Abstract
The economical production of fuels and commodity chemicals from lignocellulose requires the utilization of both the cellulose and hemicellulose fractions. Xylanase enzymes allow greater utilization of hemicellulose while also increasing cellulose hydrolysis. Recent metabolic engineering efforts have resulted in a strain of Thermoanaerobacterium saccharolyticum that can convert C5 and C6 sugars, as well as insoluble xylan, into ethanol at high yield. To better understand the process of xylan solubilization in this organism, a series of targeted deletions were constructed in the homoethanologenic T. saccharolyticum strain M0355 to characterize xylan hydrolysis and xylose utilization in this organism. While the deletion of β-xylosidase xylD slowed the growth of T. saccharolyticum on birchwood xylan and led to an accumulation of short-chain xylo-oligomers, no other single deletion, including the deletion of the previously characterized endoxylanase XynA, had a phenotype distinct from that of the wild type. This result indicates a multiplicity of xylanase enzymes which facilitate xylan degradation in T. saccharolyticum. Growth on xylan was prevented only when a previously uncharacterized endoxylanase encoded by xynC was also deleted in conjunction with xynA. Sequence analysis of xynC indicates that this enzyme, a low-molecular-weight endoxylanase with homology to glycoside hydrolase family 11 enzymes, is secreted yet untethered to the cell wall. Together, these observations expand our understanding of the enzymatic basis of xylan hydrolysis by T. saccharolyticum.
INTRODUCTION
Xylan is the second-most-abundant polysaccharide in nature after cellulose and the most-abundant component of hemicellulose, though xylans constitute a larger fraction of the hemicellulose in angiosperms than in gymnosperms (4, 6, 19, 27). Xylan-solubilizing enzymes, xylanases, have been studied for their potential use as low-cost and environmentally friendly bleaching agents in the pulp and paper industry and as a component of cost-effective lignocellulose conversion in the emerging biofuels industry. Poor removal of hemicellulose can impede cellulose hydrolysis (9), and xylo-oligomers—formed either during pretreatment or through hemicellulose hydrolysis—have been shown to have a strong inhibitory effect on cellulose hydrolysis (18). Supplementation of xylanase enzymes to the hydrolysis of pretreated lignocellulose has been shown to increase glucose yields while also increasing yields of fermentable hemicellulose-derived sugars (2, 11). Thermophilic xylanases are of particular industrial interest because of their high specific activity and increased thermostability (10, 29).
Xylans have a xylose backbone decorated with side groups, including acetic acid, arabinose, glucuronic acid, ferulic acid, and p-coumaric acid (19). These side group substituents have been linked to functionality and can be tissue specific (5). In addition, the side groups also alter the solubility and digestibility of xylan and, in turn, impact the enzymes needed for depolymerization (6). Consistent with the multiplicity of linkages involved, the collective action of a variety of saccharolytic enzymes is required to cleave hemicellulose into its constituent components. Endoxylanases cleave the internal β-1,4-glycosidic bonds of xylan to form xylo-oligomers, and β-xylosidase enzymes hydrolyze xylobiose and short xylo-oligomers to xylose. Additional accessory enzymes, such as xylan acetyl esterase and α-glucuronidase enzymes, remove the side chains, thus allowing the xylanases access to the xylose backbone (4, 19).
Thermoanaerobacterium saccharolyticum strain JW/SL-YS485 is a thermophilic anaerobe that can grow on xylan as a sole carbon source and can utilize sugars derived from both cellulose and hemicellulose. Although multiple xylanases with overlapping but distinguishable functionalities are commonly found in xylan-solubilizing microorganisms (27, 30), to date, only a single endoxylanase (XynA) has been described in T. saccharolyticum (21). This high-molecular-weight, family 10 endoxylanase has been purified from both T. saccharolyticum and a heterologous host (14, 21). Over 80% of the endoxylanase activity from cell cultures was localized to the S-layer, the outermost layer in bacterial cell walls (3). The remainder of the xylanase activity, found in the soluble protein fraction, has been presumed to be a result of cell lysis (14, 21).
Complementing the endoxylanase XynA, three distinct β-xylosidases have been identified in T. saccharolyticum JW/SL YS485 (15). Two of these β-xylosidases, XylA and XylC, have been purified as active enzymes from crude cell extract, and the sequence of XylC has been determined (24). The third β-xylosidase, XylB, has been characterized only as a recombinant enzyme (15). The gene product of xylB results in a protein smaller than both XylA and XylC with unique kinetic properties (24). XylB is a family 39 glycoside hydrolase enzyme and an ortholog of the xynB gene from T. saccharolyticum B6A-RI (13). The recently identified XylC represents a novel glycoside hydrolase family, displaying unusual kinetics and an amino acid sequence lacking similarity to any of the previously described glycoside hydrolase families with xylosidase activities (families 3, 39, 43, 52, and 54; www.cazy.org/Glycoside-Hydrolases.html) (24). While both xylB and xylC have been identified in the genome, the third β-xylosidase, XylA, has been purified to apparent homogeneity only from T. saccharolyticum. The gene for this β-xylosidase has not yet been identified (24). In addition to the primary xylanase and β-xylosidase enzymes, several accessory enzymes, including two xylan acetyl esterases (23) and an α-glucuronidase (15, 22), have been characterized.
