Abstract
The abundance of Geobacter species in contaminated aquifers in which benzene is anaerobically degraded has led to the suggestion that some Geobacter species might be capable of anaerobic benzene degradation, but this has never been documented. A strain of Geobacter, designated strain Ben, was isolated from sediments from the Fe(III)-reducing zone of a petroleum-contaminated aquifer in which there was significant capacity for anaerobic benzene oxidation. Strain Ben grew in a medium with benzene as the sole electron donor and Fe(III) oxide as the sole electron acceptor. Furthermore, additional evaluation of Geobacter metallireducens demonstrated that it could also grow in benzene-Fe(III) medium. In both strain Ben and G. metallireducens the stoichiometry of benzene metabolism and Fe(III) reduction was consistent with the oxidation of benzene to carbon dioxide with Fe(III) serving as the sole electron acceptor. With benzene as the electron donor, and Fe(III) oxide (strain Ben) or Fe(III) citrate (G. metallireducens) as the electron acceptor, the cell yields of strain Ben and G. metallireducens were 3.2 × 109 and 8.4 × 109 cells/mmol of Fe(III) reduced, respectively. Strain Ben also oxidized benzene with anthraquinone-2,6-disulfonate (AQDS) as the sole electron acceptor with cell yields of 5.9 × 109 cells/mmol of AQDS reduced. Strain Ben serves as model organism for the study of anaerobic benzene metabolism in petroleum-contaminated aquifers, and G. metallireducens is the first anaerobic benzene-degrading organism that can be genetically manipulated.
INTRODUCTION
Understanding the mechanisms for anaerobic benzene degradation may greatly aid the design and optimization of strategies for the bioremediation of subsurface environments contaminated with petroleum and is expected to reveal novel biochemistry for enzymatic attack on this highly stable molecule (9, 12, 20, 36, 38, 62). Monoaromatic hydrocarbons are often the petroleum contaminants of the greatest concern in groundwater because of their water solubility, and benzene is the most toxic of these contaminants (11). Aerobic benzene-degrading organisms are ubiquitous, and their degradation pathway and methods to detect them are well established (21, 30, 66). Many contaminated subsurface environments are, however, anoxic (29, 37, 38, 50, 54). The ability of microbial consortia to anaerobically degrade benzene has been demonstrated with a diversity of electron acceptors, and insights into potential metabolic pathways have been obtained (1, 2, 5, 8, 16, 17, 34, 45, 65). However, pure cultures are required for definitive genetic and biochemical analyses.
Strains of Dechloromonas and Azoarcus species metabolized benzene with nitrate as the electron acceptor (10, 11, 31, 32). However, analysis of the genome sequence of Dechloromonas aromatica strain RCB revealed a lack of genes for anaerobic degradation of expected intermediates in benzene degradation, such as benzoate or phenol, which in most organisms are metabolized via conserved pathways (55). Genes for aerobic benzene oxidation were present (55). These findings, and the possibility of microorganisms internally producing molecular oxygen during nitrate reduction (19, 68), has led to the suggestion that benzene degradation coupled to nitrate reduction might also involve oxygen (47, 55, 62, 64). This would be analogous to the degradation of benzene with chlorate as the electron acceptor in Alicycliphilus denitrificans in which molecular oxygen generated from (per)chlorate is used to activate benzene (63).
The one pure culture that clearly appears to anaerobically metabolize benzene is the hyperthermophile Ferroglobus placidus (27). Benzene was oxidized to carbon dioxide with Fe(III) serving as the sole electron acceptor. The use of strict anaerobic techniques and the abundance of Fe(II) in the medium ensured anoxic conditions. Furthermore, no monooxygenases were encoded in the genome. Analysis of metabolites, as well as whole-genome analysis of gene transcript abundances, suggested that benzene was first carboxylated to benzoate (27), which was then further metabolized via common pathways for anaerobic benzoate metabolism (28). However, the lack of tools for genetic manipulation has limited further study of benzene metabolism in this organism.
Geobacter species have been implicated in the degradation of aromatic hydrocarbons, including benzene, in contaminated subsurface environments (39, 53, 54) and Geobacter species capable of anaerobically oxidizing toluene and xylene with Fe(III) as the electron acceptor have been described (6, 39, 42). Stimulating Fe(III) reduction, presumably by Geobacter species, with chelators or electron shuttles is a potential mechanism for greatly accelerating anaerobic benzene degradation (40, 46). A benzene-degrading, Fe(III)-reducing Geobacter culture would be of interest not only because this environmental relevance, but also because it has been possible to develop genetic systems in Geobacter species (15, 44, 49, 59) that would be useful in further studying this unique metabolism. Here, we report on the anaerobic oxidation of benzene in two Geobacter species: a new isolate from a petroleum-contaminated aquifer and Geobacter metallireducens, which has already been shown to be genetically tractable (49, 59).
MATERIALS AND METHODS
Sediment studies.
Sediments were collected from the previously described (5, 39, 54) Fe(III) reduction zone of a petroleum-contaminated aquifer in Bemidji, MN. As previously described (5), sediment cores were obtained by drilling cores, which were transported immediately back to the laboratory, and sediment aliquots were stored anoxically in glass vessels overnight at 25°C for outlined experiments. For radiolabeled tracer studies, 20-g sediment samples were transferred to 50-ml serum bottles under N2-CO2 (95:5) and incubated at 25°C. The sediments were amended with [Ring-UL-14C]benzene (1.48 × 105 Bq 14C) (radiochemical purity = 99.7%; Moravek Biochemicals, Brea, CA).
Enrichment and isolation of strain Ben.
