Abstract
The Philadelphia-1 strain of Legionella pneumophila, the causative organism of Legionnaires' disease, contains a recently discovered noncoding RNA, lpr0035. lpr0035 straddles the 5′ chromosomal junction of a 45-kbp mobile genetic element, pLP45, which can exist as an episome or integrated in the bacterial chromosome. A 121-bp deletion was introduced in strain JR32, a Philadelphia-1 derivative. The deletion inactivated lpr0035, removed the 49-bp direct repeat at the 5′ junction of pLP45, and locked pLP45 in the chromosome. Intracellular multiplication of the deletion mutant was decreased by nearly 3 orders of magnitude in Acanthamoeba castellanii amoebae and nearly 2 orders of magnitude in J774 mouse macrophages. Entry of the deletion mutant into amoebae and macrophages was decreased by >70%. The level of entry in both hosts was restored to that in strain JR32 by plasmid copies of two open reading frames immediately downstream of the 5′ junction and plasmid lpr0035 driven by its endogenous promoter. When induced from a tac promoter, plasmid lpr0035 completely reversed the intracellular multiplication defect in macrophages but was without effect in amoebae. These data are the first evidence of a role for noncoding RNA lpr0035, which has homologs in six other Legionella genomes, in entry of L. pneumophila into amoebae and macrophages and in host-specific intracellular multiplication. The data also demonstrate that deletion of a direct-repeat sequence restricts the mobility of pLP45 and is a means of studying the role of pLP45 mobility in Legionella virulence phenotypes.
INTRODUCTION
Legionella pneumophila is the causative organism of Legionnaires' disease, a potentially fatal community- and hospital-derived pneumonia spread by aerosolization from contaminated water reservoirs (8, 25). The bacterium is widespread in aquatic niches, where it is found free-living, within biofilms, and as an intracellular parasite of aquatic amoebae (21, 33, 36, 50). The ability of Legionella to enter, replicate within, and escape from environmental amoebae and pulmonary macrophages is dependent on bacterial effector proteins, secreted into the eukaryotic host by a type 4 secretion system (T4SS) (17, 22, 29, 30, 34). Two T4SSs are associated with these virulence phenotypes in laboratory strains derived from the Legionella pneumophila Philadelphia-1 clinical isolate, named for the city (Philadelphia, PA) of the 1976 outbreak that gave the disease its name. The Dot/Icm type 4B secretion system (T4BSS) is required for invasion, intracellular multiplication, and egress from infected cells when broth-grown stationary-phase cultures are used to infect amoebae or macrophage hosts. Mutants with mutations in most Dot/Icm genes are defective in those virulence phenotypes (1, 17, 22, 38). When the aquatic and intra-amoeba niches of Legionella are mimicked in the laboratory, a second T4SS, the Lvh T4ASS, contributes to virulence phenotypes. The Lvh T4ASS is homologous to the virBD T4ASS of Agrobacterium tumefaciens and other species (11, 47). These data suggest that the Lvh T4ASS is involved in the environmental phase of the Legionella life cycle (3).
Given the central role of T4SSs and effector proteins, the mechanisms that regulate their expression have been a major focus in studies of L. pneumophila virulence phenotypes. Recently, noncoding RNAs (ncRNAs) have been implicated in those regulatory networks (18, 19, 39, 42). Initially, Legionella ncRNAs were predicted from bioinformatics or identified as Legionella homologs of ncRNAs found in other bacteria (18, 32). Homologs of Escherichia coli ncRNAs RsmY and RsmZ bind to and inhibit L. pneumophila CsrA, which negatively affects expression of Dot/Icm effector proteins in a pathway regulated by RpoS, LetA, and PmrA (28, 39, 42). A 6S RNA, homologous to RNAs known to regulate σ70 RNA polymerase, was predicted by a computational approach and shown to positively regulate expression of Dot/Icm effector proteins and to be required for optimal intracellular multiplication in Acanthamoeba castellanii and in THP-1-derived macrophages (18). With the advent of deep sequencing technologies, hundreds of additional L. pneumophila ncRNAs have been identified (43, 52), and functions for the vast majority of these are unknown. Deep sequencing of L. pneumophila strain Paris identified 713 ncRNAs, including 2 novel ncRNAs and a structural homolog of RsmY and RsmZ, which were shown to be required for optimal replication in Acanthamoeba castellanii (43).
Here we report on the results of studies of a deletion mutant of lpr0035, a previously uncharacterized ncRNA identified by RNA sequencing of the Philadelphia-1 strain (52). lpr0035 straddles the chromosomal junction of pLP45, a mobile genetic element containing the Lvh T4ASS locus (10). Loci for the Lvh and other T4ASSs in L. pneumophila genomes are frequently found on mobile genetic elements (7, 10, 24, 45). In contrast, Dot/Icm loci have been found exclusively in the chromosome. Our data establish a role for lpr0035 in entry of Legionella strain JR32, a laboratory strain derived from the Philadelphia-1 clinical isolate (41, 53), into macrophages and amoebae and in intracellular multiplication of JR32 in macrophages. The lpr0035 deletion removed a direct-repeat sequence at the 5′ end of pLP45, locking the genetic element in the chromosome and preventing its excision to an episomal form.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Parental L. pneumophila strain JR32, serotype 1, and mutants derived from it (41, 53) were plated on charcoal–N-(2-acetamido)-2-aminoethanesulfonic acid (ACES)-buffered (pH 6.9) yeast extract agar (CYE) plates and cultured in ACES-buffered yeast extract (AYE) broth (20, 27). When required, chloramphenicol and hygromycin were present at 5 and 100 μg/ml, respectively. E. coli strain DH5α, cultured in Luria-Bertani broth, was used for recombinant constructions with chloramphenicol and hygromycin at 25 and 150 μg/ml, respectively. All bacteria were cultured at 37°C. Water stress (WS)-treated cultures were prepared as previously described (3, 9), 22- to 24-h AYE broth cultures were resuspended in autoclaved deionized water, and the cultures were incubated for an additional 18 to 19 h in 16- by 150-mm glass culture tubes on a rotary drum.
Amoeba and macrophage cell line and culture conditions.
A. castellanii (ATCC 302340) was cultured at 28°C in peptone-yeast extract-glucose (PYG) medium as described previously (26, 48). The BALB/c mouse peritoneal macrophage line J774 (ATCC TIB-67) was maintained in RPMI 1640 medium with 2 mM l-glutamine, 10% heat-inactivated fetal bovine serum, and penicillin-streptomycin (5,000 U/ml) at 37°C in humidified 5% CO2 (3, 9).
Construction of lpr0035 deletion.
An unmarked, 121-bp deletion was constructed in JR32 by a two-step allelic-exchange procedure using the kanamycin-resistant, sacB-containing allelic-exchange vector pNPTS138, as described previously (4). The region deleted began 50 nucleotides from the 5′ end of lpr0035 and was entirely within the 252-bp ncRNA (Fig. 1A). The deletion removed the 49-bp direct repeat at the 5′ end of the pLP45 mobile element and an additional 72 bp from the lpg1227-lp1228 intergenic region, from 114 nucleotides (nt) downstream of the lpg1227 stop codon to 157 nt upstream of the lpg1228 start codon. The 5′ and the 3′ regions, 1.493 kb and 1.563 kb, respectively, flanking the 121-bp region were amplified by PCR with Platinum Pfx polymerase (Invitrogen, Carlsbad, CA) and JR32 cells as the template. The PCR products were restriction digested using the appropriate restriction enzymes (PstI, BamHI, and SalI; New England BioLabs, Ipswich, MA), purified using an Illustra GFX PCR DNA and gel band purification kit (GE Healthcare), and ligated into pNPTS138. After sequence confirmation, the allelic-exchange plasmid was introduced into strain JR32 by triparental mating using E. coli DH5α with pRK600 (31) as the helper strain and with the pNPTS138 construct as the donor. The exconjugants were purified and plated on CYE agar supplemented with 5% sucrose. Sucrose-resistant and kanamycin-sensitive strains were tested by PCR to identify strains containing the lpr0035 deletion and lacking the parental lpr0035 allele.
