Abstract
In turkey-derived Campylobacter coli isolates of a unique lineage (cluster II), the tetracycline resistance determinant tet(O) was chromosomal and was part of a gene cassette (transposon) interrupting a Campylobacter jejuni-associated putative citrate transporter gene. In contrast, the swine-derived C. coli strain 6461 harbored a chromosomal tet(O) in a different genomic location.
TEXT
Campylobacter spp. are leading diarrhea-causing bacterial agents in humans. Campylobacter jejuni causes the majority (>85%) of campylobacteriosis cases, while the remainder are caused primarily by C. coli (6, 21). Campylobacter is a zoonotic agent that commonly colonizes poultry and other food animals, including swine, cattle, and sheep (12). Campylobacter spp. fre- quently display resistance to antibiotics, such as tetracycline and quinolones (1, 15). Tetracycline resistance is especially widespread and is encountered even in isolates from animals raised without antibiotics (7, 11, 14, 16, 20, 24).
The gene responsible for tetracycline resistance in Campylobacter, tet(O), has been most commonly reported on plasmids (8, 9, 15, 22, 23). Plasmids harboring tet(O) frequently harbor kanamycin resistance determinants as well (22). However, there is also evidence of tet(O) being harbored on the chromosome. For instance, an estimated 33% of tetracycline-resistant C. jejuni isolates from Canada lacked plasmids (9). The prevalence of chromosomally borne tet(O) was even higher (76%) among isolates from Australia (18). Such chromosomal determinants, however, have not yet been characterized.
Tetracycline resistance is highly prevalent among C. coli and C. jejuni isolates from turkeys and swine in North Carolina and other regions (7, 10, 16). In turkey-derived C. jejuni isolates, resistance to kanamycin was typically accompanied by tetracycline resistance, suggesting the presence of plasmids harboring tetracycline and kanamycin resistance determinants (10).
Data on possible chromosomal carriage of tet(O) among turkey- or swine-derived Campylobacter spp. have not been available. Chromosomal carriage of resistance determinants has important implications, including higher stability compared with plasmids and potential to be disseminated via transformation. The latter is especially relevant for Campylobacter, which is known to be naturally competent (25).
In this study, we describe chromosomal regions harboring tet(O) in two C. coli strains, C. coli 6067 and C. coli 6461. C. coli 6067 (ST-1150) was obtained from drinking water available to turkeys inside a turkey house, while strain 6461 (ST-854) was isolated from a swine fecal sample. Animals were grown conventionally in eastern North Carolina; the farms and bacterial isolations were previously described (27). C. coli 6067 is a member of cluster II, a lineage primarily associated with turkeys and highly distinct from other animal-derived C. coli strains (17). C. coli cluster II strains constitute more than 40% of turkey-derived C. coli isolates in eastern North Carolina (26; R. M. Siletzky and S. Kathariou, unpublished findings) and exhibit several unique attributes: they harbor a C. jejuni-like aspA allele, lack intervening sequences in 23S rRNA genes, and are invariably susceptible to erythromycin (4, 5, 17).
In C. coli 6067 (cluster II), tet(O) is part of a putative IS605 element inserted in a C. jejuni-associated citrate transporter gene.
The complete genome sequence of C. coli 6067 was determined at the Genome Core Facility, Duke University, Durham, NC. Sequence annotation using the updated GAMOLA suite (2) and subsequent comparative analyses using the Functional Genome Distribution algorithm (3) revealed a chromosomal tet(O) gene with >99% identity at the nucleotide sequence level to other Campylobacter tet(O) sequences in GenBank.