To further enrich benzene metabolic capacity, the benzene-degrading sediments were incubated at 30°C under anoxic conditions as previously described (43) and amended with 100 μM benzene (Sigma-Aldrich, St. Louis, MO; Chemical purity, ≥99.9%) as the electron donor and 100 mM poorly crystalline Fe(III) oxide (43) as the electron acceptor. Fe(II) production was monitored and when sufficient Fe(II) to account for oxidation of the added benzene accumulated additional benzene was added. This was repeated three times. Sediments were then serially diluted in a bicarbonate-buffered freshwater medium (43) with benzene (100 μM) as the sole electron donor and poorly crystalline Fe(III) oxide (100 mM) as the sole electron acceptor. The liquid enrichments were transferred five times. Then enrichment cultures were inoculated (10% [vol/vol]) into an anaerobic dilution series of the same medium amended with 2% Difco noble agar (BD, Franklin, NJ) to form roll tubes. A single isolated colony was picked and further purified by repeating serial dilution in roll tubes and restreaking of single colonies on solidified Fe(III) oxide medium as described previously (48) with the modification that benzene was the sole electron donor. A colony from this extended purification, designated strain Ben, was selected for further study. The purity of the culture was confirmed by analysis of 16S rRNA gene sequence. Overall, the enrichment process took nearly 1 year.
Acetate (10 mM) served as the electron donor to evaluate whether Fe(III) oxide, Fe(III) nitrilotriacetate, Fe(III) pyrophosphate, Fe(III) citrate, or anthraquinone-2,6-disulfonate (AQDS) could serve as an electron acceptor (41, 48). AQDS (5 mM) served as the electron acceptor for evaluating whether acetate, ethanol, fumarate, H2, lactate, pyruvate, benzoate, or toluene could serve as electron donors (41, 48).
Geobacter metallireducens.
Geobacter metallireducens strain GS-15 (ATCC 53774 and DSM 7210) (41) was obtained from our laboratory culture collection and was routinely cultured as previously described (43).
Analysis of growth and metabolism on benzene.
Anaerobic pressure tubes (25 ml; 15-ml headspace) were used for benzene growth studies. Cells of strain Ben and G. metallireducens were grown for four successive transfers of a 20% inoculum in medium with 250 μM benzene as the sole electron donor, before conducting studies on the stoichiometry of growth on benzene. The electron acceptors in these studies were 5 mM AQDS or 100 mmol of Fe(III) oxide/liter for strain Ben and 55 mM Fe(III) citrate for G. metallireducens. Controls for growth experiments were live cells without donor or without acceptor and heat-killed cultures with donor and acceptor. For studies on conversion of benzene to carbon dioxide, medium containing 250 μM benzene was amended with of 3.7 × 104 Bq of [U-14C]benzene (2.78 × 109 Bq mmol−1; Radiochemical purity, 99.7%; Moravek Biochemicals, Brea, CA) from an anaerobic stock of 2.96 × 105 Bq ml−1 in water.
Additional studies of [U-14C]benzene metabolism were investigated with concentrated cells to provide for more rapid cell metabolism. Cells grown on acetate were concentrated under anaerobic conditions via centrifugation (4,400 × g for 10 min at 15°C), the pellet was washed twice with medium and then resuspended to provide a 30-fold concentration of cells. A total of 3.7 × 104 Bq [U-14C]benzene was added to aliquots (3 ml) of the cell suspensions, which were then incubated anaerobically at 30°C. Controls for radiotracer experiments were heat-killed cultures with donor and acceptor.
Statistical analysis.
Data are reported as the means and standard deviations of triplicate cultures. Statistical analysis was performed using Student t test. A P value of <0.05 was considered statistically significant.
Light and electron microscopy.
Cells were routinely examined by phase-contrast microscopy with a Nikon E600 microscope as previously described (33). The Gram type was determined using Gram staining and bright-field microscopy as previously described (48). For transmission electron microscopy, strain Ben cultures were placed on 300-mesh carbon-coated copper grids, incubated for 15 min, and then stained with 2% aqueous uranyl acetate. The cells were observed using a JEOL 100S transmission electron microscope at an accelerating voltage of 80 kV. Images were taken digitally using the MaxIm-DL software and analyzed using ImageJ (http://rsbweb.nih.giv/ij/index.html).
Phylogenetic analysis.
For analysis of the 16S rRNA gene sequence of strain Ben, 10 ml of culture were collected by centrifugation, and genomic DNA was extracted with the FastDNA spin kit (Bio 101 Inc., Vista, CA) according to the manufacturer's instructions. 16S rRNA gene sequences were amplified with primers 8 forward (18) and 1492 reverse (4) as previously described (25, 48). PCR products were agarose gel purified with a QIAquick gel extraction kit (Qiagen, Inc., Valencia, CA) and ligated into the TOPO TA cloning kit, version K2 (Invitrogen, Carlsbad, CA), according to the manufacturers' instructions. Plasmid inserts were then amplified with M13 forward and reverse primers (Invitrogen) and the 519F internal primer (35) and sequenced at the University of Massachusetts sequencing facility.
The 16S rRNA gene from strain Ben was reconstructed from the sequenced fragments and compared to the GenBank nucleotide and protein databases with the BLASTN algorithm (3). Nucleotide sequences of strain Ben and the most similar isolates found by the BLAST search were aligned using CLC sequence viewer (CLC Bio, Cambridge, MA). Aligned sequences were imported into Seaview (22), where phylogenetic trees were inferred. Identical branching orders were observed with maximum parsimony, maximum-likelihood, and distance-based algorithms when 16S rRNA gene sequences were compared (not shown). Bootstrap values were calculated by all three analyses, and 1,309 nucleotide positions were considered for 16S rRNA gene comparisons.
Analytical techniques.
Cell numbers were monitored with acridine orange staining and epifluorescence microscopy (42), and Fe(III) reduction was monitored by measuring the accumulation of Fe(II) over time as previously described (42). The reduction of AQDS was monitored by the absorbance of 2,6-anthrahydroquinone disulfonate produced at 525 nm (40).
Benzene concentrations in culture headspaces were quantified with a gas chromatograph equipped with a flame ionization detector. The samples were run through a Supelco VOCOL fused silica capillary column (60 m by 0.25 mm by 1.5 μm) held at 50°C for 0.5 min, followed by an increase to 200°C at 10°C/min. The concentrations of benzene in the aqueous phase were calculated with Henry's law using the constant at 25°C of 0.25 for benzene (57).
[14C]carbon dioxide was quantified with a gas proportional counter as previously described (14, 24).