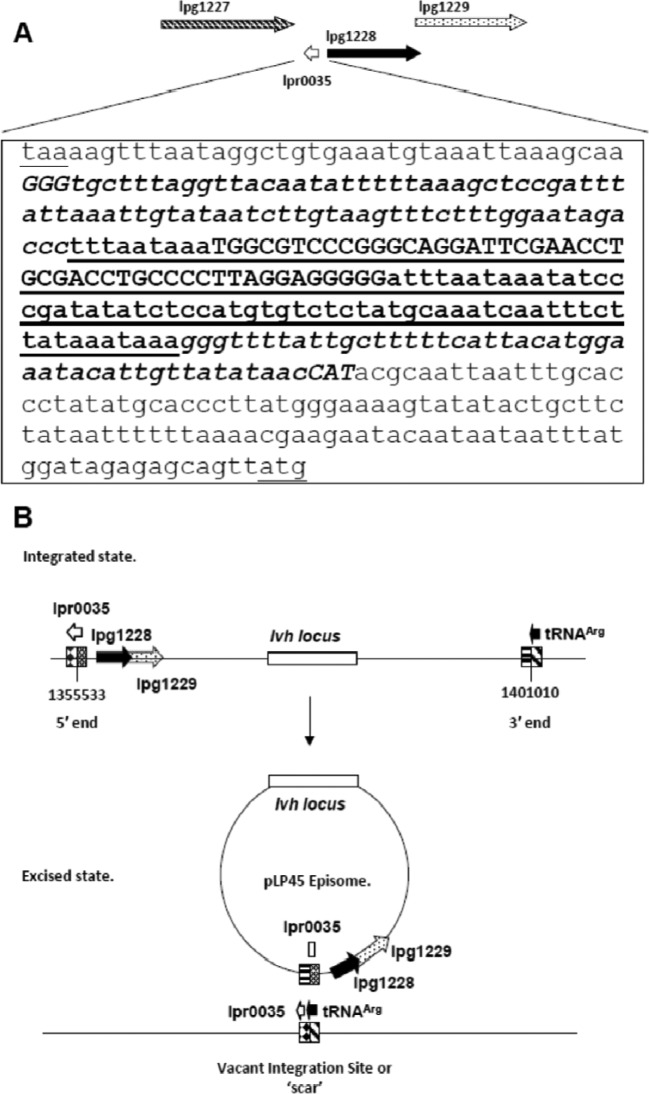
Fig 1.
Genomic region at the 5′ junction of pLP45. (A) Nucleotide sequence from coordinates 1355407 to 1355804 and genes at the 5′ junction of pLP45. The TAA stop codon for lpg1227 and the ATG start codon for lpg1228 are underlined and in lowercase letters in the first and last lines, respectively. Noncoding RNA lpr0035, which runs in the minus direction, is the reverse complement of the sequence in bold, from GGG in the second line to CAT in the eighth line. The 121-bp 5′ deletion is underlined. Within the deletion, the 49-bp direct repeat is capitalized. (B) Schematic representation of the locations of lpr0035 in the integrated and excised states of the pLP45 mobile genetic element. (Integrated state) The gene coordinates of the 5′ and 3′ ends of pLP45 are indicated, as are the location and orientation of noncoding RNAs, lpr0035, and tRNAArg and the open reading frames for the lpg1228 (hypothetical protein) and lpg1229 (site-specific recombinase) genes. Identical 49-bp direct repeats begin at nt 1355533 and 1401011. (Excised state) The sequence elements juxtaposed by excision are indicated in the episome and vacant integration site. Excision disrupts the lpr0035 noncoding RNA element, depicted as the white block in the episome and the white arrow in the vacant integration site.
Plasmids for transformation of the deletion mutant.
The following open reading frames (ORFs) were amplified by PCR using strain JR32 cells as the template: lpg1228, from 157 nt upstream of the start codon to 93 nt downstream of the stop codon; lpg1229, from 157 nt upstream of the start codon to 99 nt downstream of the stop codon; lpg1272, a tRNAArg, from 159 nt upstream of the start codon to 58 nt downstream of the stop codon; and lpg1245, Lvh VirD4, from 204 bp upstream of the start codon to 168 bp downstream of the stop codon. The open reading frames of both lpg1228 and lpg1229 were amplified from 157 nt upstream of the start codon of lpg1228 to 99 nt downstream of the stop codon of lpg1229. lpr0035 noncoding RNA (52) was amplified from 204 nt upstream of the start codon to 98 nt downstream of the stop codon. lpr0035 was also amplified from 33 nt upstream of the start codon to 98 nt downstream of the stop codon and cloned in frame with the tac promoter in pMMB207C (9). All PCR products were restriction digested with the appropriate enzymes (EcoRI, BamHI, and PstI), purified as described above, and then ligated into the pMMB207C vector. For cloning of lpg1272 tRNAArg into the pJN105 hygromycin-resistant vector (5), the region from 159 nt upstream of the start codon to 82 nt downstream of the stop codon was amplified. All plasmid constructs were confirmed by sequencing. The plasmid vector containing the lvh locus (pGS-VBD32) was described previously (47). These plasmid constructs were introduced into the lpr0035 deletion mutant and the lpr0035 deletion mutant bearing the pJN105::GFP (green fluorescent protein) plasmid by triparental mating. The pJN105::GFP plasmid used for entry experiments (5) was introduced into the lpr0035 mutant by triparental mating.
Assay of intracellular multiplication by titer of L. pneumophila.
Monolayers of A. castellanii cells at 30°C or of J774 macrophages at 37°C were infected with L. pneumophila as described previously (2, 3, 5), and aliquots were taken from the infection medium for titration on CYE plates. In all experiments, titers from replicate wells were in good agreement.
Immunofluorescence assay for entry of Legionella into A. castellanii amoebae and J774 macrophages.
Amoebae (2 × 106 cells in PYG medium) or macrophages (2 × 105 cells in the RPMI 1640 tissue culture medium cited above) were applied in 0.5 ml to 12-mm-diameter glass coverslips in 24-well microtiter dishes, as described previously (5, 26). Bacteria, grown as indicated, were resuspended in A. castellanii buffer or tissue culture medium without antibiotics, added at the indicated multiplicity of infection (MOI; 5 for A. castellanii and 100 for macrophages), incubated for 30 min at 28°C for amoebae and 1 h at 37°C for macrophages, and then fixed with formaldehyde and stained with rabbit anti-L. pneumophila serotype 1 antibody (m-Tech, Atlanta, GA), followed by Cy3-conjugated donkey anti-rabbit immunoglobulin G (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA), as previously described (5, 26). Immunofluorescence samples were observed and scored at ×60 magnification with a Zeiss Axioskop epifluorescence microscope or an Olympus IX-81 microscope, and the phagocytic index was calculated as the number of internalized Legionella bacteria per 50 host cells. The relative phagocytic indices reported here for JR32 and its dotB mutant were in good agreement with our previous data, indicating that the entry phenotype and WS reversal were reproduced and substantiating those phenotypes for the new derivatives of JR32 in this study. However, the phagocytic index values were 6- to 8-fold lower than those in our earlier studies. Using different laboratory stores of bacteria, macrophages, or amoebae or using new macrophage and amoeba samples from ATCC did not result in increased phagocytic indices.
PCR identification of integrated and episomal forms of pLP45.
The integrated form of pLP45 was detected by PCR amplification of the 5′ chromosomal junction, from 32 nt downstream of the stop codon of lpg1227 to 80 nt upstream of the start codon of lpg1228. The episomal form was detected by amplification of the juxtaposed 5′ and 3′ ends of pLP45, from 131 nt upstream of the start codon of lpg1228 to 556 nt downstream of the start codon of lpg1271. A region expected to be in the chromosome permanently, from 1,379 nt upstream of the 3′ end of lpg1227 to 10 nt upstream of the start of the integrated pLP45, was amplified as a control. All PCR experiments used Platinum Pfx polymerase with 95°C for 6 min, followed by 30 cycles of 95°C for 30 s, 55°C for 30 s, and 68°C for 2 min, with a final extension at 68°C for 6 min. The template was ∼1 × 105 cells, and products were analyzed on a 3% agarose gel.
LacZ translational and transcriptional fusions.
The first 20 nucleotides of lpg1228 and sequences beginning 392, 277, and 156 nucleotides upstream of the start codon were amplified by PCR and ligated into the EcoRI site of the LacZ translational fusion vector p-GS-lac-01 (55), using EcoRI sites in the PCR primers. Similarly, the first 29 nucleotides and the 442 nucleotides upstream of the lpg1229 start codon were cloned into p-GS-lac-01. The first 10 nucleotides of lpg1272 tRNAArg and the 293 nucleotides were PCR amplified by PCR and directionally ligated into the EcoRI and BamHI sites of the LacZ transcriptional fusion vector pMR-TV-1 (39). Constructs containing the lpg1228 and lpg1229 fusions in the forward and opposite orientation with respect to the lacZ open reading frame and the lpg1272 transcriptional construct were introduced by triparental mating into strain JR32 and the lpr0035 mutant. Activity of β-galactosidase was determined by kinetic assays of the rate of change of the absorbance at 405 nm, as described previously, using o-nitrophenyl-β-galactoside as the substrate (4). Corrections for the spontaneous rate of substrate hydrolysis were calculated from assays in the absence of bacterial extract. Corrections for endogenous β-galactosidase activity in extracts were calculated from assays of extracts of strains in which the lacZ reporter and the tested promoter were in opposite orientations. In combination, these two corrections were less than 20% of the average rate measured.