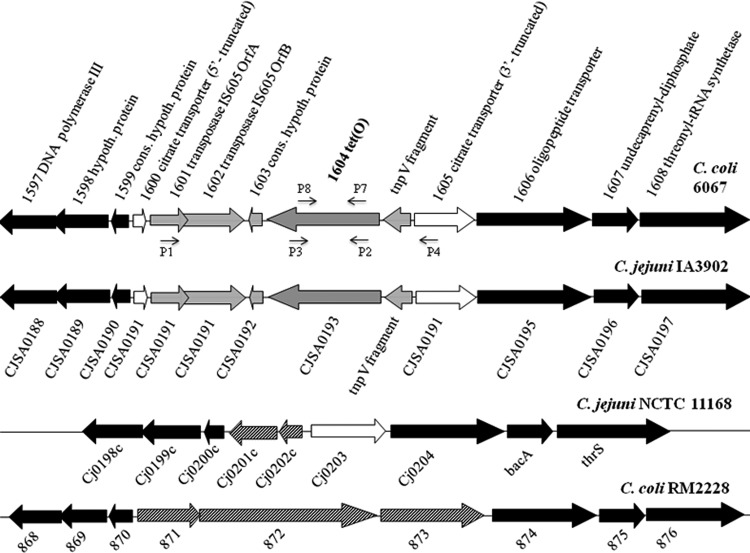
Analysis of the tet(O)-harboring region of C. coli 6067 revealed that the gene was part of a chromosomal gene cassette (transposon) that carries, in addition to tet(O), putative IS605 transposases ORF A and ORF B and the putative TnpV gene (Fig. 1). The transposon has interrupted a putative citrate transporter gene, the 5′ portion of which (167 nucleotides [nt]) is designated ORF 1600, with the remaining 3′ portion of the coding sequence designated ORF 1605 (Fig. 1). Sequence analysis suggests that the transposon has become inserted in the putative citrate transporter gene recently: there is no evidence of sequence degeneration in the insertionally disrupted gene, with ORFs 1600 and 1605 exhibiting 96 and 98% identity, respectively, at the nucleotide sequence level to the corresponding fragments of ORF Cj0203 of C. jejuni NCTC 11168. The interrupted putative citrate transporter was flanked on both sides by genes conserved among other sequenced Campylobacter genomes (e.g., C. jejuni IA3902, C. jejuni NCTC 11168, C. jejuni RM1221, and C. coli RM2228) (Fig. 1).
Fig 1.
Organization of the tet(O)-harboring region in C. coli 6067 and C. jejuni IA3902 and genomically equivalent regions in other Campylobacter genomes. Arrow orientations indicate the putative directions of transcription. ORFs flanking the tet(O) cassette and conserved (cons.) among the genomes are in black. ORFs composing the cassette are in gray, while the putative citrate transporter ORF harboring the tet(O) cassette is in white. ORFs unique to specific genomes are striped. P1, P2, P4, P5, P7, and P8 are primers, which are also listed in Table 2. hypoth., hypothetical.
Comparative genomic analysis of this region and nucleotide BLAST searches revealed that the putative citrate transporter gene was highly associated with C. jejuni; it was absent from sequenced C. coli genomes, with the sole exception of C. coli 317/04, a human clinical isolate (Fig. 1) (data not shown). In fact, this gene has been identified as one of the C. jejuni core genes that are dispensable in C. coli (13).
Surprisingly, the genomic organization of the region containing tet(O) in C. coli 6067 was identical to that of C. jejuni IA3902, a tetracycline-resistant strain representing a clonal group implicated in sheep abortions in the United States (19) (Fig. 1). The cassette harboring tet(O) was highly conserved (99% identity at the nucleotide sequence level) between C. coli 6067 and C. jejuni IA3902. However, in other C. jejuni and C. coli strains with completely sequenced genomes (e.g., C. jejuni NCTC 11168, C. jejuni RM1221, and C. coli RM2228), this region lacked tet(O) and instead harbored unrelated ORFs (Fig. 1). In C. jejuni NCTC 11168 and C. jejuni RM1221, these included the gene encoding the putative citrate transporter (also harbored by C. coli 6067 and C. jejuni IA3902, as discussed above), a putative integral membrane protein, and a hypothetical protein. In C. coli RM2228, ORFs 871 to 873 encoded two putative alpha-2 macroglobulin family proteins and a putative penicillin binding protein 1C (Fig. 1).
Nucleotide BLAST analysis revealed that in C. jejuni and C. coli, the sequences in the regions genomically equivalent to the tet(O) region of C. coli 6067 were associated with the respective species; for instance, ORFs 871 to 873 of C. coli RM2228 were harbored by diverse C. coli strains but not by any of the C. jejuni genomes in GenBank (data not shown). Similarly, ORFs Cj0201c and Cj0202c in C. jejuni NCTC 11168 were harbored by other C. jejuni strains but were absent from C. coli (with the exception of C. coli 317/04, as also described above for the putative citrate transporter gene Cj0203) (Fig. 1) (data not shown).
