Abstract
An assay to identify the common food-borne pathogens Salmonella, Escherichia coli, Shigella, and Listeria monocytogenes was developed in collaboration with Ibis Biosciences (a division of Abbott Molecular) for the Plex-ID biosensor system, a platform that uses electrospray ionization mass spectroscopy (ESI-MS) to detect the base composition of short PCR amplicons. The new food-borne pathogen (FBP) plate has been experimentally designed using four gene segments for a total of eight amplicon targets. Initial work built a DNA base count database that contains more than 140 Salmonella enterica, 139 E. coli, 11 Shigella, and 36 Listeria patterns and 18 other Enterobacteriaceae organisms. This assay was tested to determine the scope of the assay's ability to detect and differentiate the enteric pathogens and to improve the reference database associated with the assay. More than 800 bacterial isolates of S. enterica, E. coli, and Shigella species were analyzed. Overall, 100% of S. enterica, 99% of E. coli, and 73% of Shigella spp. were detected using this assay. The assay was also able to identify 30% of the S. enterica serovars to the serovar level. To further characterize the assay, spiked food matrices and food samples collected during regulatory field work were also studied. While analysis of preenrichment media was inconsistent, identification of S. enterica from selective enrichment media resulted in serovar-level identifications for 8 of 10 regulatory samples. The results of this study suggest that this high-throughput method may be useful in clinical and regulatory laboratories testing for these pathogens.
INTRODUCTION
Mass spectrometry is an established analytical technique with growing applications within microbiology. With high sensitivity and high resolution, mass spectrometry can be used to differentiate microbial species based on subcellular variations. Recently, several articles concerning the application of either matrix-assisted laser desorption ionization (MALDI) (6, 14, 36) or electrospray ionization (ESI) mass spectrometry (MS) (13, 21, 30, 37) to the detection and identification of microbes have been published. While some methods examine protein expression, this work centers on the use of nucleic acid information to identify bacteria.
MS techniques involving the analysis of DNA take advantage of the difference in mass between strands with different base compositions. In order to utilize MS for DNA-based identification of bacteria, a region of DNA that varies between species or subspecies is amplified by PCR, and the mass of this amplicon is then determined. Since the exact masses of the individual bases in DNA are known, the quantity of each of these bases within the amplified sequence can be calculated based on the exact mass of the strand. While the exact sequence is not obtained through this method, the base compositions, or base counts, can provide enough information to discriminate between species, subspecies, and even serovars depending on the organism and the assay (15, 19).
This technique is comparable to other methods that differentiate between microbes using nucleic acid information, such as 16S rRNA gene sequencing. 16S sequencing is widely used for bacterial identification and classification, and a library of data has been amassed for reference and support (11). While DNA sequencing does provide actual sequence information that can be translated to labs using other instruments and methods, it is relatively time-consuming and requires a pure sample unless PCR products are cloned prior to sequencing. Although new sequencing technology can provide results from mixed cultures, sample preparation and data analysis remain time and resource intensive. PCR-MS can analyze samples containing mixtures of bacteria with minimal sample preparation. DNA can be extracted directly from the enrichment broth, thereby eliminating the need to isolate individual colonies, and results can be obtained from extracted DNA in under 5 h and from culture in well under 8 h. While PCR-MS is similar to real-time PCR in these respects, it has the advantage of being able to provide both breadth and depth in the identification of organisms. Real-time PCR methods can often detect multiple species or can provide subspecies characterization of one species, but few methods can do both in one assay (10, 25, 26, 28, 31).
PCR-MS has been successfully applied to clinical microbial characterization (2, 29, 33), and it would also be a welcome addition to other fields, including food safety. The U.S. Food and Drug Administration (FDA) analyzed 206,723 import food lines and 25,214 domestic food facilities in 2010 for signs of adulteration or mishandling of foods, and a multitude of samples were tested for microbial contamination as a result of these inspections (16). Samples analyzed for microbial contamination at FDA are processed using culturing techniques that take days to weeks to provide isolate confirmation (1). Traditional serology testing of Salmonella enterica, for example, takes place once an isolate is confirmed as S. enterica, and it can require up to an additional month to complete the characterization. High-throughput screening methods that could provide serotype information in just a few days, including time for the initial enrichment, would greatly enhance the ability of the agency to conduct real-time monitoring and outbreak investigations.
To this end, a PCR-MS assay for use on the Plex-ID biosensor system was developed with the aim of detecting and differentiating between Salmonella spp., Escherichia coli, Shigella spp., and Listeria monocytogenes. Along with identifying these four important food-borne pathogens (FBPs) to the species level, this assay was designed to simultaneously differentiate the six subspecies of S. enterica (I, II, IIIa/IIIb, IV, and VI) and provide some serotyping of S. enterica and E. coli. Despite the promise of this assay, it is heavily reliant on a reference database, and the current database requires the inclusion of numerous important serovars. Furthermore, the strength of this assay has yet to be tested with regulatory samples.
The goal of this work was to improve the size and scope of the database while assessing the detection and identification abilities of the assay. To enlarge the database, more than 800 pure culture isolates were analyzed with the assay. The isolates consisted of S. enterica, pathogenic E. coli, and Shigella species. These species are all in the Enterobacteriaceae family, are found in foods, and cause illness in humans. Listeria species isolates were not tested. Nontarget bacterial cultures were also analyzed to test the limit of assay specificity. In addition, several foods were spiked with S. enterica, and samples of the preenrichment broth were analyzed to determine the effects of food matrices and background microflora on assay performance. Finally, enrichment broth aliquots from regulatory samples positive for S. enterica were examined to compare results obtained by PCR-MS to those obtained with serological identification methods.
MATERIALS AND METHODS
Bacterial strains and template preparation.
S. enterica isolates (n = 437) were taken from the collection at the U.S. FDA Pacific Regional Lab-Southwest (PRL-SW) in Irvine, CA. These cultures had previously been serotyped at FDA laboratories in Arkansas and Colorado and were held at −70°C in motility test media. The cultures were thawed, plated on Trypticase soy agar (TSA), and incubated for 18 to 24 h at 37°C. Two methods were used for DNA extraction of the isolated colonies. A boiling method described previously was used for the extraction of most isolates (9). An automated method was also employed for some samples, as follows. The PrepSEQ nucleic acid extraction kit (Life Technologies, Foster City, CA) was used on the MagMax sample-handling system (Life Technologies) according to the manufacturer's instructions, with two exceptions: rather than using the proteinase K in the kit, the samples were heated at 95°C for 20 min at the start of extraction. In addition, the elution volume was reduced to 150 μl. Both methods provided suitable DNA for PCR-MS analysis. During testing, two isolates were determined not to be S. enterica and removed from further data analysis. This was confirmed via Vitek 2 GN cards or API 20E strips (bioMérieux, Durham, NC) and a real-time PCR method (8, 9). In addition, two isolates identified by serology as subspecies II and one identified as subspecies IV were reclassified as subspecies I based on 16S sequencing results obtained with a MicroSEQ 500 16S rRNA gene bacterial identification kit (Applied Biosystems, Foster City, CA) combined with a 3500xl genetic analyzer (Applied Biosystems). These isolates were also excluded from the results described here.