Stress tests.
The resistance of overnight AYE cultures to sodium ion stress was quantified as the ratio of the titer on CYE plates containing 100 mM NaCl to the titer on CYE plates without added NaCl (2, 6).
Statistical analyses.
For intracellular multiplication assays, the triplicate values obtained for each strain were normalized by dividing by the average titer at day 0. Two-tailed t tests were performed to obtain P values for comparisons between normalized data for two strains. For entry experiments, the total data set from 200 to 230 host cells was divided into four subgroups, which were then used for calculation of means ± standard deviations. P values were calculated using the chi-square test available on the SPSS or PASW statistics software, as described previously (3, 5).
RESULTS
Strategy for construction of the deletion mutant in strain JR32.
The 121-bp deletion in lpr0035 was positioned to minimize effects on expression of the upstream lpg1227 and the downstream lpg1228 ORFs. The 5′ end of the deletion was 115 bp downstream of the stop codon of lpg1227, encoding a hypothetical protein. The 3′ end was 157 bp upstream of the start codon of lpg1228, also encoding a hypothetical protein (Fig. 1A). The 121-bp deletion was internal to the 252 nt of lpr0035 ncRNA and was expected to inactivate lpr0035. The deletion also removed the 49-bp direct repeat at the 5′ end of pLP45 (Fig. 1A and B). The 121-bp deletion was confirmed by sequencing a PCR amplicon of the region.
Analysis of the deletion mutant for entry into macrophages and amoebae.
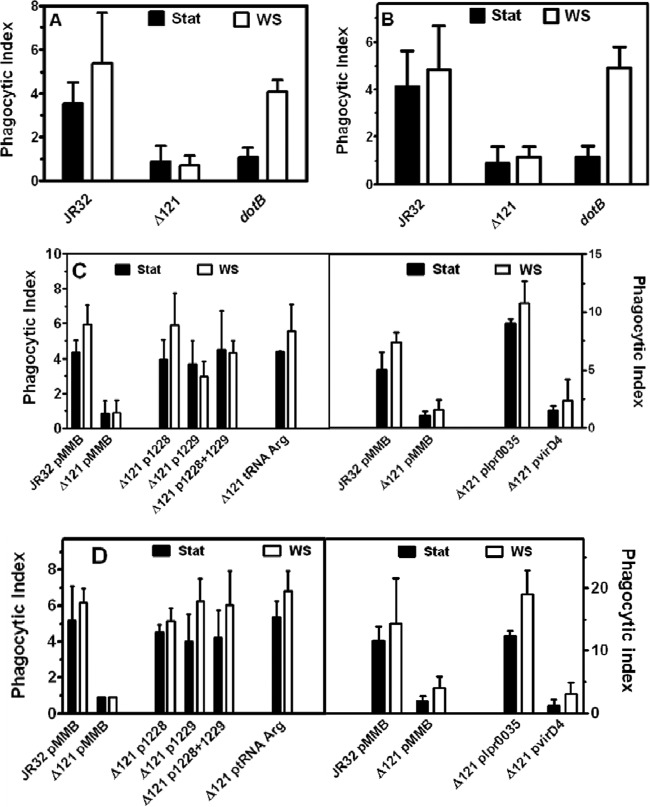
Assay for entry into host cells at 1 h and at 30 min after infection of J774 macrophages and A. castellanii amoebae showed a virulence phenotype that is a prerequisite to intracellular multiplication. As previously reported (5, 26), a dotB mutant was defective for entry following infection with broth stationary-phase cultures, showing entry at levels 31% and 28% of the level for strain JR32 for macrophages and amoebae, respectively. The deletion mutant (Δ121) showed an entry defect comparable to that for the dotB mutant: 25% and 21% of the level for JR32 for macrophages and amoebae, respectively (Fig. 2A and B).
Fig 2.
Entry phenotypes in J774 macrophages and A. castellanii amoebae. (A) The deletion mutant is defective for entry into J774 macrophages. J774 macrophages were infected at 37°C at an MOI of 100 with stationary-phase (Stat) or WS-treated bacteria expressing GFP. Means and standard deviations are plotted. For stationary-phase bacteria, P values were equal to 0.007 for JR32 versus the deletion mutant (Δ121) and 0.015 for JR32 versus dotB. For WS-treated bacteria, P values were <0.001 for JR32 versus the deletion mutant and 0.425 for JR32 versus dotB. (B) The deletion mutant is defective for entry into amoebae. A. castellanii trophozoites were infected at 28°C at an MOI of 5 with stationary-phase or WS-treated bacteria expressing GFP. Means and standard deviations are plotted. For stationary-phase bacteria, P values were 0.002 for JR32 versus the deletion mutant (Δ121) and 0.009 for JR32 versus dotB. For WS-treated bacteria, P values were 0.001 for JR32 versus the deletion mutant and 0.892 for JR32 versus dotB. (C) Defective entry of the deletion mutant into J774 macrophages is reversed with the plasmid with lpg1228 plus lpg1229, tRNAArg (lpg1272), or lpr0035 but not by plasmid virD4. To test the reversal of defective entry by plasmid genes driven by their endogenous promoters, J774 macrophages were infected at 37°C at an MOI of 100 with stationary-phase or WS-treated bacteria expressing GFP. Means and standard deviations are plotted. Δ121, the deletion mutant; pMMB, empty vector pMMB207C. For stationary-phase and WS-treated bacteria, P was 0.002 for JR32/pMMB207C versus Δ121/pMMB207C or Δ121/pvirD4. For stationary-phase and WS-treated bacteria, P values ranged from 0.101 to 0.923 for JR32/pMMB207C versus Δ121 with plasmids with lpg1228 (p1228), lpg1229 (p1229), lpg1228 and lpg1229 (p1228 + p1229), or tRNAArg (ptRNA Arg). Phagocytic indices for the right panel are higher because a different batch of macrophages was used. (D) Defective entry of the deletion mutant into A. castellanii amoebae is reversed with plasmid lpg1228 plus lpg1229, tRNAArg (lpg1272), or lpr0035 but not by plasmid virD4. To test the reversal of defective entry by plasmid genes driven by their endogenous promoters, A. castellanii trophozoites were infected at 28°C at an MOI of 5 with stationary-phase or WS-treated bacteria expressing GFP. Means and standard deviations are plotted. For stationary-phase and WS-treated bacteria, P was < 0.001 for JR32/pMMB207C versus Δ121/pMMB207C or Δ121 with plasmid virD4. For stationary-phase and WS-treated bacteria, P values ranged from 0.277 to 0.949 for JR32/pMMB207C versus Δ121 with plasmids with lpg1228, lpg1229, lpg1228 and lpg1229, or tRNAArg. The phagocytic indices for the right panel are higher because a different batch of amoebae was used.
Previous work from this laboratory demonstrated that defective entry of dotB and other dot/icm mutants was reversed following incubation of broth stationary-phase cultures in deionized water, a treatment termed water stress (WS), which mimicked the Legionella aquatic environment (3, 5). WS reversal of the entry defect in dotB JR32 was reproduced here for macrophages and amoebae (Fig. 2A and B). However, WS treatment failed to reverse the entry defect in the deletion mutant in either host.
Reversal of defective entry in the deletion mutant by lpr0035 and by genes near the 5′ and 3′ junctions of pLP45.
Plasmid lpr0035 restored entry to that of parental strain JR32 with J774 macrophages and A. castellanii amoebae, following infection with stationary-phase or with WS-treated cultures (Fig. 2C and D, right). This complementation established that lpr0035 plays a role in the Legionella entry phenotype. Reversal of the entry defect was also tested with plasmid copies of genes near the 5′ and 3′ junctions of the pLP45 mobile genetic element with the chromosome, because expression of those genes could be affected by the 121-bp deletion itself and/or by changes in the mobility of pLP45. Plasmid lpg1228, plasmid lpg1229, both plasmids lpg1228 and lpg1229, and plasmid tRNAArg (Fig. 1B) all strongly reversed the entry defect in both macrophage and amoeba hosts following infection with either stationary-phase or WS-treated cultures (Fig. 2C and D, left).
