Abstract
Tamavidins are fungal biotin-binding proteins (BBPs) displaying antifungal activity against phytopathogens. Here we show high toxicity of tamavidins toward nematodes, insects, and amoebae. As these organisms represent important phyla of fungal predators and parasites, we propose that BBPs are part of the chemical defense system of fungi.
TEXT
Biotin is a vitamin required by all prokaryotes and eukaryotes as an essential cofactor of several carboxylases involved in central metabolic pathways (2, 22). The biosynthesis of biotin is restricted to plants and some microorganisms; animals and other organisms are dependent on uptake from the environment, diet, or intestinal flora (1, 16, 22).
Biotin-binding proteins (BBPs) have been identified from different organisms, including bacteria, birds, amphibians, and recently, fungi also (see Table S2 in the supplemental material). Some of these proteins exhibit one of the strongest noncovalent bonds known in nature between a protein and a small ligand (Kd [dissociation constant] = 10−14 to 10−16 M) (12), making biotin binding irreversible and complete at equimolar concentrations (10). BBPs have been suggested as broad-range antimicrobial agents by forming a biotin-free zone. Avidin, for example, has been proposed as an antimicrobial host defense factor against pathogen infections in chicken, as it inhibits the growth of some microorganisms, and is induced in different tissues in chicken upon injury and bacterial and viral infection (11, 14, 18). Similarly, streptavidins are suggested to be part of a synergistic antibiotic complex in the filamentous bacterium Streptomyces (4, 8, 9), and bradavidin from Bradyrhizobium japonicum is proposed to protect the host plant from microbes, insects, and other herbivores (19). Finally, avidin and streptavidin added to diet-based bioassays and expressed transgenically in plants have shown to be highly toxic to many insect species (6, 10).
On the basis of the broad antimicrobial and insecticidal activity of avidin and streptavidin and the recent evidence for a protein-mediated defense of fungi against predators and parasites (5, 20), we tested whether the recently identified BBPs from the edible mushroom Pleurotus cornucopiae, tamavidins 1 and 2 (24), may serve as effector proteins of fungal defense. Both proteins were previously reported to inhibit the growth of the phytopathogenic fungus Magnaporthe grisea in culture medium (24) and to confer resistance to the blast fungus Magnaporthe oryzae in transgenic rice (23). In this study, we assayed the toxicity of tamavidins toward the nematode Caenorhabditis elegans, the amoeba Acanthamoeba sp., and the insect Drosophila melanogaster as these organisms represent three of the most important groups of fungal antagonists in nature (17).
Tamavidins 1 and 2 were expressed as soluble proteins in the cytoplasm of Escherichia coli BL21 as described previously (24). Biotoxicity of the fungal BBPs was assessed by feeding the recombinant E. coli cells to C. elegans and Acanthamoeba sp. as previously described (15, 20) and by adding purified recombinant tamavidin 2 to the rearing medium of D. melanogaster as described in Method S3 in the supplemental material and in reference 21.