E. coli isolates (n = 234) were obtained from the Orange County Public Health Lab (OCPHL) in Santa Ana, CA, and the U.S. FDA Center for Food Safety and Applied Nutrition (CFSAN). Shigella species (n = 207) isolates were obtained from OCPHL, CFSAN, the Los Angeles County Public Health Lab (LACPHL), and the American Type Culture Collection (ATCC). Serology for E. coli was performed at the originating labs. Species-level identifications of Shigella species isolates were obtained by real-time PCR and/or microarray analysis. DNA was extracted using the boiling method described by Cheng et al. (8).
Nontarget bacterial strains analyzed for exclusivity were obtained from ATCC. These included Enterobacter aerogenes ATCC 13048, Klebsiella pneumoniae ATCC 13883, Pseudomonas aeruginosa ATCC 9027, P. aeruginosa ATCC 27853, Proteus hauseri ATCC 13315, Rhodococcus equi ATCC 6939, Staphylococcus aureus ATCC 6538, S. aureus ATCC 25923, and Staphylococcus epidermidis ATCC 14990. In addition, two Citrobacter freundii isolates were obtained from the culture collection at PRL-SW. The control cultures were plated on TSA and incubated for 24 h at 37°C, and DNA was extracted via the boiling protocol used for S. enterica isolates.
Template preparation for regulatory samples.
Regulatory samples were analyzed to determine the ability of the instrument to detect the target organisms in food samples containing normal background microflora. As part of the microbiological workflow at the FDA, samples are incubated in a preenrichment broth for 24 h, followed by selective enrichment in both Rappaport-Vassiliadis (RV) medium and tetrathionate (TT) broth (1). After preenrichment, a 1-ml aliquot of the preenrichment broth was set aside and held either at −70°C or 4°C. A 1-ml aliquot was also taken following selective enrichment with RV and TT broths. The aliquots from RV/TT broths were combined in a 1:1 ratio for each sample and held either at −70°C or 4°C. If samples were found to be presumptive positive for S. enterica using the Vidas assay (bioMérieux), the enrichment broths were frozen at −70°C. Broths were held at 4°C for a maximum of 1 week. Prior to PCR-MS, the broths were thawed and DNA was extracted directly from the broths using a modified boiling protocol (9). No enumeration was performed on regulatory samples.
Template preparation for spiked food matrices.
Food matrices spiked with S. enterica were also analyzed with PCR-MS. S. enterica was spiked at two levels: 2 to 3.8 CFU/25 g of food and 20 to 38 CFU/25 g of food. Samples were spiked in replicates of six subsamples for each level. Other samples were unspiked, and the samples were all blinded. The spiking levels were chosen to be consistent with other evaluations of rapid detection methods (12, 32, 34). After PCR-MS results were obtained and interpreted, the identities of the spiked samples were revealed. The foods and their unspiked controls were sampled and incubated as described in the Bacteriological Analytical Manual (BAM) (1) except that the preenrichment broth for all foods was modified buffered peptone water (mBPW). For these samples, only the mBPW preenrichment broth and not the RV or TT enrichment broth was analyzed by PCR-MS. The boiling protocol described above for regulatory samples was used to extract DNA from the mBPW postincubation.
FBP plate design.
PCR primer sets that were capable of amplifying all known species, subspecies, and serotypes of Salmonella and their close relatives in E. coli and Shigella were developed. Primers were chosen that would be able to distinguish essential molecular lineages using base composition signatures. The Gram-negative enteric primer designs were based upon analysis of previous sequence alignments of the mutS and mdh genes across a large selection of Salmonella diversity (4). A surveillance panel of eight primer pairs, comprising three Gram-negative enteric primer pairs targeting variable mutS gene fragments and three primer pairs targeting variable mdh gene fragments, was selected. Two additional gene targets, to detect L. monocytogenes, were designed, the invasion-associated secreted endopeptidase gene (iap) for the p60 protein gene (27) and the prfA gene, which positively regulates the expression of listeriolysin for use in the identification of Listeria samples (35) (Fig. 1). The eight sets of forward/reverse primer sequences (1 set in each well) are listed in Table 1. All primers used in this study had a thymine nucleotide at the 5′ end to minimize addition of nontemplated adenosines during amplification using Taq polymerase (5). The sensitivity of each PCR primer pair was determined using known quantities of a synthetic calibrant DNA template as described previously (22). Each of the primer pairs was sensitive to as little as 20 copies of the calibrant DNA, and several primers were sensitive to 5 copies. Additionally, an ultraclean DNA polymerase, Immolase, was used for amplification due to the ability of these primers to pick up residual E. coli DNA commonly found in some commercial preparations of polymerase (data not shown). The food-borne pathogen (FBP) plate comes preloaded with 0.2 mM deoxynucleoside triphosphates (dNTPs), 0.5 μM PCR primers, and 100 copies of calibrant in each well. The DNA base count database to which sample results are matched includes more than 140 Salmonella enterica, 139 E. coli, 11 Shigella, and 36 Listeria patterns and 18 other Enterobacteriaceae organisms.
Fig 1.
Layout of the FBP assay plate. The 96-well plate can accommodate 12 samples.
Table 1.
Primer sequences on FBP plate
| Well | Primer (5′–3′) |
|
|---|---|---|
| Forward | Reverse | |
| 1 | TAT CAC CGA AGG TCG CCA CCC | TCG CAT ATA GGT ACT TTT ACC GCC CA |
| 2 | TGG GCG GTA AAA GTA CCT ATA TGC G | TCCGACGCGGGTAAAAATACGGTC |
| 3 | TAT TTT TAC CCG CGT CGG CGC AGC | TAC CGC GTC CGA TTT CAT CCA TCA |
| 4 | TAC GAC AAA AAC AAA CTG TTT GGC GT | TCA GAA TAG TCA CGC CGG AGT G |
| 5 | TCG GAT CGG CAA CCC TTT CTA TG | TGA GAG AAG AAG CGG GCA TAC TG |
| 6 | TGC ACT TGA AGG CGC TGA CG | TCG GGC AGG TTT TAG CAA TCT G |
| 7 | TCA ATG GGA GCC ACA CGA ATA TTG T | TGA AAG CGC CTT TGT AGT ATT GTA AAT TCA |
| 8 | TCG TGG AAT AAT TTA TCT GCT TCT TCT ATT TAT GT | TTG TTT TTC AGC TGC TGG AGC TT |
PCR/ESI-MS.
