Abstract
The exploitation of soil ecosystem services by agricultural management strategies requires knowledge of microbial communities in different management regimes. Crop cover by no-till management protects the soil surface, reducing the risk of erosion and nutrient leaching, but might increase straw residue-borne and soilborne plant-pathogenic fungi. A cross-site study of soil microbial communities and Fusarium fungistasis was conducted on six long-term agricultural fields with no-till and moldboard-plowed treatments. Microbial communities were studied at the topsoil surface (0 to 5 cm) and bottom (10 to 20 cm) by general bacterial and actinobacterial terminal restriction fragment length polymorphism (T-RFLP) and phospholipid fatty acid (PLFA) analyses. Fusarium culmorum soil fungistasis describing soil receptivity to plant-pathogenic fungi was explored by using the surface layer method. Soil depth had a significant impact on general bacterial as well as actinobacterial communities and PLFA profiles in no-till treatment, with a clear spatial distinction of communities (P < 0.05), whereas the depth-related separation of microbial communities was not observed in plowed fields. The fungal biomass was higher in no-till surface soil than in plowed soil (P < 0.07). Soil total microbial biomass and fungal biomass correlated with fungistasis (P < 0.02 for the sum of PLFAs; P < 0.001 for PLFA 18:2ω6). Our cross-site study demonstrated that agricultural management strategies can have a major impact on soil microbial community structures, indicating that it is possible to influence the soil processes with management decisions. The interactions between plant-pathogenic fungi and soil microbial communities are multifaceted, and a high level of fungistasis could be linked to the high microbial biomass in soil but not to the specific management strategy.
INTRODUCTION
Arable land is an exceptional ecosystem where the soil functional processes are altered by human management such as tillage. Unbalanced soil processes accelerate environmental problems like erosion, the leaching of nutrients (43, 51), and environmental chemicalization due to excess chemical plant disease control (7). On a global scale, no-till management has increased more than 230% during the last 10 years, being 111 million ha in 2009 (49). Increased interest in no-till management, in which crop is sown without prior tillage (leaving 30 to 100% of the soil surface crop covered), results from its advantages in the control of erosion and nutrient leaching and the reduction in fossil fuel consumption and labor hours of cultivation (49). One objective of traditional moldboard plowing and other primary tillage managements is to mix the crop residues into the tillage layer of 10 to 30 cm. In no-till management, mechanical disturbance of the soil is minimized, and crop residues are left on the soil surface, enhancing carbon stratification on the uppermost 0- to 5-cm soil layer (e.g., see reference 3). The adoption of no-till management creates gradual changes in soil physical properties relevant to soil ecological processes. No-till soil has also been reported to retain more soil moisture (e.g., see reference 41) and to dry and warm more slowly in the spring than plowed soil. On the other hand, crop residue-covered zero-tilled soil has been found to stay warmer during autumn and winter.
However, changes in soil carbon allocation, mixing intensity, and soil moisture and temperature conditions affect the distribution and living conditions of microbial communities in soil. No-till management increases soil fungal hypha density and plant arbuscular mycorrhizal fungus colonization in comparison to plowing (26, 35). Microbial biomass and enzyme activities have been found to be higher in no-till soil (54). No-till fields also have larger amounts of cultivated microorganisms and higher denitrification rates in the soil surface layer (0 to 7.5 cm) than plowed soils, but the opposite seems to be true for deeper layers (7.5 to 30 cm) (20). Overwintering crop residues at the soil-air interface might function as a source of plant fungal pathogens infecting new crops sown directly into the stubble (4), causing losses in yield and increasing the undesirable use of fungicides. The emergence of soilborne plant diseases is a result of relationships between and among microorganisms, pathogens, and plants interfering with soil properties.
In suppressive soils, disease incidence or severity remains low regardless of the presence of the pathogen, the host plant, and favorable environmental conditions for disease development (1). One form of soil suppressiveness is fungistasis, defined as the attribute of the soil that restricts the germination and growth of fungi (29, 38). In theory, soil-resident microorganisms mediate suppressiveness and fungistasis by competing for nutrients, producing antagonistic compounds and occupying the potential niches of pathogens (46). Fungistasis was suggested previously to be important for the general suppressiveness of fungal pathogens (52). Microbial community composition can affect soil fungistasis (19). When agricultural management changes the soil general microbial activities and biomass (39), it can potentially also affect soil suppressiveness. The management of tillage intensity, which may affect microbial activities and microbial biomass, has been hypothesized to have an effect on soil fungistasis. The relationship of the general microbial community structure and specific bacterial groups to fungistasis is not understood, and it is not known if certain microbial populations are important for the establishment of fungistasis.
The knowledge of how soil management changes microbial community structures is a prerequisite for optimized management practices, since soil microbial communities constitute a central factor controlling soil processes (9, 14). These communities offer important ecosystem services, e.g., the decomposing of organic matter, the recycling of nutrients essential for plant growth, and soil suppressivity. However, tillage management practices have rarely been studied by modern microbiological methods, including microbial community analysis, to show the structural and functional diversity of microorganisms (15, 55). Previous microbial ecology studies that recognized the diversity of microbes were performed mostly within single fields, describing treatment effects at the site level (8, 15, 23). The generalization of treatment-specific changes from single-field studies into a larger ecological context is difficult, because microbial communities are largely affected by environmental variables at the site, e.g., cultivation history, soil type, weather parameters, and management practices performed at the site (30, 32, 53). Cross-site studies are needed for the identification of treatment-specific changes that have significance on regional and global scales (24).
The hypothesis of the present study is that long-term tillage management has predictable effects on soil microbial structures that can be noted regardless of cross-site variation. In no-till agroecosystems, the location of crop residue on the surface favors fungi as the primary decomposers, and bacteria are expected to be more important in conventional tillage (26, 34). These management-borne shifts in detritus food webs are expected to alter interactions between soil fungi and bacteria, and they are hypothesized to have an impact on the soil fungistasis of Fusarium culmorum, a pathogen of barley (Hordeum vulgare) and wheat (Triticum aestivum). Furthermore, we aim to link specific microbial markers and soil fungistasis to gain insight into the microbial community structures contributing to soil fungistatic properties.
MATERIALS AND METHODS
Field sites and sampling.
Six independent field sites with their distinctive cultivation histories were used in the study. Sites 1 to 3 were located in Jokioinen, and site 4 was located in Vihti, in southern Finland (Table 1). Säkylä (5) is located in southwest Finland, and Ylistaro (6) is located in western Finland. The exact coordinates of each site are shown in Table 1. The fields represent the typical arable soils in Finland. Soils 1 to 4 were classified as Vertic Cambisol, and soils 5 and 6 were classified as Eutric Regosols (22).
Table 1.
