulatory elements. Despite the proven utility of cis-regulatory analysis to understand gene regulation during development [31], this is one of the first studies applying cis-regulatory analysis to understand an equally complex phenomenon: social behaviour [6], and the first to do so in a vertebrate. It lays the stepping-stones to the ultimate characterisation of the neurogenomic states underlying complex decision-making in response to social challenges.
2. Material and methods
Males were collected from a freshwater population and maintained in the laboratory on a 16 L : 8 D photoperiod and at 18°C. Males were provided with nesting material; only males with completed nests were used in the experiment (n = 15). We randomly divided the males into three groups with n = 5 males per group. One group consisted of territorial males that were confronted by an intruder (experimental group), the second group comprised territorial males that were not confronted by an intruder (control group) and the third group comprised males that served as intruders. Pairs of fish in the control and experimental groups were matched for size and euthanized at the same time; intruders were always smaller than the experimental individual. Control, experimental and intruder males were in the same stage of the nesting cycle [24]; they vigorously defended their territories and would court females. Therefore, whatever differences that were observed between treatments reflects the response of experimental males to an intrusion, as opposed to more general differences in reproductive maturity or nesting stage.
At the start of the experiment, a single intruder was introduced into an experimental male's tank. The intruder was removed after 15 mins. The interaction between the experimental male and the intruder was video-recorded and the videos were later scored for the following behaviours by the experimental male that were directed to the intruder: latency to orient, number of orients, time spent orienting, number of bites, number of chases and time spent chasing.
Thirty minutes after the intruder was introduced, the experimental male and their matched (paired) control male were netted and quickly euthanized by decapitation within seconds following an Institutional Animal Care and Use Committee approved protocol (#06178) of the University of Illinois at Urbana-Champaign. We elected to sample at 30 min because that is when the stress response to intrusion in sticklebacks is high [21], and when differences in gene expression, including immediate early genes (IEGs) [32], and other socially responsive genes are likely to be detected [33,34]. We dissected four brain regions on dry ice: telencephalon, diencephalon, cerebellum and brain stem. The telencephalon was dissected out first by cutting along the natural commissure between the telencephalon and diencephalon. The diencephalon was dissected by cutting the two lobes away from the cerebellum and removing the entire structure from the skull. The cerebellum was removed by cutting off the structure from the brain stem. The anterior midbrain, which is usually considered part of the brain stem, was co-dissected with the cerebellum, potentially leading to similarities between the cerebellum and the brain stem in our results. Finally, the brainstem was designated as everything that remained before the spinal cord began.
The brain regions were placed individually in Eppendorf tubes containing 500 µl of TRIzol Reagent (Invitrogen, Carlsbad, CA). Total RNA was isolated as described in [22].
For microarray labelling and hybridization, up to 1 µg of total RNA from each sample was labelled using the Agilent Two-Colour Microarray-Based Gene Expression Analysis (Quick Amp Labelling) following the manufacturer's instructions (see the electronic supplementary material). We used a ‘balanced’ design in the microarray experiment and controlled for dye effects by performing dye swaps on biological replicates (individuals). For hybridization, 900 ng cRNA of samples with different dyes were mixed, fragmented and hybridized onto an Agilent 4 × 44 K oligonucleotide microarray following the manufacturer's recommendations. Hybridization was performed within brain regions and between control and experimental replicates. A total of 20 microarrays were processed; the experimental design is in electronic supplementary material, table S1.
Microarray validation by qPCR was conducted on the Applied Biosystems 7900HT (Applied Biosystems, Foster City, CA) using the FastStart Universal SYBR Green Master (ROX) (Roche Diagnostics GmbH, Mennheim, Germany) following the manufacturer's recommendations. Because different genes were differentially expressed (DE) between control and experimental males in the four brain regions, we conducted validation on each brain region separately by selecting the top 5–8 DE genes in each brain region based upon their p-values and fold change. qPCR was conducted as in [22]. A list of primers used in qPCR and their amplification efficiencies are in electronic supplementary material, table S2. Validation was on the same samples as in the microarray.
To identify the DE genes in the four brain regions in control relative to experimental fish, we used the Rank Product implemented in R Bioconductor, an analysis especially suited for experiments with small sample sizes and samples with data that have high variation [35]. Rank product is a non-parametric statistical method based on the mean rank of fold change of each gene under the null hypothesis of no differential expression. A total of 1000 permutations were conducted. Transcripts with false discovery rate (FDR) ≤ 0.05 were considered DE.
The annotation of the DE genes was performed using the function BiomaRt [36], as implemented in R Bioconductor. We conducted gene ontology (GO) enrichment analysis in R Bioconductor as described in [22].
(a). Detection of cis-regulatory elements
We detected cis-regulatory elements by searching for common TF-binding sites (‘motifs’) upstream of a set of DE genes. If the same motif is consistently present upstream of a set of genes, it suggests that the TF that binds to that motif regulates the set of genes. In that case, we refer to the motif as ‘enriched in’ or ‘associated with’ the gene set.
Genomic annotation files were obtained from UCSC Genome Browser (Stickleback, gasAcu1) and used to define the promoter region for each gene as the 5000 bp upstream from its transcription start site. There were 17 121 annotated genes in the universe of genes for our analysis. For each TF binding motif, the Stubb algorithm [37] was used to score every 500-bp genomic window with 250-bp shifts. A ‘motif target gene set’ for a given motif was defined as the genes that contain within their promoter regions a window that scores in the top 0.1 per cent of the motif's genomic distribution of scores. We considered all the DE gene sets in each of the four regions that passed the p-value threshold of 0.01 without FDR, considering upregulated and downregulated gene sets separately (in total eight gene sets). The threshold for the analysis of cis-regulatory elements is less strict than the one used for other analyses in this paper (FDR < 0.05) because larger gene sets allow for the identification of more robust, reliable associations between motifs and DE genes. Associations were tested between each of the 661 motif target gene sets from the JASPAR and TRANSFAC databases (more information in the electronic supplementary material) and each of the eight DE gene sets with a one-sided Fisher's exact test and the association p-values were recorded.