Reversal of entry defects by plasmid copies of genes at the 5′ and 3′ junctions of pLP45 suggested that expression of those genes was diminished in the deletion mutant. To test this hypothesis, strain JR32 and the deletion mutant were transformed with plasmid LacZ translational and transcriptional fusions to genes at the 5′ and 3′ junctions of pLP45. Translational fusions were made for the two genes downstream of the 5′ junction. For lpg1228, the first downstream gene, fusions with 392, 277, and 156 nucleotides preceding the lpg1228 start codon were tested. For the next downstream gene, lpg1229, one LacZ fusion with the 442 nucleotides preceding the start codon was tested. A 50 to 80% reduction in LacZ activity for the fusions was consistent with decreased expression of lpg1228 and lpg1229 in the deletion mutant (Table 1). The low values for β-galactosidase activity, which are comparable to the values reported for LacZ translational fusions to certain L. pneumophila dot/icm genes (23), may reflect low levels of ORF expression or instability of the LacZ fusion protein. Expression from the promoter region of lpg1272, encoding a tRNAArg outside the 3′ junction of pLP45 (Fig. 1B), was tested by a LacZ transcriptional fusion to the 293 nt upstream of the tRNA start. The LacZ activity of the transcriptional fusion was not significantly different in JR32 and the deletion mutant. These results were consistent with control of tRNAArg expression by stability of a transcriptional termination stem-loop, described below in the Discussion.
Table 1.
LacZ fusions to lpg1228, lpg1229, and lpg1272
| Fusion type, gene | No. of nucleotides upstream of start codon | β-Galactosidase activitya |
|
|---|---|---|---|
| JR32 | Deletion mutant | ||
| Translational fusions | |||
| lpg1228 | 392 | 5.6 ± 1.1 | 2.2 ± 0.19 |
| 277 | 5.1 ± 0.42 | 1.2 ± 0.42 | |
| 156 | 17 ± 2.1 | 7.5 ± 1.3 | |
| lpg1229 | 442 | 7.1 ± 1.0 | 3.5 ± 0.64 |
| Transcriptional fusion, lpg1272 tRNAArg | 293 | 413 ± 114 | 372 ± 27 |
β-Galactosidase activity was measured with o-nitrophenyl-β-galactoside as the substrate and is expressed as Miller units. Data are means and standard deviations of two independent experiments. The P value for the lpg1228 and lpg1229 translational fusions in JR32 versus the deletion mutant is <0.001 for all constructs. The P value for the lpg1272 tRNAArg transcriptional fusion in JR32 versus the deletion mutant is 0.43.
To determine if entry defects could be reversed by a gene within pLP45 but distant from the junctions of pLP45 with the chromosome, plasmid virD4 was tested. virD4 lies in the lvh locus, 19 kbp from the 5′ junction and 25 kbp from the 3′ junction of pLP45 (Fig. 1B). Plasmid virD4 failed to reverse the entry defect in either macrophage or amoeba hosts following infection with either stationary-phase or WS-treated cultures, suggesting that the deletion preferentially affected expression of genes at or near the 5′ and 3′ junctions of pLP45 (Fig. 2C and D, right). The empty complementation vector pMMB207C had no significant effect on the entry defect of the deletion mutant (Fig. 2C and D).
Analysis of the distribution of pLP45 between integrated and excised states in the deletion mutant.
PCR was used to diagnose the effect of the 121-bp deletion on pLP45 mobility by amplifying fragments across the chromosomal, the episomal, and the vacant site/scar regions (Fig. 1B). PCR amplicons from strain JR32 containing the 5′ junction and containing the vacant site were sequenced and found to be identical to those predicted from the Philadelphia-1 genome (data not shown). These data confirmed that the 5′ and 3′ junction sites in the JR32 strain were identical to those in the genome of the Philadelphia-1 clinical isolate (10) and that laboratory culturing had not changed the integration sites of pLP45.
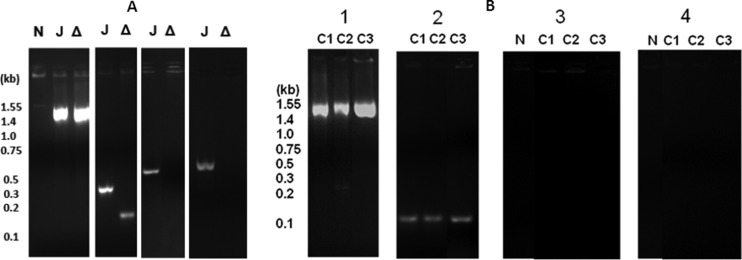
PCR amplification across the 5′ junction of pLP45 with the chromosome produced a smaller product for the deletion mutant than for strain JR32, as expected from the 121-bp deletion (Fig. 3A, second panel). PCR amplification across the episomal junction, in which 5′ and 3′ ends of pLP45 are joined (Fig. 1B), gave a product of the expected size from JR32 but no product from the deletion mutant (Fig. 3A, third panel), even after increasing the sensitivity of detection by increasing the number of PCR cycles from 30 to 45. PCR amplification across the chromosomal vacant site, or scar, of pLP45 (Fig. 1B) gave a band of the expected size for strain JR32 but no product for the deletion mutant (Fig. 3A, fourth panel). These results demonstrated that, within the limits of PCR detection, the episomal form and the vacant site on the chromosome were absent in the deletion mutant and the pLP45 element was locked in the chromosome.
Fig 3.
PCR identification of chromosomal and episomal forms of pLP45. (A) Deletion of the 5′ junction region locks pLP45 in the chromosome. Amplification of a region outside pLP45 (first panel), of the 5′ junction (second panel), of the episomal junction (third panel), and of the vacant site/scar region (fourth panel) for JR32 (J), the deletion mutant (Δ), and a no-template control (N). Expected sizes of the amplicons are 1.493 kb (first panel), 0.281 and 0.161 kb (second panel), 0.581 and 0.469 kb (third panel), and 0.598 and 0.589 kb (fourth panel). (B) pLP45 remains locked in the chromosome after transformation of the deletion mutant with plasmids lpg1228 plus lpg1229 and lpr0035. Amplification of a region outside pLP45 (panel 1), of the 5′ junction (panel 2), of the episomal junction (panel 3), and of the scar or vacant integration site (panel 4) for the deletion mutant cultured in AYE and transformed with pMMB207C containing lpg1228 and lpg1229 (lanes C1) and lpr0035 (lanes C2) or cultured in AYE with 0.1 mM IPTG and transformed with pMMB207C with Ptac driving lpr0035 (lanes C3) and for no-template controls (lanes N). The expected sizes of the amplicons are 1.493 kb (panel 1), 0.161 kb (panel 2), 0.581 kb (panel 3), and 0.598 kb (panel 4).
PCR analysis was performed to determine if pLP45 became unlocked from the chromosome when the entry defect was reversed. An amplicon indicative of integrated pLP45 was seen in the deletion mutant transformed with plasmid lpg1228 and lpg1229 or with plasmid lpr0035 (Fig. 3B, lanes C1 and C2 in panel 2), all of which reversed the entry defect. However, no amplicons indicative of the episomal state or the vacant integration site were detected in those strains (Fig. 3B, lanes C1 and C2 in panels 3 and 4). Amplicons of the sizes expected for the episomal state and the vacant site were seen for strain JR32 (data not shown). These data indicate that reversal of the entry defect occurred without unlocking pLP45 from the integrated state.
Analysis of intracellular multiplication of the deletion mutant in macrophages.
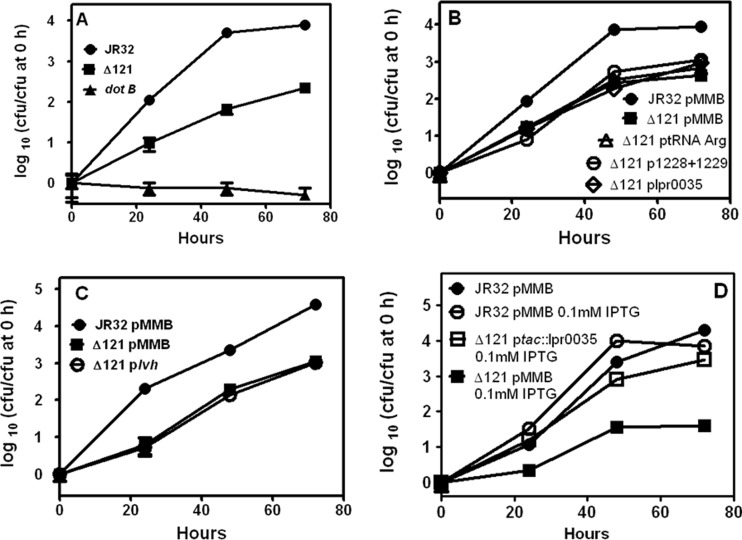
Intracellular multiplication of the deletion mutant in J774 macrophages was decreased by 1 to 2 orders of magnitude compared to that for strain JR32 throughout the 3-day experiment (Fig. 4A). After 3 days, the titer of JR32 in the infection medium had increased by nearly 4 orders of magnitude, whereas it increased approximately 2 orders of magnitude for the deletion mutant. The doubling times of the mutant (2.0 ± 0.3 h) and strain JR32 (2.0 h) were identical in AYE broth medium, indicating that differences in intracellular multiplication were not attributable to differences in in vitro growth. Consistent with previous reports, a mutant with a mutation in dotB, lacking an essential ATPase of the Dot/Icm T4BSS (49), was nonreplicative.