Description of experimental fields with no-till and plowed management plotsd
| Parameter | Description or value for field site: |
|||||
|---|---|---|---|---|---|---|
| 1 (Jokioinen) | 2 (Jokioinen) | 3 (Jokioinen) | 4 (Vihti) | 5 (Säkylä) | 6 (Ylistaro) | |
| Coordinates | 60°49′N, 23°28′E | 60°49′N, 23°28′E | 60°49′N, 23°28′E | 60°21′N, 24°22′E | 60°58′N, 22°31′E | 62°46′N, 22°50′E |
| Time in no-till regime (yr) | 9 | 10 | 8 | 9 | 11 | 11 |
| Crop type in 2009 | Spring barley | Spring barley | Crop rotationc | Spring barley | Spring barley/wheat | Spring barley |
| Climatea | ||||||
| Annual precipitation (mm) | 607 | 607 | 607 | 626 | 618 | 527 |
| Annual mean air temp (°C) | 4.30 | 4.30 | 4.30 | 4.50 | 4.20 | 3.60 |
| No. of days with ground minimum temp of <0.0°C | 190 | 190 | 190 | 204 | 206 | 211 |
| Total N in plowed soil/total N in no-till soil (% dry wt) at soil depth (cm) of: | ||||||
| 0–5 | 0.20/0.24 | 0.16/0.19 | 0.15/0.19 | 0.27/0.27 | 0.09/0.19 | 0.15/0.22 |
| 5–10 | 0.20/0.2 | 0.16/0.17 | 0.16/0.16 | 0.28/0.19 | 0.09/0.15 | 0.14/0.17 |
| 10–20 | 0.20/0.19 | 0.16/0.16 | 0.15/0.08 | 0.27/0.20 | 0.09/0.13 | 0.15/0.16 |
| Total C in plowed soil/total C in no-till soil (% dry wt) at soil depth (cm) ofb: | ||||||
| 0–5 | 2.64/3.18 | 2.57/2.94 | 2.50/2.91 | 3.53/3.35 | 1.87/2.63 | 1.81/2.69 |
| 5–10 | 2.6/2.68 | 2.56/2.6 | 2.50/2.47 | 3.70/2.37 | 1.92/2.10 | 1.76/2.00 |
| 10–20 | 2.6/2.54 | 2.57/2.5 | 2.40/2.46 | 3.62/2.42 | 1.95/1.94 | 1.83/1.91 |
| pH in plowed soil/pH in no-till soil at soil depth (cm) ofb: | ||||||
| 0–5 | 5.86/5.8 | 6.80/6.4 | 5.84/5.65 | 5.78/5.98 | 6.16/6.02 | 5.28/6.18 |
| 5–10 | 6.08/5.96 | 6.11/6.51 | 5.73/5.77 | 5.80/5.80 | 6.33/6.39 | 5.13/5.26 |
| 10–20 | 6.13/6.09 | 6.03/6.54 | 5.94/5.90 | 5.79/5.75 | 6.28/6.77 | 5.85/5.31 |
| Dry bulk density in plowed soil/dry bulk density in no-till soil (g/cm3)b | 1.14/1.17 | 1.13/1.16 | ND | 1.23/1.31 | 1.22/1.33 | 1.15/1.37 |
| Texture (g/g)b | ||||||
| Clay (<2 μm) | 0.46 | 0.617 | 0.595 | 0.476 | 0.188 | 0.211 |
| Silt (2–20 μm) | 0.287 | 0.19 | 0.195 | 0.342 | 0.295 | 0.35 |
| Fine sand (20–200 μm) | 0.143 | 0.112 | 0.127 | 0.131 | 0.345 | 0.334 |
| Sand (>200 μm) | 0.109 | 0.081 | 0.083 | 0.052 | 0.199 | 0.117 |
| Porosity of plowed soil/porosity of no-till soil (m3/m3)b | ||||||
| Macropores (>0.03 mm) | 0.119/0.102 | 0.064/0.091 | 0.046/0.022 | 0.097/0.059 | 0.136/0.093 | 0.176/0.094 |
| Micropores (<0.0002 mm) | 0.263/0.252 | 0.276/0.259 | ND | 0.245/0.266 | 0.169/0.209 | 0.182/0.213 |
| No. of biopores ≥2 mm (m2) in plowed soil/no. of biopores ≥2 mm (m2) in no-till soil | 37/358 | 19/170 | 151/264 | 132/150 | 113/434 | 0/151 |
| Water-holding capacity in plowed soil/water-holding capacity in no-till soil (ml H2O/g soil [dry wt]) | 0.52/0.58 | 0.52/0.64 | 0.54/0.64 | 0.56/0.53 | 0.42/0.49 | 0.41/0.53 |
Reference period of 1971 to 2000 (21).
Number of biopores at a 20-cm depth in the 0- to 20-cm layer, as described previously by Regina and Alakukku (45), except for site 3.
The crop rotation was spring wheat, turnip rape, barley, and pea. Turnip rape was cultivated during the sampling season.
For each table entry that has two values (e.g., 95/80), the first value refers to the “plowed” condition and the second pertains to the “no-till” condition. ND, not determined.
All fields contained pairs of long-term (8 to 11 years) no-till and plowed soil treatments. Plowed soil was moldboard plowed in autumn to a 20- to 25-cm depth. In spring, the plowed treatment was seedbed prepared to a 5-cm depth before sowing with a combined drill with shoe coulters. No-till treatment was directly sown to a 3- to 5-cm depth with a combined drill with single- or double-disk coulters. The fields were cultivated primarily with spring cereals. In 2009, spring barley (Hordeum vulgare) was cultivated, except for one no-till area, where spring wheat (Triticum aestivum) was grown, and one field, where spring turnip rape (Brassica rapa subsp. oleifera) was grown (Table 1). Composite soil samples (minimum of 20 soil samplings with augers with a diameter of 2 cm) from each field site and treatment plot were randomly collected in October 2009 before conventional tillage (within the 2-week sampling period). Long-term treatment plots (not tilled and tilled, with a size of 100 to 250 m2) or fields (in Säkylä) were located nearby within the experimental field (maximum of 30 m away from each other). The soils were sampled at depths of 0 to 5 and 10 to 20 cm and manually homogenized on site. DNA samples were stored at −20°C until used for analysis, phospholipid fatty acid (PLFA) samples were freeze-dried, and fungistasis samples taken from a depth of 0 to 10 cm were stored in the dark at +4°C and sieved (0.2 cm) before analysis.
Analysis of soil parameters.
Soil chemical and physical parameters were analyzed as described previously by Regina and Alakukku (45). Briefly, total carbon and nitrogen were analyzed by a using CN analyzer (CN-2000; Leco Corp., St. Joseph, MI). Soil pH was measured from a soil-water suspension at a dilution of 1:2.5 (vol/vol). The water-holding capacity of sieved soil was measured by wetting the soil samples in 100 ml water for 30 min and letting them drain for 0.5 to 2 h. The soil porosity was measured for each plot in 2006 from three replicate soil cores (15-cm diameter and 0- to 20-cm depth). Biopores that were ≥2 mm (m2) were counted at the soil surface. Macro- and micropore volumes were determined by water hanging (−10 kPa) and osmotic (−1,500 kPa) methods, respectively (5). Soils were oven dried (105°C at 48 h) to obtain the dry bulk density.
DNA isolation and amplification of marker genes.
Total DNA for analyses of bacterial communities was extracted from 0.27 ± 0.02 g soil from field sites with the PowerSoil DNA isolation kit (Mo Bio Laboratories, Inc.). The bead-beating step was performed by using Precellys24-Dual tissue homogenizers (Berting Technologies, France), by two 15-s beatings and a 15-s pause between the bead beatings. The quantity and quality of the DNA were assessed by using a NanoDrop 2000 spectrophotometer (Thermo Scientific) and agarose gel electrophoresis. On average, the DNA yield was 4.7 μg/gram of soil (fresh weight) (standard deviation, 1.8), and the A260/A280 ratio was 1.8 (standard deviation, 0.2).
The general 16S rRNA bacterial marker gene was amplified by using primer pair 27f (AGAGTTTGATCCTGGCTCAG) and 1492r (ACGGCTACCTTGTTACGACTT) (56). Primer 27f was labeled with 5′-carboxyfluorescein. Actinobacterium-specific 16S rRNA genes were amplified with primers S-C-Act-0235-a-S-20 (CGCGGCCTATCAGCTTGTTG) and S-C-Act-0878-a-A-19 (CCGTACTCCCCAGGCGGGG) (50). The 5′ end of primer S-C-Act-0235-a-S-20 was labeled by using 5′-carboxyfluorescein.
The 25-μl PCR mixtures for the amplification of the 16S rRNA genes of both universal bacteria and actinobacteria contained 0.2 μM each primer, 0.2 mM deoxynucleoside triphosphate (dNTP), 1.25 units of Dreamtaq DNA polymerase (Fermentas GmbH, Germany), and 1× reaction buffer containing 2 mM MgCl2. The PCR protocol for universal bacteria was 80°C for 5 min followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 56°C for 30 s, and elongation at 72°C for 90 s, followed by a final elongation step at 72°C for 10 min. The PCR protocol for actinobacteria was 80°C for 3 min, followed by 28 cycles of denaturation at 95°C for 20 s, annealing at 65°C for 45 s, and elongation at 72°C for 60 s, followed by a final elongation step at 72°C for 10 min. The quantity of PCR products were measured by using a Qubit fluorometer (Invitrogen).
T-RFLP analysis.
The universal bacterial 16S rRNA PCR product (20 ng) was digested by using 3 units of the HaeIII restriction enzyme (Promega Corporation, Madison, WI), and 10 ng actinobacterium-specific 16S rRNA PCR products was digested with the HhaI restriction enzyme (Fermentas GmbH, Germany, and Promega), using restriction buffer as recommended by the manufacturer. Digestion was performed overnight at 37°C to ensure complete restriction in a 20-μl volume. Digested fragments were ethanol precipitated and dissolved in formamide (15 μl). A Gene Scan 500 standard (Applied Biosystems, Foster City, CA) was used for capillary electrophoresis (ABI 310 DNA sequencer; Applied Biosystems).
