Abstract
Genetic factors that regulate the pathogenesis of pneumonia caused by the fungus Cryptococcus neoformans are poorly understood. Through a phenotypic strain survey we observed that inbred C3H/HeN mice develop a significantly greater lung fungal burden than mice of the resistant CBA/J strain 4 weeks following intratracheal infection with C. neoformans ATCC 24067. The aim of the present study was to characterize the inflammatory response of C3H/HeN mice following C. neoformans pulmonary infection and to identify genetic loci that regulate host defense. Following cryptococcal infection, C3H/HeN mice demonstrated a Th2 immune response with heightened airway and tissue eosinophilia, goblet cell metaplasia, and significantly higher lung interleukin-5 (IL-5) and IL-13 protein expression relative to CBA/J mice. Conversely, CBA/J mice exhibited greater airway and tissue neutrophilia that was associated with significantly higher pulmonary expression of gamma interferon, CXCL10, and IL-17 proteins than C3H/HeN mice. Using the fungal burden at 4 weeks postinfection as a phenotype, genome-wide quantitative trait locus (QTL) analysis among 435 segregating (C3H/HeN × CBA/J)F2 (C3HCBAF2) hybrids identified two significant QTLs on chromosomes 1 (Cnes4) and 9 (Cnes5) that control susceptibility to cryptococcal pneumonia in an additive manner. Susceptible C3H/HeN mice carry a resistance allele at Cnes4 and a susceptibility allele at Cnes5. These studies reveal additional genetic complexity of the host response to C. neoformans that is associated with divergent patterns of pulmonary inflammation.
INTRODUCTION
Cryptococcus neoformans is a ubiquitous environmental fungus that primarily causes pneumonia and meningitis among immune-deficient populations (3, 11) and is being increasingly recognized in immunocompetent individuals (28, 30, 36). The spectrum of illness ranges from latent or subclinical infection to severe and potentially lethal disease and is consistent with the notion that susceptibility to C. neoformans infection is controlled by multiple genetic and environmental factors (14, 16). Nevertheless, due to a paucity of reports that describe familial clustering of infection, as well as the frequent confounding effects of HIV infection or iatrogenic immune suppression, clinical and epidemiological studies have not identified these susceptibility factors.
Experimental analysis of mouse models has been a highly effective strategy to identify the cellular and molecular basis of the host immune response to cryptococcal infection (50). Data from mice that harbor natural or genetically engineered immune defects have identified a role for several components of the innate immune system as well as the complexity of antibody (Ab)- and cell-mediated anticryptococcal responses that generate protective immunity (55). Interestingly, immunocompetent inbred mouse strains also show remarkable variation in their susceptibility to experimental C. neoformans lung infection (10, 17, 21, 43, 57). As a consequence of the evolutionary origins and distinct breeding histories (4, 41), inbred strains carry extensive genomic sequence diversity and can serve as a valuable resource for identification of underlying susceptibility loci, genes, and biochemical pathways through complex trait analysis (25). Despite this potential, a current limitation to this approach for investigation of cryptococcal pneumonia is that careful characterization of host susceptibility phenotypes has been limited to relatively few inbred strains.
The overall aim of this study was to advance the understanding of genetically regulated host defense against cryptococcal pneumonia. To achieve this goal, we first phenotyped a panel of 10 widely used inbred mouse strains using a well-established infection model that recapitulates clinical disease (22, 41). We defined relative susceptibility among inbred mouse strains on the basis of the lung fungal burden at 28 days postinfection. From this survey we observed that the previously uncharacterized C3H/HeN inbred strain is highly susceptible to C. neoformans infection, despite the fact that it is considered to be immunocompetent and shares recent common ancestry with the resistant CBA/J strain. To investigate the immunological and genetic basis for differential host resistance between the closely related C3H/HeN and CBA/J inbred strains, we then performed a comparative study of lung inflammatory responses and a genome-wide quantitative trait locus (QTL) analysis following intratracheal C. neoformans infection.
MATERIALS AND METHODS
Mouse strains and crosses.
Inbred C3H/HeN and CBA/JCrHsd (CBA/J) mice were obtained from Harlan Laboratories or bred in our specific-pathogen-free facility. (C3H/HeN × CBA/JCrHsd)F1 (C3HCBAF1) and (C3HCBAF1 × C3HCBAF1)F2 (C3HCBAF2) mice were bred and maintained in our specific-pathogen-free facility. All animals were maintained in compliance with the Canadian Council on Animal Care, as approved by McGill University.