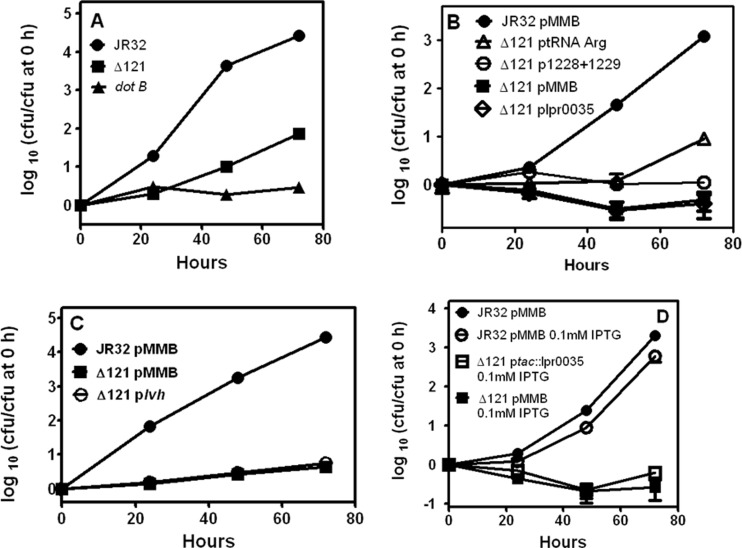
Fig 4.
Intracellular multiplication phenotypes in J774 macrophages. (A) The deletion mutant is defective in intracellular multiplication in J774 macrophages. J774 macrophages were infected at 37°C at an MOI of 1 with stationary-phase bacteria. At the indicated times, the titers in aliquots from the infection medium were determined to count the numbers of CFU. Means and standard deviations are plotted. The P value for JR32 versus the deletion mutant (Δ121) at 72 h is 2.9E−06. (B) Defective intracellular multiplication of the deletion mutant in J774 macrophages is not substantially reversed by transformation with lpg1228 plus lpg1229, tRNAArg (lpg1272), or lpr0035. To test the reversal of defective intracellular multiplication by plasmid genes driven by their endogenous promoters, J774 macrophages were infected at 37°C at an MOI of 1 with stationary-phase bacteria. At the indicated times, the titers in aliquots from the infection medium were determined to count the numbers of CFU. Means and standard deviations are plotted. Δ121, the deletion mutant; pMMB, empty vector pMMB207C. (C) Defective intracellular multiplication of the deletion mutant in J774 macrophages is not substantially reversed by transformation with a plasmid with the lvh locus. To test the reversal of defective intracellular multiplication by plasmid lvh driven by its endogenous promoters, J774 macrophages were infected at 37°C at an MOI of 1 with stationary-phase bacteria. At the indicated times, the titers in aliquots from the infection medium were determined to count the numbers of CFU. Means and standard deviations are plotted. (D) Defective intracellular multiplication of the deletion mutant on J774 macrophages is reversed by IPTG induction of lpr0035 from the tac promoter. To test the reversal of defective intracellular multiplication by plasmid lpr0035 driven by the tac promoter without the endogenous lpr0035 promoter, J774 macrophages were infected at 37°C at an MOI of 1 with stationary-phase bacteria. As indicated, IPTG was present throughout the experiment, and at the indicated times titers were determined in aliquots taken from the infection medium to count the numbers of CFU. Means and standard deviations are plotted. The P values for the 0.1 mM IPTG culture of the deletion mutant containing pMMB207C versus the mutant containing ptac::lpr0035 were ≤0.0002 at 24, 48, and 72 h.
The same plasmids that reversed defective entry were tested for reversal of the intracellular multiplication defect. The empty complementation plasmid pMMB207C did not affect the difference in intracellular multiplication between JR32 and the deletion mutant (Fig. 4B). No significant restoration of intracellular multiplication was observed for the deletion mutant containing pMMB207C plasmids with both lpg1228 and lpg1229 or with lpr0035, or with the tRNAArg (Fig. 4B) or with plasmids containing lpg1228 alone or lpg1229 alone or for a mutant with two plasmids, one expressing lpr0035 and the other expressing lpg1228 and lpg1229 (data not shown). Excision of mobile elements containing Legionella T4SS loci has been proposed as a means of increasing expression through multiple copies of the episomal form (16). Since the episomal form of pLP45 was absent in the deletion mutant, the mutant was transformed with a plasmid expressing the Lvh T4ASS locus, which lies in pLP45. However, no reversal of the defect in intracellular multiplication in J774 macrophages was observed (Fig. 4C). The preceding experiments used genes under the control of their natural promoters.
The Legionella literature contains reports where a gene under Ptac control was induced by isopropyl-β-d-thiogalactopyranoside (IPTG) to complement intracellular multiplication defects in mutants for noncoding RNAs RsmY and RsmZ (28, 42), the RNA binding protein CsrA (35), and the PmrA response regulator (54). Since ncRNAs may have regulatory functions influencing multiple downstream genes, this approach was tested with ncRNA lpr0035 by placing it under Ptac control. When 0.1 mM IPTG was present throughout the infection, the defect was significantly and nearly completely reversed in the deletion mutant (Fig. 4D). No significant reversal was observed with the Ptac-lpr0035 plasmid when IPTG was omitted from the experiment (data not shown).
To determine if pLP45 became unlocked from the chromosome when the intracellular multiplication defect was reversed, PCR analysis was performed as described above for reversal of the entry defect. An amplicon indicative of integrated pLP45 was seen in the deletion mutant containing the Ptac-driven lpr0035 and cultured with IPTG; however, no amplicons indicative of episomal state or vacant integration sites were detected (lanes C3 in Fig. 3B). These data indicated that reversal of the intracellular multiplication defect occurred without unlocking pLP45 from its integrated state and established a role for lpr0035 in intracellular multiplication of strain JR32 in J774 macrophages.
Analysis of intracellular multiplication of the deletion mutant in A. castellanii amoebae.
In A. castellanii, the deletion mutant showed an even stronger defect in intracellular multiplication: nearly 3 orders of magnitude less than that for strain JR32 at 2 to 3 days after infection (Fig. 5A). Similar to results with J774 macrophages, the intracellular multiplication defect in amoebae was not significantly reversed by plasmid genes lpg1228 and lpg1229, lpr0035, or the tRNAArg (Fig. 5B), a plasmid with the lvh locus (Fig. 5C), or plasmids containing lpg1228 alone, lpg1229 alone, or two plasmids, one expressing lpr0035 and the other expressing lpg1228 and lpg1229 (data not shown). In contrast to the macrophage phenotype, the intracellular multiplication defect in amoebae was not reversed by IPTG-induced expression of lpr0035 under Ptac control (Fig. 5D). These data suggested that factors other than lpr0035 contribute to the defect in A. castellanii and indicated host specificity in the intracellular multiplication defect of the deletion mutant. An independent isolate of the deletion mutant showed an identical intracellular multiplication defect in macrophages and amoebae (data not shown), evidence that the defect was attributable to the 121-bp deletion and unlikely to spontaneous mutation in the two isolates.
Fig 5.