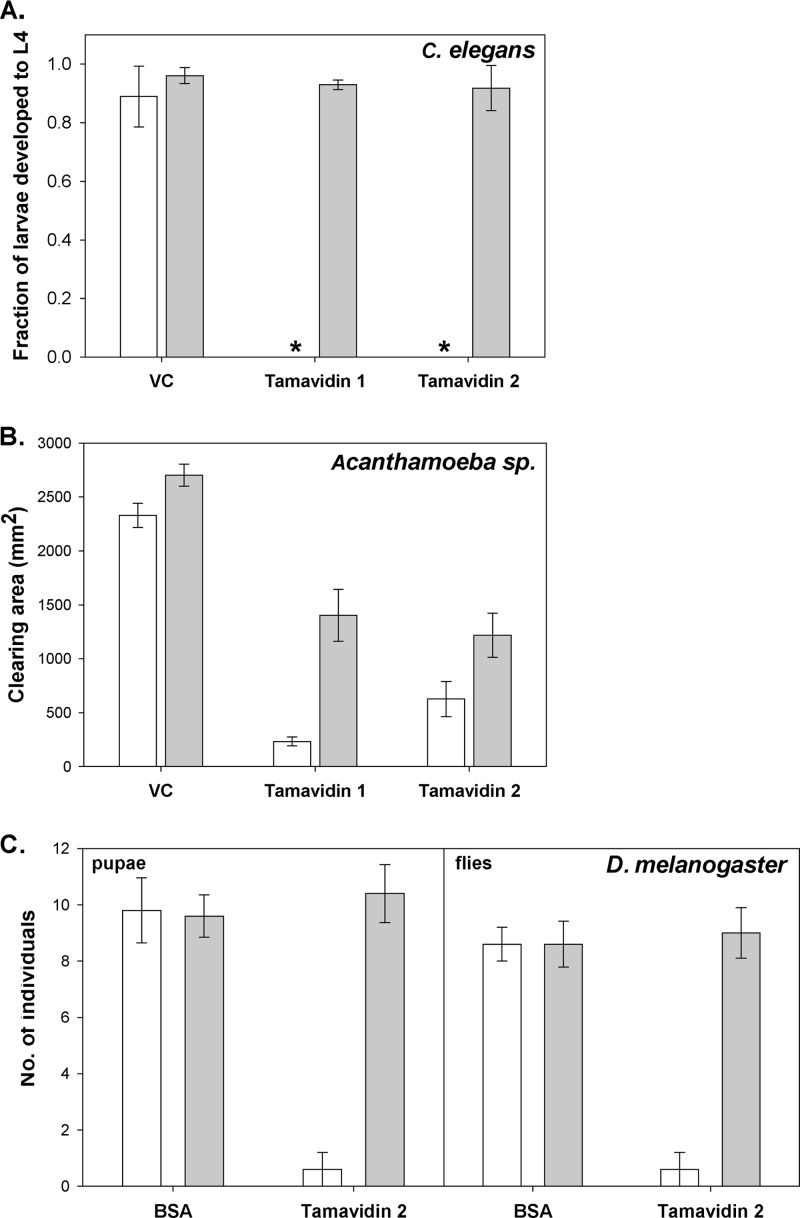
E. coli cells expressing tamavidin 1 and tamavidin 2 were highly toxic to both C. elegans and Acanthamoeba sp. fed on these E. coli cells. When exogenous biotin was added to the C. elegans bacterial suspension (20 μg/ml), the antinutritional effect of the tamavidins was completely abolished, and all larvae developed normally (Fig. 1A). In the case of Acanthamoeba sp., the addition of exogenous biotin (10 μg/ml) partially abolished the antinutritional effect of the tamavidins by increasing approximately five and two times, respectively, the clearing area of amoebae feeding on tamavidins 1 and 2 (Fig. 1B). Tamavidin 2 was toxic to D. melanogaster when added to the rearing medium, significantly reducing the number of pupae and flies in comparison to the bovine serum albumin (BSA) control. The addition of exogenous biotin to the medium (26 μg/ml) completely rescued the development of pupae and flies (Fig. 1C).
Fig 1.
Toxicity of tamavidins. (A to C) Effect of feeding E. coli expressing tamavidin 1 and tamavidin 2 to C. elegans (A) and to Acanthamoeba sp. (B) and E. coli expressing purified tamavidin 2 to D. melanogaster (C) without (white bars) and with (gray bars) the addition of exogenous biotin. Asterisks show cases where all data were 0. E. coli containing empty vector (VC) was used as a control in panels A and B, and BSA was used as a control in panel C. Values are means ± standard errors of the means (error bars). L4, larval stage 4.
In general, the toxicity of BBPs is thought to be based on their high affinity and low dissociation for biotin, which makes this essential nutrient unavailable for the antagonists (18). When a BBP is, for example, ingested by insects like D. melanogaster at an equimolar concentration or above the concentration of coingested biotin, no biotin remains for absorption, and it cannot be released from the complex, as BBPs are resistant to proteolysis (10). As expected and suggested by our results, this antinutritional effect can be at least partially abolished by addition of an excess of biotin to the food containing BBP. In the case of amoebae, it was previously shown that several soil amoebae require biotin for growth and that the addition of avidin to the medium affects the growth of amoebae in culture (3). In our bacterium-based assays, it can be assumed that the nematodes and amoebae obtain biotin from the ingested E. coli, as no other source of vitamins is available in the assays. However, when tamavidin-producing E. coli cells are ingested, tamavidins probably bind to all biotin available, affecting growth and survival of the feeding organisms. Similar to the experiments with D. melanogaster, the addition of extra biotin to the medium reduced the toxic effect of tamavidins in these cases. To our knowledge, the toxic effect of tamavidins toward C. elegans is the first demonstration of the susceptibility of nematodes to BBPs.