To discover significant, higher level ‘meta-associations’ between one or more motif target sets with more than one DE gene set, we performed an analysis with cis-Metalysis [15]. Cis-Metalysis systematically combines association p-values into a single statistic and then analytically calculates the significance (‘meta p-value’) of the combined statistics. To assess the significance of a given ‘meta p-value’ while accounting for multiple hypothesis testing, an estimated FDR statistic, ‘eFDR’, was calculated as the proportion of the number of results at that significance level from shuffled data compared with real data (more details in the electronic supplementary material). We examined the data with two different settings of the cis-Metalysis program. First, to determine single motifs and motif pairs that were enriched in the promoter regions of DE genes across all regions of the brain, we ran the ‘flexible’ mode of cis-Metalysis where the DE genes could be either the upregulated or the downregulated genes in each of the brain regions. That is, a reported meta-association could involve overrepresentation of the motif in the upregulated genes in one region and downregulated genes in another region. Second, we investigated gene regulation patterns in the diencephalon by using the ‘pattern’ mode of Metalysis, explained next. One of the most intriguing results of the analysis of DE genes was that there were many genes that were upregulated in diencephalon and downregulated in other regions (see §3). To identify possible gene regulatory causes of this pattern, we restricted the program to only search the space of meta-associations (i) that contained a significant association in the diencephalon region and (ii) where the associations in the other regions were in the opposite direction as in the diencephalon.
(b). Correlations between behaviour and gene expression
The behaviour of each individual experimental male (n = 5) was recorded during the territorial intrusion. Because cRNA of individual brains were hybridized to the arrays, we were able to extract individual-specific values of expression of each gene for the five experimental fish, and to correlate individual levels of gene expression with individual levels of behaviour by the experimental males using Spearman correlations. Individual levels of gene expression in the brain were estimated using the normalized, log-transformed expression values of the DE genes.
To assess whether the observed correlations between behaviour and gene expression of the top 20 DE genes in each region were tighter than expected by chance, we compared the mean correlation coefficients between the top 20 DE genes in each region and behaviour with the mean correlation coefficients of 100 000 random samples of 20 genes from throughout the genome. For each of the five behaviours that were recorded for the experimental males, we compared the observed absolute average value of the correlation coefficients between behaviour and gene expression of the top 20 genes with the 95% CIs surrounding the expected mean correlations between behaviour and gene expression from the random samples and their associated p-values.
3. Results
Upon introduction to the resident male's tank, all of the intruders behaved submissively; they froze, hid and attempted to swim away from the experimental males. By contrast, the experimental males made frequent trips to the nest while constantly driving away the intruder and patrolling the territory. For example, on average (±s.e.), the resident males oriented to the intruder within 46 s (±12.7), and then chased (18.8 ± 4.6 s) and bit at the intruder (10.2 ± 7.2 times) (see the electronic supplementary material, video S1).
Analysis of the microarray gene expression data revealed hundreds of genes that were DE (FDR = 0.05) between males that were confronted by an intruder compared with those that were not. The greatest number of DE genes was in the diencephalon (n = 266) and cerebellum (n = 225). In the diencephalon, there were more upregulated (n = 168) than downregulated (n = 98) transcripts, whereas we observed the opposite pattern in the cerebellum (106 up- and 119 downregulated) and the brain stem (24 up- and 49 downregulated). ‘Upregulation’ indicates higher expression in experimental males compared with controls. Contrary to our prediction, relatively few genes were DE in the telencephalon (27 up- and 23 downregulated). On the basis of fold change, the glycoprotein hormone, alpha polypeptide (CGA) was both the most highly upregulated transcript (FC = 40.3, diencephalon), and the most highly downregulated transcript (FC = −9.9, cerebellum).
Comparing the list of genes DE between control and experimental males revealed both similarities and differences between brain regions (see the electronic supplementary material, figure S1 and table S3). For example, four transcripts were DE between the experimental and the control fish in all four brain regions. There were a large (n = 65) number of genes that were DE in both the cerebellum and diencephalon (28% and 24% of the total DE genes, respectively; electronic supplementary material, figure S1). However, the heat map (figure 1) shows that many of these genes were regulated in opposite directions in the diencephalon and the cerebellum, a finding validated with qPCR (see the electronic supplementary material, figure S2). Expression of the genes selected for validation by qPCR was consistent with gene expression observed by microarray in the diencephalon (100%, n = 8). Validation was also successful in cerebellum and brain stem; however, one gene in cerebellum (n = 6) and two genes in brain stem (n = 6) that showed a similar trend did not pass the p = 0.05 cutoff, and one gene in the telencephalon showed the opposite trend. Further details on microarray validation by qPCR are in electronic supplementary material.
Figure 1.
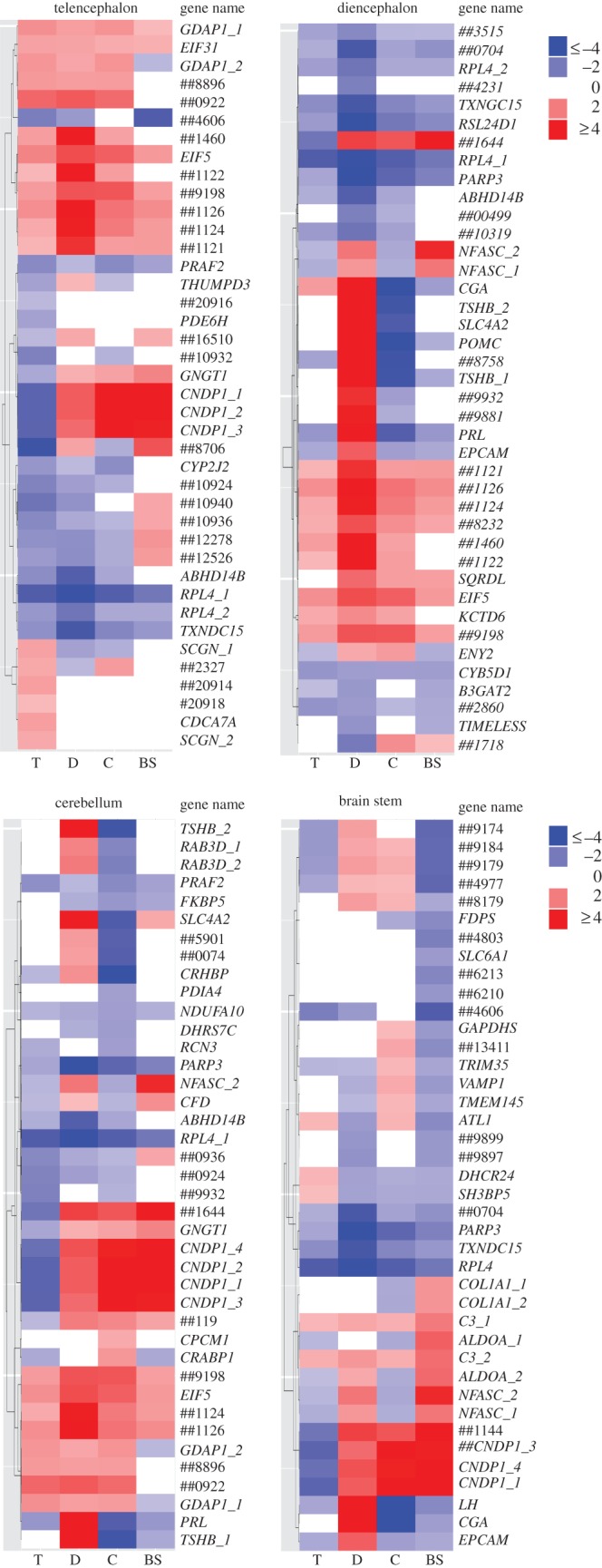
Heat maps of the top 40 genes differentially expressed (FDR ≤ 0.05) between males that experienced a territorial intrusion (experimental) compared with males that did not (control). Shown are separate heat maps for the top DE genes in each region. The columns within each heat map show the extent of differential expression in the four regions. The heat maps were generated using the package Neatmap [38]. Gene names that begin with ‘##’ include the last non-zero digits of the Ensembl Gene IDs of novel unannotated genes. Transcripts with the same names are putative transcript variants. Red in the heat map signifies upregulation, blue signifies downregulation, where ‘up’ refers to higher expression in experimental males. White indicates that the genes were not DE in the specific brain region at raw p = 0.01 cut-off. T, telencephalon; D, diencephalon; C, cerebellum; BS, brain stem. The colour scale in the legend corresponds to the intensity of fold change.