Intracellular multiplication phenotypes in A. castellanii amoebae. (A) The deletion mutant is defective in intracellular multiplication in A. castellanii amoebae. A. castellanii amoebae were infected at 30°C at an MOI of 1 with stationary-phase bacteria. At the indicated times, the titers in aliquots from the infection medium were determined to count the numbers of CFU. Means and standard deviations are plotted. The P value for JR32 versus the deletion mutant (Δ121) at 72 h was 4.8E−05. (B) Defective intracellular multiplication of the deletion mutant in A. castellanii amoebae is not substantially reversed by transformation with lpg1228 plus lpg1229, tRNAArg (lpg1272), or lpr0035. To test the reversal of defective intracellular multiplication by plasmid genes driven by their endogenous promoters, A. castellanii amoebae were infected at 30°C at an MOI of 1 with stationary-phase bacteria. At the indicated times, the titers in aliquots from the infection medium were determined to count the numbers of CFU. Means and standard deviations are plotted. Δ121, the deletion mutant; pMMB, empty vector pMMB207C. (C) Defective intracellular multiplication of the deletion mutant in A. castellanii amoebae is not substantially reversed by transformation with a plasmid with the lvh locus. To test the reversal of defective intracellular multiplication by the plasmid with lvh driven by its endogenous promoters, A. castellanii amoebae were infected at 30°C at an MOI of 1 with stationary-phase bacteria. At the indicated times, the titers in aliquots from the infection medium were determined to count the numbers of CFU. Means and standard deviations are plotted. (D) Defective intracellular multiplication of the deletion mutant in A. castellanii amoebae is not substantially reversed by IPTG induction of lpr0035 from the tac promoter. To test the reversal of defective intracellular multiplication by plasmid lpr0035 driven by the tac promoter without the endogenous lpr0035 promoter, A. castellanii amoebae were infected at 30°C at an MOI of 1 with stationary-phase bacteria. As indicated, IPTG was present throughout the experiment, and at the indicated times titers were determined in aliquots taken from the infection medium to count the numbers of CFU. Means and standard deviations are plotted.
Sensitivity of the deletion mutant to sodium ion.
Wild-type L. pneumophila is sensitive to sodium ion, but mutants with mutations in dot/icm become sodium resistant, showing increased plating efficiency in the presence of 100 mM sodium ion (46). The plating efficiency of the deletion mutant was identical to that of strain JR32, 39% and 34%, respectively, and significantly less than the 45% previously reported for a dotB mutant of JR32 (2). These results suggested that the macrophage and amoeba intracellular multiplication defects were not directly attributable to a nonfunctional Dot/Icm T4BSS in the mutant, similar to the inference from wild-type sodium sensitivity in a pmrA mutant of strain JR32 (54).
DISCUSSION
The experiments reported here were begun to study the contribution of pLP45 mobility to virulence phenotypes of L. pneumophila strain JR32. When strain JR32 is cultured under WS conditions, which mimic the aquatic environment of the pathogen, the Lvh T4ASS can functionally substitute for the Dot/Icm T4BSS. The Lvh locus lies within pLP45. In other L. pneumophila strains, the mobility of genetic elements containing Lvh or other T4ASSs has been correlated with virulence phenotypes. In Legionella strain Paris, pP36, a 36-kbp mobile genetic element containing an Lvh T4ASS locus, moves from episomal to chromosomal states during the transition from lag to stationary phase in broth, when Legionella virulence phenotypes are known to change (16). In strain Corby, excision of a mobile element containing a Trb-1 T4ASS is increased in Legionella after intracellular multiplication in A. castellanii amoebae (24). In other Legionella strains, virulence phenotypes are enhanced following intracellular growth in amoebae (12, 13), supporting a correlation between mobility and virulence phenotypes.
To more directly address the virulence contribution of pLP45 mobility in strain JR32, a 121-bp deletion was introduced at the 5′ chromosomal junction of pLP45, removing the 49-bp direct repeat at that junction. This deletion restricted pLP45 mobility and locked the genetic element in the chromosome with no detectable episomal form. During our characterization of the deletion mutant, RNA sequencing identified a novel noncoding RNA, lpr0035, which spanned the 5′ junction of pLP45 in the Philadelphia-1 clinical isolate, from which JR32 is derived (52). The 121-bp deletion lay entirely within the 252-bp ncRNA and was expected to inactivate lpr0035. Thus, our deletion mutant had two genetic alterations: an immobile genetic element and an inactivated ncRNA of unknown function and not antisense to any L. pneumophila gene. To dissect out the contributions of these two alterations to defects in the entry and intracellular multiplication phenotypes of the deletion mutant, we tested if defective phenotypes were reversed by plasmid copies of lpr0035 and of genes adjacent to the 5′ and 3′ junctions, whose expression might be altered by restricted pLP45 mobility or loss of lpr0035 function. As discussed below, our results definitively established a role for lpr0035 in entry and intracellular multiplication and revealed that further studies are required for definitive assignment of a virulence role for pLP45 mobility.
Defective entry of the deletion mutant.
Complementation of defective entry into J774 macrophages and A. castellanii amoebae implicated new genes in the entry phenotype of strain JR32. Complementation of this defect by plasmid lpr0035 indicated that the 121-bp deletion inactivated the ncRNA and lpr0035 plays a role in entry into both hosts. However, inactivation of lpr0035 by locking pLP45 in the chromosome could affect expression of other genes at the 5′ and 3′ junctions of pLP45. Mobility of pLP45 could change gene expression by changing upstream regulatory sequences or downstream termination sequences or by interrupting and rejoining genes or regulatory elements that span 5′ or 3′ chromosomal junctions of the element or through a copy number effect, if the episome replicates independently of the chromosome (Fig. 1B). In fact, entry defects were reversed by plasmid copies of lpg1228 and lpg1229, just inside the 5′ end of pLP45, and of the tRNAArg gene, just outside the 3′ end, implicating those genes in the entry phenotype and suggesting that expression of those genes was decreased in the deletion mutant. Reduced reporter activity in the deletion mutant for three LacZ translational fusions to lpg1228 and one fusion to lpg1229 supported decreased expression in the mutant. Decreased expression could arise from elimination of a positive regulatory element in the 121-bp deletion, by inactivation of lpr0035, or from decreased expression related to the restricted mobility of pLP45 by mechanisms described above.
The appropriate LacZ reporter for the tRNAArg gene at the 3′ end of pLP45 was a transcriptional fusion. Comparable LacZ reporter activity in strain JR32 and the deletion mutant indicated comparable promoter activity and suggested a promoter-independent mechanism for the inferred decrease in expression of the tRNAArg in the mutant. Regulation of translational termination was investigated using the mfold/RNA website (http://mfold.rna.albany.edu/?q=mfold/RNA-Folding-Form) to predict the stability of a 25-nt stem-loop downstream of lpg1272 (Fig. 1B). rG values of +0.3 kcal/mol and −4.4 kcal/mol were predicted for the stem-loop in the integrated and episomal states of the tRNAArg, respectively. These should be compared to a mean ΔG of −4.3 ± 1.1 kcal/mol for the three other tRNAArg genes in the L. pneumophila Philadelphia-1 genome. These calculations predict a decreased expression of the chromosomally locked tRNAArg gene in the mutant, attributable to a less stable transcription termination loop. Alternatively, if lpr0035 ncRNA stabilized the tRNAArg transcriptional termination stem-loop, then decreased expression of the tRNAArg could be attributed to inactivation of lpr0035. Distinguishing between the contributions of lpr0035 and pLP45 mobility to the entry phenotype could be studied with a mutant that lacks functional lpr0035 but retains pLP45 mobility.
The entry defect was reversed by plasmid copies of the tRNAArg gene, and plasmids lpg1228 and lpg1229 annotated as a hypothetical protein and a site-specific recombinase, respectively. This result was not entirely unexpected. Defective entry into mammalian and amoeba host cells has been reported for Legionella mutants with mutations in proteins with a variety of functions and structural motifs. Among these are an ecto-triphosphate diphosphohydrolase (44), the tetratricopeptide repeat-containing proteins LpnE and EnhC (14, 37), the repeats of structure toxin protein RtxA (15), and LvhB2, a putative pilin protein (40). While the mechanisms by which these diverse proteins contribute to entry are poorly understood, the mutant phenotypes clearly indicate redundant contributions to the entry process. Our data were therefore consistent with this literature of diverse and redundant contributions to entry. Of interest was that plasmid clones of each gene reversed defective entry without unlocking pLP45 from the chromosome and without restoring lpr0035.
Lastly, we tested for WS reversal of defective entry in the deletion mutant. Mutants in dot/icm genes are generally defective in entry, phagosome acidification, and intracellular multiplication virulence phenotypes (1, 17, 22, 26, 38). We previously showed that those virulence defects could be reversed by WS treatment of broth stationary-phase cultures of dot/icm mutants used for infection, indicating that the Dot/Icm T4BSS was conditionally required for those virulence phenotypes. We demonstrated that double mutants—with mutations in the dot/icm gene and the lvh T4ASS locus—did not show WS reversal, implying that the Lvh T4ASS was required for WS reversal and that the Lvh T4ASS could substitute for the Dot/Icm T4BSS following WS treatment (3, 5). In the current study, defective entry of the 121-bp deletion mutant could not be reversed by WS treatment, even though the Lvh T4ASS locus was present, locked in the chromosome. Inability to reverse the entry defect suggests that WS reversal may require the Lvh locus in episomal or mobile pLP45. Alternatively, the entry defect in the deletion mutant may not involve a T4SS or may involve aspects of entry not reversible by WS.