RESULTS AND DISCUSSION
Isolation and characterization of Geobacter strain Ben.
[U-14C]benzene was rapidly oxidized to 14CO2 in sediments from a zone in the petroleum-contaminated study site, which previous molecular studies (67) suggested were enriched in Geobacter species involved in the degradation of monoaromatic compounds (Fig. 1). An isolate was obtained from these sediments by (i) further enriching the benzene-degrading activity in the sediments with the addition of Fe(III) oxide and repeated benzene amendments, (ii) inoculation of enriched sediments into defined medium with benzene as the sole electron donor and Fe(III) oxide as the sole electron acceptor, (iii) repeated transfer of the enrichment culture, and (iv) isolation and purification with solidified benzene-Fe(III) oxide medium. The isolate was designated strain Ben.
Fig 1.
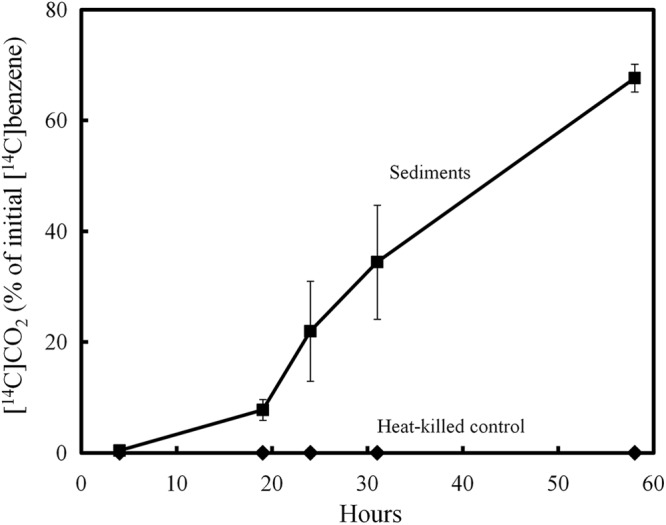
14CO2 production from [14C]benzene in Bemidji sediments. The results are means and standard deviations for triplicate incubations.
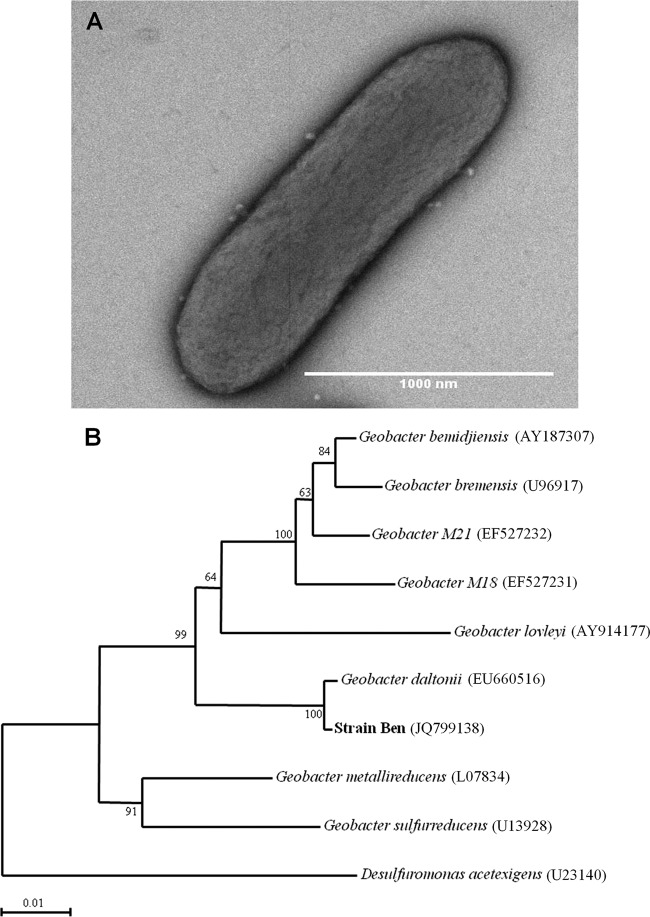
Cells of strain Ben, were Gram-negative, slightly curved rods 1.5 to 2.5 μm by 0.5 μm with round ends (Fig. 2A). Analysis of the 16S rRNA gene of strain Ben revealed that it belongs to the genus Geobacter within the family Geobacteraceae (Fig. 2B). Strain Ben is most closely related to Geobacter daltonii (99% sequence similarity), an Fe(III)- and U(VI)-reducing bacterium isolated from the Oak Ridge Field Research Center (52). This phylogenetic placement of strain Ben was consistent in distance, parsimony algorithms, and maximum-likelihood analyses. The GenBank accession number for the 16S rRNA of strain Ben is JQ799138.
Fig 2.
Morphology and phylogenetic analysis of Geobacter strain Ben. (A) Transmission electron micrograph of strain Ben grown on medium with AQDS (5 mM) provided as the electron acceptor and acetate (5 mM) as the electron donor. (B) Neighbor-joining phylogenetic tree of the 16S rRNA sequences of strain Ben and closest relatives supported by bootstrap analysis. The same topology was obtained from parsimony and maximum-likelihood analyses.
Strain Ben grew with Fe(III) oxide, Fe(III) nitrilotriacetic acid, Fe(III) pyrophosphate, or anthraquinone-2,6-disulfonate (AQDS), but not Fe(III) citrate, as electron acceptors. Various organic electron donors such as acetate (5 mM), ethanol (10 mM), fumarate (10 mM), H2 (130 kPa with 100 μM acetate as the carbon source), lactate (10 mM), pyruvate (10 mM), benzoate (1 mM), or toluene (250 μM) served as sole electron donors.
Benzene oxidation coupled to the reduction of Fe(III) oxide or AQDS by strain Ben.
Strain Ben grew when inoculated (20% inoculum) into a medium with benzene as the sole electron donor and poorly crystalline Fe(III) oxide or AQDS as the sole electron acceptor (Fig. 3). The results shown here for all studies are those for the fourth successive transfer (20% inoculum) on benzene medium. Benzene-dependent growth was slow, and between-tube differences in growth rates resulted in high replicate variability during growth on Fe(III) oxide, but there was a statistically significant increase in cell numbers and the accumulation of the reduced products of electron acceptor reduction (Fig. 3).