Swine-derived C. coli 6461 harbors a tet(O) cassette in a different chromosomal location.
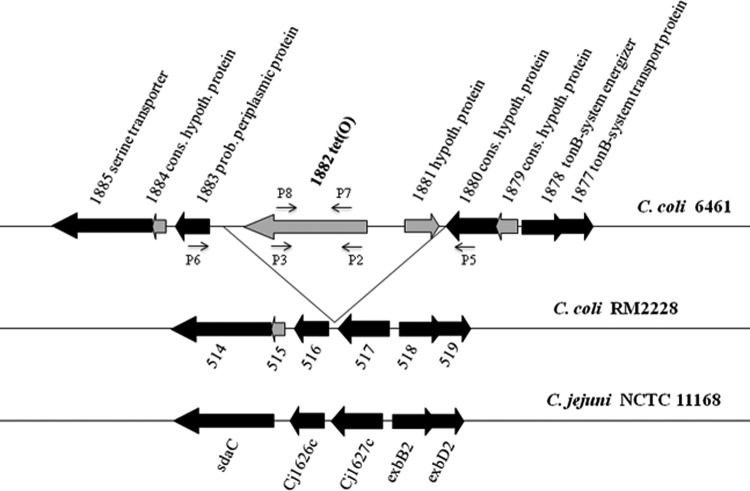
In the swine-derived C. coli strain 6461, a chromosomal tet(O) was identified in a different location from that in C. coli 6067 and was accompanied by an additional ORF (ORF 1881) encoding a hypothetical protein. The sequences flanking tet(O) and ORF 1881 in C. coli 6461 were generally highly conserved among other genomes of C. coli and C. jejuni (Fig. 2). Nucleotide BLAST analysis of ORF 1881 revealed a homologous ORF (100% identity) in the tetracycline resistance plasmid pTet of a C. jejuni strain implicated in a Guillain-Barré syndrome outbreak (28). In this plasmid, the ORF 1881 homolog was also adjacent to tet(O), suggesting the possibility that the tet(O) cassette in C. coli 6461 may have resulted from the chromosomal integration of a plasmid.
Fig 2.
Genomic organization of the tet(O)-harboring-region in C. coli 6461 and homologous regions in other Campylobacter genomes. Arrow orientations indicate putative directions of transcription. ORFs flanking the tet(O) cassette and conserved among the genomes are in black. C. coli-specific ORFs in the flanking regions are in gray. P2 to P8 are primers, which are also listed in Table 2.
Conservation of chromosomal tet(O) cassette in cluster II C. coli.
As mentioned above, C. coli 6067 belongs to the distinct clonal group cluster II, which is primarily associated with turkeys (17). To determine whether the chromosomal tet(O) genomic organization observed in this strain was conserved in other cluster II strains, we analyzed a panel of strains (Table 1) using PCR with primers derived from tet(O) and the sequences flanking the tet(O) cassette in C. coli 6067 (Table 2). The results showed that chromosomal tet(O) found in C. coli 6067 was indeed harbored in the same region by all other tested cluster II C. coli strains (Fig. 3). Two strains (6100 and 6252) produced weak amplicons with both sets of flanking primers, while their tet(O) amplicon was of normal intensity (Fig. 3). This was reproducibly observed and may reflect diversity in the primer sequences of these strains.
Table 1.