The resulting terminal restriction fragment (T-RF) length polymorphism (T-RFLP) profiles were manually inspected by using Peak Scanner software (Applied Biosystems). The relative abundance of each T-RF was calculated by using the total fluorescence area of the profile. The T-RFLP profiles from general bacterial communities were analyzed according to two schemes: in the 1%-cutoff scheme, all the peaks with a relative abundance of over 1% of the total fluorescence were accepted in the comparison of community structures, and in the “most abundant” scheme, the five largest T-RFs (relative abundance) from each profile and their corresponding T-RFs from all the profiles with a relative abundance higher than 0.5% were included in the analysis. The T-RFs with a relative abundance lower than 0.5% were excluded from the analysis. In both schemes, peaks within a size range of ±1 bp were accepted as the same T-RFs.
PLFA analysis.
PLFA (phospholipid fatty acid) and NLFA (neutral lipid fatty acid) analyses were performed according to methods described previously by Frostegard et al. (27), with minor modifications. Briefly, 3 g of freeze-dried soil was extracted with a chloroform-methanol-citrate buffer mixture (1:2:0.8) (“Bligh and Dyer mixture”). The lipids were separated into neutral lipids, glycolipids, and phospholipids in a silicic acid column (Varian Bond Elut Silica SI, 500 mg; Varian, Palo Alto, CA). The phospholipids and neutral lipids were subjected to mild alkaline methanolysis to recover fatty acid methyl esters. Samples were analyzed by using a gas chromatograph with a mass selective detector (gas chromatography-mass spectrometry [GC-MS]) (5890 series II gas chromatograph and 5971 series mass selective detector; Hewlett Packard, Palo Alto, CA) equipped with a 50-m-long, nonpolar, phenyl-methyl silicone capillary column (HP-5; 0.2 mm by 0.33 μm). Helium was used as the carrier gas at a flow rate of 0.9 ml min−1. The temperature of the injector was 280°C. The time-temperature program for the oven was as follows: an initial temperature of 90°C for 2 min, an increase of 30°C min−1 until 160°C was reached, an increase of 3°C min−1 until 280°C was reached, and a final temperature of 280°C for 10 min. The identification and response factors of the different PLFA compounds were based on fatty acid methyl ester (FAME) standards (from Sigma-Aldrich, Nu-Chek-Prep, and Larodan Fine Chemicals AB, Sweden). Altogether, 36 different PLFAs were identified.
The sum of PLFAs (PLFAtot) (nmol g−1 dry soil) was used as an indicator of total microbial biomass, and PLFA 18:2ω6 was used as an indicator of saprophytic fungal biomass in soil (6). PLFA 16:1ω5, NLFA 16:1ω5 and the ratio of NLFA 16:1ω5 to PLFA 16:1ω5 indicated the presence of arbuscular mycorrhiza (42). The sum of PLFA p10-17:0 and p10-19:0 was used as an indicator of actinobacterial biomass (13). The bacterial indicators used were those described previously by Bååth and Anderson (6).
Fungistasis experiments.
Fungistasis biotests were performed by using a surface method described previously by De Boer et al. (17). Three replicate platings were used in the experiments, and when differences between treatments were observed, the experiment was repeated with four replicates. Fusarium culmorum was used as an inoculum in fungistasis experiment because of its relevance as a common pathogen of barley and wheat with soil and plant residue-related dispersion (36). Fungistasis plates were prepared with either diluted soil (ratio of 20% [soil]/80% [diluent]) or 100% soil. Sterile montmorillonite clay (Sigma-Aldrich) and sand (0.5 to 1 mm) (Maxit; Maxit PLC, France) (ratio, 50%/50%) were used as a diluent and in the control petri dishes included in each assay. The soil 20%/80% dilution ratio was selected on the basis of a preliminary experiment with different soil/montmorillonite clay-sand weight percentage ratios (20%/80%, 10%/90%, 5%/95%, and 1%/99%). The selected dilution ratio of 20% (soil)/80% (diluent) allowed the growth of F. culmorum but still showed growth inhibition in comparison to the control (100% sterile montmorillonite clay-sand) (data not shown). The water content of each plate was adjusted to 85% of the water-holding capacity of the soil mixture. Freshly grown F. culmorum (from the growth margin) was used in the biotest as a 1.5-cm-diameter circle and inoculated onto sterile cover glass (2.2 by 2.2 cm) placed onto the test soil (50 g) in a petri dish (9 cm). The petri dishes were closed by double wrapping with Parafilm and incubated for 14 days at 25°C. The area (cm2) of fungal growth was measured after 2 weeks.
Statistical analysis.
Statistical analysis was performed by using the Past v. 1.73c program (31) and the R FactoMineR package (37). Shannon diversity indices, calculated as H′ = −∑Si = 1(ni/N)ln(ni/N) (47), and Simpson diversity indices, calculated as (1 − D) = ∑Si = 1(ni/N)2 (48), take into consideration both the species richness and species dominance. Shannon (H′) and Simpson (1 − D) diversity indices increase with T-RF richness and evenness, but the highest possible value for the Simpson diversity index is 1.
A nonparametric analysis of similarity (ANOSIM) was used to test the significance of the differences between bacterial communities (44). ANOSIM produces R statistics that can range from −1 to 1. Objects that are more dissimilar between groups than within groups will be indicated by an R statistic greater than zero. An R value of zero indicates that the null hypothesis is true. Microbial communities were subjected to principal-components analysis (PCA) with the FactoMineR package to sum up and to simplify the data by reducing the dimensionality of the community structures. The PCA plots were calculated with a scaled correlation matrix. To identify if the treatments are significantly different from each other, a confidence ellipse (95%) based to the mean variance of replicas in dimensions 1 and 2 was drawn for the each plot.
Soil characteristics, fungistasis, and bacterial community diversity indices were statistically analyzed by using paired t tests using a two-tailed distribution. Pearson correlation coefficients between microbial markers and soil fungistasis were calculated by using Microsoft Excel 2007 (Microsoft Corporation).
RESULTS
Field sites and soils.
Six fields, representing two different soil types with long-term no-till and plowed soil treatments, were included in the study (Vertic Cambisol and Eutric Regosols). Long-term no-till practice decreased the macroporosity and increased the soil bulk density and the water-holding capacity compared to plowed topsoils (0 to 20 cm) (Table 1). In no-till treatments, soil total carbon and nitrogen contents decreased by depth and formed a resource gradient for soil microorganisms. The gradient was especially steep at the soil surface (P < 0.01 for 0 to 5 cm versus 5 to 10 cm). On average, higher concentrations of total carbon and nitrogen were detected on no-till surfaces (0 to 5 cm) (2.95% C and 0.22% N) than on plowed surfaces (2.49% C and 0.17% N) (P < 0.05). Sites 5 and 6 contained clearly lower clay and higher fine-sand contents than the other sites. Sites 1 to 3, located in the same area in Jokioinen, showed some notable internal differences. Field 1 had lower soil clay and higher silt contents than the two other sites displaying different cultivation properties. Field 3 differed from sites 1 and 2 by the crop rotation practices performed (spring wheat [Triticum aestivum], turnip rape [Brassica rapa], barley [Hordeum vulgare], and pea [Pisum sativum]).
T-RFLP of 16S rRNA bacterial communities.
Bacterial communities derived from composite samples were depicted as terminal restriction fragments (T-RFs) in the range of 48 to 453 bp displaying a total of 30 peaks (T-RFs) constituting bacterial phylotypes (see Table S1 in the supplemental material). The Shannon and Simpson diversity indices were similar for all treatments (H′ of 2.7 ± 0.03 and 1 − D of 0.9 ± 0.006, calculated from data which include all the T-RFs above the 1% cutoff limit).
The treatment effects on microbial communities were compared by using ANOSIM. The overall differences in communities with global R statistics between tillage treatments and depths were small but significant (R = 0.2; P < 0.05). A significant differentiation of microbial communities was detected within no-till treatments with sampling depth (R = 0.6; P < 0.01), demonstrating the establishment of a depth gradient in all experimental fields (Table 2). Depth-related changes in microbial communities were less obvious in plowed sites (R = 0.03). No statistically significant difference could be observed between no-till and plowed treatments at either depth. The possible differences were most likely masked by the overall differences between fields (see below).
Table 2.