Fig 3.
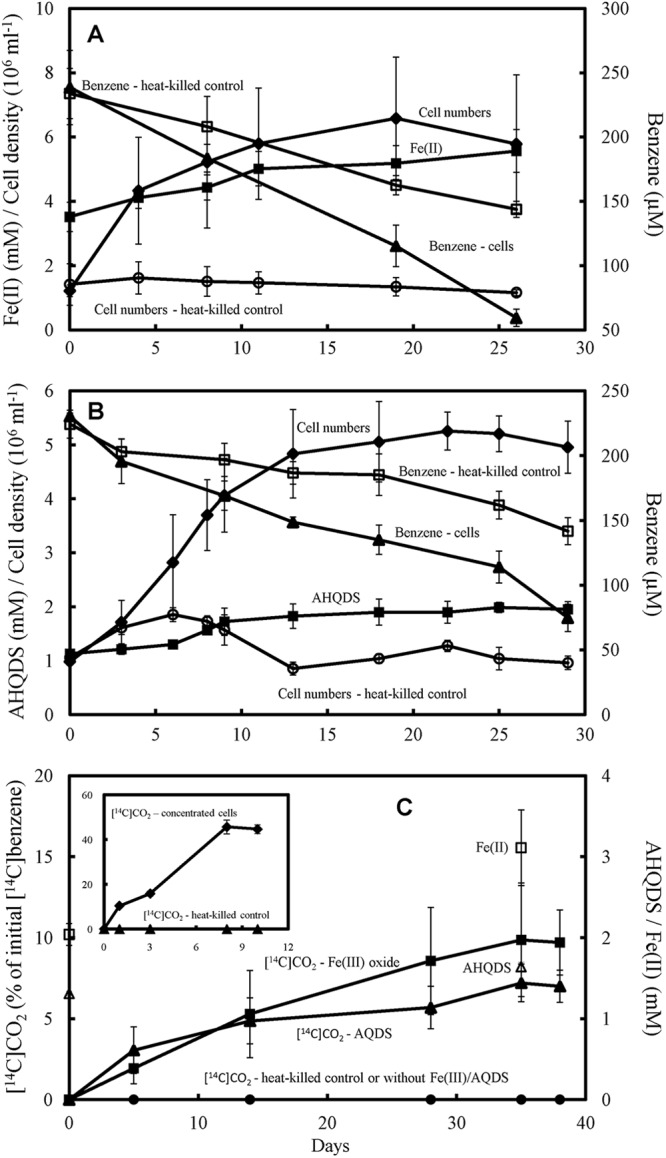
Anaerobic oxidation of benzene by strain Ben. (A) Cell growth, accumulation of Fe(II) from Fe(III) reduction, and benzene loss in medium with benzene as the sole electron donor and Fe(III) oxide as the sole electron acceptor. (B) Cell growth, accumulation of AHQDS from AQDS reduction, and benzene loss in medium with benzene as the sole electron donor and AQDS as the sole electron acceptor. (C) Production of 14CO2 from [14C]benzene and associated Fe(II) or AHQDS accumulation. Production of 14CO2 from [14C]benzene by concentrated cell with AQDS as electron acceptor is shown as an inset in panel C. As outlined in Materials and Methods, the results presented in panel C are from different sets of experiments than those shown in panels A and B. The results are means and standard deviations for triplicate cultures. The P values versus time zero for cell number, Fe(III) reduction, AQDS reduction, and benzene loss data collected during the stationary phase (panel A, day 8 to day 26; panel B, day 13 to day 29) are <0.05.
There was no growth in heat-killed controls (Fig. 3) or controls that contained benzene but no electron acceptor or contained an electron acceptor, but no benzene (see Fig. S1 in the supplemental material). There was some loss of benzene in heat-killed controls (Fig. 3), presumably due to adsorption into the butyl rubber stopper (11, 27). In live Fe(III) oxide cultures there was an increase in cell density over time that was accompanied by an accumulation of Fe(II) and a loss of benzene that was greater than the loss of benzene in the heat-killed controls (Fig. 3A). When this abiotic loss is considered, benzene depletion that could be attributed to strain Ben was 89 ± 15.9 μM benzene (mean ± the standard deviation, n = 3), which could provide electrons sufficient to reduce 2.7 mM Fe(III) oxide if benzene was completely oxidized to carbon dioxide according to the reaction:
| (1) |
The observed accumulation of Fe(II) of 2.1 ± 0.2 mM was consistent with the fact that some of the substrate would be required for biomass formation. During the active growth phase (days 0 to 18) the cell yield per mol of Fe(II) produced [(3.23 ± 0.15) × 109 cells/mmol Fe(II)] with benzene as the sole electron donor was comparable to the yields previously reported (42) for the growth of G. metallireducens with toluene [4.74 × 109 cells/mmol Fe(II)] or phenol [5.11 × 109 cells/mmol Fe(II)] as the sole electron donor and poorly crystalline Fe(III) oxide as the electron acceptor.
In a similar manner, in medium with AQDS as the electron acceptor, cell growth was associated with an accumulation of reduced electron acceptor (anthrahydroquinone-2,6-disulfonate [AHQDS]) and loss of benzene (Fig. 3B). When abiotic benzene loss was considered, the loss of benzene that could be attributed to the activity of strain Ben (73 ± 6 μM, n = 3) could provide electrons sufficient to reduce 1.1 ± 0.1 mM AQDS if benzene was completely oxidized to carbon dioxide:
| (2) |
The observed accumulation of AHQDS was 0.9 ± 0.1 mM, suggesting that AQDS was the sole electron acceptor for benzene oxidation. During the active growth phase (days 0 to 11) the cell yield per mol AHQDS produced (5.92 ± 0.27 109 cells/mmol AHQDS) was comparable to that for growth on benzene with Fe(III) oxide as the electron acceptor when it is considered that two electrons are required to reduce AQDS versus one electron to reduce Fe(III).
