Abstract
Streptococcus iniae causes severe septicemia and meningitis in farmed fish and is also occasionally zoonotic. Vaccination against S. iniae is problematic, with frequent breakdown of protection in vaccinated fish. The major protective antigens in S. iniae are the polysaccharides of the capsule, which are essential for virulence. Capsular biosynthesis is driven and regulated by a 21-kb operon comprising up to 20 genes. In a long-term study, we have sequenced the capsular operon of strains that have been used in autogenous vaccines across Australia and compared it with the capsular operon sequences of strains subsequently isolated from infected vaccinated fish. Intriguingly, strains isolated from vaccinated fish that subsequently become infected have coding mutations that are confined to a limited number of genes in the cps operon, with the remainder of the genes in the operon remaining stable. Mutations in strains in diseased vaccinated fish occur in key genes in the capsular operon that are associated with polysaccharide configuration (cpsG) and with regulation of biosynthesis (cpsD and cpsE). This, along with high ratios of nonsynonymous to synonymous mutations within the cps genes, suggests that immune response directed predominantly against capsular polysaccharide may be driving evolution in a very specific set of genes in the operon. From these data, it may be possible to design a simple polyvalent vaccine with a greater operational life span than the current monovalent killed bacterins.
INTRODUCTION
Evasion of the adaptive immune response through genetic evolution of a pathogen is a major factor governing long-term vaccine success (15). This is dependent on the nature of the antigens targeted by the adaptive immune response: if they are highly conserved, the diversity among the population will be low, or the population may be eliminated altogether, with reemergence of disease in vaccinated stock rarely occurring (15, 16). In contrast, adaptive immune selection against highly polymorphic antigenic determinants may result in pathogen populations restructuring into antigenic types or serotypes, with certain types predominating in vaccinated stock and fluctuations in the genetic structure of the population dictated by the serotype or types included in the vaccine (15).
Vaccinations against Streptococcus iniae infections in barramundi farms in Australia provide an ideal model for exploring evolution of pathogen populations. First, it is almost impossible to eliminate the source of infection from the farm. In ponds and marine cage systems, reinfection from sediments, wild fish, and other aquatic inhabitants is unavoidable (1, 7, 8). Even in completely enclosed recirculating systems, economics dictate that the system cannot be shut down and disinfected completely; thus, populations of the pathogen are likely to remain in biofilms on the pipework, tanks, and filters. Second, the infectious agent S. iniae is highly variable (6, 12, 26), with its major antigenic determinant, polysaccharide capsule (6, 18, 21), being highly polymorphic: novel capsular serotypes have already led to vaccine failure in fish farms in several parts of the world, including Australia (1, 4, 11, 26). Finally, in Australia, there is no licensed generic vaccine against S. iniae, so autogenous vaccines (“custom” vaccines prepared from an isolate taken directly from the original farm and used only on that farm) are routinely employed to prevent streptococcosis. This creates a closed cycle whereby a pathogenic strain isolated from a particular farm is used to prepare a vaccine that is then used to vaccinate stocks that are reintroduced onto the same farm.
The capsular polysaccharide (CPS) is located on the outermost layer of bacterial cells and is ubiquitous across several bacterial species (28). For pathogenic Streptococcus species, CPS varies depending on serotype and is a recognized virulence factor that contributes to immune evasion (10, 18). In group B Streptococcus (Streptococcus agalactiae), variations in the polysaccharide capsule have been implicated in reduced complement C3b binding and, ultimately, avoidance of phagocytosis (19). Similarly, research on Streptococcus pneumoniae revealed that morphological changes in the capsule occur in order to adapt to the host's environment (13). In Streptococcus iniae, as with the other streptococcal species, production of the capsular polysaccharide appears to be necessary for infection (21). Locke and colleagues (17) elegantly demonstrated that cpsD, a homologue of a gene shown to be required for CPS production in group B Streptococcus and S. pneumoniae, was required for capsule formation and export in S. iniae. Although a few studies have investigated virulence factors in S. iniae, the function of the genes in the capsular operon are less well known, and how these translate into multiple serotypes in this species is yet to be elucidated.
In the present study, we investigated molecular evolution of the capsular operon of S. iniae from diverse origins in order to determine which genes were most variable. We found that five genes out of the approximately 21-kb cps operon were highly variable, and when we investigated case studies of repeated autogenous vaccination and vaccine failure at Australian barramundi farms for links between the highly mutable genes and reinfection of vaccinated stock, we found a direct correlation with the variable cps genes and vaccination failure. Surprisingly, no capsule was formed in some of our isolates, yet the pathogen was still able to infect the host, albeit with a completely different pathology. To further understand these findings on the effect of vaccination against S. iniae, we examined serological cross-reactivity using antibodies raised in fish between isolates with differing cps gene sequences from several of the case studies in which vaccinated stock became reinfected. Our results suggest that polyvalent vaccines comprising different cps sequence types are partially effective but that future generic vaccines may need to target alternative antigens that are less polymorphic than CPS.
MATERIALS AND METHODS
Bacterial strains and culturing.
Bacterial isolates received from culture collections or direct from fish farms or veterinarians were stored at −80°C in Todd-Hewitt broth (THB) containing 20% glycerol until required and are listed in Table S1 in the supplemental material. Strains were recovered from stock without defrosting and grown on Columbia agar base containing 5% defibrinated sheep blood (Oxoid, Australia) at 28°C for 24 to 48 h. Where isolates were obtained from farm cases through veterinarians, histopathology was performed by the veterinary laboratories using standard techniques. Identity of strains was confirmed by diagnostic PCR as previously described (20, 27) and by sequencing the 16S rRNA gene (26, 27).
DNA extraction, primer design, PCR, and sequencing.
S. iniae genomic DNA was extracted from freshly grown cells using an enzymatic lysis method as previously described (27a). Primers for PCR and sequencing were designed using the S. iniae capsule operon sequence available on GenBank (sequence accession number AY904444). The primers used were as described in Barnes (5) and are listed in Tables S2 and S3 in the supplemental material.