Comparison of microbial community structures by three analysis methods, 16S rRNA gene T-RFLP, actinobacterial 16S rRNA gene T-RFLP, and phospholipid fatty acid analysis, with no-till and plowed soil treatments at depths of 0 to 5 and 10 to 20 cm by one-way ANOSIMa
| Treatment | Soil depth (cm) |
R value |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No till |
Plowed |
||||||||||||
| 0–5 cm |
10–20 cm |
0–5 cm |
10–20 cm |
||||||||||
| 16S rRNA | ACT 16S rRNA | PLFA | 16S rRNA | ACT 16S rRNA | PLFA | 16S rRNA | ACT 16S rRNA | PLFA | 16S rRNA | ACT 16S rRNA | PLFA | ||
| No till | 0–5 | 0.6* | 0.5* | 0.3* | −0.1 | 0 | 0.1 | 0.3* | 0.3* | 0.1 | |||
| No till | 10–20 | 0.2* | 0.1 | 0.0 | −0.0 | 0.3* | 0.1 | ||||||
| Plowed | 0–5 | 0.03 | 0.2 | 0.12 | |||||||||
| Plowed | 10–20 | ||||||||||||
The received R values between each treatment and soil depth are shown. Statistical significance at a P value of <0.05 is indicated by an asterisk. 16S rRNA, 16S rRNA gene T-RFLP; ACT 16S rRNA, actinobacterial 16S rRNA gene T-RFLP.
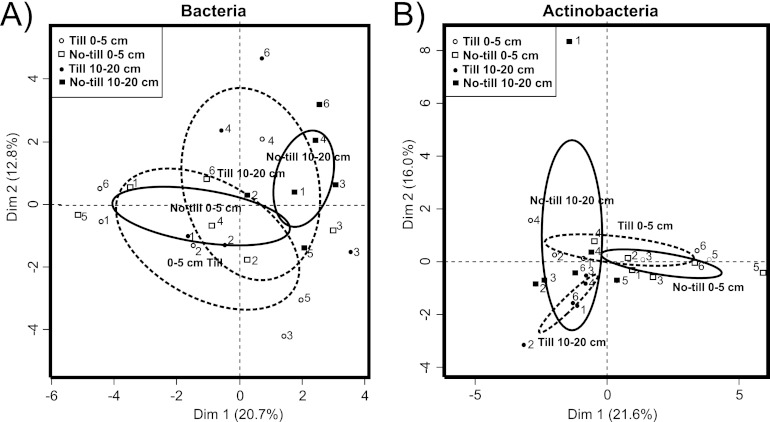
PCA plots were calculated from the T-RFLP profiles showing a clustering of microbial communities in two dimensions (33.5% of variance) (Fig. 1A). The no-till bottom (10 to 20 cm) demonstrated the most compact clustering by PCA. Subclustering occurred between bottom bacterial communities from fields 1 and 2 in both plowed and no-till treatments. Fields 1 and 2 are located near each other, with similar chemical/physical compositions (total carbon and dry bulk density) (Table 1) and cultivation histories (long-term spring barley monoculture), explaining the subclusters shown in Fig. 1A. Surface soil (0 to 5 cm) communities from both treatments were largely dispersed but were different from bottom soils (10 to 20 cm), separated in dimension 1 (20.7% of variance). Confidence ellipses of no-till treatment showed a separation of bacterial communities with depth versus plowed treatment with overlapping ellipse areas. The observed field-specific clustering of site 3 (Fig. 1A) might be due to the crop rotation practice at the site and the rhizosphere effect of turnip rape cultivated in summer 2009. Although bacterial community structures had considerable cross-site variation, it was still possible to detect differences between no-till and plowed soil treatments on the vertical stratification of microbial community structures.
Fig 1.
Principal-components analysis of microbial communities from six field sites with no-till and plowed soil treatments at sampling depths of 0 to 5 and 10 to 20 cm. Confidence ellipses (95% confidence) are drawn around the barycenters of treatments, including till treatment (broken line) and no-till treatment (black line). (A) General 16S rRNA bacterial communities. The two dimensions explain 33.5% of the variance. (B) Actinobacterial communities. The dimensions explain 37.6% of the variance. The numbers 1 to 6 represent the experimental fields described in Table 1.
T-RFLP analysis of actinobacterial communities.
Analysis of actinobacterial communities revealed 25 T-RFs (0.5% cutoff; analysis range, 32 to 622 bp), and the 135-bp peak (phylotype) dominated all communities (65% of the total fluorescence) (see Table S2 in the supplemental material). A higher level of diversity of actinobacterial communities was found at the surface (0 to 5 cm) than at the bottom (10 to 20 cm) (H′ of 1.5 versus 0.9; P < 0.05 by t test) for the no-till treatment, but depth-related diversity differences could not be detected in plowed plots (H′ of 1.2 versus 1.0).
Global R statistics showed that treatments had only a moderate effect on actinobacterial community structures (R = 0.2; P < 0.05). The pairwise comparison of communities showed the largest difference between the no-till surface (0 to 5 cm) and bottom (10 to 20 cm) soils (R = 0.5; P < 0.01) (Table 2). In lowed plots, the depth-related changes were minor (R = 0.2; P < 0.08). Bottom soil (10 to 20 cm) actinobacterial communities were different in no-till and plowed treatments (R = 0.3; P < 0.05), but this treatment effect was not detected for surface soils (R = 0.0; P < 0.4). Surface microbial communities contained more field-specific variation, which masks potential tillage treatment effects (Fig. 1B).
In PCA, the actinobacterial communities from bottom soil (10 to 20 cm) clustered closely, with the exception of no-till field 1, but the clustering was treatment specific (Fig. 1B). The no-till treatment showed depth-related separation supported by the confidence ellipses and evidently explaining the variation in dimension 1 (21.6%). Some depth-related separation was also detected in plowed plots, partly explaining the variance in dimension 2 (16.0%). The surface soils (0 to 5 cm) from no-till treatments were also tightly clustered in the plot, with the exception of soil from field 5. The clustering of actinobacteria was mostly treatment specific, with the exception of field 1. Surface soils from fields 5 and 6 clustered together and separated somewhat from other surface soil samples, reflecting the different soil type of these fields (Table 1).
PLFA analysis of microbial communities.
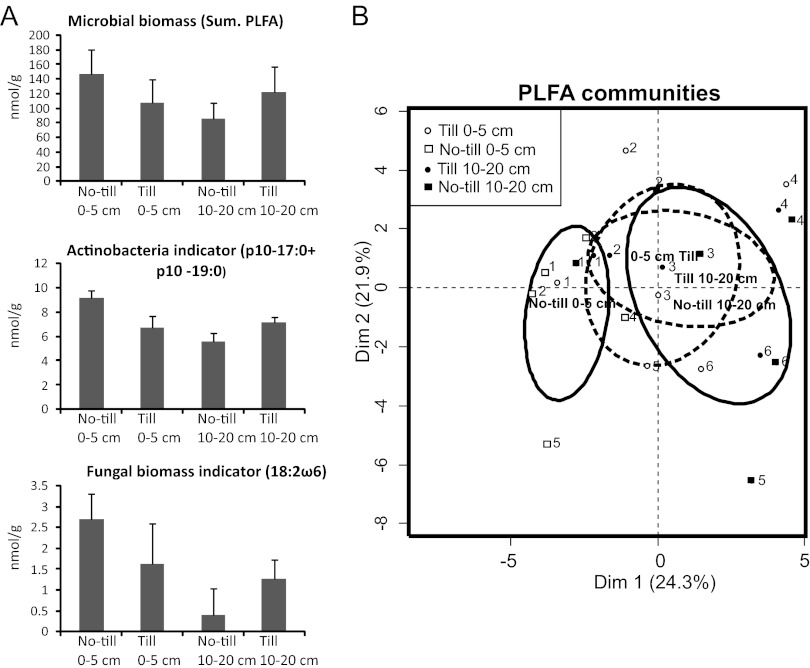
The microbial biomass was higher in no-till surface soils (0 to 5 cm) (146.2 nmol g−1) than in no-till bottom soils (10 to 20 cm) (85.2 nmol g−1), and this was also evident for the actinobacterial indicators (P < 0.05 by pairwise t test) (Fig. 2A). The actinobacterial biomass correlated with the total microbial biomass at each site (Pearson correlation r = 0.89; P < 0.001). The fungal biomass (18:2ω6) was highest in no-till surface soils (2.7 nmol g−1) and lowest in no-till bottom (10 to 20 cm) soil (0.4 nmol g−1) (P < 0.01). No-till surface soils showed higher fungal biomasses than soils from plowed treatment (P < 0.07); the situation was reversed in bottom soil (P < 0.05). The total microbial biomass was thus depth dependent in no-till but not in plowed fields. The PLFA results were in good agreement with the T-RFLP results regarding the general bacterial community.
Fig 2.