After DNA extraction, template DNA was diluted 1:10 with PCR-grade water. The template was distributed onto the FBP assay plate (Abbott Molecular, Abbott Park, IL) using an automated liquid handler (Abbott Molecular). The liquid handler added 5 μl of diluted template DNA and 1 U of Immolase DNA polymerase in PCR buffer to each well of the plate. The plate was then heat sealed with foil on a Thermo Scientific Alps microplate heat sealer (Rockford, IL). Each sealed plate was loaded onto a Mastercycler Pro thermocycler (Eppendorf, Hauppauge, NY) and PCR amplified under the following conditions: 95°C for 10 min, and then 8 cycles of 95°C for 30 s, 48°C for 30 s, and 72°C for 30 s, followed by 37 cycles of 95°C for 15 s, 56°C for 20 s, and 72°C for 20 s, followed by 72°C for 2 min, and then 99°C for 20 min. The plate was then loaded onto the Plex-ID system (Abbott Molecular) for amplicon desalting and analysis. The Plex-ID system has a desalting carousel and a dual-sprayer ESI quadrupole time-of-flight mass spectrometer. Analysis of each plate took about an hour.
Data analysis.
The Plex-ID instrument software analyzed the spectra for each sample and determined the base counts of the peaks in the spectra. For the software to confirm a specific base count, both cDNA strands had to be detected. Spectra were manually analyzed to confirm the results. The software then compared these base counts to known organisms in the Plex-ID FBP database containing base compositions from more than 700 validated isolates from across a diversity of genera. To consider an isolate as validated, the identity must have been confirmed using an alternative to standard serology, including pulsed-field gel electrophoresis (PFGE), Vitek (bioMérieux), molecular serotyping, multilocus sequence typing (MLST), or whole-genome sequencing.
For the S. enterica isolates studied, identifications made by PCR-MS were compared to those made by serology. If the serovar determined by serology was one of the top identifications in the software, the serology and PCR-MS results were called concordant for that isolate. It should be noted that for some serovars, the top identification resulted in an identical match to more than one serovar. If the correct serovar was included in this group, the isolate was still called concordant. If all isolates with the same serovar were found to be concordant, the serovar was considered concordant. In some cases, different isolates with the same serovar had different PCR-MS identifications. For these serovars, if more than one isolate was found to be concordant the serovar was described as semiconcordant. Isolates that did not match serological results were described as discordant. E. coli isolates were compared to serological results in a manner similar to that for the S. enterica isolates; however, the isolates were compared only at the serogroup level (O antigens) rather than the serovar level (O and H antigens).
RESULTS
Common disease-causing S. enterica.
While there are more than 2,500 serovars of S. enterica, 85% of human illness caused by Salmonella is associated with only 40 serovars (Table 2) (7). Of those 40 serovars, 32 were represented in the collection at PRL-SW and were analyzed on the assay plate (n = 178 isolates). Up to nine isolates of each serovar were analyzed. For 18 of the 32 serovars, all isolates had identifications concordant with serology. An additional 10 serovars were semiconcordant. For the semiconcordant serovars, discordant isolates often had single nucleotide polymorphisms (SNPs) or differences in one or two amplicons (out of six) that resulted in a different identification. Only 4 of the top 40 serovars tested did not have concordant identifications for any isolates. Some of the discordant serovars, such as Hartford, appear to be missing from the database, while others, like Mbandaka, do not match the database strains. A full list of serovars tested can be found in Table S1 in the supplemental material.
Table 2.
The 40 most common disease-causing S. enterica serovars from the United States in 1996 to 2006a
| Rankb | % human disease | S. enterica serovarc | Concordanced |
|---|---|---|---|
| 1 | 19.2 | Typhimurium (+1) | Concordant |
| 2 | 17.8 | Enteritidis | Semiconcordant |
| 3 | 8.4 | Newport | Concordant |
| 4 | 5.2 | Heidelberg | Concordant |
| 5 | 3.4 | Javiana (+1) | Concordant |
| 6 | 2.4 | Typhimurium var. 5- (+1) | Concordant |
| 7 | 2.4 | Montevideo (+3) | Concordant |
| 8 | 2.0 | Muenchen (+1) | Concordant |
| 9 | 1.7 | Oranienburg | Semiconcordant |
| 10 | 1.6 | Saintpaul | Semiconcordant |
| 11 | 1.5 | Infantis | Semiconcordant |
| 12 | 1.5 | Thompson | Semiconcordant |
| 13 | 1.5 | Braenderup | Concordant |
| 14 | 1.5 | Agona (+1) | Concordant |
| 16 | 1.1 | Hadar (+1) | Concordant |
| 19 | 1.0 | Paratyphi B var. Java (+1) | Concordant |
| 20 | 0.8 | Poona | Concordant |
| 22 | 0.6 | Stanley (+1) | Concordant |
| 23 | 0.6 | Anatum (+1) | Concordant |
| 24 | 0.5 | Bareilly | Concordant |
| 25 | 0.5 | Mbandaka | Discordant |
| 26 | 0.5 | Paratyphi B | Semiconcordant |
| 27 | 0.5 | Hartford | Discordant |
| 28 | 0.4 | Panama | Semiconcordant |
| 29 | 0.4 | Derby | Semiconcordant |
| 30 | 0.4 | Litchfield | Discordant |
| 31 | 0.4 | Schwarzengrund | Semiconcordant |
| 32 | 0.4 | Senftenberg (+10) | Concordant |
| 33 | 0.4 | Brandenburg (+10) | Concordant |
| 34 | 0.3 | Sandiego | Discordant |
| 36 | 0.3 | Give | Semiconcordant |
| 38 | 0.2 | Rubislaw | Semiconcordant |
See reference 7.
The following serovars, with their corresponding ranks, were not available for analysis: rank 15, I 4,[5],12:i:-; rank 17, Mississippi; rank 18, Typhi; rank 21, Berta; rank 35, Paratyphi A; rank 37, Reading; rank 39, Norwich; and rank 40, Miami.
Numbers in parentheses are the number of other serovars identified at the top confidence level along with the concordant serovar.
Concordant, the serology and PCR-MS results matched for all isolates; semiconcordant, the serology and PCR-MS results matched for some but not all isolates; discordant, the serology and PCR-MS results did not match for any isolates.
Less-common S. enterica subspecies I isolates.
To determine the range of the identification capabilities of the assay, a collection of Salmonella serovars that are uncommon agents of human disease was also investigated. The isolates (n = 232) were from 116 different subspecies I serovars. While all of these serovars were isolated from food samples, not all of them have been implicated in human illness. Although these isolates are less clinically relevant than those discussed above, detecting and identifying these S. enterica bacteria is an important food safety effort since these serovars still have the potential to cause illness and may be agents in future outbreaks.