Capsular operon genes were amplified individually or in blocks of several genes (see Table S2 in the supplemental material) with a proofreading DNA polymerase (PrimeStar HS DNA polymerase; TaKaRa, Japan) in 25-μl reactions composed of 5 μl of 5× PCR buffer, 0.5 μl of each deoxynucleoside triphosphate (dNTP) (2.5 mM), 100 ng of each primer, 0.15 μl of PrimeStar DNA HS DNA polymerase, 100 to 200 ng of extracted bacterial DNA, and the balance made up of sterile Milli-Q water. Reactions were carried out at annealing temperatures appropriate to each primer pair (see Table S2 in the supplemental material) (5). A “hot start” technique was employed to reduce the likelihood of nonspecific amplification products prior to cycling as follows: 2 min at 94°C for one cycle, followed by 35 cycles of denaturation for 15 s at 94°C, 30 s at an appropriate annealing temperature, and extension at 72°C of 1 min for every expected kb of the amplicon. Reaction mixtures were analyzed by agarose gel electrophoresis.
Where a single amplicon was produced, 0.6 μl of PCR product was added to 0.3 μl of a 1:2 mixture of exonuclease I and shrimp alkaline phosphatase and 3.1 μl of sterile Milli-Q water and incubated at 37°C for 30 min, followed by heating at 85°C for 15 min before being sent for DNA sequencing by the Australian Genome Research Facility (AGRF; Brisbane, Australia). Where more than one amplicon was observed (rarely), they were excised from the gel with a sterile scalpel blade and then extracted from the gel slice using a commercial gel purification kit for subsequent sequencing (MEGA-Spin gel extraction kit; Intron Biotechnology, South Korea).
Chromatograms were analyzed using Sequencher v. 4.9 (Genecodes). Contigs were assembled using equivalent genes from the published genome as references, and the assembled complete cDNA sequences were compared to the reference sequences using both nucleotide and translated variance tables in Sequencher 4.9. Sequence types (STs) were assigned on an ad hoc basis as discovered, with the sequence types for the type strain assigned ST1. Changes resulting in an amino acid change were assigned ST numbers, while synonymous nucleotide changes were denoted with a letter appended to the identical amino acid sequence type. Sequences for each of the STs of cps genes were uploaded to GenBank as follows: cpsY ST1 and ST2, GenBank accession numbers JX164243 and JX164242, respectively; cpsD ST1, ST2, ST3, ST3A, and ST4, GenBank accession numbers JX164238, JX164245, JX164246, JX164239, and JX164247, respectively; cpsE ST1, ST2, ST3, ST4, ST5A, ST5, ST6, ST7, and ST8, GenBank accession numbers JX164231, JX164248, JX164232, JX164233, JX164249, JX164234, JX164235, JX164236, JX164237, and JX164250, respectively; cpsG ST1, ST2, and ST3, GenBank accession numbers JX164240, JX14244, and JX164241, respectively; and cpsH ST1, GenBank accession number JX181784.
Buoyant density assays.
As an estimate of quantity of CPS expressed by differing strains, buoyant density was determined in continuous Percoll gradients as described previously (17). Briefly, a standard isotonic Percoll solution was prepared by mixing 9 parts Percoll with 1 part 1.5 M NaCl. Mid-exponential-phase and stationary-phase THB cultures of each strain were washed in phosphate-buffered saline (PBS) and resuspended to an optical density at 600 nm (OD600) of ∼2.5, and a 0.5-ml aliquot was layered onto the Percoll. All gradients were centrifuged simultaneously in an Eppendorf 5518E refrigerated centrifuge at 4,000 × g for 90 min with no brake. The experiment was repeated using new cultures at least twice.
Serological cross-reactivity by a whole-cell enzyme-linked immunosorbent assay (ELISA).
Lates calcarifer barramundi weighing 35 g were obtained from a commercial farm and held in 300-liter round plastic tanks with aerated brackish (5-ppt) water at 30 ± 1°C that were connected to a recirculating system. Water quality was maintained with mechanical and biological filtration and checked daily, and water changes were conducted when required. The fish were acclimated for 7 days and were fed to satiation with a commercial pelleted diet (Ridley Aqua Feeds, Narangba, Australia).
Fish were anesthetized with Aqui-S (Aqui-S, Lower Hutt, New Zealand) in accordance with the manufacturer's instructions and then vaccinated by intraperitoneal (i.p.) injection with 100 μl of oil-adjuvanted (1:1 emulsion in Freund's incomplete adjuvant) formalin-killed bacterin. Control fish were injected i.p. with PBS/adjuvant emulsion. Fish were allowed to recover in clean aquarium water before being returned to their respective tanks.
A total of 900 degree days postvaccination (i.e., 30 days at 30°C), the fish were euthanized by lethal overdose of anesthetic (Aqui-S) and bled by caudal venipuncture. Blood samples were allowed to clot at 4°C overnight and were centrifuged at 6,000 × g for 15 min to collect serum that was then stored at −20°C for subsequent analysis.