In this study, we constructed a series of deletion strains in order to better understand the suite of enzymes that contribute to xylan solubilization by T. saccharolyticum. In addition, we characterized the effect of these enzymes on the xylo-oligomer pool during xylan hydrolysis as the starting point for future studies on the effects of xylo-oligomers on cellulase activity.
MATERIALS AND METHODS
Strains, media, and culturing conditions.
MTC medium, used for fermentation experiments involving T. saccharolyticum, was prepared as described in reference 31 with modifications (17) and with 5 g/liter birchwood xylan (batch number 010M0169; Sigma) as the carbon source for fermentation. MTC medium with 5 g/liter cellobiose was used for inoculum cultures grown from freezer stocks. TSC1 medium, used for molecular work involving T. saccharolyticum with kanamycin selection, contained 1.85 g (NH4)2SO4, 0.05 g FeSO4, 1 g KH2PO4, 1.0 g MgSO4, 0.1 g CaCl2 · 2H2O, 2 g trisodium citrate · 2H2O, 8.5 g yeast extract, and 0.5 g l-cysteine · HCl per liter and was adjusted to pH 6.7 prior to inoculation (25). Defined M122 medium, used for uracil autotrophy selection, contained 1.3 g (NH4)2SO4, 1.43 g KH2PO4, 1.8 g K2HPO4 · 3H2O, 2.6 g MgCl2 · 6H2O, 0.13 g CaCl2 · 2H2O, 6.0 g glycerol-2-phosphate disodium, 5.0 g cellobiose, 1.1 mg FeSO4 · 7H2O, and 0.5 g l-cysteine · HCl per liter and was adjusted to pH 5.0 prior to inoculation. After autoclaving, 2× RPMI 1640 vitamins (Sigma R7256) and 1× minimal essential medium (MEM) amino acids (Sigma M5550) were added (25).
Saccharomyces cerevisiae InvSc1 was grown on YPD medium for normal growth and SD-Ura medium (Sunrise Science Products, San Diego, CA) when selecting for URA3+ plasmids. Escherichia coli was maintained in LB medium and supplemented with kanamycin (50 μg/ml) when selecting for the presence of plasmids. The parent strain T. saccharolyticum M0355 (25) and its subsequent deletion strains were grown inside an anaerobic chamber (Coy Laboratory Products, Grass Lake, MI) at 55°C on TSC1 medium solidified with 1.2% agar, supplemented with kanamycin (200 μg/ml) when selecting for gene replacement. When selecting for removal of the marker system, strains were grown on defined M122 medium supplemented with 0.2 mM sodium chloroacetate. Markerless strains were restreaked to obtain single colonies. After verification of deletion by PCR and sequencing, strains were maintained at −80°C in 5% dimethyl sulfoxide (DMSO).
Fermentations on xylan were performed in anaerobic serum bottles. Xylan in water was sterilized by autoclaving at 121°C. MTC components were added as described above, using 5 g/liter MES (morpholinoethanesulfonic acid) as a buffer. A 4% (vol/vol) inoculum, grown overnight from freezer stocks in MTC medium with 5 g/liter cellobiose, was used to seed the fermentation. Bottles were incubated at 50°C, and samples were withdrawn every 24 h for analysis.
Vector construction.
Plasmid pYC2/CT (Invitrogen, Carlsbad, CA) was digested with BamHI (New England BioLabs, Ipswich, MA). The resulting linearized vector was purified by gel extraction (Zymo Research, Orange, CA). The upstream, downstream, and internal fragments for each target were amplified by PCR from T. saccharolyticum cells using the primers listed in Table 1. Primers were designed from the T. saccharolyticum genome sequence from Mascoma Corporation (GenBank accession number CP003184). The pta, ack, and kanamycin (Kan) resistance genes were amplified as a single selection cassette from plasmid pMU433, a derivative of pMU424 that was provided by Mascoma Corporation (25). The cut pYC2/CT backbone, the upstream, downstream, and internal fragments, and the pta/ack/Kan selection cassette were combined using yeast-mediated gap repair cloning (20). A representative plasmid is shown in Fig. S1 in the supplemental material. Yeast plasmids were extracted using a Zymoprep yeast plasmid miniprep II kit (Zymo Research, Orange, CA) and transformed into E. coli Top10 (Invitrogen, Carlsbad, CA) via electroporation. E. coli plasmids were purified using a Qiagen miniprep kit (Qiagen Inc., Germantown, MD). All regions PCR amplified during cloning were confirmed by DNA sequencing.
Table 1.