C. coli strains employed in this study
| Strain | Sourcea | Date (mo/yr) | Antibiotic susceptibility profileb | STc |
|---|---|---|---|---|
| 6067 | Water, turkey house | 11/2003 | TSQ | 1150 |
| 6077 | Turkey | 12/2003 | TSQ | 1161 |
| 6100 | Turkey | 12/2003 | T | 1161 |
| 6252 | Turkey | 03/2004 | TKQ | 1487 |
| 6690 | Turkey | 05/2004 | T | 1833 |
| 6890 | Turkey | 06/2004 | TQ | 1161 |
| 6979 | Turkey | 06/2004 | TKS | 1150 |
| 7725 | Turkey | 08/2004 | T | 1150 |
| 8023 | Turkey | 08/2004 | TQ | 1192 |
| 8901 | Turkey | 10/2004 | TQ | 1192 |
| WP145 | Swine | 12/2002 | T | ND |
| 5973 | Swine | 10/2003 | TK | 1142 |
| 5974 | Swine | 10/2003 | TSE | 1151 |
| 5979 | Swine | 10/2003 | TSK | 1153 |
| 5997 | Swine | 11/2003 | TSEK | 1246 |
| 6008 | Swine | 11/2003 | TSEK | 1246 |
| 6022 | Swine | 11/2003 | TEK | 1142 |
| 6029 | Swine | 11/2003 | TSE | 1151 |
| 6084 | Swine | 12/2003 | TE | 829 |
| 6087 | Swine | 12/2003 | TEK | 828 |
| 6093 | Swine | 12/2003 | TEK | 1157 |
| 6094 | Swine | 12/2003 | TE | 1157 |
| 6123 | Swine | 11/2003 | TEK | 1164 |
| 6461 | Swine | 04/2004 | TSE | 854 |
With the exception of C. coli 6067 (isolated from water in the turkey house), bacteria were obtained as described previously (27) from fecal samples from the indicated animal source.
Antimicrobial susceptibility profiles were determined as described previously (10) for a panel of antibiotics, including tetracycline (T), streptomycin (S), erythromycin (E), kanamycin (K), and (fluoro)quinolones (nalidixic acid and ciprofloxacin [Q]).
Sequence types (STs) were determined by multilocus sequence typing as described previously (17). ND, not determined.
Table 2.
Primers employed in the studya
| Primer (alternative name) | Sequence (5′→3′) |
|---|---|
| 6067_1601F (P1) | GCTAGACTTTATGGCTCACG |
| 6461_1882F (P2) | ATGGAGGGGGTTCTTTATGG |
| 6461_1882R (P3) | ATGCCATCCTTTGCAGAAAC |
| 6067_1605R (P4) | GTATCCGTTACGCTTTTGAC |
| 6461_1880F (P5) | GCTTGAGGTTTGTGATGCAA |
| 6461_1883R (P6) | CACCCAATAAAGCCGCTAAA |
| Tet(O) F (P7) | CAAAGGGGAATCACTATCC |
| Tet(O) R (P8) | AACCTGCCCGCATAGTTC |
Fig 3.
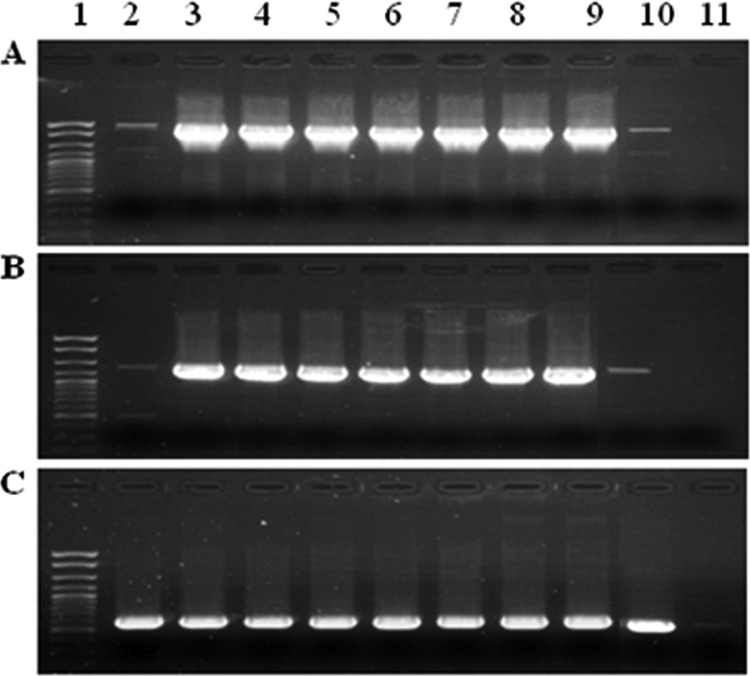
Conservation of the genomic location of the tet(O) cassette among cluster II C. coli strains. The PCR employed primers derived from the tet(O) cassette of C. coli 6067. (A) PCR using primers for region downstream of tet(O) (P1 and P2); (B) PCR using primers for the region upstream of tet(O) (primers P3 and P4); (C) PCR using tet(O) primers (internal primers P7 and P8). Lane 1, molecular weight markers (exACTGene cloning DNA ladder; Fisher Scientific International, Inc.); lanes 2 to 10, cluster II C. coli strains 6100, 6077, 6690, 6890, 6979, 7725, 8023, 8901, and 6252, respectively; lane 11, negative control (no DNA).