(a). Identification of brain-region-specific enriched gene ontology processes
In order to gain insight into the various processes involved in the brain's response to territorial intrusion, we tested for overrepresentation of biological, molecular and cellular processes in the DE genes from different brain regions. GO analysis confirmed that different biological processes were enriched in each brain region in response to a territorial intrusion (see the electronic supplementary material, figure S3), and that territorial intrusion elicited diverse molecular and cellular processes from immune defence to hormonal, peptide and lipid metabolism and homeostasis (see the electronic supplementary material, figures S4 and S5). Biological processes overrepresented in the set of genes shared between the diencephalon and the cerebellum, the two brain regions with the most DE genes, included peptide hormone processing, maternal aggressive behaviour, female mating behaviour, social behaviour, female pregnancy, male mating behaviour and adult feeding behaviour (see the electronic supplementary material, figure S6).
(b). Gene functional annotation
The DE genes in the diencephalon point to the importance of GnRH-controlled pituitary hormones and the pro-opiomelanocortin (POMC) neuronal system in responding to a territorial intrusion. The most upregulated transcripts were the genes encoding the glycoprotein hormone CGA and its associated hormones, luteinizing hormone (LH) precursor and thyroid stimulating hormone, beta subunit (TSHB). Another study on song sparrows also implicated CGA and LH with territorial behaviour [10], and POMC has also been associated with territorial dominance in fish [27]. Consistent with previous findings [27,39], the gene encoding prolactin (PRL), a pituitary hormone that is often associated with parental care [40], was upregulated.
Our study also revealed a large number of genes that have not heretofore been implicated with aggression or territorial defence. In particular, we identified several TFs that were DE between control and experimental males in the diencephalon, including ENY2, SIX2, SCRIB, EIF5, NR5A1, SF1 and ATF4 (see the electronic supplementary material, table S5). Particular TFs were also DE in the cerebellum (STAT3, EIF5, EIF3I) and the telencephalon (EIF5, CDCA7L). These findings are consistent with the hypothesis that TFs within transcription regulatory networks mediate shifts in neurogenomic state.
The vast majority of the downregulated transcripts in the diencephalon were genes associated with metabolic processes (e.g. the ribosomal protein RPL24D1 and RPL4), genome integrity and chromatin remodelling (e.g. the poly (ADP-ribose) polymerase family, member 3 (PARP3)) immune response (e.g. the protein tyrosine phosphatase (PTPN7)) and feeding (e.g. the cocaine and amphetamine regulated transcript prepropeptide (CARTPT)); electronic supplementary material, table S4. These results suggest some of the molecular mechanisms that might underlie the costs of aggression [41] for survival and self-maintenance, including immunity and feeding.
Some of the most upregulated genes in the cerebellum encoded for pituitary hormones (CGA, TSHB, POMC) and corticotropin releasing hormone-binding protein (CRHBP). There is growing evidence that these genes are expressed outside the pituitary [42], suggesting that POMC neuronal projections might modulate gene expression in response to territorial intrusion.
The telencephalon is often a key area involved in aggression [43]. Therefore, it was surprising that we observed the fewest DE genes in response to territorial intrusion in the telencephalon (figure 1). However, the genes and biological processes identified in this region are nonetheless relevant to territorial defence (see the electronic supplementary material, figure S3). A complete list of the top DE genes in each region is provided in electronic supplementary material, tables S4–S7.
(b). Transcriptional regulation in response to territorial intrusion
We identified TF motifs associated with up- and downregulated genes within each brain region using cis-Metalysis [15] (figures 2 and 3; electronic supplementary material, figure S7). In the following discussion, we refer to a motif by the name of the TF whose experimentally determined binding specificity is represented by the motif. However, we note that our statistical findings pertain to the motif itself and not necessarily to the corresponding TF; multiple TFs may have nearly identical motifs, which implies that an association involving a TF's motif may in fact reflect a regulatory role for a different TF that has very similar binding specificity.
Figure 2.