Defective intracellular multiplication of the deletion mutant.
In addition to showing defective entry into J774 macrophages and A. castellanii amoebae, the deletion mutant was defective in intracellular multiplication in both hosts by 1 to 3 orders of magnitude over 2 to 3 days following infection. This intracellular multiplication defect was not reversed by the above-described plasmids, which reversed defective entry. Plasmids with the lvh locus or two plasmids, one with lpg1228 and lpg1229 and one with the tRNAArg gene, were also ineffective in reversing the defect in intracellular multiplication.
Although the function of lpr0035 RNA is unknown, a role in virulence is suggested by a 16-fold decrease in expression during transition from broth exponential to postexponential phase (52), because virulence phenotypes related to invasion and intracellular multiplication increase dramatically during that transition in growth (6, 34, 51). When cloned behind a Ptac promoter and induced by IPTG present throughout infection, plasmid lpr0035 reversed the intracellular multiplication defect in J774 macrophages but not in amoebae and did not unlock pLP45 from the chromosome. Driven by its endogenous promoter, plasmid lpr0035 failed to reverse intracellular multiplication defects in either host.
These results are similar to complementation data seen with Legionella mutants with mutations in rsmY and rsmZ. Mutants with mutations in those ncRNAs are attenuated in intracellular multiplication in A. castellanii and macrophages derived from the THP-1 monocyte line (42). Complementation of the multiplication defect was accomplished by IPTG-induced expression of rsmY and rsmZ throughout infection. Similarly, intracellular multiplication defects of the CsrA protein (35) and of the PmrA response regulator (54) were complemented by IPTG-induced expression. PCR analysis showed that reversal of the intracellular multiplication defect by lpr0035 occurred without excision of pLP45. This result indicated that pLP45 mobility was not required for reversal and suggested a regulatory role for lpr0035 in Legionella virulence phenotypes.
The intracellular multiplication defect of the deletion mutant was more pronounced in A. castellanii amoebae than in J774 macrophages and was not reversed by IPTG-induced expression of lpr0035. A JR32 mutant of the PmrA regulatory protein also displayed a host-specific attenuation, showing a more pronounced intracellular multiplication defect in A. castellanii than macrophages derived from HL-60 cells (54). Host-specific differences such as these quantitative differences in virulence phenotypes have been attributed to host-specific factors or to differences between primary and cultured cells. Alternatively, the host-specific phenotype may reflect bacterial factors required for replication in amoebae but not in macrophage hosts. For example, in addition to lpr0035, replication in amoebae may require episomal pLP45 and/or mobility of pLP45. In strain Corby, growth in A. castellanii is accompanied by an increase in the episomal form of a mobile genetic element that harbors a T4ASS locus. Since our deletion mutant contains no detectable pLP45 episome, the intracellular multiplication defect in amoebae may be related to locking pLP45 in the chromosome, restricting the mobility of that genetic element and eliminating the episomal form.
Summary.
The studies reported here establish that a previously uncharacterized ncRNA in the Philadelphia-1 strain, lpr0035, plays a role in the entry of Legionella into macrophages and amoebae and in intracellular multiplication in macrophages. The presence of lpr0035 homologs in six other L. pneumophila genomes, those of strains 130b, Alcoy, Corby, Lens, Lorraine, and Paris, but no other microbial species, suggests that this ncRNA may play similar roles in other Legionella strains. Genes adjacent to the 5′ and 3′ junctions of pLP45 were also implicated in entry into macrophages and amoebae. Our results demonstrate that mobility of the pLP45 genetic element in the JR32 strain of Legionella pneumophila can be restricted and the mobile element can be locked in the chromosome by deleting the direct-repeat sequence at the 5′ chromosomal junction. This strategy may be applicable for studying the mobility of genetic elements in other strains of L. pneumophila and in other bacterial species. In the present study, inferences about the virulence role of lpr0035 were complicated by overlap of lpr0035 and the 5′ direct repeat of pLP45. Although lpr0035 is not antisense to any ORF in the L. pneumophila Philadelphia-1 genome, it is antisense to the direct repeats at the 5′ and 3′ ends of pLP45. In that way, pLP45 itself could influence the mobility of pLP45. To further dissect the contributions of lpr0035 and pLP45 mobility to the virulence phenotypes of strain JR32, it will be necessary to construct mutants in which lpr0035 is inactivated but mobility is not impaired and, conversely, in which lpr0035 is functional but mobility is restricted. Such studies and proteomic analyses of the deletion mutant are under way in our laboratory.
ACKNOWLEDGMENTS
We thank the Albert Einstein Analytical Imaging Facility for use of epifluorescence microscopes and Gil Segal (Tel Aviv University) for the p-GS-lac-01 translational and pMR-TV-1 lacZ transcriptional fusion vectors.
This work was supported by Public Health Service grant AI 072487 from the National Institutes of Health to H.M.S.
Footnotes
Published ahead of print 10 September 2012
REFERENCES
- 1. Al-Khodor S, Price CT, Habyarimana F, Kalia A, Abu Kwaik Y. 2008. A Dot/Icm-translocated ankyrin protein of Legionella pneumophila is required for intracellular proliferation within human macrophages and protozoa. Mol. Microbiol. 70:908–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bandyopadhyay P, Byrne B, Chan Y, Swanson MS, Steinman HM. 2003. Legionella pneumophila catalase-peroxidases are required for proper trafficking and growth in primary macrophages. Infect. Immun. 71:4526–4535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bandyopadhyay P, Liu S, Gabbai CB, Venitelli Z, Steinman HM. 2007. Environmental mimics and the Lvh type IVA secretion system contribute to virulence-related phenotypes of Legionella pneumophila. Infect. Immun. 75:723–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bandyopadhyay P, Steinman HM. 2000. Catalase-peroxidases of Legionella pneumophila: cloning of the katA gene and studies of KatA function. J. Bacteriol. 182:6679–6686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bandyopadhyay P, Xiao H, Coleman HA, Price-Whelan A, Steinman HM. 2004. Icm/Dot-independent entry of Legionella pneumophila into amoeba and macrophage hosts. Infect. Immun. 72:4541–4551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Byrne B, Swanson MS. 1998. Expression of Legionella pneumophila virulence traits in response to growth conditions. Infect. Immun. 66:3029–3034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cazalet C, et al. 2004. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat. Genet. 36:1165–1173 [DOI] [PubMed] [Google Scholar]
- 8. Centers for Disease Control and Prevention 2011. Legionellosis—United States, 2000-2009. MMWR Morb. Mortal. Wkly. Rep. 60:1083–1086 [PubMed] [Google Scholar]
- 9. Chen J, et al. 2004. Legionella effectors that promote nonlytic release from protozoa. Science 303:1358–1361 [DOI] [PubMed] [Google Scholar]
- 10. Chien M, et al. 2004. The genomic sequence of the accidental pathogen Legionella pneumophila. Science 305:1966–1968 [DOI] [PubMed] [Google Scholar]
- 11. Christie PJ, Vogel JP. 2000. Bacterial type IV secretion: conjugation systems adapted to deliver effector molecules to host cells. Trends Microbiol. 8:354–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cirillo JD, et al. 1999. Intracellular growth in Acanthamoeba castellanii affects monocyte entry mechanisms and enhances virulence of Legionella pneumophila. Infect. Immun. 67:4427–4434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cirillo JD, Falkow S, Tompkins LS. 1994. Growth of Legionella pneumophila in Acanthamoeba castellanii enhances invasion. Infect. Immun. 62:3254–3261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cirillo SL, Lum J, Cirillo JD. 2000. Identification of novel loci involved in entry by Legionella pneumophila. Microbiology 146:1345–1359 [DOI] [PubMed] [Google Scholar]
- 15. Cirillo SL, Yan L, Littman M, Samrakandi MM, Cirillo JD. 2002. Role of the Legionella pneumophila rtxA gene in amoebae. Microbiology 148:1667–1677 [DOI] [PubMed] [Google Scholar]