Primers for upstream, downstream, and internal regions used to construct deletion strainsa
| Target | Forward primer (3′→5′) | Reverse primer (5′→3′) |
|---|---|---|
| or0897 | ||
| Upstream | AGGAGAATTTGCAAGAATGA AGG | CAATTTACCTCCTCTCCGCT G |
| Downstream | GAAGGCATTTAATGGTTTTC AGC | CCAACACCATTCTCTTACAT TACCC |
| Internal | GATGTTAAACTTTAAGAGAA TTTTTACGTTAATTTG | CTCACCAAACTGTTACATTA GCATAACC |
| or1451 | ||
| Upstream | CGATGGAGAATTTAAATTTG CAGCC | TTTAAAACACCTCTTCTTAA ATAAATTTCATTCAACC |
| Downstream | ACTTTGGGGGTCTTAACATG ATTAAAG | CCATGAAATTGCGATCGC |
| Internal | GGAGCTTTTAAAGAGAGACG ATGG | ACACCTGTTGCCACATTGC |
| or1452 | ||
| Upstream | GTCTTCTGGGAAATGAGGTT CC | AAAGTCTATTTCATCCACAA TACACTTGTC |
| Downstream | TATGCATTTGGAGGGATTGA TATG | TAATGTCGTAGGCATACAAG ACCC |
| Internal | CAGGCAGACTTGGCCTTG | CATATGCTCATCTCTATAAA GCATTTCC |
| or1456 | ||
| Upstream | CAAGCCTATCATACCATTTA GCTGTC | ACAAGATATTAAAAAATTCA GAAAATATCAGAAATATTT |
| Downstream | TGAATTAATATAATTAAAAA CTTTCTGTAATGAAGTACG | AAGAAACAACTGAGTTAAAT ATACCGGC |
| Internal | AAAACTTTCTGAATTGACAG GTGTG | GAATAATCTTTACCTTCAAT TCCCCAG |
| or1457 | ||
| Upstream | TAATAAAGCACATCCATTTG CAATAAG | AAATAATATTTTAAGCGTAA TGAAGTACGGAAC |
| Downstream | TCTAATTTACAAGATTTCAG GGGGTTATAAAGTGGC | TGTCTAATAAGTATTTTTAA ATTTCCCCCC |
| Internal | CATGGCTGTTACAGATGCTA GAGTAG | TTAAAATGCACTTTCACCAC TAAGTG |
| or1458 | ||
| Upstream | GATGCTAGAGTAGAAGTTTT ATCAGAAGGG | TTTATAACCCCCTGAAATCT TGTAAATTAG |
| Downstream | TACATCTTTGGTTTTTTATA AATGCTGAAAG | CCATTCGCCTTTATTTACAT TTGC |
| Internal | GTGGCTAAAAATAATAGAAT AAGGTTGAGC | CTAACCCTTAACACTGCCAA TGG |
| or1459 | ||
| Upstream | AGCACAGAAGATATTGTATT TGACACG | TTCTTACTTCCTCCCTCAGT AAATTTAATTTATTG |
| Downstream | AAAAAACAAATAATCTTTAA GTAAAAAGGCAG | AAAAAGGCTTCAGAATGGCT TG |
| Internal | GAACATTATATAGTTAAGAG GGATGGTACAGG | CACAACTTTCTGTTGTCCTT CACC |
| Endoxylanase region | ||
| Upstream | CATGATTCCGGCAGGC | CGCAACATATCTACCCATAT ATACCATC |
| Downstream | GCCTGTAAATTCGAATTCAC CTG | GCCAATGGTGAATTGAAAGA AAATG |
| Internal | CAATGCTTTTTTGTTCCATG AATC | CTTGTATTGGATTTTGTGGT GTCACTAC |
| β-Xylosidase region | ||
| Upstream | AAAGTCTGTGGCGATGGAGA | GCATTTGTTTGAGCTTCCA |
| Downstream | GGAGATGCTTTAAGCATCTC CT | TGGGTGAAGAATCTCGCTTG |
| Internal | GGGACTTTTCGACATGCCAT TAC | CCAACGCACATTAAGACATC G |
| Xylose catabolism region | ||
| Upstream | CACCGTTTTCTACGTAGCTT CTGGCT | GGCATGTAAGTCCTTCCTCG C |
| Downstream | AGCTTCCTCCTTCTATTCGT TTGTCTG | GGTCCTGCTGGAATGTCTGT TG |
| Internal | ATCCAACTTCCCAATTTCTT CAAAGG | CCAGAGGACACAGGAAGAAT GC |
| pta/ack/kan cassette | CGTGCCCATTGTGAAGTGG | GCGCCTACGAGGAATTTGTA TCG |
Overlapping sequences used in vector construction, matching an adjacent region, are not shown.
Transformation.
Xylanase mutant strains were constructed in T. saccharolyticum strain M0355, which lacks lactate dehydrogenase (ldh), phosphotransacetylase (pta), and acetate kinase (ack) and does not produce significant amounts of lactate or acetate (25). DNA for transformation was PCR amplified from E. coli plasmids and purified prior to transformation using the Zymo Clean and Concentrator kit (Zymo Research, Irvine, CA). DNA was introduced using natural competence as described previously with the initial selection (25). A diagram of the selections and predicted crossover events is shown in Fig. S2 in the supplemental material. Strains constructed in the T. saccharolyticum M0355 background are listed in Table 2, and the predicted functions of the open reading frames contained within are described in Table S1 in the supplemental material.
Table 2.
Deletion strains constructed in the T. saccharolyticum M0355 background
| Strain name | Deleted region(s) | Description |
|---|---|---|
| ΔB | Δor1451–Δor1453 | Deleted for the β-xylosidase region |
| ΔE | Δor1456–Δor1459 | Deleted for the endoxylanase region |
| ΔX | Δor0271–Δor0277 | Deleted for the xylose catabolism region |
| ΔBEX | Δor1451–Δor1453, Δor1456–Δor1459, Δor0271–Δor0277 | Deleted for the combined β-xylosidase, endoxylanase, and xylose utilization regions |
| ΔxylD | Δor1451 | Deleted for the putative β-xylosidase xylD |
| ΔxylB | Δor1452 | Deleted for the β-xylosidase xylB |
| ΔxynA | Δor1459 | Deleted for the endoxylanase xynA |
| ΔxynC | Δor0897 | Deleted for the putative endoxylanase xynC |
| ΔxynA ΔxynC | Δor1458 Δor1459 | Deleted for the endoxylanases xynA and xynC |
Analytical methods.