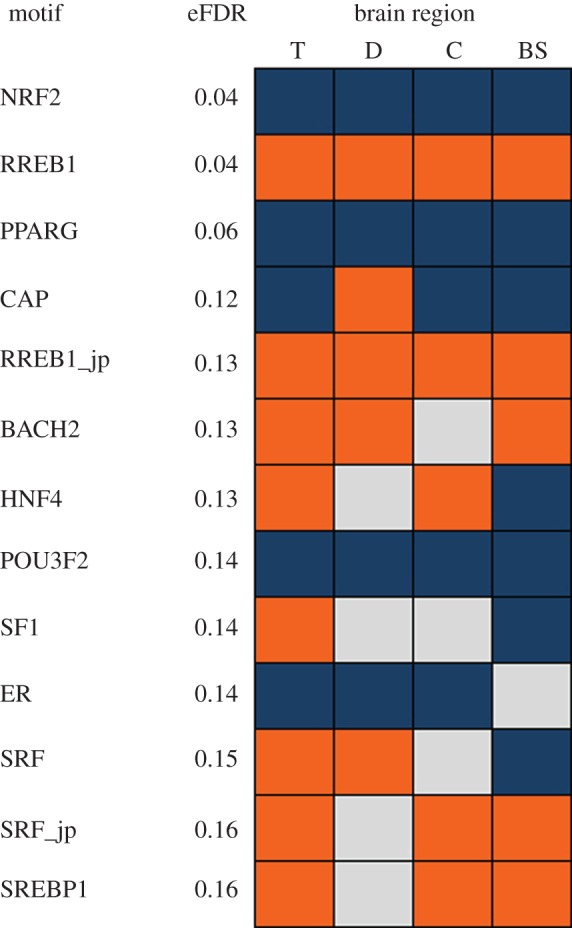
Transcription factor motifs enriched in genes that were differentially expressed in different brain regions of stickleback following a territorial intrusion. T, telencephalon; D, diencephalon; C, cerebellum; BS, brain stem. Orange indicates TF motifs that were associated with upregulated genes, blue indicates TF motifs that were associated with downregulated genes and grey indicates TF motifs whose association with gene expression was not significant (not included in cis-Metalysis). eFDR (estimated FDR) represents the probability of finding a meta-association as strong as the result in randomly generated data. All TF motifs are from the Transfac database except those indicated by ‘–jp’, which are from Jaspar database.
Figure 3.
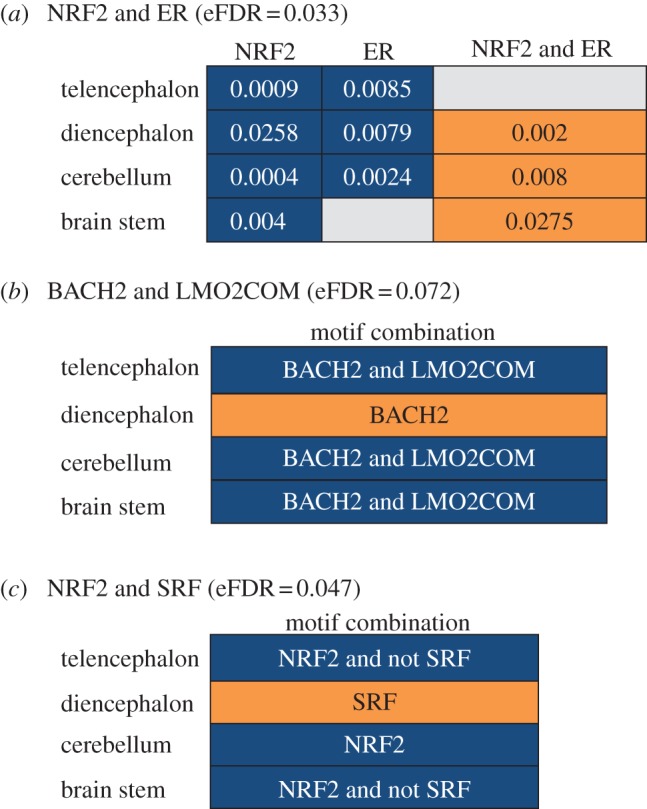
Motif pairs found in significant meta-associations with flexible, patterned cis-Metalysis. Orange cells indicate that the strongest association in each brain region was with the upregulated genes, and blue cells indicate that the strongest association was with the downregulated genes. Grey cells represent associations that were not significant and were not included in the computation by cis-Metalysis. eFDR (estimated FDR) are provided for each meta-association. (a) P-values of association between only NRF2, only ER, and NRF2 and ER motif target sets and the DE genes in the four brain regions. (b,c) Meta-association involving the motif pairs BACH2/LMO2COM and NRF2/SRF with the cell values being the motif combination of the strongest association in the brain region. BACH2 was most strongly associated with upregulated genes in diencephalon. However, in combination with LMO2COM, BACH2 was most strongly associated with downregulated genes in other brain regions.
In agreement with the gene expression analysis, some TF motifs were enriched in upregulated gene sets in all four regions (e.g. the RAS responsive element binding protein 1 (RREB1)), and some were consistently enriched in downregulated gene sets (e.g. peroxisome proliferator-activated receptor gamma (PPARG), nuclear respiratory factor 2 (NRF2) and oestrogen receptor (ER)) (figure 2). Some motifs (e.g. the catabolite activator protein (CAP) and the hepatocyte nuclear factor 4 (HNF4)) were enriched in gene sets that exhibited different directions of regulation in different brain regions (figure 2). It is noteworthy that several of the TFs whose motifs were associated with DE genes by cis-Metalysis were also themselves DE (FDR ≤ 0.05), including NR5A1 (SF1), STAT3 and CREB2 (ATF4), a master regulator of neural and behavioural plasticity [17]. The TF motif analysis also points to the involvement of the POMC neuronal system in the regulation of the brain's response to a territorial intrusion. For example, another study implicated RREB1 (identified above as enriched in upregulated genes across regions) in the transcription of NEUROD1, a gene that regulates the cell-specific transcription of POMC gene in synergy with Ptx1 in the anterior pituitary [44]. Two other TFs identified in our study (POU3F2 and ER) have also been shown to regulate POMC expression [45]. When we more closely examined the location of binding sites of one of the motif CAP in the stickleback genome, we discovered that they were preferentially positioned within approximately 125 bp upstream and downstream of the transcription initiation site, as has been reported previously [46]. This finding strongly suggests that the predicted motif for CAP reflects the true transcription binding site for CAP. Taken together, these results support the hypothesis that behavioural response to territorial intrusion is modulated via transcription regulatory networks.
One of the most striking patterns in the gene expression data is that there were a large number of genes upregulated in the diencephalon and downregulated in the other regions, especially cerebellum and brain stem (figure 1). The motif analysis identified several meta-associations that corroborated this pattern. For example, the single motif CAP was significantly associated with upregulated genes in diencephalon and downregulated genes in cerebellum, while the motif pair ER/TIFF1 was associated with downregulated genes in the diencephalon and upregulated genes in the cerebellum (figure 3, electronic supplementary material, figure S7).