Of the 116 non-top-40 serovars tested, only 8 were concordant. An additional 4 serovars were semiconcordant, and the remaining 104 serovars were discordant. While the results were discordant at the serovar level, all isolates but one were classified as subspecies I, indicating that the assay can identify to the subspecies level well. The rate of discordance is strikingly different from the results obtained with clinically common S. enterica. This discrepancy highlights the large number of validated isolates already populating the Plex-ID database for the top 40 Salmonella serotypes causing food-borne illness and the need for a stronger reference database for many of the remaining serovars. Adding the isolates under investigation herein to the database should improve serovar-level identification for S. enterica serovars that are found in foods yet might not have been implicated in prior human illness.
S. enterica subspecies II to VI.
S. enterica from five other subspecies were also investigated with this technique (n = 22): subspecies II (n = 4), subspecies IIIa (n = 2), subspecies IIIb (n = 7), subspecies IV (n = 8), and subspecies VI (n = 1). While subspecies IV bacteria occasionally cause human illness—often through handling of reptiles and other carrier animals—S. enterica from other subspecies are infrequent disease agents. While the assay was not able to provide serovar-level identifications with these isolates, it was capable of providing resolution at the subspecies level. All isolates tested were concordant to the subspecies identifications (data not shown). Although the sample size is small, the results show that the assay is capable of classifying all six subspecies.
Since the majority of serovars analyzed were uncommon and were not found in the database, the rate of concordance was low when all S. enterica results are considered (n = 432 isolates). In total, 108 serovars were discordant, 14 serovars were semiconcordant, and 31 serovars were fully concordant with conventional serology. As discussed above, the difference in rates of concordance between common and uncommon S. enterica serovars suggests that increasing the database size will improve the serovar-level identifications.
Diarrheagenic E. coli isolates.
The PRL-SW E. coli isolate collection included 11 enteropathogenic E. coli, 33 general diarrheagenic E. coli, 72 non-O157 shiga-toxin producing E. coli, and 112 O157 enterohemorrhagic E. coli isolates (Table 3). Of the 228 isolates analyzed, 226 were identified by this assay. The majority of isolates in the collection have the serotype O157:H7 or O157:H unknown. The O157:H7 isolates (n = 66) were all concordantly identified. Among the O157:H unknown isolates, one was not identified by the instrument and the remaining isolates (n = 45) were identified as O157:H7. E. coli O55:H7 (n = 7), an evolutionary precursor to E. coli O157:H7, could not be separated from E. coli O157:H7 with this assay and showed equivalent matches to O55:H7 and O157:H7.
Table 3.
PCR-MS identifications for diarrheagenic E. coli isolates
| Serotype(s) (na) | No. of isolates analyzed | PCR-MS IDb |
|---|---|---|
| O157:H7 (66) | 66 | O157:H7 (and O55:H7) |
| O157:H unknown (46) | 45 | O157:H7 (and O55:H7) |
| 1 | Unidentified | |
| O55:H7 (7) | 7 | O55:H7 (and O157:H7) |
| O45:H2 or O45:HND (5) | 5 | O45:H2 (and O103, O111, O121) |
| O26:H11 or O26:HND (32) | 11 | Shigella spp., O111 |
| 10 | O26:H2 and O26:H11 | |
| 6 | O26:H11 and O111 | |
| 4 | Others | |
| 1 | Unidentified | |
| O103:H2, O103:H6, O103:H(25) (11) | 6 | O103 (and O45, O111, O139, O128, O91) |
| 4 | O103, O45 | |
| 1 | O104:H21 | |
| O104:H4 (1) | 1 | O111/EAEC and Shigella spp. |
| O111:H8, O111:H11, O111:HNM (47) | 17 | O26:H2 and O26:H11 |
| 8 | O103 | |
| 8 | O111 (and O26, O103) | |
| 5 | O113:H21 | |
| 5 | O111:H8, S. flexneri | |
| 4 | O45 | |
| O113:H21 (1) | 1 | O113:H21 |
| O118:H16 (1) | 1 | O26:H2 and O26:H11 |
| O121:H19, O121:HND (5) | 1 | O121 (and O45, O143) |
| 1 | O26:H2 and O26:H11 | |
| 1 | O157:not H7 | |
| 2 | E. coli (serotype unknown) | |
| O143:H4 (1) | 1 | O138 |
| O145:H(25), O145:HND (6) | 3 | O157:H7 |
| 1 | O145:HND | |
| 1 | O111 | |
| 1 | E. coli (serotype unknown) |
n, total no. of isolates analyzed.
Bold results indicate PCR-MS identification (ID) was concordant with serology at the serogroup level (O antigen).
The results for non-O157 diarrheagenic E. coli are included in Table 3. While isolates of several serogroups were concordant at the serogroup level, isolates from other serogroups were inconsistently identified. For example, all five O45 isolates and 10 of 11 O103 isolates were concordant at the serogroup level. However, identifications of only 16 of 32 O26 isolates and 12 of 47 O111 isolates were concordant with serology.
Shigella species and EIEC isolates.
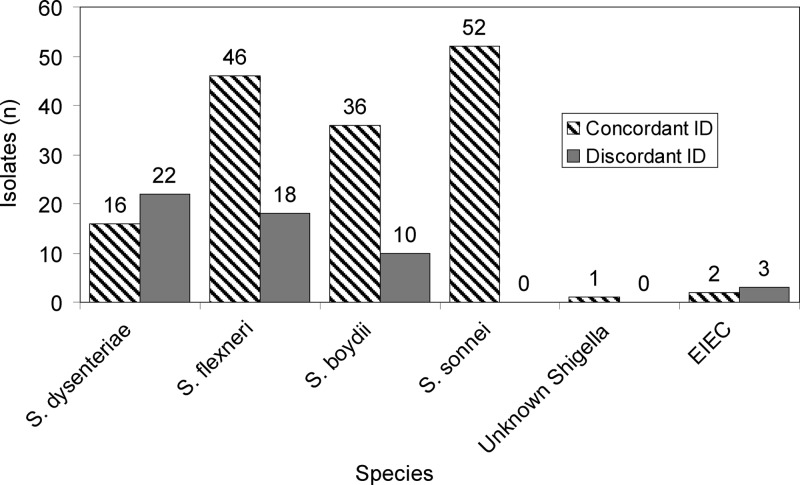
Shigella species isolates obtained from local public health laboratories as well as national collections at FDA/CFSAN and ATCC were analyzed using PCR-MS (n = 201) (Fig. 2). Shigella sonnei isolates had the strongest correlation between species identification data and PCR-MS results: all 52 isolates were concordant. Identification of other Shigella spp. was less successful. Of 64 Shigella flexneri isolates, 46 were concordant at the species level, and 36 of 46 Shigella boydii isolates were concordant. Only 16 of 38 Shigella dysenteriae isolates were concordant, with discordant isolates being identified as E. coli or other Shigella species. One Shigella isolate, the species of which could not be determined by other methods, was identified as either S. flexneri or S. boydii by PCR-MS, but that identification was also observed with several S. dysenteriae isolates, thereby preventing a conclusive species identification. Shigella species isolates that were not identified to the species level were frequently identified as various serotypes of E. coli. One isolate of S. boydii was identified as Escherichia albertii. E. albertii, which causes diarrhea, is a recent addition to the Escherichia genus and has been shown to be closely related to several strains of S. boydii (23).