The specific antibody response of each individual vaccinated fish was determined using an enzyme-linked immunosorbent assay (ELISA) as described previously (9) with modifications. Microlon flat-bottom 96-well ELISA plates (Greiner) were coated by evaporating 100 μl/well of formalin-inactivated suspension (OD600 = 1.0) of QMA0177 and QMA0191 in coating buffer (carbonate-bicarbonate buffer [Sigma, Australia]) with a hair dryer. Coated plates were washed 3 times with Tris-buffered saline containing 0.1% Tween 20 (TBST) and blocked with 2% normal goat serum in TBST at room temperature for 1 h. The plates were washed 3 times with TBST, and 100 μl primary antisera from vaccinated fish and control serum from sham-vaccinated siblings was diluted 1:4 in TBST and incubated for 2 h at room temperature. Wells were washed 3 times with TBST, and secondary antibody (monoclonal mouse anti-barramundi IgM, diluted 1:32-fold in TBST [Aquatic Diagnostics, Stirling, United Kingdom]) was added and incubated for 1 h at room temperature. Wells were washed three times with TBST before addition of tertiary antibody conjugate (polyclonal goat anti-mouse IgG alkaline phosphatase conjugate, diluted 1:30,000 in TBST [Sigma-Aldrich]) and incubated for 1 h at room temperature. Wells were washed twice in 0.1% TBST and once in TBS, and then color was developed for 1 h using p-nitrophenyl phosphate liquid substrate (50 μl/well) (Sigma-Aldrich). Absorbance was measured at 405 nm with a FLUOstar Optima spectrophotometer/fluorimeter/luminometer (BMG LabTech).
RESULTS
Variation in capsular genotype is confined to a few genes in the operon.
Capsular biosynthesis in S. iniae is under the control of a 21-kb operon containing around 20 genes. The operon has been fully sequenced (21) and characterized to some degree using site-directed mutagenesis (18). To determine whether genetic changes in the capsular operon correlate with infection in vaccinated animals and virulence, we initially sequenced all genes within the capsular operon of 10 S. iniae isolates. These strains came from diverse origins and included strains from documented vaccine failures (4, 5, 26).
Sequences of the genes from the capsular operon of these 10 isolates resulted in a number of surprising findings (Table 1). First, the number of synonymous (noncoding) mutations across the 21-kb operon was incredibly low, compared to coding mutations, which occurred frequently. Coding mutations, resulting in amino acid changes in the expressed proteins, were restricted to a limited set of genes within the operon. That is, the mutations were not evenly distributed across all of the 21 kb but were confined to a limited set of 5 genes, cpsY, cpsD, cpsE, cpsG, and cpsH (Table 1).
Table 1.
Sequence types of genes from the capsular operon of 10 isolates of S. iniae separated either geographically or serologicallya
| Gene | ST for each strain no. (origin) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| QMA0072 (QLD) | QMA0076 (QLD) | QMA0083 (WA) | QMA0140 (USA) | QMA0155 (NSW) | QMA0165 (QLD) | QMA0177 (NT) | QMA0191 (NT) | QMA0188 (ISR) | QMA0186 (ISR) | |
| cpsY | 2 | 2 | ||||||||
| cpsD | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3A | 2 | |
| cpsE | 3A | 4 | 3 | 3 | 3A | 3 | 3 | 2 | 2 | |
| cpsG | 2 | 2 | 3 | X | X | |||||
| cpsH | 2 | |||||||||
Gene sequences from the type strain (QMA0140) were designated ST1. As coding mutations were discovered, they were allocated a new sequence type number arbitrarily in the order of discovery. Noncoding mutations are given the ST number of the identical expressed sequence and identified as variants with a letter (e.g., ST3A expresses the same protein as ST3 but has one or more noncoding single nucleotide polymorphisms [SNPs]). Genes are ST1 if not otherwise indicated. X indicates a deleted gene. Only genes that differ from the type strain are included in the table. Strain geographic origin abbreviations: QLD, Queensland, Australia; WA, Western Australia; NSW, New South Wales, Australia; NT, Northern Territory, Australia; USA, United States of America; ISR, Israel.
Subsequently, we extended our study to an additional 29 isolates and sequenced blocks of genes encompassing these regions (Table 2). We found that cpsE had the largest diversity (that is, the largest number of variant sequence types [STs]). However, cpsE was not the most frequently modified gene in the operon; variance in cpsG occurred more frequently in Australian isolates (see Table S1 in the supplemental material) (5). cpsD was also variable, with 4 different coding sequence types found among the isolates sequenced. In contrast, diversity of cpsD was lower in Australia, with only two sequence types found. Indeed, ST3 was universal across all states in Australia, and ST1 was identical to the type strain from dolphins, arising only under high selective pressure in two cases in New South Wales and South Australia (see Table S1 in the supplemental material; see also the case histories described below). The cpsY gene also had low variability; all isolates examined carried ST1, regardless of global geographic origin, with the exception of the isolates from Northern Territory, Australia, that, without exception, carried the ST2 cpsY variant (Table 2).
Table 2.
Nucleotide and amino acid changes associated with differing cps gene sequence types
| Gene and position | Nucleotides/amino acids ina: |
|||||||
|---|---|---|---|---|---|---|---|---|
| ST1 | ST2 | ST3 | ST4 | ST5b | ST6b | ST7 | ST8 | |
| cpsY | ||||||||
| 535/179 | TTC/F | TCC/S | ||||||
| cpsD | ||||||||
| 367/123 | CCA/P | CTA/L | ||||||
| 385/129 | TTA/L | |||||||
| 475/175 | GTG/V | TTG/L | TTG/L | |||||
| cpsE | ||||||||
| 119/40 | GAA/E | GGA/G | GGA/G | GGA/G | GGA/G | GGA/G | GGA/G | |
| 136/46 | TTT/F | GTT/V | ||||||
| 1426/476 | CGT/R | CTT/L | ||||||
| 1081/361 | CAT/H | Del C→ | Del C→ | |||||
| 1108/370 | TTA/L | TAA/stop | TAA/stop | |||||
| 1096/366 | AAG/K | ACG/T | ||||||
| 1315/439 | GCA/A | GTA/V | ||||||
| cpsG | ||||||||
| 495/165 | TTA/L | Del TTA/L | ||||||
| 498/166 | TCA/S | Del TCA/S | ||||||
| 501/167 | AAG/K | Del AAG/K | ||||||
| 504/167.1 | Ins TTA/L | |||||||
| 507/167.2 | Ins TCA/S | |||||||
| 510/167.3 | Ins AAG/K | |||||||
| cpsH | ||||||||
| 669/223 | TTA/L | TTT/F | ||||||
Underlining indicates the changed nucleotide relative to the type strain (ST1). Del, deletion; ins, insertion.