Our analysis also suggests that some transcription motifs interact with other TFs to differently regulate gene expression in different brain regions. For example, the motif for NFR2 was associated with downregulated genes in all brain regions, but in combination with another TF motif, ER, NRF2 was associated with upregulated genes in the diencephalon and cerebellum (figure 3). Interestingly, while the motif pair ER, NRF2 was associated with upregulation of genes in the diencephalon, the combination of ER and TIFF resulted in opposing direction of regulation in the same brain region (see the electronic supplementary material, figure S7). Similarly, BACH2 was associated with upregulated genes in the diencephalon, while in combination with LMO2COM, BACH2 was associated with downregulated genes in other regions (figure 3). This pattern is distinct from the pattern we observed with the motif pair NRF2 and SRF: SRF was associated with upregulated genes in the diencephalon, but its binding sites explicitly avoided downregulated genes in brain stem and telencephalon (figure 3). An example of meta-association between TF motifs (here CAP) and gene target sets, including the numbers of genes implicated in the meta-association in each brain region, is provided in electronic supplementary material, figure S8. These TFs and pairs of TFs are good candidates for causing the dramatic differential gene regulation specific to the diencephalon.
(d). Gene expression—behaviour correlations
We observed significant associations between behaviour and gene expression in all four brain regions. In particular, many (n = 14) of the top 20 genes in the diencephalon (the region where territorial intrusion provoked the largest gene expression response) were significantly (p < 0.05) correlated with the behaviour of experimental males (see the electronic supplementary material, figure S9, table S9). The males that spent more time chasing the intruder (Time chasing) had higher levels of expression of the transcripts encoding EPCAM, NFASC and lower levels of PRL in diencephalon. It is interesting to note that PRL was upregulated in males that were confronted by an intruder, but was actually negatively correlated with aggression (see the electronic supplementary material, table S9). The strong behaviour-gene expression correlations offer further validation that our DE genes are unlikely to be false positives and are consistent with a growing number of studies showing a remarkably close quantitative relationship between gene expression and behaviour [34,47].
To evaluate whether the most DE genes were, in general, more tightly correlated with behaviour than other genes in the genome, we examined the average correlation coefficients between gene expression and behaviour for the top 20 DE genes compared with the rest of the genome. For example, the average correlation coefficient between Number of chases and the top 20 DE genes in diencephalon was r = 0.675, which is outside the 95% CI surrounding the expected correlation (0.395–0.645, p = 0.016). For other behaviours, see electronic supplementary material, table S10. In other words, the top 20 DE genes in diencephalon were more strongly correlated with behaviour than genes in the rest of the genome, offering further evidence that our DE genes are directly involved in the behavioural response to a territorial intrusion. Results for the other brain regions are in electronic supplementary material, table S10.
4. Discussion
In this study, we took a systems biology approach encompassing the transcriptome and the regulome to gain insights into the molecular processes that modulate the behavioural response of territorial animals to a social challenge. We showed that a territorial intrusion by a conspecific male elicited strong aggressive behaviour, and induced the differential expression of hundreds of genes in the brain. Consistent with the functional and anatomical compartmentalization of the brain [48], our study showed that distinct genes and biological processes were expressed in each of the four brain regions. While many of the GO terms overrepresented in the list of the DE genes in control compared with experimental fish were related to behaviour, some of these terms such as female pregnancy or maternal behaviour did not necessarily reflect the behaviour of a territorial male fish. This points to the limitations of GO analysis, which is based upon annotation in model organisms and calls for a more ecological annotation [49]. We detected the most differential expression of genes in the diencephalon and the cerebellum, and showed that aggressive behaviour was correlated with gene expression at the individual level. Previous studies on cichlids showed that sex steroid hormone receptors are expressed primarily in the diencephalon, the telencephalon and the mesencephalic structures of the brain [29] and that sex hormones regulate social behaviour [50]. In agreement with these findings, our study identified the most DE genes in the diencephalon; however, fewer genes were identified in the telencephalon. Steroid hormone receptors that were identified in these studies were not among the top DE genes detected in our study, but some of these genes (especially ER) were implicated by the cis-regulatory motif analysis. These differences may reflect the differences in the social-context (territorial intrusion versus social dominance) and the timing of sampling in these experiments.
Intriguingly, a large number of DE genes that were shared between brain regions showed opposite directions of regulation in different brain regions, suggesting the role of neural gene transcription regulation networks in the expression of aggressive behaviour. In support of these observations, cis-regulatory network analysis identified TF motifs that were enriched in the promoters of genes that were upregulated in one region and downregulated in others. Our motif results highlight the importance of TFs operating together to regulate gene expression in different directions.
Our transcriptome analysis reveals some of the molecular mechanisms underlying the rich social life of nesting male sticklebacks. For example, territorial males engage in energetically costly and risky interactions with their neighbours and intruders. Our gene expression data highlight some of the molecular mechanisms underlying the costs of these activities, e.g. downregulation in the diencephalon of immunity-related genes (PTPN7), genes associated with feeding (CARTPT) and the ribosomal proteins (RPL24D1, RPL4), known for their associations with brain plasticity.
The strong correlations between behaviour and gene expression at the individual level strongly implicate our list of DE genes with aggressive behaviour. However, we do not know whether the genes that were upregulated in the diencephalon and correlated with aggression were causally related to aggressive behaviour, or if they were DE as a consequence of aggressive behaviour. Manipulative experiments that change the expression of particular genes can help disentangle cause from consequence. However, it is becoming increasingly evident that for complex traits such as aggression, perturbations to single candidate genes might not be fruitful because the behaviour reflects the coordinated action of an entire network of genes.
Indeed, the motif analysis strongly implicates the involvement of complex transcription regulatory networks in the behavioural response of territorial animals to an intrusion, and offers insights into why there were such different patterns in the different brain regions. The pattern of predicted regulation of gene expression by TFs mirrored that of gene expression in the different brain regions. For example, certain TFs motifs were associated with genes that were consistently upregulated (e.g. RREB1) or downregulated (e.g. PPARG, NRF2, POU3F2) in all brain regions and other TF motifs were associated with genes that were differentially regulated in different brain regions (e.g. CAP, HNF4) (figure 2). In parallel, some genes were consistently upregulated (e.g. EIF5) or downregulated (e.g. RPL4) in all brain regions, while other genes exhibited different directions of regulation across brain regions (e.g. CGA, TSHB, PRL, POMC). Of particular interest was the identification, by both microarray and qPCR gene expression profiling, of the pituitary glycopeptide hormones (CGA, TSHB, LH) and polypeptide hormones (POMC, CRHBP and PRL) as upregulated in the diencephalon and downregulated in the cerebellum and the brain stem. These results suggest that territorial intrusion evokes varying transcriptomic responses in different brain regions, often with the same genes being regulated in opposite directions in different regions. This finding strongly suggests the involvement of transcription regulatory networks in response to an intruder. This is consistent with studies that have shown that the production of GnRH and its receptors is under the control of a ‘transcription code’ involving IEGs [51] and supports the hypothesis that POMC and GnRH-controlled neurons modulate behaviour via transcription regulatory networks operating within and across brain regions.