PLFA analysis of microbial communities from six field sites (fields 1 to 6) with no-till and plowed soil (till) treatments. (A) Microbial biomass averages (sum of PLFAs) and actinobacterial (p10-17:0 plus p10-19:0) and fungal biomass (18:2ω6) indicators from the treatments (nmol g−1 ± standard deviation) (field sites 1 to 6 as replicates). (B) Principal-components analysis of PLFAs from six field sites. Confidence ellipses are drawn around the barycenters of till treatments (broken line) and no-till treatments (black line). The two dimensions explain 46.3% of the variance. The numbers in plots 1 to 6 represent the experimental fields described in Table 1.
The relative percentages (mol%) of each phospholipid fatty acid (34 peaks were detected) were used for community-level PLFA analysis (see Table S3 in the supplemental material). The ANOSIM showed a small depth-related differentiation of microbial communities for no-till treatments (R = 0.3; P < 0.05), but no differentiation was detected for plowed treatments. The PCA of PLFA profiles revealed a field-specific separation of microbial communities (Fig. 2B), but the confidence ellipses showed a depth-related separation of microbial communities in no-till plots, evidently explaining the variance in dimension 1 (24.4%). The second dimension is probably the soil effect, as Vertic Cambisol (fields 1 to 4) and Eutric Regosols (fields 5 and 6) are separated by it (21.9% variance). The field effect was further shown by one-way ANOSIM, where samples (all depths and treatments) from the same field site were set as a replicate. This analysis revealed differences in microbial communities at the field level (R = 0.6; P < 0.0001). The exceptions were fields 1 and 2 (from same region), which had similar microbial communities (R = 0.24; P < 0.10), as already shown by T-RFLP bacterial community analysis.
Fusarium culmorum fungistasis in arable soils.
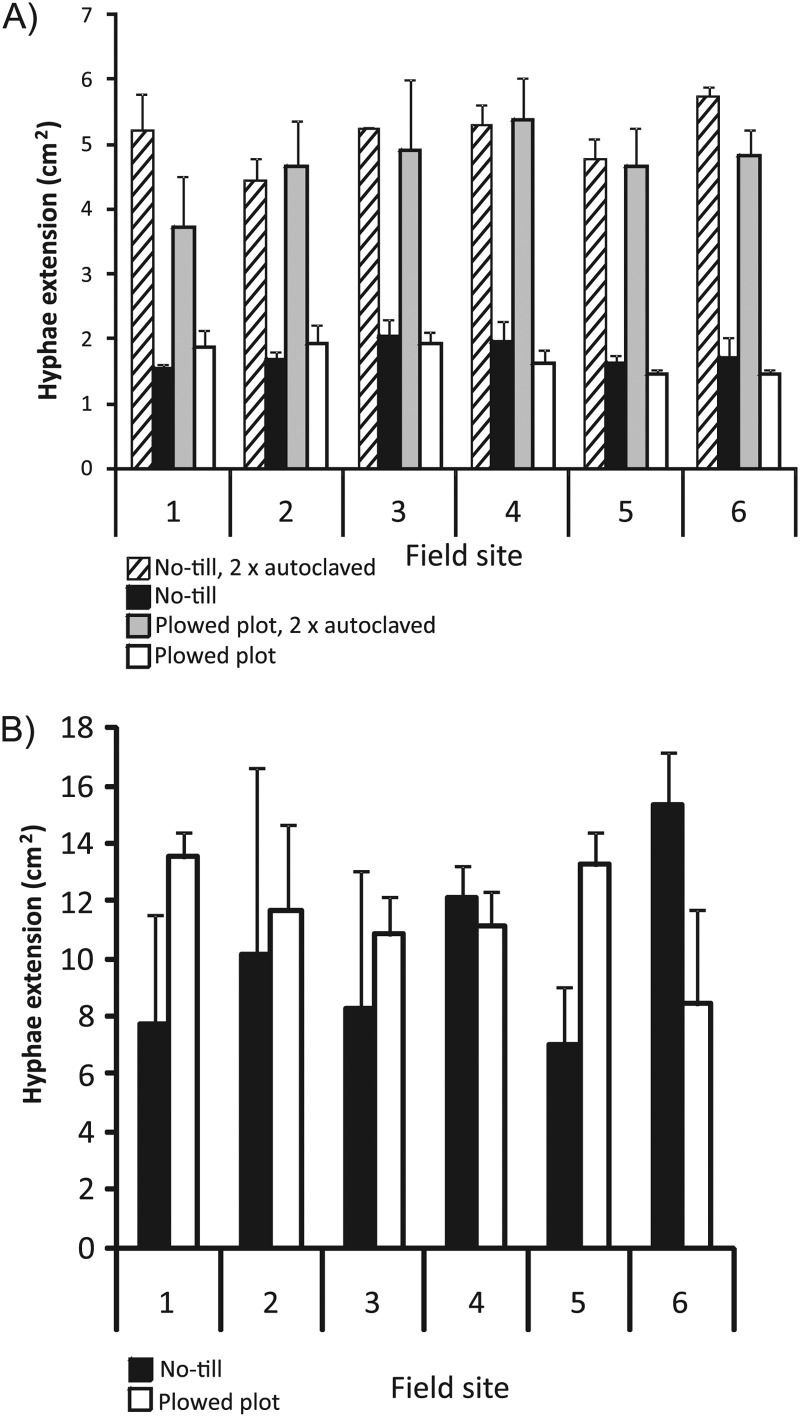
Fungistasis is an inherent property of natural soil mediated mostly by soil microorganisms, and it is interlinked to soil plant pathogen suppressiveness. Fungistasis was detected in all soils, and it was released by autoclaving (Fig. 3A). The average growth areas of F. culmorum in arable soil were very small (1.7 cm2 [standard deviation, 0.2 cm2]) in comparison to the growth in autoclaved soils (4.9 cm2 [standard deviation, 0.5 cm2]), showing growth inhibition. No differences between treatments could initially be detected in nontreated or autoclaved soils. To improve the resolution of the fungistasis assay, the soil was diluted with sterile montmorillonite clay. With these soil dilutions, different strengths of fungistasis were shown at the field and treatment levels (Fig. 3B). Tillage treatment did not have an overall effect (all fields included) on F. culmorum fungistasis (pairwise t test). The growth of F. culmorum in the biotest correlated negatively with the surface soil C/N ratio (0 to 5 cm) (r = −0.55; P < 0.08).
Fig 3.
Fusarium culmorum fungistasis measured with soil from six field sites with no-till and plowed soil treatments using the surface layer method. The growth areas (cm2) on plates were determined after 2 weeks of growth at 25°C. Standard deviations from three replicate analyses are shown. (A) F. culmorum hyphal extensions in nondiluted soils, including both natural soils and soils autoclaved twice (121°C for 15 min). (B) F. culmorum hyphal extensions in diluted soils (dilution, 20%/80%). Sterile montmorillonite clay and sand (0.5 to 1 mm) (dilution, 50%/50%) were used as the diluent. The average extension in sterile montmorillonite clay and sand was 12.3 cm2, with a standard deviation of 0.83 cm2.
Microbial markers and fungistasis.