Fig 2.
Chart of PCR-MS identifications for Shigella species and EIEC isolates.
The pathogenic E. coli subgroup that is the most closely related to Shigella is enteroinvasive E. coli (EIEC). EIEC and Shigella spp. cause similar illnesses in humans. Four of five EIEC isolates analyzed were identified as E. coli. The fifth isolate was identified as S. dysenteriae. Only one of the five isolates, an EIEC isolate from the O111 serogroup, was concordant at the serogroup level. Two O124 EIEC isolates were identified as belonging to the O78 serogroup, while the final isolate was from an unknown serogroup.
S. enterica in spiked food samples.
In addition to bacterial isolates, DNA from the preenrichment broth of 72 blind food samples spiked with S. enterica was analyzed by PCR-MS (Table 4). Results from spiked tomato and chili powder samples were relatively poor. Only 4 of the 12 spiked chili powder samples and 6 of the 12 spiked tomato samples were identified with high confidence as S. enterica. While all 4 of the chili powder S. enterica identifications listed the correct serovar, Weltevreden, only 4 of the 6 tomato S. enterica identifications listed the correct serovar (i.e., Newport). Cheese samples spiked with S. enterica serovar Typhimurium were more promising: the serovar was identified with high confidence in all 12 of the spiked samples. S. enterica was also identified in all 12 spiked fish samples; however, the serovar used for spiking, Senftenberg, is one in a cluster of more than 15 serovars that cannot be distinguished in this assay. In general, unspiked foods resulted in no S. enterica identification. However, 5 of the 24 controls did have low-quality matches to S. enterica in which only one of six primer pairs matched. While not all of the spiked food samples were well characterized by PCR-MS, results from the regulatory samples described below indicate that results from the tomato and chili powder samples may have improved if the selective enrichments rather than the preenrichments were analyzed.
Table 4.
Results from PCR-MS analysis of spiked foodsa
| Food | Spike (CFU/25 g) | S. enterica serovar | PCR-MS resultsb |
|---|---|---|---|
| Chili powder | 3 | Weltevreden | |
| 3 | Weltevreden | LQ | |
| 3 | Weltevreden | LQ | |
| 3 | Weltevreden | LQ | |
| 3 | Weltevreden | LQ | |
| 3 | Weltevreden | LQ | |
| 30 | Weltevreden | LQ | |
| 30 | Weltevreden | LQ | |
| 30 | Weltevreden | Weltevreden (+8) | |
| 30 | Weltevreden | Weltevreden (+8) | |
| 30 | Weltevreden | Weltevreden (+2) | |
| 30 | Weltevreden | Weltevreden (+2) | |
| Tomato | 2.4 | Newport | |
| 2.4 | Newport | ||
| 2.4 | Newport | ||
| 2.4 | Newport | LQ | |
| 2.4 | Newport | Mgulani (+1) | |
| 2.4 | Newport | Newport (+7) | |
| 24 | Newport | ||
| 24 | Newport | ||
| 24 | Newport | Telaviv (+2) | |
| 24 | Newport | Newport (+1) | |
| 24 | Newport | Newport | |
| 24 | Newport | Newport | |
| Soft cheese | 3.8 | Typhimurium | Typhimurium (+1) |
| 3.8 | Typhimurium | Typhimurium (+1) | |
| 3.8 | Typhimurium | Typhimurium (+1) | |
| 3.8 | Typhimurium | Typhimurium (+1) | |
| 3.8 | Typhimurium | Typhimurium (+1) | |
| 3.8 | Typhimurium | Typhimurium (+1) | |
| 38 | Typhimurium | Typhimurium (+1) | |
| 38 | Typhimurium | Typhimurium (+1) | |
| 38 | Typhimurium | Typhimurium (+1) | |
| 38 | Typhimurium | Typhimurium (+1) | |
| 38 | Typhimurium | Typhimurium (+1) | |
| 38 | Typhimurium | Typhimurium (+1) | |
| Fish | 2 | Senftenberg | LQ |
| 2 | Senftenberg | LQ | |
| 2 | Senftenberg | Butantan | |
| 2 | Senftenberg | Senftenberg (+22) | |
| 2 | Senftenberg | Senftenberg (+16) | |
| 2 | Senftenberg | Senftenberg (+15) | |
| 20 | Senftenberg | Senftenberg (+16) | |
| 20 | Senftenberg | Senftenberg (+16) | |
| 20 | Senftenberg | Senftenberg (+15) | |
| 20 | Senftenberg | Senftenberg (+15) | |
| 20 | Senftenberg | Senftenberg (+15) | |
| 20 | Senftenberg | Senftenberg (+15) |
Only the preenrichment broth (mBPW) was tested for each sample. Results from unspiked samples are not shown.
Values in parentheses indicate the number of other serovars listed at the same confidence level. LQ, low-quality match to S. enterica (match in 1 of 6 wells).
S. enterica in regulatory samples.
Aliquots of preenrichment and selective enrichment broths were taken from food samples during the normal regulatory workflow. Only samples that were confirmed to be positive for S. enterica were analyzed for this project. Results were poor when analyzing the preenrichment broth (n = 16) (Table 5). While identifications were made from several samples, they were of low quality, with amplification at 2 of 6 primer sites or less.
Table 5.