PCR using primers P6 and P2, as well as P3 and P5 (Table 2 and Fig. 2), was also employed to determine whether the chromosomal tet(O) found in the swine-derived C. coli strain 6461 was harbored in the same region in other tetracycline-resistant C. coli strains from swine (Table 1). None of the 13 strains tested yielded evidence of a tet(O) cassette in the same region as C. coli 6461 (data not shown).
In conclusion, our findings suggest that a chromosomal transposable unit (IS605) harboring tet(O) has become inserted in a C. jejuni-associated citrate transporter gene and has been acquired by cluster II C. coli, a unique lineage commonly associated with turkeys, and possibly by additional C. coli strains. Remarkable conservation was noted in the sequence content, genomic location, and organization of this tet(O) cassette in cluster II C. coli strains and in a C. jejuni strain (IA3902) responsible for outbreaks of sheep abortions (19). One may speculate that the chromosomal cassette became transferred, likely via transformation, between strains such as C. jejuni IA3902 and the cluster II C. coli strains. Further studies are needed to characterize the distribution and potential impact on fitness (including the animal colonization potential) of this cassette, as well as the chromosomal tet(O) cassette identified in C. coli 6461.
Nucleotide sequence accession numbers.
The nucleotide sequences of the tet(O) regions in C. coli 6067 and 6461 have been submitted to GenBank under accession no. JQ613155 and JQ613156, respectively.
ACKNOWLEDGMENTS
This research was partially supported by USDA-NRI competitive grant 2008-35201-04664.
We are grateful to Gregory Wray and the Genome Sequencing and Analysis Core Resource at Duke University for carrying out the sequencing. We thank all members of our laboratory for feedback and support.
Footnotes
Published ahead of print 28 September 2012
REFERENCES
- 1. Alfredson DA, Korolik V. 2007. Antibiotic resistance and resistance mechanisms in Campylobacter jejuni and Campylobacter coli. FEMS Microbiol. Lett. 277:123–132 [DOI] [PubMed] [Google Scholar]
- 2. Altermann E, Klaenhammer TR. 2003. GAMOLA: a new local solution for sequence annotation and analyzing draft and finished prokaryotic genomes. OMICS 7:161–169 [DOI] [PubMed] [Google Scholar]
- 3. Altermann E. 2012. Tracing lifestyle adaptation in prokaryotic genomes. Front. Microbiol. 3:48 doi:10.3389/fmicb.2012.00048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chan K, Elhanafi D, Kathariou S. 2008. Genomic evidence for interspecies acquisition of chromosomal DNA from Campylobacter jejuni by Campylobacter coli strains of a turkey-associated clonal group (cluster II). Foodborne Pathog. Dis. 5:387–398 [DOI] [PubMed] [Google Scholar]
- 5. Chan K, Miller WG, Mandrell RE, Kathariou S. 2007. The absence of intervening sequences in 23S rRNA genes of Campylobacter coli isolates from turkeys is a unique attribute of a cluster of related strains which also lack resistance to erythromycin. Appl. Environ. Microbiol. 73:1208–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Friedman CR, Neimann J, Wegener HC, Tauxe RV. 2000. Epidemiology of Campylobacter jejuni infections in the United States and other industrialized nations, p 121–138 In Nachamkin I, Blaser M. (ed), Campylobacter, 2nd ed American Society for Microbiology, Washington, DC [Google Scholar]
- 7. Gebreyes WA, Thakur S, Morrow WE. 2005. Campylobacter coli: prevalence and antimicrobial resistance in antimicrobial-free (ABF) swine production systems. J. Antimicrob. Chemother. 56:765–768 [DOI] [PubMed] [Google Scholar]