To determine the fermentation product concentrations, high-performance liquid chromatography (HPLC) analysis was performed using a Waters HPLC at 60°C using an Aminex HPX-87H column (Bio-Rad Laboratories, Hercules, CA) with a refractive index (RI) detector and 2.5 mM sulfuric acid as the mobile phase. Xylo-oligomers were analyzed by separation on an ICS 300 Dionex system with a PA-100 column (Dionex Corp., Sunnyvale, CA). The method described by Qing et al. was followed (18) except that the gradient was established with 100 mM NaOH and 500 mM NaAc. Xylose and xylo-oligomer standards (degrees of polymerization [DP] 2 to 6; Megazyme Inc., Wicklow, Ireland) were used to manually calculate the oligomer concentration based on the correlation between oligomer length and peak area. The concentration of oligomers larger than DP10 (approximately DP10 to DP15) was estimated based on the total combined area relative to the DP10 area. Residual carbohydrate was measured by scaling down (500-μl sample, 18 μl acid) the method described by Sluiter et al. (26); oligomers in the samples were hydrolyzed to monomers using 4% sulfuric acid at 121°C for 1 h.
RESULTS
In order to systematically characterize the suite of enzymes used by T. saccharolyticum during hemicellulose utilization, a series of deletion strains were constructed to remove genes known or expected to be involved in xylan degradation.
Deletion of gene clusters encoding xylose utilization, β-xylosidases, and endoxylanases.
Several genes known or expected to be involved in xylan hydrolysis are colocated on the T. saccharolyticum chromosome (Fig. 1A). A separate region of the genome that encodes proteins involved in xylose utilization was identified (Fig. 1B). Using the marker removal system described by Shaw et al. (25), strains were constructed with deletions of the entire xylose utilization (called ΔX), β-xylosidase (called ΔB), and endoxylanase (called ΔE) regions, both individually and in combination (ΔBEX). Subsequently, individual genes were deleted in order to assess their effect on the growth of T. saccharolyticum on xylan. A list of the constructed strains is presented in Table 2. The putative function of the targeted regions is described in Table S1 in the supplemental material.
Fig 1.
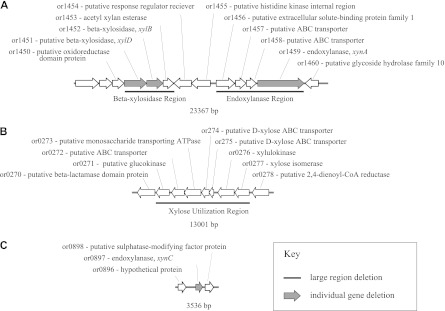
Regions of the T. saccharolyticum genome targeted for deletion. (A) β-xylosidase and endoxylanase regions. Regions encoding multiple genes thought to contribute to xylan hydrolysis. (B) Xylose utilization region. (C) Family 11 endoxylanase, or0897. Underlined regions indicate large region deletions. Solid gray genes indicate genes deleted individually.
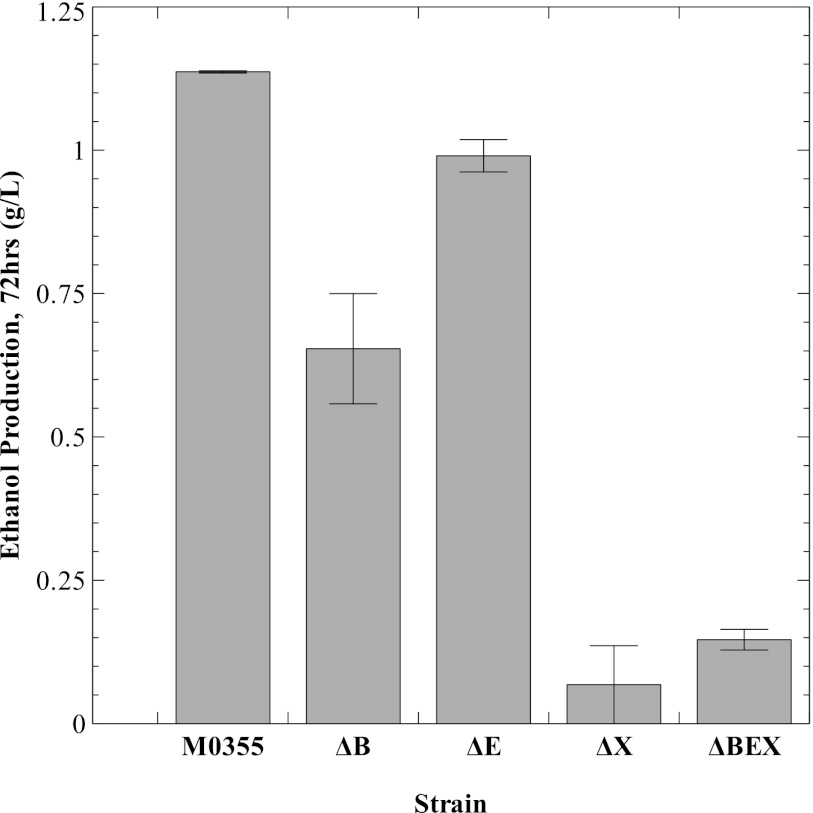
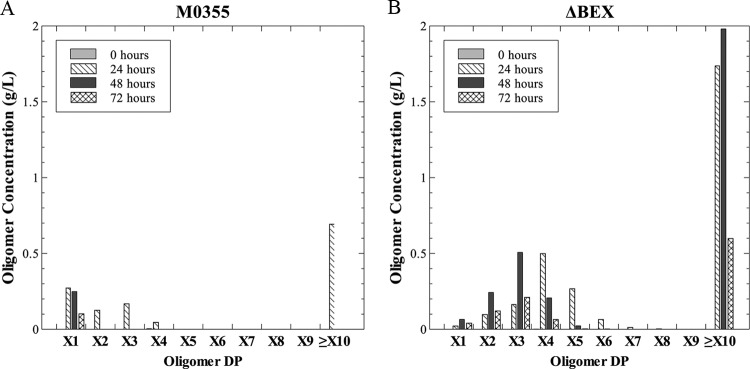
These strains were characterized by growth on 5 g/liter birchwood xylan using the production of ethanol, the main fermentation product, as a metric for growth. Interestingly, the ΔE strain retained the ability to grow on xylan (Fig. 2), suggesting the presence of an additional endoxylanase. Likewise, strain ΔB, deleted for the β-xylosidase region, retained the ability to grow on xylan, though it produced less ethanol at 72 h than the parent strain and the ΔE strain. Only strains deleted for the xylose utilization region (strains ΔX and ΔBEX) showed substantially impaired xylan fermentation. Further analysis of fermentation samples from strain ΔBEX revealed the accumulation of xylo-oligomers (Fig. 3). After 24 h, the fermentation samples from the ΔBEX strain contained high concentrations of oligomers with a DP value of ≥10 and also contained substantial quantities of xylotriose. The concentration of oligomers over DP10 increased at incubation times up to 48 h before decreasing in the last 24 h. In contrast, after 24 h, the concentrations of short-chain oligomers (xylose to xylotriose) and oligomers over DP10 observed in fermentation samples from the parent strain M0355 were low. Only low levels of xylose were detected in the last 48 h of the fermentation with strain M0355.