In order to further evaluate the stoichiometry of benzene oxidation in a manner that alleviated concerns about adsorption of benzene into the rubber stoppers, a set of separate incubations were conducted in which [14C]benzene was added along with the 250 μM unlabeled benzene (Fig. 3C). 14CO2 was produced with either Fe(III) or AQDS as electron acceptors (Fig. 3C), but there was no 14CO2 production if electron acceptors were omitted. With Fe(III) oxide as the electron acceptor 13.2% ± 2.4% of the added [14C]benzene was recovered as 14CO2. Oxidation of this proportion of the added benzene would be expected to result in the reduction of 1 ± 0.2 mM Fe(III), which compared well with the observed accumulation of 1.3 ± 0.6 mM Fe(II). With AQDS as the electron acceptor, 8.4% ± 0.5% of the added benzene (mean ± the standard deviation, n = 3) was recovered as 14CO2 which would be expected to result in the reduction of 316 ± 33 μM AQDS. The amount of AQDS reduced was 450 ± 50 μM.
In order to evaluate whether strain Ben could anaerobically oxidize a higher proportion of [14C]benzene to 14CO2, studies were also conducted with dense cell suspensions. With AQDS as the electron acceptor 46% ± 3% of the added [14C]benzene was recovered as 14CO2 (Fig. 3C, inset). This level of recovery is a clear indication that the observed benzene oxidation activity is not due to a possible nonbenzene contaminant in the provided radiolabeled benzene, which had a radiochemical purity of 99.7%.
Benzene oxidation coupled to the reduction of Fe(III) by G. metallireducens.
The metabolism of aromatic compounds in Geobacter species has been studied most thoroughly in G. metallireducens (44). It was previously reported that G. metallireducens did not metabolize benzene when evaluated with an initial benzene concentration of 500 μM and Fe(III) oxide as the electron acceptor (41). However, when Fe(III) was supplied as Fe(III) citrate, which promotes faster growth of G. metallireducens, and the initial benzene concentration was lowered to 250 μM, G. metallireducens metabolized benzene with the reduction of Fe(III) (Fig. 4A). The increase in cell density over time was accompanied by an accumulation of Fe(II) and loss of benzene (Fig. 4A). There was no growth in the absence of added benzene or if no Fe(III) was provided (see Fig. S2 in the supplemental material). When the abiotic loss of benzene in heat-killed controls was considered, the benzene depletion that could be attributed to metabolism of G. metallireducens was 84.2 ± 2.4 μM, which could provide electrons sufficient to reduce 2.5 ± 0.1 mM Fe(III) if benzene was completely oxidized to carbon dioxide. The observed accumulation of Fe(II) was 2.2 ± 0.3 mM. The cell yield per mol of Fe(II) produced [(8.41 ± 0.19) × 109 cells/mmol Fe(II)] was higher than for growth of strain Ben on benzene, which may be attributed to Fe(III) citrate rather than Fe(III) oxide serving as the electron acceptor. When [14C]benzene was added along with the unlabeled benzene (250 μM), 14CO2 was produced when Fe(III) was provided as the sole electron acceptor, but there was no 14CO2 production if the Fe(III) was omitted (Fig. 4B). 28.0% ± 8.8% of the added [14C]benzene was recovered as 14CO2. Oxidation of this fraction of the added benzene would be expected to lead to the reduction of 2.1 ± 0.7 mM Fe(II) and 2.4 ± 0.3 mM Fe(II) was generated. Concentrated cell suspension of G. metallireducens oxidized 80% ± 5% of the added [14C]benzene to 14CO2 with Fe(III) as the electron acceptor (Fig. 4B, inset).
Fig 4.
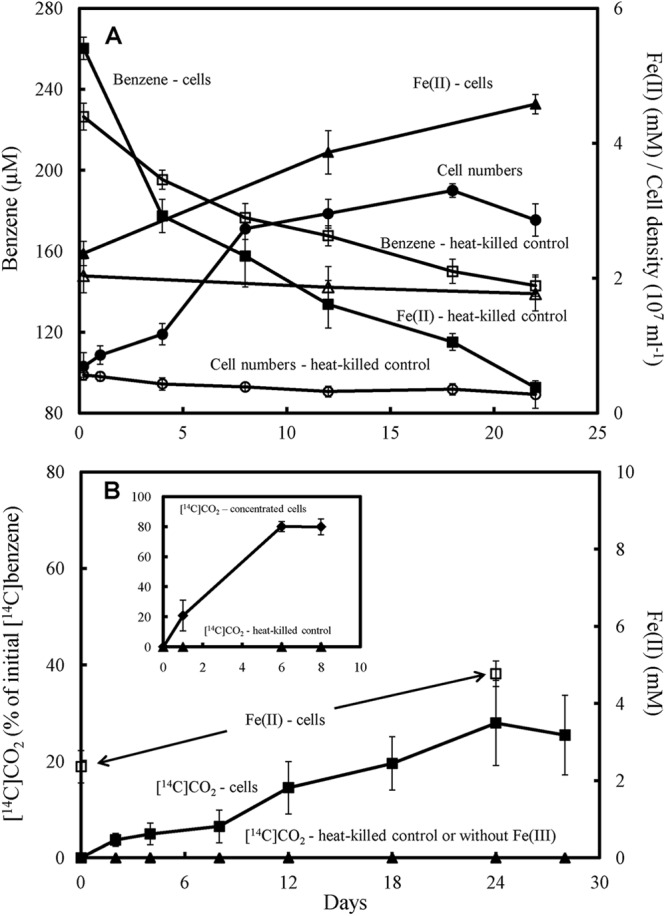
Anaerobic oxidation of benzene by G. metallireducens. (A) Cell growth, accumulation of Fe(II) from Fe(III) reduction, and benzene loss in medium with benzene as the sole electron donor and Fe(III) citrate as the sole electron acceptor. (B) Production of 14CO2 from [14C]benzene and associated Fe(II) accumulation. The production of 14CO2 from [14C]benzene by concentrated cell is shown as an inset in panel B. The results are the mean and standard deviation for triplicate cultures. The P values versus time zero for cell number, Fe(III) reduction, and benzene loss data collected during the stationary phase (panel A, day 8 to day 22) are <0.005.