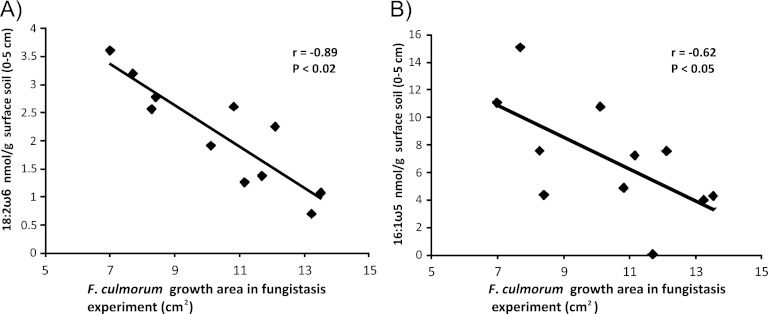
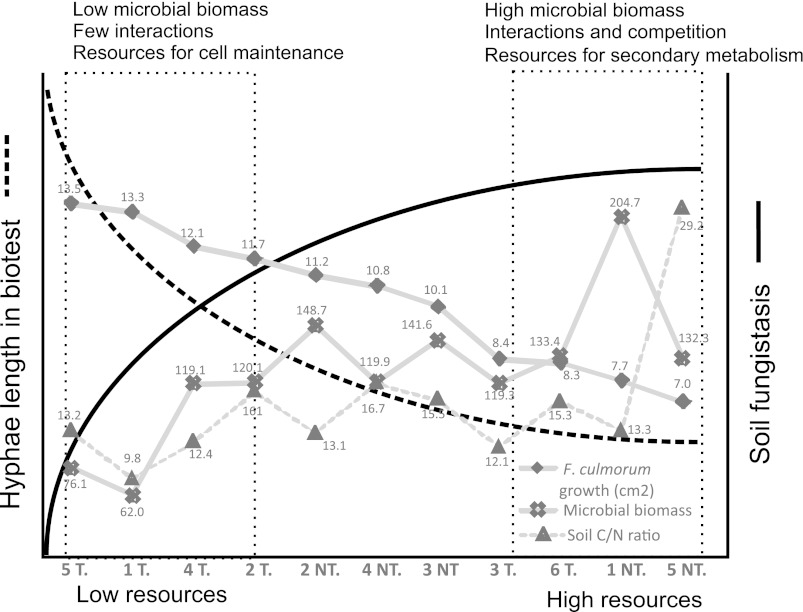
The simultaneous analysis of microbial markers (16S rRNA T-RFLP, Actinobacteria-specific T-RFLP, and PLFA) and fungistasis allowed an elucidation of relationships between microbial community groups and fungistasis by Pearson correlation coefficients (Table 3). The surface soil (0 to 5 cm) microbial biomass correlated negatively (r = −0.70; P < 0.02) with F. culmorum growth in fungistasis assays, supporting the soil sterilization results and the notion that soil microbial activity mediates fungistasis (Table 3). High negative correlations were found with fungally derived (18:2ω6) and arbuscular mycorrhizal fungus (16:1ω5)-derived PLFAs (r = − 0.89 to −0.60; P < 0.02) (Table 3 and Fig. 4). PLFAs associated with bacteria (Table 3) showed only a modest correlation with fungistasis (r = 0.02 to −0.54). The actinobacterial PLFA (p10-19:0) showed the most significant negative correlation to fungistasis (r = −0.54; P < 0.07). The correlations between fungistasis and PLFA markers in bottom soil (10 to 20 cm) were negligible. The soil properties C/N ratio, microbial biomass, and fungistasis were connected in the conceptual model, showing that fungistasis is related to high microbial biomass and C/N ratios (Fig. 5).
Table 3.
Pearson correlations between F. culmorum growth in a fungistasis bioassaya and microbial markers (general bacterial and actinobacterial 16S rRNA T-RFs and PLFAs) from six experimental field sites with no-till and conventional tillage treatmentsb
| Microbial marker | Soil depth (cm) | Pearson correlation coefficient (r) |
|---|---|---|
| Actinobacterial 16S rRNA HhaI T-RFs (bp) | ||
| 218 | 0–5 | −0.51 |
| 447 | 0–5 | −0.42 |
| 449 | 0–5 | −0.48 |
| Bacterial 16S rRNA HaeIII T-RFs (bp) | ||
| 54 | 0–5 | −0.42 |
| 55 | 0–5 | −0.42 |
| 75 | 0–5 | −0.57* |
| 77 | 0–5 | −0.44 |
| 192 | 0–5 | 0.43 |
| 239 | 0–5 | −0.45 |
| 297 | 0–5 | 0.41 |
| Phospholipid fatty acid marker | ||
| Originating from bacteria | ||
| i14:0 | 0–5 | −0.43 |
| i15:0 | 0–5 | −0.50 |
| i16:0 | 0–5 | −0.49 |
| 16:1ω7t | 0–5 | −0.41 |
| ai17:0 | 0–5 | −0.48 |
| p10-19:0 | 0–5 | −0.54* |
| Sum p10-17:0 + p10-19:0 (Actinobacteria) | 0–5 | −0.51* |
| Originating from fungi | ||
| 18:2ω6 | 0–5 | −0.89*** |
| 16:1ω5(AMF) | 0–5 | −0.63*** |
| Others | ||
| 14:0 | 0–5 | −0.43 |
| 15:0 | 0–5 | −0.50 |
| 16:1 | 0–5 | −0.65** |
| 16:0 | 0–5 | −0.66* |
| 17:1 | 0–5 | −0.45 |
| 17:0 | 0–5 | −0.62** |
| 18:0 | 0–5 | −0.75*** |
| Sum PLFAs (biomass indicator) | 0–5 | −0.70*** |
See Fig. 3B.
Correlations higher than 0.4 or lower than −0.4 are shown.
, significance value of <0.02;
, significance value of <0.05;
, significance value of <0.10.
Fig 4.
Correlation between fungal PLFA markers and fungistasis in surface soils of six field sites with no-till and plowed soil treatments. Shown are scatter plots with a linear trend line of the PLFA 18:2ω6 fungal indicator (A) and the 16:1ω5 arbuscular mycorrhizal fungal indicator (B) and Fusarium culmorum growth areas in fungistasis assays (Fig. 3B).
Fig 5.
Relationships between fungistasis, microbial biomass, and soil C/N ratios in a conceptual model describing interconnections between hyphal biotest, soil fungistasis, and soil properties. The measured values are drawn in gray, the sites (fields 1 to 6) and treatments (T, plowed soil; NT, no-till soil) in the x axis are organized from the highest hyphal extension value to the lowest value, and values of microbial biomass and the soil C/N ratio are set accordingly. Measured values are shown for F. culmorum growth by biotest (cm2), microbial biomass (sum PLFAs) (nmol g−1), and the soil C/N ratio.
Most general bacterial T-RFs (54, 55, 75, 77, and 239 bp) from surface soils (0 to 5 cm) displayed a negative correlation (r ≤ −0.4) (Table 3) or no correlation with F. culmorum growth, but T-RFs of 297 and 192 bp, however, correlated positively with F. culmorum growth in the assay. The abundance of actinobacterial T-RF phylotypes 218, 247, 449, 451, and 617 were negatively correlated with the F. culmorum growth area; e.g., when these phylotypes were more abundant, F. culmorum hyphal growth was reduced, showing a connection to a high level of fungistasis. No actinobacterial groups displayed positive correlations (r > 0.4) with fungal growth. T-RFLP correlations were mostly negative (12 out of 18) and modest (r = −0.4 to −0.57), which is in line with the theory that fungistasis is known to be mediated by several bacterial groups together and that no single bacterial marker could explain the whole phenomenon (18).
DISCUSSION
Agricultural management had wide-ranging impacts on microbial communities, including six different fields with a history of long-term tillage intensity management. The no-till treatment structured the soil so that a depth-related bacterial community gradient emerged, which was shown by multiple assays, including general and actinobacterial fingerprinting as well as phospholipid fatty acid community analyses. Soil fungistasis could not be linked to a specific management strategy but could be linked to high microbial and fungal biomasses in soil, indicating that the general microbial activity at the site is an important determinant of soil fungistasis. This demonstrated that human management alters soil microbial structures, supporting the notion that optimized management strategies have the potential to change soil processes. The detailed microbial community analysis (T-RFLP and PLFA) gave insight into interactions between bacteria and fungi in agricultural soil.
Microbial ecology studies of agricultural systems are traditionally performed in a single field experiment with replication plots according to the selected experimental design (8, 15, 23). The generalization of such results is often difficult, since one single field is rarely representative of the diversity of soils and the agricultural and climatic conditions abundant on the regional or even on the global scale. From these studies, it is not possible to predict how management practices generally impact the microbial communities (in different fields and soils and under different environmental conditions). To the best of our knowledge, the only study with a cross-site experimental setup and microbial community analysis was reported recently (33), which included both no-till and conventional till managements. In that study, however, tillage management did not affect the community composition of bacteria and fungi but affected only biomass, in contrast to our study, where no-till management led to a similar stratification of microbial communities regardless of the soil properties or location. This clear discrepancy could best be explained by the low-resolution capacity of the denaturing gradient gel electrophoresis (DGGE) analysis of diverse community compositions deduced from dense banding patterns.
Within a single field, Bissett et al. (11) performed an extensive study to analyze communities of Bacteria, Archaea, and Fungi in both conventional and no-till soils in a long-term experiment. In that study, tillage treatment had only a modest effect on microbial communities, probably due to the sampling strategy, which unfortunately overlooked the depth gradient in no-till treatment by composite sampling (depth of 0 to 15 cm). It should be taken into account that in no-till management, the organic matter resources (crop residues) are located in the surface soil and decline with the depth of the soil. In plowed soil, on the other hand, the incoming resources are turned down to a soil depth of 20 to 25 cm, and the soil structure is under mechanical disturbance. Depth-related changes in soil microbial communities have been described for several natural undisturbed environments (25, 28), and it was hypothesized previously that resource availability is the main driving force in the depth-related changes of microbial communities (25). Our results showing the depth-related separation of microbial communities in no-till treatments support the resource availability hypothesis. Several years of no-till management of fields led to the development of a higher biomass in the surface layer and, consequently, the lowest biomass in the bottom layer. In this respect, no-till management more resembles the natural situation in pristine soil. Also, the differences in soil physical parameters, such as soil contraction or oxygen level, and mechanical disturbances can explain the observed changes in microbial community structures between managements.