Fig 2.
Ethanol production at 72 h from strains with large regions deleted, ΔB, ΔE, ΔX, and ΔBEX. Production of ethanol from the fermentation of 5 g/liter birchwood xylan. Ethanol, the main fermentation product of the parent strain M0355, was used as a metric for growth because insoluble xylan prevents optical density from being used to monitor growth. Values are the averages of duplicate runs, with error bars indicating the standard errors of the mean.
Fig 3.
Xylo-oligomer distribution of T. saccharolyticum strains M0355 and ΔBEX. Xylo-oligomer concentration from the fermentation supernatant of 5 g/liter birchwood xylan of strains M0355 (A) and ΔBEX (B) at 0 (gray), 24 (diagonal), 48 (black), and 72 (hatched) hours. X1 to X10 indicate the oligomer degree of polymerization.
Deletion of individual genes involved in xylan utilization.
Growth on xylan and the accumulation of xylo-oligomers in the ΔBEX strain indicated residual endoxylanase activity despite the removal of the endoxylanase region, which included the only previously characterized endoxylanase-encoding gene, xynA.
Further examination of the genome revealed an additional putative endoxylanase (Fig. 1C). This putative endoxylanase, encoded by or0897 and here referred to as XynC, shares homology with conserved domains in family 11 glycoside hydrolase enzymes, with similarity to several Bacillus and Clostridium endoxylanases using BLASTP (http://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins; accession numbers of top hits include NP_389765.1 and YP_001559210.1). Further analysis of the sequence reveals that XynC is likely a secreted protein, with a signal peptide probability of 0.998 using SignalP (http://www.cbs.dtu.dk/services/SignalP/). However, xynC encodes a much smaller protein than XynA and does not have any predicted S-layer homology (SLH) regions or carbohydrate binding module (CBM) regions. As such, xynC likely encodes an untethered endoxylanase.
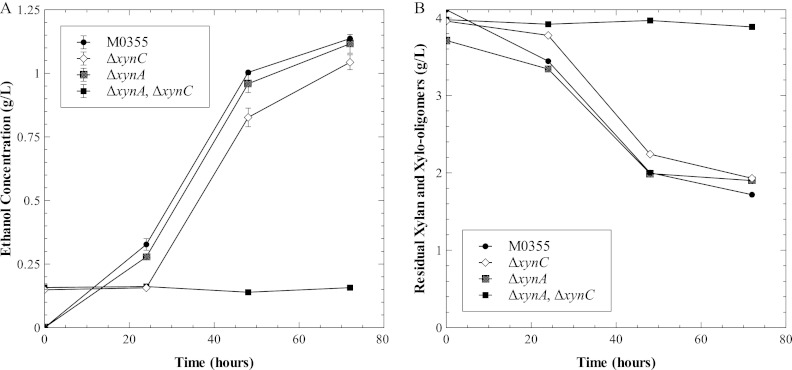
To examine the role of this putative endoxylanase xynC and the previously characterized xynA, these genes were deleted individually and in combination. These strains, ΔxynA, ΔxynC, ΔxynA ΔxynC, and the parent strain M0355, were characterized by monitoring the production of ethanol to assess their roles while growing on xylan (Fig. 4A). Neither single endoxylanase gene deletion prevented growth on xylan. However, in the xynC xynA double mutant, growth on xylan was eliminated. Identification of XynC as a T. saccharolyticum xylanase was further confirmed by measurement of residual xylan and xylo-oligomers in all four strains (Fig. 4B). The individual deletion of either xynA or xynC showed utilization of xylan over the fermentation equivalent to that of the parent strain M0355. In contrast, the ΔxynA ΔxynC strain did not consume xylan, as the total xylan concentration remained constant over the 72-h time course. In addition, no detectable xylo-oligomers were observed for this double mutant strain at 24, 48, or 72 h (data not shown).