Implications.
The Geobacter species described here provide additional model organisms for the study of anaerobic benzene degradation. The abundance of Fe(II) during growth on Fe(III) as the electron acceptor ensured anaerobic conditions, and there is no possibility of producing molecular oxygen intracellularly from Fe(III) as there is with nitrate. Further evidence for a lack of degradation of benzene via an aerobic pathway is the lack of monooxygenase genes in the genome of G. metallireducens (49, 59) or in a draft genome of strain Ben (unpublished data).
The three primary mechanisms for anaerobic activation of benzene have been hypothesized to be hydroxylation to phenol (10, 23, 60, 61), carboxylation to benzoate (1, 2, 34, 51), or methylation to toluene (60). The G. metallireducens genome encodes pathways for anaerobic benzoate, toluene, and phenol complete oxidation (7), and previous studies have shown that G. metallireducens can anaerobically metabolize the hydrocarbon, toluene, and other monoaromatic compounds such as benzoate, phenol, and p-cresol (39, 41, 42). Pathways for the anaerobic degradation of benzoate, phenol, toluene and p-cresol are also encoded in the genome of Geobacter strain Ben (unpublished data).
The finding that there are Geobacter species that can anaerobically degrade benzene supports previous suggestions that Geobacter species play an important role in the removal of benzene and other aromatic hydrocarbon contaminants from polluted groundwater (6, 13, 26, 44, 53, 56, 58). Methods for genetically manipulating G. metallireducens have recently been developed (49, 59), making it the first genetically tractable organism known to be capable of anoxic benzene degradation under strict anaerobic conditions. This opens the possibility of definitively elucidating the pathway for anaerobic benzene degradation in this organism, identifying molecular signals that can be useful for monitoring rates of anaerobic benzene degradation in the subsurface, and developing new strategies for stimulating anaerobic benzene degradation from a better understanding of this metabolism.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dale Callaham of the University of Massachusetts Amherst Microscopy facility for transmission electron microscopy.
This research was supported by the Office of Naval Research, grant N00014-09-1-0190.
Footnotes
Published ahead of print 21 September 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Abu Laban N, Selesi D, Jobelius C, Meckenstock RU. 2009. Anaerobic benzene degradation by Gram-positive sulfate-reducing bacteria. FEMS Microbiol. Ecol. 68:300–311 [DOI] [PubMed] [Google Scholar]
- 2. Abu Laban N, Selesi D, Rattei T, Tischler P, Meckenstock R. 2010. Identification of enzymes involved in anaerobic benzene degradation by a strictly anaerobic iron-reducing enrichment culture. Environ. Microbiol. 12:2783–2796 [DOI] [PubMed] [Google Scholar]
- 3. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 4. Amann RI, et al. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Anderson RT, Rooney-Varga JN, Gaw CV, Lovley DR. 1998. Anaerobic benzene oxidation in the Fe(III) reduction zone of petroleum contaminated aquifers. Environ. Sci. Technol. 32:1222–1229 [Google Scholar]
- 6. Botton S, van Harmelen M, Braster M, Parsons JR, Roling WF. 2007. Dominance of Geobacteraceae in BTX-degrading enrichments from an iron-reducing aquifer. FEMS Microbiol. Ecol. 62:118–130 [DOI] [PubMed] [Google Scholar]
- 7. Butler JE, et al. 2007. Genomic and microarray analysis of aromatics degradation in Geobacter metallireducens and comparison to a Geobacter isolate from a contaminated field site. BMC Genomics 8:180 doi:10.1186/1471-2164-8-180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Caldwell ME, Suflita JM. 2000. Detection of phenol and benzoate as intermediates of anaerobic benzene biodegradation under different terminal electron-accepting conditions. Environ. Sci. Technol. 34:1216–1220 [Google Scholar]
- 9. Chakraborty R, Coates JD. 2004. Anaerobic degradation of monoaromatic hydrocarbons. Appl. Microbiol. Biotechnol. 64:437–446 [DOI] [PubMed] [Google Scholar]
- 10. Chakraborty R, Coates JD. 2005. Hydroxylation and carboxylation-two crucial steps of anaerobic benzene degradation by Dechloromonas strain RCB. Appl. Environ. Microbiol. 71:5427–5432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Coates JD, et al. 2001. Anaerobic benzene oxidation coupled to nitrate reduction in pure culture by two strains of Dechloromonas. Nature 411:1039–1043 [DOI] [PubMed] [Google Scholar]
- 12. Coates JD, Chakraborty R, McInerney MJ. 2002. Anaerobic benzene biodegradation-a new era. Res. Microbiol. 153:621–628 [DOI] [PubMed] [Google Scholar]
- 13. Coates JD, Phillips EJ, Lonergan DJ, Jenter H, Lovley DR. 1996. Isolation of Geobacter species from diverse sedimentary environments. Appl. Environ. Microbiol. 62:1531–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Coates JD, Woodward J, Allen J, Philp P, Lovley DR. 1997. Anaerobic degradation of polycyclic aromatic hydrocarbons and alkanes in petroleum-contaminated marine harbor sediments. Appl. Environ. Microbiol. 63:3589–3593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Coppi MV, Leang C, Sandler SJ, Lovley DR. 2001. Development of a genetic system for Geobacter sulfurreducens. Appl. Environ. Microbiol. 67:3180–3187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Da Silva ML, Ruiz-Aguilar GM, Alvarez PJ. 2005. Enhanced anaerobic biodegradation of BTEX-ethanol mixtures in aquifer columns amended with sulfate, chelated ferric iron or nitrate. Biodegradation 16:105–114 [DOI] [PubMed] [Google Scholar]