A single nucleotide deletion leads to a changed amino acid sequence from aa 361 to an early termination at aa 370.
Coding mutations in the capsular operon correlate with autogenous vaccine failure.
Reoccurrence of infection by S. iniae in vaccinated fish has been reported previously in Israel (4), and this appeared to be a result of changes in capsular polysaccharide and secreted exopolysaccharide (11). We sequenced the complete capsular operon in two postvaccine strains from Israel that had differing random amplified polymorphic DNA (RAPD) profiles (4) but that were apparently serologically cross-reactive (4, 11) and found a single coding mutation in cpsD (Table 1) resulting in a change from proline to leucine at amino acid (aa) position 123.
Northern Territory, Australia.
In April 2005, streptococcosis emerged as a cause of high mortalities in farmed barramundi in Port Hurd, Northern Territory (NT), Australia. In July 2005, a cohort of fingerlings was vaccinated with an autogenous vaccine prepared from the isolate obtained from the index case and designated strain QMA0191 in the UQ strain collection (Table 3) and introduced onto the site. Further cases of clinical streptococcosis were recorded in the fish at Port Hurd in July, August, and September as well as isolated from brain and kidney in subclinically infected fish. All these cases were in nonvaccinated fish and are represented by the strain numbers QMA0142, QMA0150, and QMA0153 (Table 3). In July 2006, an outbreak of clinical streptococcosis occurred in multiple cohorts of fish across the farm, including fish vaccinated with the autogenous vaccine containing strain QMA0191. An isolate taken at this time was also deposited in the UQ collection and designated QMA0177.
Table 3.
Case histories of strain isolation and vaccination in three farms, a sea farm in Northern Territory and recirculating aquaculture farms in South Australia and New South Walesa
| Location | Strain | ST of: |
Date of isolation (mo/yr) | Dates used in vaccines | ||
|---|---|---|---|---|---|---|
| cpsD | cpsE | cpsG | ||||
| Northern Territory | QMA0191 (v) | 3 | 3 | 1 | 04/2005 | 2005-2006 |
| QMA0142 | 3 | 3 | 1 | 07/2005 | ||
| QMA0150 | 3 | 3 | 1 | 08/2005 | ||
| QMA0153 | 3 | 3 | 1 | 09/2005 | ||
| QMA0177 | 3 | 3 | 3 | 07/2006 | ||
| South Australia | QMA0160 (v) | 3 | 3 | 1 | 12/1999 | 2004–2009 |
| QMA0159 | 3 | 3 | 2 | 03/2006 | ||
| QMA0243 | 3 | 3 | 2 | 05/2006 | ||
| QMA0244, QMA0245 | 3 | 3 | 2 | 10/2008 | ||
| QMA0246+, QMA0247, QMA0248 | 3 | 3 | 2 | 03/2009 | March/April 2009 divalent with QMA0160 | |
| QMA0249 | 1 | 5* | X | 05/2009 | ||
| New South Wales | QMA0155 (v) | 3 | 3 | 2 | 12/2005 | Early 2006 monovalent |
| QMA0220+ | 3 | 3 | 2 | 08/2006 | 2007-2008 | |
| QMA0250+ | 3 | 3 | 2 | 11/2007 | January 2008 | |
| QMA0251+, QMA0252+ | 3 | 3 | 2 | 06/2008 | July 2008 divalent | |
| QMA0233 | 1 | 5A* | X | 11/2008 | ||
| QMA0253+, QMA0254+ | 1 | 5A* | X | 01/2009 | March 2009 divalent | |
| QMA0236 | 1 | 5A* | X | 03/2009 | ||
Numbers indicate sequence types of indicated cps genes. X indicates a gene deletion. All other genes in the cps operon were identical among strains in the same table unless otherwise indicated.
, strain had genes cpsF-cpsM deleted; (v), strain used in initial autogenous vaccine;
, isolate used in subsequent vaccinations.
Analysis of the capsular genotypes of the strains recovered from unvaccinated fish during 2005 indicated a consistent molecular serotype identical to that of the strain (QMA0191) used to produce the autogenous vaccine and explains the efficacy of the vaccine (Table 3). When disease arose across the site in July 2006, more than 6 months after vaccination, mortality resulted from strains with a coding change in cpsG, a 6-amino-acid (2× LSK repeat) insert.
South Australia.
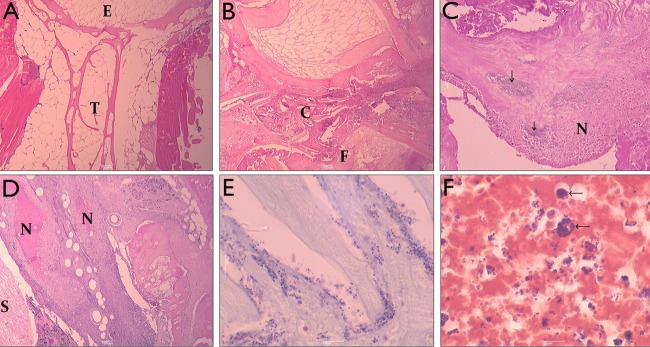
Vaccination of barramundi in a tank-based aquaculture facility in South Australia (SA) commenced in late 2004 using an autogenous vaccine prepared from an isolate from an outbreak in December 1999 (QMA0160). A subsequent outbreak resulted from a new strain in May 2006 (QMA0243) and again in October 2008 (QMA0244, QMA0255), continuing through the Southern-hemisphere summer to March 2009 (QMA0246 to QMA248), with all isolates exhibiting a shift in amino acid composition in the cpsG (ST1 and ST2) genes (Table 3). A new autogenous vaccine incorporating both strain types was used to vaccinate new stock in March to April 2009. In May 2009, low-level mortality was detected in vaccinated stock, but symptoms were atypical; barramundi developed spinal fractures. In some vaccinated fish, these spinal fractures were colonized by Gram-positive cocci, perpetuating spinal inflammation (Fig. 1). The brain and head-kidney (pronephros) of nine fish were sampled for bacteria, but no S. iniae was recovered, nor was S. iniae observed in the kidney during histology. S. iniae was observed in, and recovered from, only the bone lesions and the resulting exudate (Fig. 1). The isolates that caused this unusual pathology were found to have a frameshift mutation (single nucleotide deletion at nucleotide [nt] position 1081) in the cpsE gene that resulted in an early stop codon at amino acid 370, resulting in a severely truncated protein. Moreover, all of the capsular operon genes from cpsF-cpsM were deleted in these isolates (Table 3).