The observed toxicities suggest that the fungal BBPs play a role as effectors in the defense of fungi against insect, nematode, and amoebal predators and parasites. A similar role has recently been proposed for fungal lectins (5) and protease inhibitors (20). Besides their toxicity, these proteins share other features, e.g., the lack of a signal sequence for classical secretion, their low molecular weight, their resistance to oxidation, high temperature, and proteolysis, and their abundant expression in fruiting bodies. The lack of a secretory signal in the tamavidins contrasts with other members of the avidin superfamily but is in agreement with the proposed mechanism of protein-mediated defense in fungi (5). According to this model, the tamavidins would be kept innocuous by being sequestered to the cytoplasm (see Discussion S5 in the supplemental material). Upon disruption of the fungal cell during fungivory, the tamavidins would be released and bind to the biotin that is available in the digestive tract of the fungivore before it is absorbed by the intestinal epithelium. Such an antinutritional mechanism has also been proposed for protease inhibitors functioning as fungal defense proteins. These proteins are believed to inhibit gut proteases of fungivores necessary to digest ingested proteins (20). Fungal defense lectins, on the other hand, function in a more direct way. Although the exact mechanism of lectin-mediated toxicity is not well understood, the toxicity is known to be dependent on binding of the lectins to specific glycoepitopes displayed on the surfaces of epithelial cells of the fungivore (7, 25). This binding of toxic lectins to epithelial cells causes damage of the epithelial cells and expansion of the intestinal lumen (7, 21). In contrast, nematotoxicity caused by the antinutritional effect of the tamavidins does not result in morphological changes of the intestine (Fig. S4). In conclusion, BBPs would be the only fungal defense protein identified so far that do not directly interact with a cell or molecule of the target organism but act indirectly by sequestration of an essential component of the food.
Large-scale sequencing of fungal genomes has revealed the presence of genes coding for BBPs homologous to tamavidins in other basidiomycetes besides P. cornucopiae (13) (see Fig. S1 in the supplemental material). The phylogenetic distribution of these genes is random and does not follow an obvious pattern (data not shown). Such a “patchy” distribution is typical for genes that are not involved in conserved physiological processes but rather function in biotic and abiotic interactions. Accordingly, the same type of distribution is observed for the genes coding for fungal defense lectins and protease inhibitors (5, 20). In conclusion, we propose that cytoplasmic biotin-binding proteins constitute a novel class of effector proteins in fungal resistance and/or defense against antagonists.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Swiss National Science Foundation (grant 31003A-130671) and by ETH Zürich.
We thank W. Rudin and P. Müller (Swiss Tropical and Public Health Institute, Basel, Switzerland), E. Hafen and H. Stocker (Institute of Molecular Systems Biology, ETH Zürich, Switzerland), and M. O. Hengartner (Institute of Molecular Life Sciences, University of Zürich, Switzerland) for supplying A. aegypti eggs, D. melanogaster eggs, and C. elegans worms, respectively. We are grateful to Y. Takakura (Japan Tobacco Inc.) for providing the plasmids for expression of tamavidins in E. coli and for critically reading the manuscript.
Footnotes
Published ahead of print 21 September 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Alban C, Job D, Douce R. 2000. Biotin metabolism in plants. Annu. Rev. Plant Biol. 51:17–47 [DOI] [PubMed] [Google Scholar]
- 2. Attwood PV, Wallace JC. 2002. Chemical and catalytic mechanisms of carboxyl transfer reactions in biotin-dependent enzymes. Acc. Chem. Res. 35:113–120 [DOI] [PubMed] [Google Scholar]
- 3. Band RN. 1961. Biotin, a growth requirement for four soil amoebae. Nature 192:674. [DOI] [PubMed] [Google Scholar]
- 4. Bayer EA, Kulik T, Adar R, Wilchek M. 1995. Close similarity among streptavidin-like, biotin-binding proteins from Streptomyces. Biochim. Biophys. Acta 1263:60–66 [DOI] [PubMed] [Google Scholar]