- 8. Gibreel A, et al. 2005. Macrolide resistance in Campylobacter jejuni and Campylobacter coli: molecular mechanism and stability of the resistance phenotype. Antimicrob. Agents Chemother. 49:2753–2759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gibreel A, et al. 2004. Incidence of antibiotic resistance in Campylobacter jejuni isolated in Alberta, Canada, from 1999 to 2002, with special reference to tet(O)-mediated tetracycline resistance. Antimicrob. Agents Chemother. 48:3442–3450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gu W, Siletzky RM, Wright S, Islam M, Kathariou S. 2009. Antimicrobial susceptibility profiles and strain type diversity of Campylobacter jejuni isolates from turkeys in eastern North Carolina. Appl. Environ. Microbiol. 75:474–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Halbert LW, et al. 2006. Genetic mechanisms contributing to reduced tetracycline susceptibility of Campylobacter isolated from organic and conventional dairy farms in the midwestern and northeastern United States. J. Food Prot. 69:482–488 [DOI] [PubMed] [Google Scholar]
- 12. Jacobs-Reitsma W. 2000. Campylobacter in the food supply, p 467–481 In Nachamkin I, Blaser MJ. (ed), Campylobacter, 2nd ed American Society for Microbiology, Washington, DC [Google Scholar]
- 13. Lefébure T, Bitar PD, Suzuki H, Stanhope MJ. 2010. Evolutionary dynamics of complete Campylobacter pan-genomes and the bacterial species concept. Genome Biol. Evol. 2:646–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Luangtongkum T, et al. 2008. Prevalence of tetracycline-resistant Campylobacter in organic broilers during a production cycle. Avian Dis. 52:487–490 [DOI] [PubMed] [Google Scholar]
- 15. Luangtongkum T, et al. 2009. Antibiotic resistance in Campylobacter: emergence, transmission and persistence. Future Microbiol. 4:189–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Luangtongkum T, et al. 2006. Effect of conventional and organic production practices on the prevalence and antimicrobial resistance of Campylobacter spp. in poultry. Appl. Environ. Microbiol. 72:3600–3607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Miller WG, et al. 2006. Identification of host-associated alleles by multilocus sequence typing of Campylobacter coli strains from food animals. Microbiology 152:245–255 [DOI] [PubMed] [Google Scholar]
- 18. Pratt A, Korolik V. 2005. Tetracycline resistance of Australian Campylobacter jejuni and Campylobacter coli isolates. J. Antimicrob. Chemother. 55:452–460 [DOI] [PubMed] [Google Scholar]
- 19. Sahin O, et al. 2008. Emergence of a tetracycline-resistant Campylobacter jejuni clone associated with outbreaks of ovine abortion in the United States. J. Clin. Microbiol. 46:1663–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sato K, Bartlett PC, Kaneene JB, Downes FP. 2004. Comparison of prevalence and antimicrobial susceptibilities of Campylobacter spp. isolates from organic and conventional dairy herds in Wisconsin. Appl. Environ. Microbiol. 70:1442–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Scallan E, et al. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg. Infect. Dis. 17:7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Taylor DE, Garner RS, Allan BJ. 1983. Characterization of tetracycline resistance plasmids from Campylobacter jejuni and Campylobacter coli. Antimicrob. Agents Chemother. 24:930–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tenover FC, Williams S, Gordon KP, Nolan C, Plorde JJ. 1985. Survey of plasmids and resistance factors in Campylobacter jejuni and Campylobacter coli. Antimicrob. Agents Chemother. 27:37–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thibodeau A, et al. 2011. Presence and characterization of Campylobacter jejuni in organically raised chickens in Quebec. Can. J. Vet. Res. 75:298–307 [PMC free article] [PubMed] [Google Scholar]
- 25. Wang Y, Taylor DE. 1990. Natural transformation in Campylobacter species. J. Bacteriol. 172:949–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wright SL. 2007. Potential transfer of Campylobacter between turkeys and swine produced in close proximity, in eastern North Carolina. M.S. thesis North Carolina State University, Raleigh, NC [Google Scholar]
- 27. Wright SL, et al. 2008. Longitudinal study of prevalence of Campylobacter jejuni and Campylobacter coli from turkeys and swine grown in close proximity. J. Food Prot. 71:1791–1796 [DOI] [PubMed] [Google Scholar]
- 28. Zhang M, et al. 2010. Association study between an outbreak of Guillain-Barré syndrome in Jilin, China, and preceding Campylobacter jejuni infection. Foodborne Pathog. Dis. 7:913–919 [DOI] [PubMed] [Google Scholar]