- 16. Doleans-Jordheim A, et al. 2006. Growth-phase-dependent mobility of the lvh-encoding region in Legionella pneumophila strain Paris. Microbiology 152:3561–3568 [DOI] [PubMed] [Google Scholar]
- 17. Ensminger AW, Isberg RR. 2009. Legionella pneumophila Dot/Icm translocated substrates: a sum of parts. Curr. Opin. Microbiol. 12:67–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Faucher SP, Friedlander G, Livny J, Margalit H, Shuman HA. 2010. Legionella pneumophila 6S RNA optimizes intracellular multiplication. Proc. Natl. Acad. Sci. U. S. A. 107:7533–7538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Faucher SP, Shuman HA. 2011. Small regulatory RNA and Legionella pneumophila. Front. Microbiol. 2:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Feeley JC, et al. 1979. Charcoal-yeast extract agar: primary isolation medium for Legionella pneumophila. J. Clin. Microbiol. 10:437–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fields BS, Benson RF, Besser RE. 2002. Legionella and Legionnaires' disease: 25 years of investigation. Clin. Microbiol. Rev. 15:506–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Franco IS, Shuman HA, Charpentier X. 2009. The perplexing functions and surprising origins of Legionella pneumophila type IV secretion effectors. Cell. Microbiol. 11:1435–1443 [DOI] [PubMed] [Google Scholar]
- 23. Gal-Mor O, Zusman T, Segal G. 2002. Analysis of DNA regulatory elements required for expression of the Legionella pneumophila icm and dot virulence genes. J. Bacteriol. 184:3823–3833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Glockner G, et al. 2008. Identification and characterization of a new conjugation/type IVA secretion system (trb/tra) of Legionella pneumophila Corby localized on two mobile genomic islands. Int. J. Med. Microbiol. 298:411–428 [DOI] [PubMed] [Google Scholar]
- 25. Hicks LA, Garrison LE, Nelson GE, Hampton LM. 2012. Legionellosis—United States, 2000-2009. Am. J. Transplant. 12:250–253 [DOI] [PubMed] [Google Scholar]
- 26. Hilbi H, Segal G, Shuman HA. 2001. Icm/Dot-dependent upregulation of phagocytosis by Legionella pneumophila. Mol. Microbiol. 42:603–617 [DOI] [PubMed] [Google Scholar]
- 27. Horwitz MA, Silverstein SC. 1980. Legionnaires' disease bacterium (Legionella pneumophila) multiples intracellularly in human monocytes. J. Clin. Invest. 66:441–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hovel-Miner G, et al. 2009. SigmaS controls multiple pathways associated with intracellular multiplication of Legionella pneumophila. J. Bacteriol. 191:2461–2473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hubber A, Roy CR. 2010. Modulation of host cell function by Legionella pneumophila type IV effectors. Annu. Rev. Cell Dev. Biol. 26:261–283 [DOI] [PubMed] [Google Scholar]
- 30. Isberg RR, O'Connor TJ, Heidtman M. 2009. The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat. Rev. Microbiol. 7:13–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kessler B, de Lorenzo V, Timmis KN. 1992. A general system to integrate lacZ fusions into the chromosomes of gram-negative eubacteria: regulation of the Pm promoter of the TOL plasmid studied with all controlling elements in monocopy. Mol. Gen. Genet. 233:293–301 [DOI] [PubMed] [Google Scholar]
- 32. Kulkarni PR, Cui X, Williams JW, Stevens AM, Kulkarni RV. 2006. Prediction of CsrA-regulating small RNAs in bacteria and their experimental verification in Vibrio fischeri. Nucleic Acids Res. 34:3361–3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Molmeret M, Horn M, Wagner M, Santic M, Abu Kwaik Y. 2005. Amoebae as training grounds for intracellular bacterial pathogens. Appl. Environ. Microbiol. 71:20–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Molofsky AB, Swanson MS. 2004. Differentiate to thrive: lessons from the Legionella pneumophila life cycle. Mol. Microbiol. 53:29–40 [DOI] [PubMed] [Google Scholar]
- 35. Molofsky AB, Swanson MS. 2003. Legionella pneumophila CsrA is a pivotal repressor of transmission traits and activator of replication. Mol. Microbiol. 50:445–461 [DOI] [PubMed] [Google Scholar]
- 36. Newton HJ, Ang DK, van Driel IR, Hartland EL. 2010. Molecular pathogenesis of infections caused by Legionella pneumophila. Clin. Microbiol. Rev. 23:274–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Newton HJ, Sansom FM, Bennett-Wood V, Hartland EL. 2006. Identification of Legionella pneumophila-specific genes by genomic subtractive hybridization with Legionella micdadei and identification of lpnE, a gene required for efficient host cell entry. Infect. Immun. 74:1683–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ninio S, Roy CR. 2007. Effector proteins translocated by Legionella pneumophila: strength in numbers. Trends Microbiol. 15:372–380 [DOI] [PubMed] [Google Scholar]
- 39. Rasis M, Segal G. 2009. The LetA-RsmYZ-CsrA regulatory cascade, together with RpoS and PmrA, post-transcriptionally regulates stationary phase activation of Legionella pneumophila Icm/Dot effectors. Mol. Microbiol. 72:995–1010 [DOI] [PubMed] [Google Scholar]
- 40. Ridenour DA, Cirillo SL, Feng S, Samrakandi MM, Cirillo JD. 2003. Identification of a gene that affects the efficiency of host cell infection by Legionella pneumophila in a temperature-dependent fashion. Infect. Immun. 71:6256–6263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sadosky AB, Wiater LA, Shuman HA. 1993. Identification of Legionella pneumophila genes required for growth within and killing of human macrophages. Infect. Immun. 61:5361–5373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sahr T, et al. 2009. Two small ncRNAs jointly govern virulence and transmission in Legionella pneumophila. Mol. Microbiol. 72:741–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sahr T, et al. 2012. Deep sequencing defines the transcriptional map of L. pneumophila and identifies growth phase-dependent regulated ncRNAs implicated in virulence. RNA Biol. 9:503–519 [DOI] [PubMed] [Google Scholar]
- 44. Sansom FM, et al. 2007. A bacterial ecto-triphosphate diphosphohydrolase similar to human CD39 is essential for intracellular multiplication of Legionella pneumophila. Cell. Microbiol. 9:1922–1935 [DOI] [PubMed] [Google Scholar]
- 45. Schroeder GN, et al. 2010. Legionella pneumophila strain 130b possesses a unique combination of type IV secretion systems and novel Dot/Icm secretion system effector proteins. J. Bacteriol. 192:6001–6016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Segal G, Feldman M, Zusman T. 2005. The Icm/Dot type-IV secretion systems of Legionella pneumophila and Coxiella burnetii. FEMS Microbiol. Rev. 29:65–81 [DOI] [PubMed] [Google Scholar]
- 47. Segal G, Russo JJ, Shuman HA. 1999. Relationships between a new type IV secretion system and the icm/dot virulence system of Legionella pneumophila. Mol. Microbiol. 34:799–809 [DOI] [PubMed] [Google Scholar]
- 48. Segal G, Shuman HA. 1999. Legionella pneumophila utilizes the same genes to multiply within Acanthamoeba castellanii and human macrophages. Infect. Immun. 67:2117–2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sexton JA, Yeo HJ, Vogel JP. 2005. Genetic analysis of the Legionella pneumophila DotB ATPase reveals a role in type IV secretion system protein export. Mol. Microbiol. 57:70–84 [DOI] [PubMed] [Google Scholar]
- 50. Steinert M, Hentschel U, Hacker J. 2002. Legionella pneumophila: an aquatic microbe goes astray. FEMS Microbiol. Rev. 26:149–162 [DOI] [PubMed] [Google Scholar]
- 51. Swanson MS, Hammer BK. 2000. Legionella pneumophila pathogenesis: a fateful journey from amoebae to macrophages. Annu. Rev. Microbiol. 54:567–613 [DOI] [PubMed] [Google Scholar]
- 52. Weissenmayer BA, Prendergast JG, Lohan AJ, Loftus BJ. 2011. Sequencing illustrates the transcriptional response of Legionella pneumophila during infection and identifies seventy novel small non-coding RNAs. PLoS One 6:e17570 doi:10.1371/journal.pone.0017570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wiater LA, Sadosky AB, Shuman HA. 1994. Mutagenesis of Legionella pneumophila using Tn903 dlllacZ: identification of a growth-phase-regulated pigmentation gene. Mol. Microbiol. 11:641–653 [DOI] [PubMed] [Google Scholar]
- 54. Zusman T, et al. 2007. The response regulator PmrA is a major regulator of the icm/dot type IV secretion system in Legionella pneumophila and Coxiella burnetii. Mol. Microbiol. 63:1508–1523 [DOI] [PubMed] [Google Scholar]
- 55. Zusman T, Gal-Mor O, Segal G. 2002. Characterization of a Legionella pneumophila relA insertion mutant and roles of RelA and RpoS in virulence gene expression. J. Bacteriol. 184:67–75 [DOI] [PMC free article] [PubMed] [Google Scholar]