Comparison of serology and PCR-MS results for regulatory food samples
| Samplea | Mediumb | Serology result | PCR-MS resultsc,e | PCR-MS/PRLSWd |
|---|---|---|---|---|
| Anise seeds I | TSB | Montevideo | No ID | No ID |
| Anise seeds II | TSB | Montevideo | Montevideo (+3 others) (2 of 6) | No ID |
| Fennel seeds I | TSB | Emek | Poona (1 of 6) | No ID |
| Fennel seeds II | TSB | Emek | II/IIIa/Chester (1 of 6) | Poona/II/IIIa/Sandiego |
| Papaya I | LB | Duesseldorf | No ID | No ID |
| Papaya II | LB | Mbandaka | No ID | No ID |
| Papaya III | LB | Mbandaka | No ID | No ID |
| Papaya IV | LB | Untypeable C2 | No ID | No ID |
| Papaya V | LB | Typhimurium | No ID | No ID |
| Papaya VI | LB | Mbandaka | No ID | No ID |
| Papaya VII | LB | Meleagridis | No ID | No ID |
| Shrimp I | LB | Agona | No ID | No ID |
| Silverfish | LB | Fulica | No ID | No ID |
| Tuna I | LB | Weltevreden | No ID | No ID |
| Tuna II | LB | Monophasic C2 | No ID | No ID |
| Tilapia I | LB | Albany | No ID | No ID |
| Papaya V | RV/TT | Typhimurium | Typhimurium/Typhimurium 5- | Typhimurium/Typhimurium 5- |
| Papaya VI | RV/TT | Mbandaka | Senftenberg (4 of 6) | Mbandaka |
| Papaya VII | RV/TT | Meleagridis | Pullorum/Enteritidis | Meleagridis |
| Shrimp II | RV/TT | Weltevreden | Mbandaka/Virchow | Weltevreden/Virchow |
| Shrimp III | RV/TT | Weltevreden | Mbandaka/Virchow | Weltevreden/Virchow |
| Silverfish | RV/TT | Fulica | Salamae (5 of 6) | Salamae (5 of 6) |
| Tuna I | RV/TT | Weltevreden | No ID | No ID |
| Tuna II | RV/TT | Monophasic C2 | Hadar/Blockley | Hadar (+4 others) |
| Tilapia II | RV/TT | Tennessee | Agona/Lac - | Tennessee |
| Tilapia III | RV/TT | Enteritidis | Enteritidis (+3 others) | Enteritidis/Thompson |
Samples in gray shading were extracted from preenrichment media.
TSB, Trypticase soy broth; LB, lactose broth; RV/TT, Rappaport-Vassiliadis medium and tetrathionate medium.
Results refer to identifications (IDs) made from the database.
Results refer to identifications made by comparing the base counts for the samples to those for the S. enterica isolates studied in this work.
Number of amplicons out of six that match the listed serotype is shown.
Results were more successful when the selective enrichments (n = 10) were analyzed. Nine samples showed identifications of S. enterica at a high confidence level. In two of these cases, the identification in the PCR-MS database matched the identification made by serology. In five additional cases, comparing the samples to an external database containing base count signatures from isolates studied in this work resulted in a concordant identification. An additional S. enterica isolate from silverfish was identified as a rare subspecies I serovar, Fulica, by serology. This bacterium had no perfect match in the FBP database or among the isolates studied for this work, but the closest result was a subspecies II isolate. The final sample was identified by serology as a monophasic C2 serovar. The PCR-MS identification was Hadar or Blockley, each of which is a C2 serovar. When the results for the isolates studied at PRL-SW are included, the monophasic C2 isolate matched five serovars which are all in the O:8 serogroup (either C2 or C3). While neither PCR-MS nor serotyping could identify a serovar for this isolate, they are concordant in the serogroup identification. Among the six regulatory samples for which both the preenrichment and enrichment broths were analyzed, S. enterica was identified in five of the enrichment broths but not in any of the preenrichment broths.
Nontarget bacteria.
To determine the selectivity of the assay, bacterial isolates other than S. enterica, E. coli, or Shigella spp. were analyzed (n = 11). This set included enteric bacteria, as well as other Gram-negative and Gram-positive bacteria. Ideally, any nontarget bacteria would not amplify during PCR in order to limit the effects of PCR inhibition. However, since the PCR primers were designed to broadly target enteric bacteria, some nontarget bacteria will also be amplified. When this occurs, the nontarget bacteria should not be confused with the target organisms. To help prevent false positives in the case of nontarget amplification, several common nontarget bacteria are present in the database.
As expected, most Gram-negative bacteria amplified and were detected by the instrument (Table 6). E. aerogenes, K. pneumoniae, and P. aeruginosa all amplified in at least one well and were identified to the species level. P. hauseri, R. equi, S. epidermidis, and two strains of S. aureus did not amplify. The results with P. hauseri are surprising given that it is a Gram-negative enteric bacterium. Two C. freundii isolates were identified as nonpathogenic E. coli.
Table 6.
PCR-MS results for nontarget bacteriaa
| Species (including strain) | PCR-MS IDb |
|---|---|
| C. freundii | E. coli |
| C. freundii | E. coli |
| E. aerogenes 13048 | E. aerogenes |
| K. pneumoniae 13883 | K. pneumoniae |
| P. aeruginosa 9027 | P. aeruginosa |
| P. aeruginosa 27853 | P. aeruginosa |
| P. hauseri 13315 | No detection |
| R. equi 6939 | No detection |
| S. aureus 6538 | No detection |
| S. aureus 25923 | No detection |
| S. epidermidis 14990 | No detection |
Bold font indicates a concordant identification at the species level.
Identification by PCR-MS.
DISCUSSION
Recent advances in ESI-MS using mass spectrometers have enabled analysis of PCR amplicons with sufficient mass accuracy that the nucleotide base composition (the A, G, C, and T count) of the PCR amplicon can be unambiguously determined for 120- to 140-bp fragments (15). The measured base compositions allow identification of the bacterial species with a high degree of resolution. Broad-range primers targeted to highly conserved sites within Salmonella, E. coli, Shigella, and Listeria which flank highly variable, information-rich regions were used to amplify DNA from these bacteria and their close relatives out of mixed samples in the same assay. For this FBP plate, 8 primer pairs, each targeting the variable and informative gene segments within mutS and mdh (for Gram-negative enterics) and prfA and iap (for Listeria) where the resultant base compositions provide species, subspecies, and some serotype information, were designed. While the E. coli serotype O157:H7 is the most well known type of shiga-toxin-producing E. coli (STEC), E. coli bacteria with other serotypes are increasingly found to produce toxins and cause illness (17, 20, 24). Thus, the ability to determine E. coli serotypes is a growing need for many labs. Shigella bacteria are genetically similar organisms to E. coli (3, 18), and the genetic resemblance can cause misidentification of Shigella as E. coli, especially in DNA-based testing schemes.
The plate was able to detect and identify the target organisms with various degrees of success. In analyzing pure samples, S. enterica was detected 100% of the time, and 30% of the serovars had identifications concordant with serology (95% confidence interval [CI], 23 to 37%). E. coli was detected 99% of the time and was concordant to the serogroup level for 72% of the isolates (95% CI, 65 to 77%), although the majority of those isolates were from the O157 serogroup. Since many Shigella spp. were identified as E. coli, the Shigella species isolates were detected as Shigella 81% of the time and were identified to the species level 73% of the time (95% CI, 66 to 78%).
The results of this study indicate that expanding the current reference database will enhance the potential of this assay to provide serovar-level information for S. enterica and should enhance the detection of E. coli and Shigella spp. Of 432 S. enterica isolates tested, 181 did not match an entry in the reference database at all six primer sites. These nonmatching isolates had serovar identifications that were concordant with serology only 9% of the time. In comparison, of the 251 isolates that did match a database entry at all six primer pairs, 69% had identifications concordant with serology. Most of the isolates that did not match a database entry at all six sites still had PCR products in all six wells. The presence of PCR amplicons in wells that were not used for the identification indicates that these isolates have the potential to be uniquely identified once their base counts are added to the database.