Fig 4.
Ethanol production and residual xylan for ΔxynA and ΔxynC strains. Fermentation profiles for strains M0355 (black circle), ΔxynC (open diamond), ΔxynA (gray cross), and ΔxynA ΔxynC (black square). Ethanol concentrations (A) and residual xylan and xylo-oligomers (B) from 5-g/liter xylan fermentations. Error bars indicate standard errors of replicate runs.
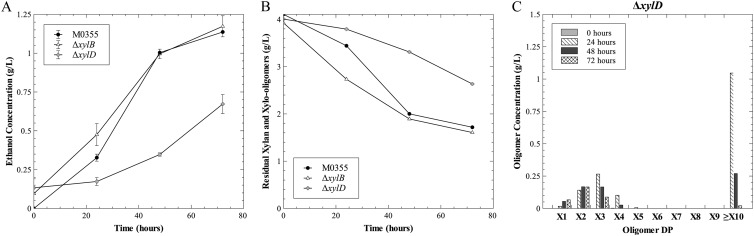
The β-xylosidase region encodes two putative β-xylosidases, xylB and or1451. Because the gene encoding XylA has not yet been identified and previous work by Lee and Zeikus (12) named the xylose isomerase xylA, or1451 will here be referred to as xylD. To examine the relative roles of the β-xylosidases within this region, each was deleted individually. The xylB deletion did not have a phenotype distinct from that of the parent strain M0355 (Fig. 5). However, the deletion of xylD led to a slowed production of ethanol from xylan (Fig. 5A). Residual carbohydrate analysis confirmed the slowed utilization of xylan (Fig. 5B). In contrast, deletion of the previously characterized β-xylosidase (xylB) did not affect the ability to hydrolyze and grow on xylan relative to that of the parent strain.
Fig 5.
Growth of β-xylosidase deletion strains on xylan. (A and B) Production of ethanol (A) and disappearance of xylan (B) by M0355 (black circle), ΔxylB (open triangle), and ΔxylD (gray cross) strains grown on xylan. (C) Xylo-oligomer profile of strain ΔxylD at 0 (gray), 24 (diagonal), 48 (black), and 72 (hatched) hours.
The xylo-oligomer pools were also measured in fermentation samples from strain ΔxylD. In contrast to the parent strain M0355 (Fig. 3A), strain ΔxylD accumulated short xylo-oligomers over time, from xylose to xylotetraose for the first 48 h, after which the oligomers were converted to xylobiose and/or consumed, thus slowing growth in this strain (Fig. 5C).
DISCUSSION
No mutant with a single targeted deletion of either a β-xylosidase or an endoxylanase gene lost the ability to grow on xylan, consistent with the notion that several enzymes depolymerize xylan. The only single gene deletion strain with a distinct phenotype during growth on xylan was the β-xylosidase xylD deletion mutant (ΔxylD), while the deletion of xylB had little effect on growth on xylan. Since XylB was not found as an active protein from cell extracts in previous studies (15, 24), XylD is likely the major β-xylosidase utilized by T. saccharolyticum during growth on birchwood xylan. This observation is also supported by the accumulation of short-chain xylo-oligomers in the absence of xylD, suggesting that in this strain, the cleavage of xylobiose to xylose is the rate-limiting step for growth on xylan. Yet since strain ΔxylD retained the ability to grow on xylan, additional β-xylosidase activity must be present in the wild type. The recent discovery of the novel T. saccharolyticum β-xylosidase XylC (24) may help explain this additional β-xylosidase activity in ΔxylD. More work is needed to clarify the relative roles of XylC and XylD.
Though microorganisms typically have more than one endoxylanase, XynA is, to date, the only endoxylanase previously reported in T. saccharolyticum. XynA is a family 10 glycoside hydrolase with three SLH repeat regions and two conserved CBM regions (14). Despite being the only characterized endoxylanase in T. saccharolyticum, the deletion of xynA did not prevent growth on xylan. Subsequently, the removal of a newly identified endoxylanase encoded by xynC (or0897) also showed no distinct phenotype. When the two genes were simultaneously deleted (ΔxynA ΔxynC), no growth was observed on xylan, as shown by both the lack of ethanol production and the high concentrations of residual xylan and xylo-oligomers (Fig. 4). Thus, we have demonstrated that T. saccharolyticum has a second endoxylanase enzyme which in the absence of XynA can facilitate solubilization of xylan. Analysis of the xylo-oligomers also supports the role of or0897 as an endoxylanase. As a secreted enzyme with no SLH-encoding regions, this newly identified endoxylanase, XynC, likely accounts for the endoxylanase activity previously detected in the soluble supernatant fraction of T. saccharolyticum cell extracts (21). Thus, XynA and XynC may have separate but partially overlapping substrate specificities and may have differing physiological roles. Future biochemical comparisons may help elucidate the differences between these xylanases.