Fig 1.
Histopathology of spinal inflammation in infected barramundi. (A) L. calcarifer sagittal spinal vertebral section showing normal structure of central vertebral spongy bone trabeculae, indicated by T, and endplate, indicated by E, at ×40 magnification (hematoxylin and eosin [H&E]). (B) L. calcarifer spinal sagittal section at the point of collapse, indicated by C, of the intervertebral disc space due to inflammatory infiltrate and fracture of the vertebra, indicated by F, at ×40 magnification (H&E). (C) L. calcarifer spinal sagittal section showing multiple colonies of Gram-positive cocci. Arrows indicate inflamed and necrotic tissue exudate, indicated by N, at the site of spinal fracture at ×200 magnification (H&E). (D) L. calcarifer spinal sagittal section showing a severe intraspinal canal infiltration with inflammatory cells, caseous necrotic foci, indicated by N, containing colonies of phagocytosed coccoid bacteria. Inflammatory exudate is in close proximity to the spinal cord, indicated by S, at ×40 magnification (H&E). (E) Gram stain of an L. calcarifer sagittal spinal section showing colonies of Gram-positive coccoid and chain-forming bacteria in close association with the vertebral bony surfaces at ×1,000 magnification. (F) Gram stain of an L. calcarifer spinal vertebral inflammatory exudate showing multiple Gram-positive coccoid bacteria. Note the potentially phagocytosed bacteria in cells (arrows), but there are also free bacteria in the exudate, at ×1,000 magnification.
New South Wales, Australia.
A similar but separate case occurred at a recirculating aquaculture farm in NSW: barramundi were vaccinated with autogenous bacterins prepared from isolates QMA0155/QMA0220 that caused an outbreak that ran from late 2005 into early 2006. In November 2008, an outbreak occurred in vaccinated stock. As with the SA case, mortalities were low and associated with spine deformities in which streptococci were able to be detected in histology and from which S. iniae was subsequently isolated in culture. These isolates had a similar frameshift mutation in cpsE, coupled with deletion of cpsF-cpsM (Table 3).
Changes in genotype associated with vaccine failure are phenotypically and antigenically relevant.
In the vaccine failure case in Northern Territory, Australia, a single mutation was found in the cpsG gene (from ST1 to ST3). A buoyant density assay was performed to assess any phenotypic variation between the cps genes. Buoyant density was different in the two isolates, with isolate QMA0177 traveling further through a continuous Percoll gradient than strain QMA0191. This occurred with multiple independently prepared cell suspensions through Percoll gradients prepared at two dilutions. In the case of the SA and NSW cases with truncated cpsE and major deletions in the operon, the differences in buoyant density were more pronounced, with the mutated isolates QMA0253, QMA0236, and QMA0249 migrating completely through the continuous gradient when prepared using diluted Percoll (Fig. 2).
Fig 2.

Percoll buoyant density assay for NT (QMA0177, QMA0191), SA (QMA0160, QMA0243, QMA0249), and NSW (QMA0155, QMA0220, QMA0236, QMA0253) isolates.
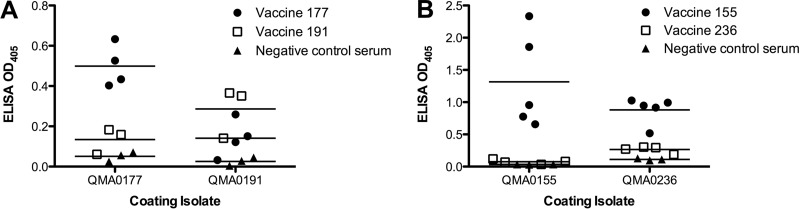
To investigate whether the changes seen in the capsular operon that coincided with reinfection of vaccinated fish were antigenically relevant, antibodies were raised in barramundi against the two isolates from the index case of reinfection in NT. Antibodies raised against QMA0191 were poorly cross-reactive with QMA0177 by a whole-cell ELISA (Fig. 3A). Similarly, antiserum raised against QMA0177 was poorly cross-reactive with QMA0191. In the case of the strains from NSW, antisera raised in barramundi against the initial vaccine isolate QMA0155 (same CPS sequence type as the subsequent vaccine strains QMA250 to QMA252) cross-reacted strongly with strain QMA0236, which occurred in vaccinated fish (Fig. 3B). When fish were vaccinated with QMA0236, a relatively low antibody response was detected in general, even against the same strain in a whole-cell ELISA, although it was higher than that for the negative-control serum. Although the response was low, it was significantly higher in a whole-cell ELISA against the homologous strain than against QMA0155 (Fig. 3B) (P < 0.05).
Fig 3.
Antibody cross-reactivity by a whole-cell ELISA for selected isolates from the index case in NT (QMA0177, QMA0191) (A) and the NSW case (QMA0155, QMA0236) (B).