- 17. Dou J, Liu X, Hu Z, Deng D. 2008. Anaerobic BTEX biodegradation linked to nitrate and sulfate reduction. J. Hazard. Mater. 151:720–729 [DOI] [PubMed] [Google Scholar]
- 18. Eden PA, Schmidt TM, Blakemore RP, Pace NR. 1991. Phylogenetic analysis of Aquaspirillum magnetotacticum using polymerase chain reaction-amplified 16S rRNA-specific DNA. Int. J. Syst. Bacteriol. 41:324–325 [DOI] [PubMed] [Google Scholar]
- 19. Ettwig KF, et al. 2010. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464:543–548 [DOI] [PubMed] [Google Scholar]
- 20. Farhadian M, Vachelard C, Duchez D, Larroche C. 2008. In situ bioremediation of monoaromatic pollutants in groundwater: a review. Bioresour. Technol. 99:5296–5308 [DOI] [PubMed] [Google Scholar]
- 21. Fischer A, et al. 2008. Combined carbon and hydrogen isotope fractionation investigations for elucidating benzene biodegradation pathways. Environ. Sci. Technol. 42:4356–4363 [DOI] [PubMed] [Google Scholar]
- 22. Gouy M, Guindon S, Gascuel O. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27:221–224 [DOI] [PubMed] [Google Scholar]
- 23. Grbic-Galic D, Vogel TM. 1987. Transformation of toluene and benzene by mixed methanogenic cultures. Appl. Environ. Microbiol. 53:254–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hayes LA, Nevin KP, Lovley DR. 1999. Role of prior exposure on anaerobic degradation of naphthalene and phenanthrene in marine harbor sediments. Org. Geochem. 30:937–945 [Google Scholar]
- 25. Holmes DE, Nevin KP, Lovley DR. 2004. Comparison of 16S rRNA, nifD, recA, gyrB, rpoB, and fusA genes within the family Geobacteraceae fam. nov. Int. J. Syst. Evol. Microbiol. 54:1591–1599 [DOI] [PubMed] [Google Scholar]
- 26. Holmes DE, et al. 2007. Subsurface clade of Geobacteraceae that predominates in a diversity of Fe(III)-reducing subsurface environments. ISME J. 1:663–677 [DOI] [PubMed] [Google Scholar]
- 27. Holmes DE, Risso C, Smith JA, Lovley DR. 2011. Anaerobic oxidation of benzene by the hyperthermophilic archaeon Ferroglobus placidus. Appl. Environ. Microbiol. 77:5926–5933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Holmes DE, Risso C, Smith JA, Lovley DR. 2012. Genome-scale analysis of anaerobic benzoate and phenol metabolism in the hyperthermophilic archaeon Ferroglobus placidus. ISME J. 6:146–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jin S, Fallgren PH, Bilgin AA, Morris JM, Barnes PW. 2007. Bioremediation of benzene, ethylbenzene, and xylenes in groundwater under iron-amended, sulfate-reducing conditions. Environ. Toxicol. Chem. 26:249–253 [DOI] [PubMed] [Google Scholar]
- 30. Jindrova E, Chocova M, Demnerova K, Brenner V. 2002. Bacterial aerobic degradation of benzene, toluene, ethylbenzene and xylene. Folia Microbiol. (Praha) 47:83–93 [DOI] [PubMed] [Google Scholar]
- 31. Kasai Y, Kodama Y, Takahata Y, Hoaki T, Watanabe K. 2007. Degradative capacities and bioaugmentation potential of an anaerobic benzene-degrading bacterium strain DN11. Environ. Sci. Technol. 41:6222–6227 [DOI] [PubMed] [Google Scholar]
- 32. Kasai Y, Takahata Y, Manefield M, Watanabe K. 2006. RNA-based stable isotope probing and isolation of anaerobic benzene-degrading bacteria from gasoline-contaminated groundwater. Appl. Environ. Microbiol. 72:3586–3592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kashefi K, Holmes DE, Baross JA, Lovley DR. 2003. Thermophily in the Geobacteraceae: Geothermobacter ehrlichii gen. nov., sp. nov., a novel thermophilic member of the Geobacteraceae from the “Bag City” hydrothermal vent. Appl. Environ. Microbiol. 69:2985–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kunapuli U, Griebler C, Beller HR, Meckenstock RU. 2008. Identification of intermediates formed during anaerobic benzene degradation by an iron-reducing enrichment culture. Environ. Microbiol. 10:1703–1712 [DOI] [PubMed] [Google Scholar]
- 35. Lane DJ, et al. 1985. Rapid determination of 16S rRNA sequences for phylogenetic analyses. Proc. Natl. Acad. Sci. U. S. A. 82:6955–6959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lin B, Van Verseveld HW, Roling WF. 2002. Microbial aspects of anaerobic BTEX degradation. Biomed. Environ. Sci. 15:130–144 [PubMed] [Google Scholar]
- 37. Lovley DR. 2000. Anaerobic benzene degradation. Biodegradation 11:107–116 [DOI] [PubMed] [Google Scholar]
- 38. Lovley DR. 1997. Potential for anaerobic bioremediation of BTEX in petroleum-contaminated aquifers. J. Ind. Microbiol. Biotechnol. 18:75–81 [Google Scholar]
- 39. Lovley DR, et al. 1989. Oxidation of aromatic contaminants coupled to microbial iron reduction. Nature 339:297–300 [Google Scholar]
- 40. Lovley DR, Coates JD, BluntHarris EL, Phillips EJP, Woodward JC. 1996. Humic substances as electron acceptors for microbial respiration. Nature 382:445–448 [Google Scholar]
- 41. Lovley DR, et al. 1993. Geobacter metallireducens gen. nov. sp. nov., a microorganism capable of coupling the complete oxidation of organic compounds to the reduction of iron and other metals. Arch. Microbiol. 159:336–344 [DOI] [PubMed] [Google Scholar]
- 42. Lovley DR, Lonergan DJ. 1990. Anaerobic oxidation of toluene, phenol, and p-cresol by the dissimilatory iron-reducing organism, GS-15. Appl. Environ. Microbiol. 56:1858–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lovley DR, Phillips EJ. 1988. Novel mode of microbial energy metabolism: organic carbon oxidation coupled to dissimilatory reduction of iron or manganese. Appl. Environ. Microbiol. 54:1472–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lovley DR, et al. 2011. Geobacter: the microbe electric's physiology, ecology, and practical applications Adv. Microb. Physiol. 59:1–100 [DOI] [PubMed] [Google Scholar]