Many organisms use both free and cell-associated enzymes to solubilize plant cell wall-derived substrates (7). The attachment of XynA to the S-layer ensures close proximity of the cell to the products of hydrolysis, a factor which may be advantageous in nature where microorganisms compete for these sugars (1, 28). For example, higher rates of cellulose solubilization by Clostridium thermocellum were observed with cells utilizing cell-bound cellulases than with cells utilizing free cellulases in controlled simultaneous saccharification and fermentation (SSF) studies (16). Cell-associated enzymes may be particularly important in an extreme environment, such as the hot spring from which T. saccharolyticum was isolated, where free enzymes would quickly diffuse away from the organism (21). However, the large size and tethered state of XynA may also prevent access to hemicellulose while being present in interior portions of biomass particles, which may be better accessed by the smaller XylC endoxylanase. Furthermore, in the absence of high substrate levels, free enzymes may be released into the environment to generate soluble xylo-oligomers, to form a chemical gradient that can be used for a chemotactic response toward additional insoluble substrate. Thus, having multiple xylanase enzymes may allow a better response to different hemicellulose substrates and to changing conditions than having only one xylanase activity.
In addition to further describing the xylanase system in T. saccharolyticum, several of the strains constructed in this study exhibited altered xylo-oligomer pools. Hydrolysis of cellulose in the presence of xylo-oligomers has indicated the inhibitory nature of these oligomers (18); however, this has not been demonstrated in a microbial system. Thus, these strains can be utilized in the future to better understand the effects of xylo-oligomers on cellulose hydrolysis. While the native strain M0355 can degrade xylan, strains ΔBEX and ΔxylD both accumulate xylo-oligomers. Using these strains, it may be possible to directly test the effect of xylo-oligomer accumulation on cellulose hydrolysis without interference from cellobiose or glucose accumulation, known inhibitors of cellulose hydrolysis (8). Thus, these strains can also be utilized to better understand the effects of xylo-oligomers on cellulose hydrolysis.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by the BioEnergy Science Center at Oak Ridge National Laboratory. The BioEnergy Science Center is a U.S. DOE Bioenergy Research Center supported by the Office of Biological and Environmental Research in the DOE Office of Science. Oak Ridge National Laboratory is managed by UT-Battelle, LLC, for the U.S. DOE under contract number DE-AC05-00OR22725. Mascoma Corporation provided additional support.
We acknowledge Phil Thorne of the Mascoma Corporation for assistance with running and processing the xylo-oligomer samples. Strain construction and growth analysis were performed at Thayer School of Engineering, Dartmouth College. Xylo-oligomer analysis was performed at Mascoma Corporation.
Footnotes
Published ahead of print 28 September 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Bayer EA, Lamed R, White BA, Ding S-Y, Himmel ME. 2010. Conversion of agricultural residues to bioethanol: the roles of cellulases and cellulosomes, p 67–96 In Blaschek HP, Ezeji TC, Scheffran J. (ed), Biofuels from agricultural wastes and byproducts. Wiley-Blackwell, Ames, IA [Google Scholar]
- 2. Berlin A, Maximenko V, Gilkes N, Saddler J. 2007. Optimization of enzyme complexes for lignocellulose hydrolysis. Biotechnol. Bioeng. 97:287–296 [DOI] [PubMed] [Google Scholar]
- 3. Beveridge TJ, et al. 1997. Functions of S-layers. FEMS Microbiol. Rev. 20:99–149 [DOI] [PubMed] [Google Scholar]
- 4. Collins T, Gerday C, Feller G. 2005. Xylanases, xylanase families and extremophilic xylanases. FEMS Microbiol. Rev. 29:3–23 [DOI] [PubMed] [Google Scholar]
- 5. Dodd D, Cann IKO. 2009. Enzymatic deconstruction of xylan for biofuel production. Glob. Change Biol. Bioenergy 1:2–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ebringerova A, Heinze T. 2000. Xylan and xylan derivatives—biopolymers with valuable properties. 1. Naturally occurring xylans structures, procedures and properties. Macromol. Rapid Commun. 21:542–556 [Google Scholar]
- 7. Himmel ME, et al. 2010. Microbial enzyme systems for biomass conversion: emerging paradigms. Biofuels 1:323–341 [Google Scholar]
- 8. Holtzapple M, Cognata M, Shu Y, Hendrickson C. 1990. Inhibition of Trichoderma reesei cellulase by sugars and solvents. Biotechnol. Bioeng. 36:275–287 [DOI] [PubMed] [Google Scholar]
- 9. Hu J, Arantes V, Saddler JN. 2011. The enhancement of enzymatic hydrolysis of lignocellulosic substrates by the addition of accessory enzymes such as xylanase: is it an additive or synergistic effect? Biotechnol. Biofuels 4:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kristjansson JK. 1989. Thermophilic organisms as sources of thermostable enzymes. Trends Biotechnol. 7:349–353 [Google Scholar]