C. neoformans culture.
C. neoformans ATCC 24067 was grown and maintained on Sabouraud dextrose agar (BD, Becton, Dickinson and Company). Cells from a single colony were grown in Sabouraud dextrose broth (BD) for 48 h with constant rotation. The stationary culture was then washed with sterile phosphate-buffered saline (PBS), counted on a hemacytometer, and diluted to 2 × 105 CFU/ml in sterile PBS. Each experimental dose was confirmed before and after infection by plating a dilution on Sabouraud dextrose agar and counting the number of CFU after 72 h of incubation at room temperature.
Intratracheal administration of C. neoformans.
Mice were anesthetized by intraperitoneal injection of ketamine (15 mg/kg of body weight; Ayerst Veterinary Laboratories) and xylazine (125 mg/kg; Bayer Inc.), and a small skin incision was made over the trachea. Subsequently, 50 μl of 2 × 105 CFU/ml of C. neoformans (104 CFU) was administered via insertion of a 22-gauge catheter into the trachea, followed immediately by 50 μl of air. The incision was then closed using a 9-mm Autoclip wound closing kit (Stoelting), and the mice were allowed to recover under a heat lamp.
Lung isolation and CFU assay.
At the indicated time points following infection, mice were euthanized by CO2 exposure and the lungs were excised and placed in 2 ml of ice-cold, sterile PBS. Lungs were then weighed and homogenized with a glass tube and pestle at 333 rpm using a tissue homogenizer (Glas-Col). The lung homogenate was then serially diluted in sterile PBS and plated on Sabouraud dextrose agar plates in duplicate. Inoculated plates were incubated at 37°C for 72 h, and then the numbers of C. neoformans CFU were counted. To detect microbial contamination arising from the infection or lung excision procedures, each homogenate was also plated on Columbia blood agar.
Lung histology.
Lungs were gently flushed with ice-cold PBS via the right ventricle and then inflated to a fixed pressure of 25 cm H2O with 10% buffered formalin acetate (Fisher Scientific) in situ, embedded in paraffin, sectioned at 5 μm, and stained with hematoxylin-eosin (H&E) or periodic acid-Schiff (PAS). Slide images were captured with a CoolSNAP-Pro cf digital capture kit (Media Cybernetics) using an Olympus BX51 light microscope (Olympus Canada Inc.).
BAL.
Bronchoalveolar lavage (BAL) fluid (BALF) was collected by flushing the airways with four 0.5-ml aliquots of ice-cold, sterile PBS via a 22-guage catheter inserted into the trachea of a euthanized mouse. The total airway cell number was determined using an automated cell counter (Beckman Coulter). Differential cell counts of macrophages, neutrophils, and eosinophils were determined by counting 300 cells following cytospin centrifugation (Shandon) and Diff-Quik staining (Dade Behring). Macrophages were identified as relatively large cells with a compact violet nucleus and abundant vacuolated cytoplasm, while neutrophils and eosinophils were identified as having segmented blue nuclei and pale or red-staining cytoplasmic granules, respectively. The absolute number of cells for each subset was determined by multiplying the percentage of each cell type by the total number of leukocytes.
Isolation of CD45+ lung leukocytes.
Individual lungs were gently flushed with ice-cold PBS via the right ventricle and immediately placed in 15 ml of sterile digestion buffer (RPMI 1640 medium supplemented with 2 mM glutamine, 100 U/ml penicillin, 100 g/ml streptomycin, 1 mg/ml collagenase D [Sigma-Aldrich], 30 μg/ml DNase I, and 10% [vol/vol] fetal bovine serum). Lungs were enzymatically digested for 90 min at 37°C, and the suspension was then drawn through a 10-ml syringe multiple times to disperse any residual fragments. Cells were then filtered through a 70-μm-sieve-size cell strainer and collected by centrifugation. Erythrocytes were lysed with ice-cold NH4Cl buffer (0.83% NH4Cl, 0.1% KHCO3, 0.037% Na2EDTA, pH 7.4) for 3 min, and the remaining leukocytes were suspended in complete medium (RPMI 1640 medium supplemented with 2 mM glutamine, 100 U/ml penicillin, 100 g/ml streptomycin). CD45+ cells were isolated from the lung cell suspension by magnetically activated cell sorting CD45+ microbeads and an AutoMACS cell sorter (Miltenyi Biotec) according to the manufacturer's protocol. The total CD45+ cells were counted using an automatic cell counter (Beckman Coulter), and differential leukocyte counts were determined as described above.
Flow cytometry.