Our results support the hypothesis that transcriptional regulatory networks modulate shifts in neurogenomic states. However, the question still remains to know how the regulatory networks modulate shifts in neurogenomic states across different brain regions. In other words, how can the same TFs regulate the same genes in opposing direction in two different brain regions? The cis-regulatory network analysis suggests that this might occur via interacting TFs. In combination with other motifs or ligands, these motifs might exert different regulatory effects on gene expression based upon the environment in the region in which they are expressed. Indeed, the promoters of many TFs, including IEGs, contain multiple regulatory elements that are responsive to different intracellular signalling pathways (hormones, receptors or ligands), and the details of these pathways will be different in different cell types and therefore in every region of the brain [52]. A good example is the oestrogen receptor (ER). The E domain of ER contains the binding site for the hormone oestrogen (ligand) as well as binding sites for other coactivator and corepressor proteins. Only in the presence of a bound ligand does ER exhibit its full gene expression regulation capacity. Our data suggest that ER acted differently in different brain regions according to its combinations with other TFs. For instance, ER motif alone was associated with downregulated genes in the diencephalon in response to territorial intrusion. However, in combination with NRF2 (i.e. the motif pair ER/NRF2), the ER motif was implicated in the upregulation of genes in the diencephalon and in the downregulation of genes in cerebellum (figure 3). The interaction between ER and NRF2 has been reported in other studies [53]. The implication of signalling molecules and cofactors in the combinatory regulation of gene expression has been reported in both vertebrates and invertebrates (see [54,55]); however, their implication in the modulation of complex behaviours has not been reported until recently [17]. A major challenge for future work is to integrate gene regulatory networks within neural circuits [52,56]. Our results offer a glimpse into the complexity of gene expression regulation involved in complex behaviours such as aggression and further demonstrate the power of emerging bioinformatic tools to discover how behaviour is modulated at the transcriptional level both within and across brain regions.
Acknowledgements
We thank Jenny Drnevich for help with statistical analysis in R Bioconductor, and Tom Newman and Gene Robinson for access to their qPCR machine. We also thank Molly Kent for help with brain dissection, and David Clayton, Gene Robinson, the Bell Laboratory, two anonymous reviewers and Associate Editor Rosemary Knapp for their valuable comments on the manuscript. This study was supported by a grant from the National Institutes of Health R01 GM082937 to A.M.B. and M.B.
References
- 1.Gesquiere L. R., Learn N. H., Simao M. C., Onyango P. O., Alberts S. C., Altmann J. 2011. Life at the top: rank and stress in wild male baboons. Science 333, 357–360 10.1126/science.1207120 (doi:10.1126/science.1207120) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huntingford F. A., Turner A. K., Downie L. M. 1987. Animal conflict. Boca Raton, FL: Chapman & Hall [Google Scholar]
- 3.Sih A., Bell A., Johnson J. C. 2004. Behavioral syndromes: an ecological and evolutionary overview. Trends Ecol. Evol. 19, 372–378 10.1016/j.tree.2004.04.009 (doi:10.1016/j.tree.2004.04.009) [DOI] [PubMed] [Google Scholar]
- 4.de Waal F. B. 2000. Primates—a natural heritage of conflict resolution. Science 289, 586–590 10.1126/science.289.5479.586 (doi:10.1126/science.289.5479.586) [DOI] [PubMed] [Google Scholar]
- 5.Dierick H. A., Greenspan R. J. 2006. Molecular analysis of flies selected for aggressive behavior. Nat. Genet. 38, 1023–1031 10.1038/ng1864 (doi:10.1038/ng1864) [DOI] [PubMed] [Google Scholar]
- 6.Alaux C., et al. 2009. Honey bee aggression supports a link between gene regulation and behavioral evolution. Proc. Natl Acad. Sci. USA 106, 15 400–15 405 10.1073/pnas.0907043106 (doi:10.1073/pnas.0907043106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edwards A. C., Rollmann S. M., Morgan T. J., Mackay T. F. 2006. Quantitative genomics of aggressive behavior in Drosophila melanogaster. PLoS Genet. 2, e154. 10.1371/journal.pgen.0020154 (doi:10.1371/journal.pgen.0020154) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feldker D. E., Datson N. A., Veenema A. H., Proutski V., Lathouwers D., De Kloet E. R., Vreugdenhil E. 2003. GeneChip analysis of hippocampal gene expression profiles of short- and long-attack-latency mice: technical and biological implications. J. Neurosci. Res. 74, 701–716 10.1002/jnr.10800 (doi:10.1002/jnr.10800) [DOI] [PubMed] [Google Scholar]
- 9.Kroes R. A., Panksepp J., Burgdorf J., Otto N. J., Moskal J. R. 2006. Modeling depression: social dominance-submission gene expression patterns in rat neocortex. Neuroscience 137, 37–49 10.1016/j.neuroscience.2005.08.076 (doi:10.1016/j.neuroscience.2005.08.076) [DOI] [PubMed] [Google Scholar]