The results for E. coli isolates did not always match serology. While E. coli O157:H7 was reliably identified, the assay inconsistently determined the serogroups of non-O157 pathogenic E. coli. Similarly, Shigella spp. were frequently identified as E. coli. Improving the size of the reference database with regard to E. coli and Shigella spp. will likely improve the detection and identification abilities of the assay. However, E. coli and Shigella spp. amplify only in five or fewer wells in this assay, while S. enterica amplifies in six. It is possible that this will limit the ability to improve differentiation of E. coli and Shigella spp.
When analyzing the regulatory and spiked food samples, it was clear that extracting DNA from preenrichment medium frequently resulted in no or low-quality identifications. Analyzing samples from selective enrichment medium was more successful. While the difficulty in identifying S. enterica in preenrichment medium does necessitate an additional day to incubate in selective medium, it indicates that the S. enterica detected are viable. Since PCR techniques amplify DNA from both living and dead organisms, it can be difficult to determine whether the detected organisms are viable and capable of reproduction. The ability to detect S. enterica only after selective enrichment suggests that the bacteria replicated in this medium, an important distinction for regulatory action.
The results with nontarget bacteria indicate that the assay is selective: Gram-positive bacteria and some Gram-negative species do not amplify, and many of the bacteria that do amplify are identified correctly. One exception was C. freundii. C. freundii can biochemically and serologically resemble atypical lactose-fermenting S. enterica but is unlikely to be confused with E. coli, the organism identified via PCR-MS. The false-positive E. coli identification would be ruled out by further testing with procedures outlined in the BAM.
While this work greatly improves the body of knowledge for the use of the FBP assay for the identification of bacteria, there are several limitations present in this study. Perhaps the biggest limitation is the small size of the pure culture exclusivity panel. Ideally, many more relevant isolates from both Gram-positive and Gram-negative species would have been tested with the assay in order to provide an adequate assessment of the false-positive rate of the assay. However, the culture collection used in this work did not support further testing. In addition, the effects of the food matrix on sensitivity would be better understood by performing spiking experiments in a wider range of foods than was attempted here. Continued research, including single-lab and multilab validation studies, will provide the best opportunity to address these concerns.
Conclusions.
This work indicates that PCR-MS in general and this assay in particular could reduce the time necessary to detect and identify food-borne pathogenic bacteria. The assay was successful at detecting all S. enterica subspecies and some serovars. This was observed in pure culture testing and in the analysis of regulatory samples and spiked food matrices. Although it was common to observe more than one serovar listed as a possible identification, reducing the list from thousands down to a few possible serovars is very useful. That information could easily be used to narrow focus during an outbreak traceback procedure. This assay has the potential to significantly reduce the time necessary to characterize bacteria in food samples and greatly improve the response time for food-borne bacterial outbreaks.
Many of the noted deficiencies can be improved upon through expansion of the current reference library. In addition, planned changes to the assay plate design are expected to improve the ability of the assay to differentiate between closely related bacteria, such as Shigella and E. coli, and to improve the serotyping resolution. New primer pairs are designed to interrogate more regions of the S. enterica genome to improve discrimination power. For E. coli, added primers will provide information on virulence factors, including the ability to express Shiga toxins and hemolysin. Due to the extensive number of serovars, for Salmonella in particular, it is unlikely that all serovars will be differentiated on one assay plate. However, changes to the database and to the assay plate design may permit the identification of many of the bacterial pathogens important to the protection of the public health. To date, hundreds of known validated bacterial cultures have been run on the FBP plates in order to incorporate the variations found in these cultures into the database and to determine the specificity and discriminating power of these particular primer pairs. The addition of new genetic targets for better serotyping will improve the accuracy of calls from the instrument. Using genetic targets other than the O and H antigen genes overcomes the limitations of directly targeting these gene segments, which require specific primer pairs for each known type. Further, since the H and O antigen gene segments evolve rapidly, newly emerging strains might not be readily detected by the traditional approach if the primer regions differentiate.
Supplementary Material
ACKNOWLEDGMENTS
Funding was provided by FDA/CFSAN and the FDA Commissioner's Fellowship Program.
We thank the microbiology analysts at PRL-SW for providing sample aliquots. We are grateful to Erik Burrows and Jeffrey Blazar for insightful comments and editing, which improved the manuscript, and to Jim Bono, Yi Chen, Rachel Binet, Keith Lampel, Thomas Hammack, Peter Feng, and Chris Keys for access to strains used to populate the original base count database. We greatly appreciate the technical support provided by Erik Burrows, Melinda McFarland, and John Callahan and all of the team at Ibis Biosciences/Abbott Molecular, including James Hannis, Sheri Manalili, Heather Matthews, Roberta Housley, and Ranga Sampath, who jointly created and designed the primers and the database upon which this study rests.