In a recent review, a correlation analysis was suggested for the identification of microbial species important for plant pathogen suppressiveness in arable soil (12), but until now, it was rarely implemented in practice. One study used a correlation analysis to detect and identify soil Burkholderia and Mitsuaria strains important in oomycete soil suppressiveness (10). We studied the correlation of fungistasis with three microbial markers to gain knowledge about their connection and importance for soil fungistasis. The conceptual model described interconnections between a hyphal biotest, soil fungistasis, and soil properties (Fig. 5.). The microbial biomass, fungal biomass, and C/N ratio correlated with the strength of fungistasis in our cross-site study, suggesting that fungistasis is not necessarily related to the specific bacterial species but rather is a result of high general activity in soil. This is in line with the notion that, overall, soil microbial activity is important for general plant pathogen suppressiveness (57). The correlation was especially strong between soil fungistasis and fungal biomass, demonstrating that the soil fungistasis level was high when fungi were abundant in soil. The most obvious explanation for this finding is that the fungal abundance in soil is important in the fungistasis process for competing with and inhibiting the growth of F. culmorum (model pathogen). Several isolated fungal strains are known to have antagonistic activities toward fungi (40). Another model suggests that soil microbial communities invest more in the inhibition of fungi (competition) when they are more abundant in soil. This model is supported by results showing that larger amounts of antagonistic bacteria were isolated from fungus-rich environments than from fungus-poor sites (16). In our cross-site study, a high level of F. culmorum fungistasis was found in soils from all studied fields that had a clay content of 18.8 to 61.7%. This finding is in good agreement with results reported previously by Amir and Alabouvette (2), who showed a high level of suppression of Fusarium oxysporum f. sp. lini in clay soil in comparison to sandy soil. The fact that fungistasis was released by autoclaving or reduced with an amendment of sterile montmorillonite clay indicated that clay minerals were not solely causing fungistasis, but the microbial populations were important in our study.
In our cross-site study, PLFA was additionally used for microbial community analysis. The fatty acid-based PLFA markers displayed more field-specific clustering than bacterial DNA fingerprints, which could be related to the different biochemical natures of these molecules. Fatty acids have high turnover rates also regulated by environmental factors such as temperature, whereas DNA markers are linked to the target species composition in soil. The group-specific actinobacterial marker analysis supported the finding that bacterial communities were changing with depth in no-till soil, but in addition to that, minor stratification was observed for till treatment. The large variation of microbial communities within treatments was due to the cross-site experimental setup. This makes the evaluation of agricultural management effects a challenging task that requires the use of parallel community analysis methods to obtain more reliable results.
The changing climate requires the adoption of new cultivation methods in the boreal zone, where it is predicted that winters will become milder, with the freezing and thawing of soils and increasing runoff. This will cause the leaching of nutrients from agricultural fields into inland watercourses and, at the end, to the Baltic Sea. No-till management is considered a way to detain erosion material and nutrients in the fields and minimize leaching (e.g., see reference 43). No-till management in crop cultivation is gaining ground among farmers, but there have been few studies that have described its effects on microbial communities at the regional level. A better comprehension of microbial communities in arable soil managements is needed for the development of agricultural strategies at the national level. Best-management practice can, for instance, reduce plant pathogen pressure, optimize nutrient inputs, and reduce the eutrophication of watercourses in Northern Europe. Treatment-specific microbial community structures could allow the development of optimized agricultural practices to alter soil processes, leading to sustainable cultivation. We conclude that long-term no-till management leads to a depth-related differentiation of microbial communities and also develops a high fungal biomass in surface soil. Indigenous fungal communities in arable soil were linked to the fungistasis phenomenon, which has a major role in the general suppression of soilborne pathogenic fungi (29). Further studies of fungistasis are needed for the development of biotechnological applications of fungal inoculants in crop residue management and biocontrol, since fungistasis is a factor hampering the use of beneficial fungi in green technology. Future research efforts will focus on the identification of fungal and bacterial species important for agricultural management connected to fungistasis.
Supplementary Material
ACKNOWLEDGMENTS
The work was funded by the University of Helsinki Centre for Environment (HENVI) and MTT Agrifood Research Finland.
We thank the technical staff at MTT Jokioinen for their invaluable assistance in the field and with the laboratory work, and we give special appreciation to Leena Mäkäräinen. We acknowledge Miriam Kellock and Sanna Vesterinen for technical assistance in the MEM Group microbiology laboratory.
Footnotes
Published ahead of print 14 September 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Alabouvette C, et al. 2006. Concepts and methods to assess the phytosanitary quality of soils, p 257–269 In Bloem J, Hopkins DW, Benedetti A. (ed), Microbiological methods for assessing soil quality. CABI Publishing, Wallingford, United Kingdom [Google Scholar]
- 2. Amir H, Alabouvette C. 1993. Involvement of soil abiotic factors in the mechanisms of soil suppressiveness to Fusarium wilts. Soil Biol. Biochem. 25:157–164 [Google Scholar]
- 3. Arshad M, Franzluebbers A, Azooz R. 1999. Components of surface soil structure under conventional and no-tillage in northwestern Canada. Soil Tillage Res. 53:41–47 [Google Scholar]
- 4. Arvidsson J. 2010. Energy use efficiency in different tillage systems for winter wheat on a clay and silt loam in Sweden. Eur. J. Agron. 33:250–256 [Google Scholar]
- 5. Aura E. 1975. Effects of soil moisture on the germination and emergence of sugar beet (Beta vulgaris L.). Ph.D. thesis University of Helsinki, Helsinki, Finland [Google Scholar]
- 6. Bååth E, Anderson TH. 2003. Comparison of soil fungal/bacterial ratios in a pH gradient using physiological and PLFA-based techniques. Soil Biol. Biochem. 35:955–963 [Google Scholar]
- 7. Bailey K, Lazarovits G. 2003. Suppressing soil-borne diseases with residue management and organic amendments. Soil Tillage Res. 72:169–180 [Google Scholar]
- 8. Balota EL, Colozzi-Filho A, Andrade DS, Dick RP. 2003. Microbial biomass in soils under different tillage and crop rotation systems. Biol. Fertil. Soils 38:15–20 [Google Scholar]
- 9. Bell T, Newman JA, Silverman BW, Turner SL, Lilley AK. 2005. The contribution of species richness and composition to bacterial services. Nature 436:1157–1160 [DOI] [PubMed] [Google Scholar]
- 10. Benitez MS, McSpadden Gardener BB. 2009. Linking sequence to function in soil bacteria: sequence-directed isolation of novel bacteria contributing to soilborne plant disease suppression. Appl. Environ. Microbiol. 75:915–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bissett A, Richardson AE, Baker G, Thrall PH. 2011. Long-term land use effects on soil microbial community structure and function. Appl. Soil Ecol. 51:66–78 [Google Scholar]
- 12. Borneman J, Becker JO. 2007. Identifying microorganisms involved in specific pathogen suppression in soil. Annu. Rev. Phytopathol. 45:153–172 [DOI] [PubMed] [Google Scholar]
- 13. Brennan P. 1988. Mycobacterium and other actinomycetes. Microb. Lipids 1:203–298 [Google Scholar]
- 14. Cavigelli MA, Robertson GP. 2000. The functional significance of denitrifier community composition in a terrestrial ecosystem. Ecology 81:1402–1414 [Google Scholar]