- 10.Mukai M., Replogle K., Drnevich J., Wang G., Wacker D., Band M., Clayton D. F., Wingfield J. C. 2009. Seasonal differences of gene expression profiles in song sparrow (Melospiza melodia) hypothalamus in relation to territorial aggression. PLoS ONE 4, e8182. 10.1371/journal.pone.0008182 (doi:10.1371/journal.pone.0008182) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sneddon L., Schmidt R., Fang Y., Cossins A. 2011. Molecular correlates of social dominance: a novel role for ependymin in aggression. PLoS ONE 6, e18181. 10.1371/journal.pone.0018181 (doi:10.1371/journal.pone.0018181) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greenwood A. K., Wark A. R., Fernald R. D., Hofmann H. A. 2008. Expression of arginine vasotocin in distinct preoptic regions is associated with dominant and subordinate behaviour in an African cichlid fish. Proc. R. Soc. B 275, 2393–2402 10.1098/rspb.2008.0622 (doi:10.1098/rspb.2008.0622) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stansberg C., Ersland K. M., van der Valk P., Steen V. M. 2011. Gene expression in the rat brain: high similarity but unique differences between frontomedial-, temporal- and occipital cortex. BMC Neurosci. 12, 15. 10.1186/1471-2202-12-15 (doi:10.1186/1471-2202-12-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sinha S., Ling X., Whitfield C. W., Zhai C., Robinson G. E. 2006. Genome scan for cis-regulatory DNA motifs associated with social behavior in honey bees. Proc. Natl Acad. Sci. USA 103, 16 352–16 357 10.1073/0607448103 (doi:10.1073/0607448103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ament S. A., et al. 2012. New meta-analysis tools reveal common transcriptional regulatory basis for multiple determinants of behavior. Proc. Natl Acad. Sci. USA 109, E1801–E1810 10.1073/pnas.1205283109 (doi:10.1073/pnas.1205283109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davidson E., Levin M. 2005. Gene regulatory networks. Proc. Natl Acad. Sci. USA 102, 4935. 10.1073/pnas.0502024102 (doi:10.1073/pnas.0502024102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chandrasekaran S., Ament S. A., Eddy J. A., Rodriguez-Zas S. L., Schatz B. R., Price N. D., Robinson G. E. 2011. Behavior-specific changes in transcriptional modules lead to distinct and predictable neurogenomic states. Proc. Natl Acad. Sci. USA 108, 18 020–18 025 10.1073/pnas.1114093108 (doi:10.1073/pnas.1114093108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vage J., Bonsdorff T. B., Arnet E., Tverdal A., Lingaas F. 2010. Differential gene expression in brain tissues of aggressive and non-aggressive dogs. BMC Vet. Res. 6, 34. 10.1186/1746-6148-6-34 (doi:10.1186/1746-6148-6-34) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bell M. A., Foster S. A. 1994. Introduction to the evolutionary biology of the threespine stickleback. In The evolutionary biology of the threespine stickleback (eds Bell M. A., Foster S. A.). Oxford, UK: Oxford University Press [Google Scholar]
- 20.Kitano J., Lema S. C., Luckenbach J. A., Mori S., Kawagishi Y., Kusakabe M., Swanson P., Peichel C. L. 2010. Adaptive divergence in the thyroid hormone signaling pathway in the stickleback radiation. Curr. Biol. 20, 2124–2130 10.1016/j.cub.2010.10.050 (doi:10.1016/j.cub.2010.10.050) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bell A. M., Backstrom T., Huntingford F. A., Pottinger T. G., Winberg S. 2007. Variable neuroendocrine responses to ecologically-relevant challenges in sticklebacks. Physiol. Behav. 91, 15–25 10.1016/j.physbeh.2007.01.012 (doi:10.1016/j.physbeh.2007.01.012) [DOI] [PubMed] [Google Scholar]
- 22.Sanogo Y. O., Hankison S., Band M., Obregon A., Bell A. M. 2011. Brain transcriptomic response of threespine sticklebacks to cues of a predator. Brain Behav. Evol. 77, 270–285 10.1159/000328221 (doi:10.1159/000328221) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aubin-Horth N., Deschenes M., Cloutier S. 2012. Natural variation in the molecular stress network correlates with a behavioural syndrome. Hormones Behav. 61, 140–146 10.1016/j.yhbeh.2011.11.008 (doi:10.1016/j.yhbeh.2011.11.008) [DOI] [PubMed] [Google Scholar]
- 24.Wootton R. J. 1976. The biology of sticklebacks. London, UK: Academic Press [Google Scholar]
- 25.Goodson J. L. 2005. The vertebrate social behavior network: evolutionary themes and variations. Hormones Behav. 48, 11–22 10.1016/j.yhbeh.2005.02.003 (doi:10.1016/j.yhbeh.2005.02.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Newman S. W. 1999. The medial extended amygdala in male reproductive behavior—a node in the mammalian social behavior network. Ann. NY Acad. Sci. 877, 242–257 10.1111/j.1749-6632.1999.tb09271.x (doi:10.1111/j.1749-6632.1999.tb09271.x) [DOI] [PubMed] [Google Scholar]
- 27.Renn S. C., Aubin-Horth N., Hofmann H. A. 2008. Fish and chips: functional genomics of social plasticity in an African cichlid fish. J. Exp. Biol. 211, 3041–3056 10.1242/jeb.018242 (doi:10.1242/jeb.018242) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oliveira R. F. 2009. Social behavior in context: hormonal modulation of behavioral plasticity and social competence. Integr. Comp. Biol. 49, 423–440 10.1093/icb/icp055 (doi:10.1093/icb/icp055) [DOI] [PubMed] [Google Scholar]
- 29.Munchrath L. A., Hofmann H. A. 2010. Distribution of sex steroid hormone receptors in the brain of an African cichlid fish, Astatotilapia burtoni. J. Comp. Neurobiol. 518, 3302–3326 10.1002/cne.22401 (doi:10.1002/cne.22401) [DOI] [PubMed] [Google Scholar]
- 30.Burmeister S. S., Jarvis E. D., Fernald R. D. 2005. Rapid behavioral and genomic responses to social opportunity. PLoS Biol. 3, e363. 10.1371/journal.pbio.0030363 (doi:10.1371/journal.pbio.0030363) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davidson E. 2001. Genomic regulatory systems: development and evolution, p. 261 Academic Press [Google Scholar]
- 32.Clayton D. F. 2000. The genomic action potential. Neurobiol. Learn. Mem. 74, 185–216 10.1006/nlme.2000.3967 (doi:10.1006/nlme.2000.3967) [DOI] [PubMed] [Google Scholar]
- 33.Ellis L. L., Carney G. E. 2011. Socially-responsive gene expression in male Drosophila melanogaster is influenced by the sex of the interacting partner. Genetics 187, 157–169 10.1534/genetics.110.122754 (doi:10.1534/genetics.110.122754) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cummings M. E., Larkins-Ford J., Reilly C. R. L., Wong R. Y., Ramsey M., Hofmann H. A. 2008. Sexual and social stimuli elicit rapid and contrasting genomic responses. Proc. R. Soc. B 275, 393–402 10.1098/rspb.2007.1454 (doi:10.1098/rspb.2007.1454) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Breitling R. 2004. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 573, 83. 10.1016/j.febslet.2004.07.055 (doi:10.1016/j.febslet.2004.07.055) [DOI] [PubMed] [Google Scholar]