- 45. Lovley DR, Woodward JC, Chapelle FH. 1996. Rapid anaerobic benzene oxidation with a variety of chelated Fe(III) forms. Appl. Environ. Microbiol. 62:288–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lovley DR, Woodward JC, Chapelle FH. 1994. Stimulated anoxic biodegradation of aromatic hydrocarbons using Fe(III) ligands. Nature 370:128–131 [DOI] [PubMed] [Google Scholar]
- 47. Meckenstock RU, Mouttaki H. 2011. Anaerobic degradation of non-substituted aromatic hydrocarbons. Curr. Opin. Biotechnol. 22:406–414 [DOI] [PubMed] [Google Scholar]
- 48. Nevin KP, et al. 2005. Geobacter bemidjiensis sp. nov. and Geobacter psychrophilus sp. nov., two novel Fe(III)-reducing subsurface isolates. Int. J. Syst. Evol. Microbiol. 55:1667–1674 [DOI] [PubMed] [Google Scholar]
- 49. Oberender J, Kung J, Seifert J, von Bergen M, Boll M. 2012. Identification and characterization of a succinyl-CoA: benzoate CoA transferase in Geobacter metallireducens. J. Bacteriol. 194:2501–2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Phelps CD, Young LY. 1999. Anaerobic biodegradation of BTEX and gasoline in various aquatic sediments. Biodegradation. 10:15–25 [DOI] [PubMed] [Google Scholar]
- 51. Phelps CD, Zhang X, Young LY. 2001. Use of stable isotopes to identify benzoate as a metabolite of benzene degradation in a sulphidogenic consortium. Environ. Microbiol. 3:600–603 [DOI] [PubMed] [Google Scholar]
- 52. Prakash O, et al. 2010. Geobacter daltonii sp. nov., an Fe(III)- and uranium(VI)-reducing bacterium isolated from a shallow subsurface exposed to mixed heavy metal and hydrocarbon contamination. Int. J. Syst. Evol. Microbiol. 60:546–553 [DOI] [PubMed] [Google Scholar]
- 53. Röling WF, van Breukelen BM, Braster M, Lin B, van Verseveld HW. 2001. Relationships between microbial community structure and hydrochemistry in a landfill leachate-polluted aquifer. Appl. Environ. Microbiol. 67:4619–4629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rooney-Varga JN, Anderson RT, Fraga JL, Ringelberg D, Lovley DR. 1999. Microbial communities associated with anaerobic benzene degradation in a petroleum-contaminated aquifer. Appl. Environ. Microbiol. 65:3056–3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Salinero KK, et al. 2009. Metabolic analysis of the soil microbe Dechloromonas aromatica str. RCB: indications of a surprisingly complex life-style and cryptic anaerobic pathways for aromatic degradation. BMC Genomics 10:351 doi:10.1186/1471-2164-10-351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Staats M, Braster M, Rölling WF. 2011. Molecular diversity and distribution of aromatic hydrocarbon-degrading anaerobes across a landfill leachate plume. Environ. Microbiol. 13:1216–1227 [DOI] [PubMed] [Google Scholar]
- 57. Staudinger J, Roberts PV. 1996. A critical review of Henry's law constants for environmental applications. Crit. Rev. Environ. Sci. Technol. 26:205–297 [Google Scholar]
- 58. Tobler NB, Hofstetter TB, Straub KL, Fontana D, Schwarzenbach RP. 2007. Iron-mediated microbial oxidation and abiotic reduction of organic contaminants under anoxic conditions. Environ. Sci. Technol. 41:7765–7772 [DOI] [PubMed] [Google Scholar]
- 59. Tremblay PL, Aklujkar M, Leang C, Nevin KP, Lovley DR. 2011. A genetic system for Geobacter metallireducens: role of the flagellin and pilin in the reduction of Fe(III) oxide. Environ. Microbiol. Rep. 4:82–88 [DOI] [PubMed] [Google Scholar]
- 60. Ulrich AC, Beller HR, Edwards EA. 2005. Metabolites detected during biodegradation of 13C6-benzene in nitrate-reducing and methanogenic enrichment cultures. Environ. Sci. Technol. 39:6681–6691 [DOI] [PubMed] [Google Scholar]
- 61. Vogel TM, Grbic-Galic D. 1986. Incorporation of oxygen from water into toluene and benzene during anaerobic fermentative transformation. Appl. Environ. Microbiol. 52:200–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Vogt C, Kleinsteuber S, Richnow HH. 2011. Anaerobic benzene degradation by bacteria. Microb. Biotechnol. 4:710–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Weelink SA, et al. 2008. Isolation and characterization of Alicycliphilus denitrificans strain BC, which grows on benzene with chlorate as the electron acceptor. Appl. Environ. Microbiol. 74:6672–6681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Weelink SAB, van Eekert MHA, Stams AJM. 2010. Degradation of BTEX by anaerobic bacteria: physiology and application. Rev. Environ. Sci. Biotechnol. 9:359–385 [Google Scholar]
- 65. Weiner JM, Lovley DR. 1998. Anaerobic benzene degradation in petroleum-contaminated aquifer sediments after inoculation with a benzene-oxidizing enrichment. Appl. Environ. Microbiol. 64:775–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wilson LP, Bouwer EJ. 1997. Biodegradation of aromatic compounds under mixed oxygen/denitrifying conditions: a review. J. Ind. Microbiol. Biotechnol. 18:116–130 [DOI] [PubMed] [Google Scholar]
- 67. Yun J, Ueki T, Miletto M, Lovley DR. 2011. Monitoring the metabolic status of Geobacter species in contaminated groundwater by quantifying key metabolic proteins with Geobacter-specific antibodies. Appl. Environ. Microbiol. 77:4597–4602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zedelius J, et al. 2011. Alkane degradation under anoxic conditions by a nitrate-reducing bacterium with possible involvement of the electron acceptor in substrate activation. Environ. Microbiol. Rep. 3:125–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.