- 15. Ceja-Navarro JA, et al. 2010. Phylogenetic and multivariate analyses to determine the effects of different tillage and residue management practices on soil bacterial communities. Appl. Environ. Microbiol. 76:3685–3691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. De Boer W, de Ridder-Duine AS, Klein Gunnewiek PJA, Smant W, Van Veen JA. 2008. Rhizosphere bacteria from sites with higher fungal densities exhibit greater levels of potential antifungal properties. Soil Biol. Biochem. 40:1542–1544 [Google Scholar]
- 17. De Boer W, Gunnewiek PJ, Woldendorp JW. 1998. Suppression of hyphal growth of soil-borne fungi by dune soils from vigorous and declining stands of Ammophila arenaria. New Phytol. 138:107–116 [Google Scholar]
- 18. De Boer W, Wagenaar AM, Klein Gunnewiek PJ, Van Veen JA. 2007. In vitro suppression of fungi caused by combinations of apparently non-antagonistic soil bacteria. FEMS Microbiol. Ecol. 59:177–185 [DOI] [PubMed] [Google Scholar]
- 19. De Boer W, Verheggen P, Gunnewiek PJ, Kowalchuk GA, Van Veen JA. 2003. Microbial community composition affects soil fungistasis. Appl. Environ. Microbiol. 69:835–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Doran JW. 1980. Soil microbial and biochemical changes associated with reduced tillage. Soil Sci. Soc. Am. J. 44:765–771 [Google Scholar]
- 21. Drebs A, Nordlund A, Karlsson P, Helminen J, Rissanen P. 2002. Climatological statistics of Finland 1971–2000. Finnish Meteorological Institute, Helsinki, Finland [Google Scholar]
- 22. FAO 1988. FAO/UNESCO soil map of the world. Revised legend with corrections. World soil resources report 60. FAO, Rome, Italy [Google Scholar]
- 23. Feng Y, et al. 2003. Soil microbial communities under conventional-till and no-till continuous cotton systems. Soil Biol. Biochem. 35:1693–1703 [Google Scholar]
- 24. Fierer N, Grandy AS, Six J, Paul EA. 2009. Searching for unifying principles in soil ecology. Soil Biol. Biochem. 41:2249–2256 [Google Scholar]
- 25. Fierer N, Schimel JP, Holden PA. 2003. Variations in microbial community composition through two soil depth profiles. Soil Biol. Biochem. 35:167–176 [Google Scholar]
- 26. Frey S, Elliott E, Paustian K. 1999. Bacterial and fungal abundance and biomass in conventional and no-tillage agroecosystems along two climatic gradients. Soil Biol. Biochem. 31:573–585 [Google Scholar]
- 27. Frostegård Å, Tunlid A, Bååth E. 1993. Phospholipid fatty acid composition, biomass, and activity of microbial communities from two soil types experimentally exposed to different heavy metals. Appl. Environ. Microbiol. 59:3605–3617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Galand PE, Saarnio S, Fritze H, Yrjala K. 2002. Depth related diversity of methanogen Archaea in Finnish oligotrophic fen. FEMS Microbiol. Ecol. 42:441–449 [DOI] [PubMed] [Google Scholar]
- 29. Garbeva P, Hol W, Termorshuizen AJ, Kowalchuk GA, de Boer W. 2010. Fungistasis and general soil biostasis—a new synthesis. Soil Biol. Biochem. 43:469–477 [Google Scholar]
- 30. Girvan MS, Bullimore J, Pretty JN, Osborn AM, Ball AS. 2003. Soil type is the primary determinant of the composition of the total and active bacterial communities in arable soils. Appl. Environ. Microbiol. 69:1800–1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hammer O, Harper DAT, Ryan PD. 2001. PAST: Paleontological Statistics software package for education and data analysis. Paleontol. Electron. 4:1–9 [Google Scholar]
- 32. Hartmann M, Fliessbach A, Oberholzer HR, Widmer F. 2006. Ranking the magnitude of crop and farming system effects on soil microbial biomass and genetic structure of bacterial communities. FEMS Microbiol. Ecol. 57:378–388 [DOI] [PubMed] [Google Scholar]
- 33. Helgason B, Walley F, Germida J. 2010. Long-term no-till management affects microbial biomass but not community composition in Canadian prairie agroecosytems. Soil Biol. Biochem. 42:2192–2202 [Google Scholar]
- 34. Hendrix PF, et al. 1986. Detritus food webs in conventional and no-tillage agroecosystems. Bioscience 36:374–380 [Google Scholar]
- 35. Kabir Z, O'Halloran I, Fyles J, Hamel C. 1997. Seasonal changes of arbuscular mycorrhizal fungi as affected by tillage practices and fertilization: hyphal density and mycorrhizal root colonization. Plant Soil 192:285–293 [Google Scholar]
- 36. Knudsen I, Debosz K, Hockenhull J, Jensen DF, Elmholt S. 1999. Suppressiveness of organically and conventionally managed soils towards brown foot rot of barley. Appl. Soil Ecol. 12:61–72 [Google Scholar]
- 37. Lê S, Josse J, Husson F. 2008. FactoMineR: an R package for multivariate analysis. J. Stat. Softw. 25:1–18 [Google Scholar]
- 38. Lockwood J. 1977. Fungistasis in soils. Biol. Rev. 52:1–43 [Google Scholar]
- 39. McCarty G, Meisinger J, Jenniskens F. 1995. Relationships between total-N, biomass-N and active-N in soil under different tillage and N fertilizer treatments. Soil Biol. Biochem. 27:1245–1250 [Google Scholar]
- 40. Minerdi D, Bossi S, Gullino ML, Garibaldi A. 2009. Volatile organic compounds: a potential direct long-distance mechanism for antagonistic action of Fusarium oxysporum strain MSA 35. Environ. Microbiol. 11:844–854 [DOI] [PubMed] [Google Scholar]
- 41. Nyborg M, Malhi S. 1989. Effect of zero and conventional tillage on barley yield and nitrate nitrogen content, moisture and temperature of soil in north-central Alberta. Soil Tillage Res. 15:1–9 [Google Scholar]
- 42. Olsson PA. 1999. Signature fatty acids provide tools for determination of the distribution and interactions of mycorrhizal fungi in soil. FEMS Microbiol. Ecol. 29:303–310 [Google Scholar]
- 43. Puustinen M, Koskiaho J, Peltonen K. 2005. Influence of cultivation methods on suspended solids and phosphorus concentrations in surface runoff on clayey sloped fields in boreal climate. Agric. Ecosyst. Environ. 105:565–579 [Google Scholar]
- 44. Rees GN, Baldwin DS, Watson GO, Perryman S, Nielsen DL. 2004. Ordination and significance testing of microbial community composition derived from terminal restriction fragment length polymorphisms: application of multivariate statistics. Antonie Van Leeuwenhoek 86:339–347 [DOI] [PubMed] [Google Scholar]
- 45. Regina K, Alakukku L. 2010. Greenhouse gas fluxes in varying soils types under conventional and no-tillage practices. Soil Tillage Res. 109:144–152 [Google Scholar]
- 46. Schroth MN, Hancock JG. 1982. Disease-suppressive soil and root-colonizing bacteria. Science 216:1376–1381 [DOI] [PubMed] [Google Scholar]
- 47. Shannon C, Weaver W. 1963. The mathematical theory of communication. University of Illinois Press, Urbana, IL [Google Scholar]
- 48. Simpson EH. 1949. Measurement of diversity. Nature 163:688 [Google Scholar]
- 49. Soane BD, et al. 2011. No-till in northern, western and south-western Europe: a review of problems and opportunities for crop production and the environment. Soil Tillage Res. 118:66–87 [Google Scholar]
- 50. Stach JEM, Maldonado LA, Ward AC, Goodfellow M, Bull AT. 2003. New primers for the class Actinobacteria: application to marine and terrestrial environments. Environ. Microbiol. 5:828–841 [DOI] [PubMed] [Google Scholar]
- 51. Syswerda S, Basso B, Hamilton S, Tausig J, Robertson G. 2012. Long-term nitrate loss along an agricultural intensity gradient in the Upper Midwest USA. Agric. Ecosyst. Environ. 149:10–19 [Google Scholar]
- 52. Termorshuizen A, Jeger M. 2008. Strategies of soilborne plant pathogenic fungi in relation to disease suppression. Fungal Ecol. 1:108–114 [Google Scholar]
- 53. Ulrich A, Becker R. 2006. Soil parent material is a key determinant of the bacterial community structure in arable soils. FEMS Microbiol. Ecol. 56:430–443 [DOI] [PubMed] [Google Scholar]
- 54. Van Den Bossche A, De Bolle S, De Neve S, Hofman G. 2009. Effect of tillage intensity on N mineralization of different crop residues in a temperate climate. Soil Tillage Res. 103:316–324 [Google Scholar]
- 55. Vargas Gil S, et al. 2010. Response of soil microbial communities to different management practices in surface soils of a soybean agroecosystem in Argentina. Eur. J. Soil Biol. 47:55–60 [Google Scholar]
- 56. Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Weller DM, Raaijmakers JM, Gardener BBMS, Thomashow LS. 2002. Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annu. Rev. Phytopathol. 40:309–348 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.