DISCUSSION
Vaccination of finfish against bacterial pathogens has proven extremely effective at preventing mass mortality in farmed animals (29). However, it is this very efficacy that places a high selective pressure on pathogens, and this may lead to dominance by new serotypes less well recognized by the resultant specific immune response to the original vaccine. In the present study, we investigated genetic changes in S. iniae, a fish pathogen with a track record of new serotype emergence in vaccinated animals (4). We found mutations in the capsular operon that coincided with infection in previously vaccinated stocks. These genetic changes likely lead to changes in polysaccharide biosynthesis (6, 11). Intriguingly, changes were restricted to a limited suite of genes within the ∼20-gene operon. Firstly, two sequence types were found in cpsY. Of 39 isolate sequences, only strains from Northern Territory, Australia, contained the cpsY ST2 variant, with all other isolates being identical to the type strain. It is intriguing that a new sequence type should be found only in NT, especially when movements of fingerlings interstate are common. This suggests that S. iniae infections arose locally in the fish after they were shipped and that the bacterium was not cotransported with the fingerlings from the Darwin hatchery. cpsY ST2 would currently appear to be a robust diagnostic indicator for NT origin of the strain. cpsY is a transcriptional regulator that has an important role in intracellular survival in neutrophils and is also essential for successful dissemination to the brain (2, 3). The functional role of the mutation in the present case is unknown, but isolates were recovered from the brain in both outbreaks.
cpsA through cpsC were always identical to the type strain, regardless of the country of origin. In S. pneumoniae, cpsD is a self-phosphorylating tyrosine kinase, acting with cpsB and cpsC to regulate activity of cpsE (22–25). cpsD was found to vary in S. iniae and was the only coding mutation in the cps operon that was different between the two postvaccine isolates from the Israeli case of vaccine failure, with a switch in the amino acid at position 123 from proline to leucine. While the effect of this mutation at the functional level remains to be determined, a proline-leucine switch at this position will substantially change the shape of the protein, removing a bend at this residue. The Israeli team reported that these two isolates (named KPF177 [QMA0188] and KPF404 [QMA0186]) were serologically similar when tested with rabbit antisera using Ouchterlony immunodiffusion, but they differ in RAPD profiles (4). Each of these strains has been used in autogenous vaccines, and a novel serotype which differed in EPS production arose (11). It would be interesting to investigate the CPS operon of the newly emerged strain to determine whether coding mutations in the capsular biosynthesis machinery that led to increased polysaccharide overexpression or failure to anchor the membrane have arisen. In Australia, a coding mutation in cpsD was never found as the sole mutation in the cps operon, with most isolates analyzed having the ST3 cpsD sequence type. The only other cpsD variant found in Australia isolates was cpsD ST1, and this was found only in association with a frameshift mutation in cpsE. Once again, this cpsD mutation (ST3 to ST1) involved the proline-leucine switch at aa 123, derived from an identical single nucleotide mutation. In Australia, however, the proline-leucine switch was also accompanied by a valine-leucine substitution at aa 175. The effects of cpsD mutations will be difficult to interpret in Australian isolates due to the universal accompaniment of the change with a frameshift in cpsE. In S. iniae, cpsE encodes a UDP-glucose-dependent glycosyl transferase that initiates synthesis and export of the capsular polymer. There was more diversity of this gene than of any of the other cps genes, with 6 different sequence types found among the S. iniae isolates recovered in Australia and 8 in total among the 39 isolates analyzed. It is likely that mutations in this gene change the amount of capsular polysaccharide produced, and this can have profound effects on the tissue distribution of the organism during disease (18). Indeed, the frameshift mutations that resulted in early termination of the cpsE gene that occurred in strains infecting vaccinated fish in NSW and SA substantially increased the buoyant density, suggesting reduced/eliminated capsular polysaccharide production. This was associated with lower mortality and completely transformed pathology and symptoms. While these mutations often cooccurred with deletion of genes cpsF through cpsM (isolates QMA0233 to QMA0236; isolate QMA0249), it appears that the frameshift mutation in cpsE alone is sufficient to cause the phenotypic changes, as isolate QMA0158 (frameshift in cpsE, but remainder of the operon intact) exhibited an increase in buoyant density similar to that of the strains carrying the deletion of cpsF-cpsM. Deletion of cpsF-cpsM has been reported previously in a “commensal” strain of S. iniae (18). Our research suggests that these strains are not commensal, as they still cause pathology and mortality in a farm situation, but are somewhat attenuated (data not shown). This association with spinal fractures in barramundi is interesting, as human spinal osteomyelitis caused by Streptococcus dysgalactiae subsp. equisimilis has been reported (14). The structure of teleost fish bone is without a marrow cavity and Haversian system, which means that osteomyelitis cannot occur. However, inflammation can develop in avascular tissue by extension from the adjacent periosteal vessels and generally takes the form of rarefaction (demineralization) rather than frank necrosis. Teleost bone usually cannot be immobilized in a fracture situation and thus will heal in a deformed fashion, as observed in the cases reported here (data not shown).
cpsG is a putative UDP-glucose 4 epimerase that converts UDP-glucose to UDP-galactose. Three variants of cpsG, involving an insertion or deletion of three amino acids, LSK, at aa positions 165, 166, and 167 in the ST1 type strain, were found. In ST2, these three amino acids were deleted, while in ST3, they were repeated, resulting in a protein that was three amino acids longer than the type strain. The phenotypic effects of these mutations remain to be determined, but mutations in this gene that alter enzyme efficiency may change the rate of conversion and thus change the ratio of glucose/galactose in the final capsular polymer, with resulting effects on surface epitopes. Mutation from cpsG ST1 to ST3 was the only change in the index case of vaccine failure in Australia. This involves a 3-aa LSK repeat insertion in ST3 compared to ST1. How this may affect function is yet to be elucidated, but the two isolates involved were serologically different, as determined by an ELISA using fish antiserum. Moreover, there was a small change in the buoyant density of the cells, suggesting a change in CPS structure. Future work will focus on expressing these variants as recombinant proteins and measuring their epimerase activity. This, coupled with liquid chromatography-mass spectrometry (LC-MS) analysis of CPS from these two isolates, may elucidate how these mutations translate into antigenic variation. cpsG was deleted in a number of isolates, including the Israeli and Thai isolates, suggesting that it may not be essential for cps biosynthesis but may play an important role in the final molecular shape of the polysaccharides.