CD45+ lung leukocytes were incubated with Fc blocking anti-mouse CD16/32 Ab (eBioscience). Aliquots of 106 cells were then incubated with peridinin chlorophyll protein-conjugated anti-CD4 Ab (BD Pharmingen), phycoerythrin-conjugated anti-CD8 Ab (eBioscience), fluorescein isothiocyanate-conjugated anti-CD19 Ab (BD Pharmingen), or a corresponding anti-rat IgG isotype control Ab. Propidium iodide staining was used to assess cell death (eBioscience). Data were collected with a FACSCalibur flow cytometer and CellQuest software (BD Bioscience) and analyzed by FlowJo software (Tree Star Inc.). A total of 50,000 events were captured per sample after setting a gate based on forward- and side-scatter patterns and CD45+ expression. The absolute number of each cell subset was determined by multiplying the percentage of each cell type by the total number of leukocytes.
Genotyping and QTL analysis.
DNA was collected from parental strains and F2 progeny using a DNeasy tissue kit (Qiagen). Informative single nucleotide polymorphisms (SNPs) between C3H/HeN and CBA/JCrHsd parental strains were identified using a JAX mouse diversity genotyping array (The Jackson Laboratory). Eleven genomic regions greater than 40 Mb did not contain an informative SNP between C3H/HeN and CBA/JCrHsd mice. A total of 94 informative SNPs were chosen at an average distance of 24.1 Mb and used to genotype all 435 C3HCBAF2 mice (218 males, 217 females). Genotyping was carried out at the Centre Hospitalier de l'Université Laval Research Centre. QTL analysis was performed using the package R/qtl in the statistical software R (5). A linear regression analysis was conducted to determine if sex was a significant covariate in the F2 population. Subsequently, single-marker linkage analysis was done in R by testing for linkage using three models with an additive, dominant, or recessive mode of inheritance. Analysis to test for all pairwise locus interactions was carried out by regression at the markers. The genome-wide significance of a locus was estimated by permutation with 10,000 resamples and took into consideration not only the total number of markers but also, at the same time, the number of models tested. A P value of 0.01 was our criterion for statistical significance.
Total lung cytokine production.
Mice were euthanized and lungs were perfused via the right ventricle with 10 ml of ice-cold PBS. Whole lungs were excised and placed in 2 ml PBS with Halt protease and phosphatase inhibitor cocktail (Fisher Scientific). Lungs were subsequently homogenized using a sterilized glass tube and pestle attached to a mechanical tissue homogenizer (Glas-Col). Homogenates were centrifuged at 12,000 rpm for 20 min, and supernatants were collected and stored at −80°C until analysis. Supernatants were assayed in 96-well plates (Greiner Bio One) using DuoSet enzyme-linked immunosorbent assay (ELISA) kits (R&D Systems) for interleukin-5 (IL-5), IL-13, IL-17, gamma interferon (IFN-γ), and CXCL10 protein per the manufacturer's protocol. Sample optical densities were determined using a microplate reader (Bio-Rad) and interpolated against a suitable standard curve.
Statistical analysis.
All fungal burden data are expressed as the mean ± standard error of the mean (SEM) unless otherwise indicated. Standard statistical analyses and graphics were done in R, version 2.13.1 (http://www.R-project.org), or Prism, version 4 (GraphPad). Pairwise comparisons were done with an unpaired t test; Tukey post hoc tests were applied to multiple comparisons.
RESULTS
Inbred mice exhibit differential susceptibility to experimental C. neoformans infection.
To survey the natural variation in host susceptibility to cryptococcal pneumonia, we phenotyped a panel of 10 inbred mouse strains from the initial priority classification of the mouse phenome project (41). We observed a wide spectrum of lung fungal burden among six previously uncharacterized strains and established their susceptibility relative to the susceptibilities of C57BL/6J, BALB/cJ, CBA/J, and SJL/J mice (Table 1). In four cases (FVB/NJ, A/J, DBA/2J, and SWR/J mice), the susceptible phenotype is likely attributable to known complement C5 deficiency (the C5 genotype has not been determined in 129S6 mice). The most notable finding from the phenotypic survey was that C5-sufficient C3H/HeN mice were highly susceptible to C. neoformans infection, with a lung fungal burden that was similar to that of C57BL/6J mice (mean lung burden, 6.49 ± 0.10 log10 CFU versus 6.94 ± 0.16 log10 CFU) (22). The marked susceptibility of C3H/HeN mice compared to that of resistant CBA/J mice (mean lung burden, 4.27 ± 0.27 log10 CFU) was particularly striking since these strains are closely related and share the H-2k/k haplotype that mediates antigen presentation by the immune system (17, 18).
Table 1.