- 5. Bleuler-Martinez S, et al. 2011. A lectin-mediated resistance of higher fungi against predators and parasites. Mol. Ecol. 20:3056–3070 [DOI] [PubMed] [Google Scholar]
- 6. Bruins BG, Scharloo W, Thorig GEW. 1991. The harmful effect of light on Drosophila is diet-dependent. Insect Biochem. 21:535–539 [Google Scholar]
- 7. Butschi A, et al. 2010. Caenorhabditis elegans N-glycan core beta-galactoside confers sensitivity towards nematotoxic fungal galectin CGL2. PLoS Pathog. 6:e1000717 doi:10.1371/journal.ppat.1000717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chaiet L, Miller TW, Tausig F, Wolf FJ. 1964. Antibiotic MSD-235. II. Separation and purification of synergistic components, p 28–32 Antimicrob. Agents Chemother. 1963. [PubMed] [Google Scholar]
- 9. Chaiet L, Wolf FJ. 1964. The properties of streptavidin, a biotin-binding protein produced by Streptomycetes. Arch. Biochem. Biophys. 106:1–5 [DOI] [PubMed] [Google Scholar]
- 10. Christeller JT, Markwick NP, Burgess EP, Malone LA. 2010. The use of biotin-binding proteins for insect control. J. Econ. Entomol. 103:497–508 [DOI] [PubMed] [Google Scholar]
- 11. Elo HA, Raisanen S, Tuohimaa PJ. 1980. Induction of an antimicrobial biotin-binding egg white protein (avidin) in chick tissues in septic Escherichia coli infection. Experientia 36:312–313 [DOI] [PubMed] [Google Scholar]
- 12. Green NM. 1990. Avidin and streptavidin. Methods Enzymol. 184:51–67 [DOI] [PubMed] [Google Scholar]
- 13. Grigoriev IV, et al. 2012. The Genome Portal of the Department of Energy Joint Genome Institute. Nucleic Acids Res. 40:D26–D32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Korpela J, Kulomaa M, Tuohimaa P, Vaheri A. 1983. Avidin is induced in chicken embryo fibroblasts by viral transformation and cell damage. EMBO J. 2:1715–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kunzler M, et al. 2010. Biotoxicity assays for fruiting body lectins and other cytoplasmic proteins. Methods Enzymol. 480:141–150 [DOI] [PubMed] [Google Scholar]
- 16. Lin S, Cronan JE. 2011. Closing in on complete pathways of biotin biosynthesis. Mol. Biosyst. 7:1811–1821 [DOI] [PubMed] [Google Scholar]
- 17. McGonigle TP. 2007. Effects of animals grazing on fungi, p 201–212 In Kubicek CP, Druzhinina IS. (ed), Environmental and microbial relationships (the Mycota), 2nd ed Springer-Verlag, Berlin, Germany [Google Scholar]
- 18. Mine Y. 2000. Avidin, p 253–264 In Naidu AS. (ed), Natural food antimicrobial systems. CRC Press, LLC, London, United Kingdom [Google Scholar]
- 19. Nordlund HR, Hytonen VP, Laitinen OH, Kulomaa MS. 2005. Novel avidin-like protein from a root nodule symbiotic bacterium, Bradyrhizobium japonicum. J. Biol. Chem. 280:13250–13255 [DOI] [PubMed] [Google Scholar]
- 20. Sabotic J, et al. 2012. Structural basis of trypsin inhibition and entomotoxicity of cospin, a serine protease inhibitor involved in defence of Coprinopsis cinerea fruiting bodies. J. Biol. Chem. 287:3898–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schubert M, et al. 2012. Plasticity of the beta-trefoil protein fold in the recognition and control of invertebrate predators and parasites by a fungal defence system. PLoS Pathog. 8:e1002706 doi:10.1371/journal.ppat.1002706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Streit WR, Entcheva P. 2003. Biotin in microbes, the genes involved in its biosynthesis, its biochemical role and perspectives for biotechnological production. Appl. Microbiol. Biotechnol. 61:21–31 [DOI] [PubMed] [Google Scholar]
- 23. Takakura Y, Oka N, Suzuki J, Tsukamoto H, Ishida Y. 2012. Intercellular production of Tamavidin 1, a biotin-binding protein from Tamogitake mushroom, confers resistance to the blast fungus Magnaporthe oryzae in transgenic rice. Mol. Biotechnol. 51:9–17 [DOI] [PubMed] [Google Scholar]
- 24. Takakura Y, et al. 2009. Tamavidins–novel avidin-like biotin-binding proteins from the Tamogitake mushroom. FEBS J. 276:1383–1397 [DOI] [PubMed] [Google Scholar]
- 25. Wohlschlager T, et al. 2011. Nematotoxicity of Marasmius oreades agglutinin (MOA) depends on glycolipid binding and cysteine protease activity. J. Biol. Chem. 286:30337–30343 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.