- 36.Durinck S., Moreau Y., Kasprzyk A., Davis S., De Moor B., Brazma A., Huber W. 2005. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics 21, 3439–3440 10.1093/bioinformatics/bti525 (doi:10.1093/bioinformatics/bti525) [DOI] [PubMed] [Google Scholar]
- 37.Sinha S., van Nimwegen E., Siggia E. D. 2003. A probabilistic method to detect regulatory modules. Bioinformatics 19(Suppl. 1), i292–i301 10.1093/bioinformatics/btg1040 (doi:10.1093/bioinformatics/btg1040) [DOI] [PubMed] [Google Scholar]
- 38.Rajaram S., Oono Y. 2010. NeatMap—non-clustering heat map alternatives in R. BMC Bioinformatics 11, 45. 10.1186/1471-2105-11-45 (doi:10.1186/1471-2105-11-45) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aubin-Horth N., Landry C. R., Letcher B. H., Hofmann H. A. 2005. Alternative life histories shape brain gene expression profiles in males of the same population. Proc. R. Soc. B 272, 1655–1662 10.1098/rspb.2005.3125 (doi:10.1098/rspb.2005.3125) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schradin C., Anzenberger G. 1999. Prolactin, the hormone of paternity. News Physiol. Sci. 14, 223–231 [DOI] [PubMed] [Google Scholar]
- 41.Marler C. A., Moore M. C. 1988. Evolutionary costs of aggression revealed by testosterone manipulations in free-living male lizards. Behav. Ecol. Sociobiol. 23, 21–26 10.1007/BF00303053 (doi:10.1007/BF00303053) [DOI] [Google Scholar]
- 42.Pandolfi M., Pozzi A. G., Canepa M., Vissio P. G., Shimizu A., Maggese M. C., Lobo G. 2009. Presence of beta-follicle-stimulating hormone and beta-luteinizing hormone transcripts in the brain of Cichlasoma dimerus (Perciformes: Cichlidae) effect of brain-derived gonadotropins on pituitary hormone release. Neuroendocrinology 89, 27–37 10.1159/000152833 (doi:10.1159/000152833) [DOI] [PubMed] [Google Scholar]
- 43.Filby A. L., Paull G. C., Hickmore T. F. A., Tyler C. R. 2010. Unravelling the neurophysiological basis of aggression in a fish model. BMC Genom. 11, 498. 10.1186/1471-2164-11-498 (doi:10.1186/1471-2164-11-498) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poulin G., Turgeon B., Drouin J. 1997. NeuroD1/beta2 contributes to cell-specific transcription of the proopiomelanocortin gene. Mol. Cell. Biol. 17, 6673–6682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Souza F. S., Nasif S., Lopez-Leal R., Levi D. H., Low M. J., Rubinsten M. 2011. The estrogen receptor alpha colocalizes with proopiomelanocortin in hypothalamic neurons and binds to a conserved motif present in the neuron-specific enhancer nPE2. Eur. J. Pharmacol. 660, 181–187 10.1016/j.ejphar.2010.10.114 (doi:10.1016/j.ejphar.2010.10.114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bucher P. 1990. Weight matrix descriptions of 4 eukaryotic RNA Polymerase-II promoter elements derived from 502 unrelated promoter sequences. J. Mol. Biol. 212, 563–578 10.1016/0022-2836(90)90223-9 (doi:10.1016/0022-2836(90)90223-9) [DOI] [PubMed] [Google Scholar]
- 47.Sabatini M. J., Ebert P., Lewis D. A., Levitt P., Cameron J. L., Mirnics K. 2007. Amygdala gene expression correlates of social behavior in monkeys experiencing maternal separation. J. Neurosci. 27, 3295–3304 10.1523/JNEUROSCI.4765-06.2007 (doi:10.1523/JNEUROSCI.4765-06.2007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roeder T., Schramm G., Marquardt H., Bussmeyer I., Franz O. 2004. Differential transcription in defined parts of the insect brain: comparative study utilizing Drosophila melanogaster and Schistocerca gregaria. Inverteb. Neurosci. 5, 77–83 10.1007/s10158-004-0030-z (doi:10.1007/s10158-004-0030-z) [DOI] [PubMed] [Google Scholar]
- 49.Landry C. R., Aubin-Horth N. 2007. Ecological annotation of genes and genomes through ecological genomics. Mol. Ecol. 16, 4419–4421 10.1111/j.1365-294X.2007.03504.x (doi:10.1111/j.1365-294X.2007.03504.x) [DOI] [PubMed] [Google Scholar]
- 50.O'Connell L. A., Hofmann H. A. 2012. Social status predicts how sex steroid receptors regulate complex behavior across levels of biological organization. Endocrinology 153, 1341–1351 10.1210/en.2011-1663 (doi:10.1210/en.2011-1663) [DOI] [PubMed] [Google Scholar]
- 51.Schang A. L., et al. 2011. GnRH receptor gene expression in the developing rat hippocampus: transcriptional regulation and potential roles in neuronal plasticity. Endocrinology 152, 568–580 10.1210/en.2010-0840 (doi:10.1210/en.2010-0840) [DOI] [PubMed] [Google Scholar]
- 52.McCarthy M. M., Arnold A. P. 2011. Reframing sexual differentiation of the brain. Nat. Neurosci. 14, 677–683 10.1038/nn.2834 (doi:10.1038/nn.2834) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yao Y., Brodie A. M., Davidson N. E., Kensler T. W., Zhou Q. 2010. Inhibition of estrogen signaling activates the NRF2 pathway in breast cancer. Breast Cancer Res. Treat. 124, 585–591 10.1007/s10549-010-1023-8 (doi:10.1007/s10549-010-1023-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gerke J., Lorenz K., Cohen B. 2009. Genetic interactions between transcription factors cause natural variation in yeast. Science 323, 498–501 10.1126/science.1166426 (doi:10.1126/science.1166426) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ravasi T., et al. 2010. An atlas of combinatorial transcriptional regulation in mouse and man. Cell 140, 744–752 10.1016/j.cell.2010.01.044 (doi:10.1016/j.cell.2010.01.044) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Betancur P., Bronner-Fraser M., Sauka-Spengler T. 2010. Assembling neural crest regulatory circuits into a gene regulatory network. Annu. Rev. Cell. Dev. Biol. 26, 581–603 10.1146/annurev.cellbio.042308.113245 (doi:10.1146/annurev.cellbio.042308.113245) [DOI] [PMC free article] [PubMed] [Google Scholar]