No cases of reinfected vaccinated fish have yet been associated with changes in cpsH, although two variants were found among the Australian strains. cpsH is a putative repeat unit polymerase, and changes in efficacy may change polymer length and, therefore, the final structure and antigenic nature of the capsule.
When disease arises in enclosed tank-based aquaculture systems, theoretically, best practice would be to close down the system, disinfect it completely, dismantle and clean pipework, pumps, and filtration, and then restart with clean stock. In practice, this is not possible, as multiple size classes are generally kept within the same system to allow a continual supply of table-sized fish for customers. Thus, outbreaks of S. iniae may be controlled with careful antibiotic treatment, and vaccinated stock may be reintroduced into the system. However, this means that the reservoir of infection is never really eradicated, providing ample opportunity for reinfection should immunity become compromised. In the closed systems in NSW and SA, careful use of autogenous vaccines initially controlled disease. In the latter period of the study, divalent vaccines were formulated after sequence typing to include the cps STs from the history of infection on the farm. It is intriguing that streptococcosis once again occurred in these farms in 2008/2009 but with substantially changed pathology and greatly reduced mortality among the infected fish. Moreover, strains from both sites were typed and found to carry the frameshift mutation in cpsE (cpsE ST5/5A) and the deletion of cpsF through cpsM, although fingerlings came from different sources. The development of these very similar cases suggests the possibility of a common origin for the isolate. However, a silent mutation after the early termination signal in cpsE was found in the NSW isolates (hence cpsE ST5A), hinting that the SA case and NSW case may have arisen independently. Interestingly, the unusual truncation of cpsE was also found in a strain isolated several years earlier in Thailand. Whether this type has been introduced or whether it is a natural adaptation to host-specific immunity that occurs independently is unknown. It is likely that such a serious mutation in the capsular operon results in complete cessation of the capsular biosynthesis machinery and therefore creates a capsule-deficient mutant, as evidenced by our buoyant density results. This may explain the unusual pathology and very low mortality rate in the fish, as a capsule-deficient strain, while not recognized by anti-CPS antibodies, would be highly susceptible to phagocytic attack and may seek refuge in the bone of already-compromised fish. When antibodies raised against QMA0155, the initial sequence type found on this farm, were used in a whole-cell ELISA, they cross-reacted with strain QMA0236 but not vice versa. This may reflect a lack of CPS in QMA0236; while whole-cell bacterins prepared with QMA0155 would result in anti-cps antibodies along with a range of antibodies against other cell components, including integral membrane proteins, which are highly conserved among S. iniae (6) and would be exposed to antibody binding on CPS-deficient QMA0236, the converse would not be true. Integral membrane proteins of QMA0155 would likely be coated with CPS and unavailable for antibody binding in a whole-cell ELISA.
In summary, coding mutations in the capsular operon of S. iniae appear to correlate with reinfection of previously vaccinated fish. These mutations are restricted to a few key genes in the ∼20-gene operon. Two key questions remain to be answered. First, how do the genetic changes in the capsular operon, translated changes in the protein machinery for polysaccharide biosynthesis and export, actually lead to phenotypic and antigenic changes on the surface of the cell? Second, is vaccination driving mutation of highly mutable loci within a single strain type extant on a farm at a particular time, or is vaccination merely selecting the fittest variant from a pool of varied serotypes that are present among the population? The high ratio of nonsynonymous to synonymous mutations (dN/dS) among the genes of the CPS operon suggests that these changes are being driven under strong selective pressure. Further work aimed at answering these questions is ongoing.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the Fisheries Research Development Corporation for financial support to A.C.B. through Aquatic Animal Health Subprogram Project 2007/226 and the Centre for Marine Science, University of Queensland (UQ), for financial support of honors (C.C.) and undergrad (B.Y.) students. F.A. was supported by a Chilean government scholarship. Candice M. Millard is supported by an APA Scholarship and ARC Discovery project DP120102755.
A.C.B. conceived and designed the project, performed much of the bioinformatics analysis, and drafted the manuscript. C.M.M. reanalyzed sequence data, prepared figures, and cowrote the manuscript. J.C.F.B. conducted the sequencing, including primer design and preliminary sequence analysis. M.L., R.S.M.C., and S.B. provided the veterinary and pathology reports and supplied many of the strains. R.S.M.C. conducted the histopathology. C.C., B.Y., and F.A. conducted the antibody and phenotypic assays.