- 11. Kumar R, Wyman CE. 2009. Effects of cellulase and xylanase enzymes on the deconstruction of solids from pretreatment of poplar by leading technologies. Biotechnol. Prog. 25:302–314 [DOI] [PubMed] [Google Scholar]
- 12. Lee YE, Ramesh MV, Zeikus JG. 1993. Cloning, sequencing and biochemical characterization of xylose isomerase from Thermoanaerobacterium saccharolyticum strain B6A-RI. J. Gen. Microbiol. 139:1227–1234 [DOI] [PubMed] [Google Scholar]
- 13. Lee YE, Zeikus JG. 1993. Genetic organization, sequence and biochemical characterization of recombinant beta-xylosidase from Thermoanaerobacterium saccharolyticum strain B6A-RI. J. Gen. Microbiol. 139:1235–1243 [DOI] [PubMed] [Google Scholar]
- 14. Liu S, Gherardini F, Matuschek M, Bahl H, Wiegel J. 1996. Cloning, sequencing, and expression of the gene encoding a large S-layer-associated endoxylanase from Thermoanaerobacterium sp. strain JW/SL-YS 485 in Escherichia coli. J. Bacteriol. 178:1539–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lorenz WW, Wiegel J. 1997. Isolation, analysis, and expression of two genes from Thermoanaerobacterium sp. strain JW/SL YS485: a beta-xylosidase and a novel acetyl xylan esterase with cephalosporin C deacetylase activity. J. Bacteriol. 179:5436–5441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lu YP, Zhang YHP, Lynd LR. 2006. Enzyme-microbe synergy during cellulose hydrolysis by Clostridium thermocellum. Proc. Natl. Acad. Sci. U. S. A. 103:16165–16169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Podkaminer KK, Shao X, Hogsett DA, Lynd LR. 2011. Enzyme inactivation by ethanol and development of a kinetic model for thermophilic simultaneous saccharification and fermentation at 50 °C with Thermoanaerobacterium saccharolyticum ALK2. Biotechnol. Bioeng. 108:1268–1278 [DOI] [PubMed] [Google Scholar]
- 18. Qing Q, Yang B, Wyman CE. 2010. Xylooligomers are strong inhibitors of cellulose hydrolysis by enzymes. Bioresour. Technol. 101:9624–9630 [DOI] [PubMed] [Google Scholar]
- 19. Saha BC. 2003. Hemicellulose bioconversion. J. Ind. Microbiol. Biotechnol. 30:291. [DOI] [PubMed] [Google Scholar]
- 20. Shanks RMQ, Caiazza NC, Hinsa SM, Toutain CM, O'Toole GA. 2006. Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from gram-negative bacteria. Appl. Environ. Microbiol. 72:5027–5036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shao W, DeBlois S, Wiegel J. 1995. A high-molecular-weight, cell-associated xylanase isolated from exponentially growing Thermoanaerobacterium sp. strain JW/SL-YS485. Appl. Environ. Microbiol. 61:937–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shao W, Obi S, Puls J, Wiegel J. 1995. Purification and characterization of the α-glucuronidase from Thermoanaerobacterium sp. strain JW/SL-YS485, an important enzyme for the utilization of substituted xylans. Appl. Environ. Microbiol. 61:1077–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shao W, Wiegel J. 1995. Purification and characterization of two thermostable acetyl xylan esterases from Thermoanaerobacterium sp. strain JW/SL-YS485. Appl. Environ. Microbiol. 61:729–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shao WL, et al. 2011. Characterization of a novel beta-xylosidase, XylC, from Thermoanaerobacterium saccharolyticum JW/SL-YS485. Appl. Environ. Microbiol. 77:719–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shaw AJ, Covalla SF, Hogsett DA, Herring CD. 2011. Marker removal system for Thermoanaerobacterium saccharolyticum and development of a markerless ethanologen. Appl. Environ. Microbiol. 77:2534–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sluiter A, et al. 2008. Determination of sugars, byproducts, and degradation products in liquid fraction process samples. Laboratory analytical procedure (LAP) (NREL/TP-510-42623). National Renewable Energy Laboratory, Golden, CO [Google Scholar]
- 27. Sunna A, Antranikian G. 1997. Xylanolytic enzymes from fungi and bacteria. Crit. Rev. Biotechnol. 17:39–67 [DOI] [PubMed] [Google Scholar]
- 28. Vazana Y, Morais S, Barak Y, Lamed R, Bayer EA. 2010. Interplay between Clostridium thermocellum family 48 and family 9 cellulases in cellulosomal versus noncellulosomal states. Appl. Environ. Microbiol. 76:3236–3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ward OP, Mooyoung M. 1988. Thermostable enzymes. Biotechnol. Adv. 6:39–69 [DOI] [PubMed] [Google Scholar]
- 30. Wong KK, Tan LU, Saddler JN. 1988. Multiplicity of β-1,4-xylanase in microorganisms: functions and applications. Microbiol. Rev. 52:305–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang Y, Lynd LR. 2003. Quantification of cell and cellulase mass concentrations during anaerobic cellulose fermentation: development of an enzyme-linked immunosorbent assay-based method with application to Clostridium thermocellum batch cultures. Anal. Chem. 75:219–227 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.