Footnotes
Published ahead of print 21 September 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Andrews WH, Jacobson A, Hammack TS. 2011. Salmonella. In Bacteriological analytical manual. U.S. Food and Drug Administration, Washington, DC: http://www.fda.gov/Food/ScienceResearch/LaboratoryMethods/BacteriologicalAnalyticalManualBAM/default.htm [Google Scholar]
- 2. Blyn LB, et al. 2008. Rapid detection and molecular serotyping of adenovirus by use of PCR followed by electrospray ionization mass spectrometry. J. Clin. Microbiol. 46:644–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brenner DJ. 1984. Family I Enterobacteriaceae, p 408–420 In Krieg NR, Holt JG. (ed), Bergey's manual of systematic bacteriology, vol 1 Williams & Wilkins, Baltimore, MD [Google Scholar]
- 4. Brown EW, Kotewicz ML, Cebula TA. 2002. Detection of recombination among Salmonella enterica strains using the incongruence length difference test. Mol. Phylogenet. Evol. 24:102–120 [DOI] [PubMed] [Google Scholar]
- 5. Brownstein MJ, Carpten JD, Smith JR. 1996. Modulation of non-templated nucleotide addition by tag DNA polymerase: primer modifications that facilitate genotyping. Biotechniques 20:1004–1006, 1008–1010 [DOI] [PubMed] [Google Scholar]
- 6. Carbonnelle E, et al. 2011. MALDI-TOF mass spectrometry tools for bacterial identification in clinical microbiology laboratory. Clin. Biochem. 44:104–109 [DOI] [PubMed] [Google Scholar]
- 7. CDC 2008. Salmonella surveillance: annual summary 2006. U.S. Department of Health and Human Services, CDC, Atlanta, GA [Google Scholar]
- 8. Cheng CM, Van KT, Lin W, Ruby RM. 2009. Interlaboratory validation of a real-time PCR 24-hour rapid method for detection of Salmonella in foods. J. Food Prot. 72:945–951 [DOI] [PubMed] [Google Scholar]
- 9. Cheng CM, et al. 2008. Rapid detection of Salmonella in foods using real-time PCR. J. Food Prot. 71:2436–2441 [DOI] [PubMed] [Google Scholar]
- 10. Chiu CH, Ou JT. 1996. Rapid identification of Salmonella serovars in feces by specific detection of virulence genes, invA and spvC, by an enrichment broth culture-multiplex PCR combination assay. J. Clin. Microbiol. 34:2619–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Clarridge JE. 2004. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin. Microbiol. Rev. 17:840–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. de Freitas CG, et al. 2010. PCR multiplex for detection of Salmonella Enteritidis, Typhi and Typhimurium and occurrence in poultry meat. Int. J. Food Microbiol. 139:15–22 [DOI] [PubMed] [Google Scholar]
- 13. Deyde VM, et al. 2010. Genomic signature-based identification of Influenza A viruses using RT-PCR/electro-spray ionization mass spectrometry (ESI-MS) technology. PloS One 5:e13293 doi:10.1371/journal.pone.0013293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dieckmann R, Malorny B. 2011. Rapid screening of epidemiologically important Salmonella enterica subsp. enterica serovars by whole-cell matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl. Environ. Microbiol. 77:4136–4146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ecker DJ, et al. 2008. Ibis T5000: a universal biosensor approach for microbiology. Nat. Rev. Microbiol. 6:553–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Food and Drug Administration 2011. Annual report on food facilities, food imports, and FDA foreign offices: section 421(h)(1). Food and Drug Administration, Washington, DC [Google Scholar]
- 17. Franci T, Mariel Sanso A, Bustamante AV, Lucchesi PM, Parma AE. 2011. Genetic characterization of non-O157 verocytotoxigenic Escherichia coli isolated from raw beef products using multiple-locus variable-number tandem repeat analysis. Foodborne Pathog. Dis. 8:1019–1023 [DOI] [PubMed] [Google Scholar]
- 18. Fukushima M, Kakinuma K, Kawaguchi R. 2002. Phylogenetic analysis of Salmonella, Shigella, and Escherichia coli strains on the basis of the gyrB gene sequence. J. Clin. Microbiol. 40:2779–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Grant RJ, et al. 2010. Application of the Ibis-T5000 pan-orthopoxvirus assay to quantitatively detect monkeypox viral loads in clinical specimens from macaques experimentally infected with aerosolized monkeypox virus. Am. J. Trop. Med. Hyg. 82:318–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hadler JL, et al. 2011. Ten-year trends and risk factors for non-O157 shiga toxin-producing Escherichia coli found through shiga toxin testing, Connecticut, 2000–2009. Clin. Infect. Dis. 53:269–276 [DOI] [PubMed] [Google Scholar]
- 21. Hofstadler SA. 2007. High throughput characterization of amplified nucleic acids by ESI-TOF mass spectrometry: applications in pathogen detection and characterization. Abstr. 8th Int. Symp. Mass Spectrom. Health Life Sci., abstr 7.4, Mol. Cell. Proteomics, p S66 [Google Scholar]
- 22. Hofstadler SA, et al. 2005. TIGER: the universal biosensor. Int. J. Mass Spectrom. 242:23–41 [Google Scholar]
- 23. Hyma KE, et al. 2005. Evolutionary genetics of a new pathogenic Escherichia species: Escherichia albertii and related Shigella boydii strains. J. Bacteriol. 187:619–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Joris M, Pierard D, De Zutter L. 2011. Occurrence and virulence patterns of E. coli O26, O103, O111 and O145 in slaughter cattle. Vet. Microbiol. 151:418–421 [DOI] [PubMed] [Google Scholar]
- 25. Kim JS, et al. 2007. A novel multiplex PCR assay for rapid and simultaneous detection of five pathogenic bacteria: Escherichia coli O157:H7, Salmonella, Staphylococcus aureus, Listeria monocytogenes, and Vibrio parahaemolyticus. J. Food Prot. 70:1656–1662 [DOI] [PubMed] [Google Scholar]
- 26. Kim S, et al. 2006. Multiplex PCR-based method for identification of common clinical serotypes of Salmonella enterica subsp. enterica. J. Clin. Microbiol. 44:3608–3615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kohler S, Leimeisterwachter M, Chakraborty T, Lottspeich F, Goebel W. 1990. The gene coding for protein P60 of Listeria monocytogenes and its use as a specific probe for Listeria monocytogenes. Infect. Immun. 58:1943–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kong RYC, Dung WF, Vrijmoed LLP, Wu RSS. 1995. Co-detection of three species of water-borne bacteria by multiplex PCR. Mar. Pollut. Bull. 31:317–324 [Google Scholar]
- 29. Lamson D, et al. 2006. MassTag polymerase-chain-reaction detection of respiratory pathogens, including a new rhinovirus genotype, that caused influenza-like illness in New York State during 2004–2005. J. Infect. Dis. 194:1398–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee JK, et al. 2011. Detection and identification of human papillomavirus using a PCR-restriction fragment mass polymorphism assay. Mol. Med. Rep. 4:645–650 [DOI] [PubMed] [Google Scholar]
- 31. Li Y, Mustapha A. 2004. Simultaneous detection of Escherichia coli O157:H7, Salmonella, and Shigella in apple cider and produce by a multiplex PCR. J. Food Prot. 67:27–33 [DOI] [PubMed] [Google Scholar]
- 32. Malorny B, Huehn S, Dieckmann R, Kramer N, Helmuth R. 2009. Polymerase chain reaction for the rapid detection and serovar identification of Salmonella in food and feeding stuff. Food Anal. Methods 2:81–95 [Google Scholar]
- 33. Sharma VK, Vouros P, Glick J. 2011. Mass spectrometric based analysis, characterization and applications of circulating cell free DNA isolated from human body fluids. Int. J. Mass Spectrom. 304:172–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang LX, Li Y, Mustapha A. 2007. Rapid and simultaneous quantitation of Escherichia coli O157:H7, Salmonella, and Shigella in ground beef by multiplex real-time PCR and immunomagnetic separation. J. Food Prot. 70:1366–1372 [DOI] [PubMed] [Google Scholar]
- 35. Wernars K, et al. 1992. Suitability of the prfA gene, which encodes a regulator of virulence genes in Listeria monocytogenes, in the identification of pathogenic Listeria spp. Appl. Environ. Microbiol. 58:765–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Williams TL, Andrzejewski D, Lay JO, Musser SM. 2003. Experimental factors affecting the quality and reproducibility of MALDI TOF mass spectra obtained from whole bacteria cells. J. Am. Soc. Mass. Spectrom. 14:342–351 [DOI] [PubMed] [Google Scholar]
- 37. Zhang JI, et al. 2011. Rapid direct lipid profiling of bacteria using desorption electrospray ionization mass spectrometry. Int. J. Mass Spectrom. 301:37–44 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.