Footnotes
Published ahead of print 21 September 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Agnew W, Barnes AC. 2007. Streptococcus iniae: an aquatic pathogen of global veterinary significance and a challenging candidate for reliable vaccination. Vet. Microbiol. 122:1–15 [DOI] [PubMed] [Google Scholar]
- 2. Allen JP, Neely MN. 2012. CpsY influences Streptococcus iniae cell wall adaptations important for neutrophil intracellular survival. Infect. Immun. 80:1707–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Allen JP, Neely MN. 2011. The Streptococcus iniae transcriptional regulator CpsY is required for protection from neutrophil-mediated killing and proper growth in vitro. Infect. Immun. 79:4638–4648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bachrach G, Zlotkin A, Hurvitz A, Evans DL, Eldar A. 2001. Recovery of Streptococcus iniae from diseased fish previously vaccinated with a streptococcus vaccine. Appl. Environ. Microbiol. 67:3756–3758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barnes AC. 2010. Aquatic animal health subprogram: rapid strain identification of the bacterial fish pathogen Streptococcus iniae and the development of an effective polyvalent vaccine for Australian barramundi. Fisheries Research and Development Corporation, Canberra, Australia [Google Scholar]
- 6. Barnes AC, Young FM, Horne MT, Ellis AE. 2003. Streptococcus iniae: serological differences, presence of capsule and resistance to immune serum killing. Dis. Aquat. Organ. 53:241–247 [DOI] [PubMed] [Google Scholar]
- 7. Bromage ES, Owens L. 2002. Infection of barramundi Lates calcarifer with Streptococcus iniae: effects of different routes of exposure. Dis. Aquat. Organ. 52:199–205 [DOI] [PubMed] [Google Scholar]
- 8. Colorni A, Diamant A, Eldar A, Kvitt H, Zlotkin A. 2002. Streptococcus iniae infections in Red Sea cage-cultured and wild fishes. Dis. Aquat. Organ. 49:165–170 [DOI] [PubMed] [Google Scholar]
- 9. Delamare-Deboutteville J, Wood D, Barnes AC. 2006. Response and function of cutaneous mucosal and serum antibodies in barramundi (Lates calcarifer) acclimated in seawater and freshwater. Fish Shellfish Immunol. 21:92–101 [DOI] [PubMed] [Google Scholar]
- 10. Eyngor M, et al. 2010. A pivotal role for the Streptococcus iniae extracellular polysaccharide in triggering proinflammatory cytokines transcription and inducing death in rainbow trout. FEMS Microbiol. Lett. 305:109–120 [DOI] [PubMed] [Google Scholar]
- 11. Eyngor M, et al. 2008. Emergence of novel Streptococcus iniae exopolysaccharide-producing strains following vaccination with nonproducing strains. Appl. Environ. Microbiol. 74:6892–6897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Facklam R, Elliott J, Shewmaker L, Reingold A. 2005. Identification and characterization of sporadic isolates of Streptococcus iniae isolated from humans. J. Clin. Microbiol. 43:933–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hammerschmidt S, et al. 2005. Illustration of pneumococcal polysaccharide capsule during adherence and invasion of epithelial cells. Infect. Immun. 73:4653–4667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kumar A, Sandoe J, Kumar N. 2005. Three cases of vertebral osteomyelitis caused by Streptococcus dysgalactiae subsp. equisimilis. J. Med. Microbiol. 54:1103–1105 [DOI] [PubMed] [Google Scholar]
- 15. Lipsitch M. 2001. Bacterial vaccines and serotype replacement: lessons from Haemophilus influenzae and prospects for Streptococcus pneumoniae. Emerg. Infect. Dis. 5:336–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lipsitch M. 1997. Vaccination against colonizing bacteria with multiple serotypes. Proc. Natl. Acad. Sci. U. S. A. 94:6571–6576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Locke JB, et al. 2007. Streptococcus iniae capsule impairs phagocytic clearance and contributes to virulence in fish. J. Bacteriol. 189:1279–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lowe BA, Miller JD, Neely MN. 2007. Analysis of the polysaccharide capsule of the systemic pathogen Streptococcus iniae and its implications in virulence. Infect. Immun. 75:1255–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Martins ER, Melo-Cristino J, Ramirez M. 2010. Evidence for rare capsular switching in Streptococcus agalactiae. J. Bacteriol. 192:1361–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mata AI, Blanco MM, Dominguez L, Fernandez-Garayzabal JF, Gibello A. 2004. Development of a PCR assay for Streptococcus iniae based on the lactate oxidase (lctO) gene with potential diagnostic value. Vet. Microbiol. 101:109–116 [DOI] [PubMed] [Google Scholar]
- 21. Miller JD, Neely MN. 2005. Large-scale screen highlights the importance of capsule for virulence in the zoonotic pathogen Streptococcus iniae. Infect. Immun. 73:921–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Morona JK, Miller DC, Morona R, Paton JC. 2004. The effect that mutations in the conserved capsular polysaccharide biosynthesis genes cpsA, cpsB, and cpsD have on virulence of Streptococcus pneumoniae. J. Infect. Dis. 189:1905–1913 [DOI] [PubMed] [Google Scholar]
- 23. Morona JK, Morona R, Miller DC, Paton JC. 2003. Mutational analysis of the carboxy-terminal (YGX)4 repeat domain of CpsD, an autophosphorylating tyrosine kinase required for capsule biosynthesis in Streptococcus pneumoniae. J. Bacteriol. 185:3009–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Morona JK, Morona R, Paton JC. 1999. Analysis of the 5′ portion of the type 19A capsule locus identifies two classes of cpsC, cpsD, and cpsE genes in Streptococcus pneumoniae. J. Bacteriol. 181:3599–3605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Morona JK, Paton JC, Miller DC, Morona R. 2000. Tyrosine phosphorylation of CpsD negatively regulates capsular polysaccharide biosynthesis in Streptococcus pneumoniae. Mol. Microbiol. 35:1431–1442 [DOI] [PubMed] [Google Scholar]
- 26. Nawawi RA, Baiano JCF, Barnes AC. 2008. Genetic variability amongst Streptococcus iniae isolates from Australia. J. Fish Dis. 31:305–309 [DOI] [PubMed] [Google Scholar]
- 27. Nawawi RA, Baiano JCF, Kvennefors ECE, Barnes AC. 2009. Host-directed evolution of a novel lactate oxidase in Streptococcus iniae isolates from barramundi (Lates calcarifer). Appl. Environ. Microbiol. 75:2908–2919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27a. Pruksakorn S, et al. 2000. Epidemiological analysis of non-M-typeable group A streptococcus isolates from a Thai population in northern Thailand. J. Clin. Microbiol. 38:1250–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Roberts IS. 1996. The biochemistry and genetics of capsular polysaccharide production in bacteria. Annu. Rev. Microbiol. 50:285–315 [DOI] [PubMed] [Google Scholar]
- 29. Sommerset I, Krossoy B, Biering E, Frost P. 2005. Vaccines for fish in aquaculture. Expert Rev. Vaccines 4:89–101 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.