Lung fungal burden of inbred mouse strainsa
| Strain | Log10 no. of CFUb | C5c | MHC H2 haplotyped |
|---|---|---|---|
| C57BL/6J | 6.94 ± 0.16 | + | H-2b |
| FVB/N | 6.63 ± 0.06 | − | H-2q |
| C3H/HeN | 6.49 ± 0.10 | + | H-2k |
| 129S6 | 6.06 ± 0.15 | ? | H-2b |
| SWR/J | 5.92 ± 0.11 | − | H-2q |
| DBA/2J | 5.80 ± 0.20 | − | H-2d |
| A/J | 5.13 ± 0.22 | − | H-2a |
| Balb/cJ | 4.90 ± 0.17 | + | H-2d |
| CBA/J | 4.27 ± 0.27 | + | H-2k |
| SJL/J | 3.25 ± 0.20 | + | H-2s |
Ten different inbred strains of 7-week-old mice (n > 7) were intratracheally infected with 104 CFU of C. neoformans ATCC 24067. The lungs were harvested at 28 days postinfection, and the numbers of CFU were determined.
Mean number of CFU in the lung ± SEM.
MHC, major histocompatibility complex. H2 haplotype data were obtained from http://jaxmice.jax.org/jaxnotes.
C3H/HeN is susceptible to progressive C. neoformans infection.
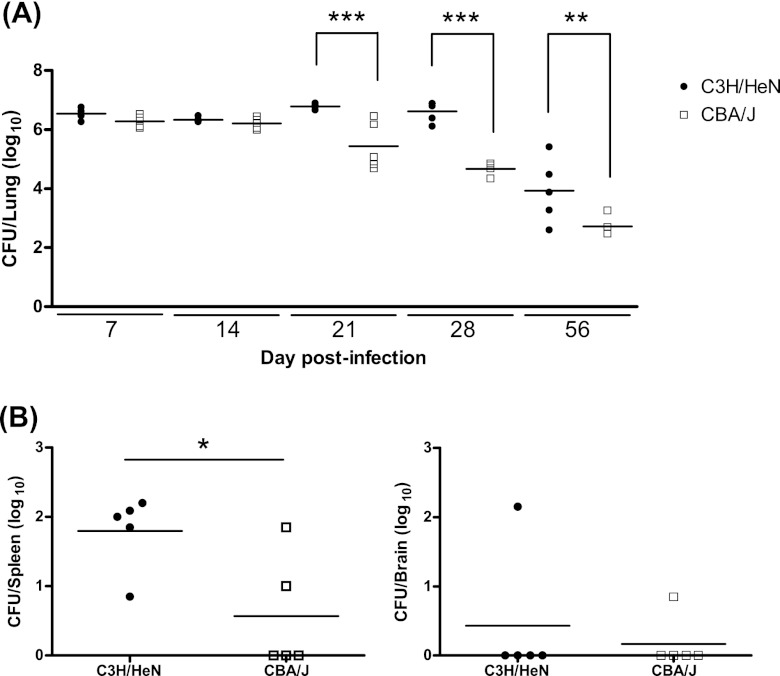
To determine the kinetics of differential fungal growth between C3H/HeN and CBA/J mice, each inbred strain was intratracheally infected with 104 CFU of C. neoformans ATCC 24067 and lung fungal burden was followed at serial time points (Fig. 1A). At day 21, C3H/HeN mice presented a significantly greater lung fungal burden than CBA/J mice (6.79 ± 0.04 log10 CFU for C3H/HeN mice versus 5.45 ± 0.37 log10 CFU for CBA/J mice; P < 0.001) that was also observed at day 28 (6.61 ± 0.16 log10 CFU for C3H/HeN mice versus 4.67 ± 0.11 log10 CFU for CBA/J mice; P < 0.001) and day 56 (3.94 ± 0.49 log10 CFU for C3H/HeN mice versus 2.73 ± 0.18 log10 CFU for CBA/J mice; P < 0.01). C3H/HeN mice also demonstrated significantly greater fungal dissemination to the spleen (1.80 ± 0.24 log10 CFU for C3H/HeN mice versus 0.57 ± 0.37 log10 CFU for CBA/J mice; P = 0.01) at day 28 than the CBA/J inbred strain (Fig. 1B). No significant difference in fungal burden was observed between male and female C3H/HeN mice or CBA/J mice up to day 56 (data not shown) (10).
Fig 1.
Fungal burden and dissemination following C. neoformans infection. Inbred 7-week-old female C3H/HeN and CBA/J mice underwent intratracheal infection with 104 CFU of C. neoformans ATCC 24067. (A) Fungal burden in the lung was determined at 7, 14, 21, 28, and 56 days after infection. (B) Fungal burden in the spleen and brain was determined 28 days after infection. Each filled circle represents an individual mouse; horizontal bars represent the mean number of CFU/organ (n = 4 or 5 mice/strain/day; *, P = 0.01; **, P < 0.01; ***, P < 0.001).
C3H/HeN and CBA/J mount differential immune responses to experimental C. neoformans infection.
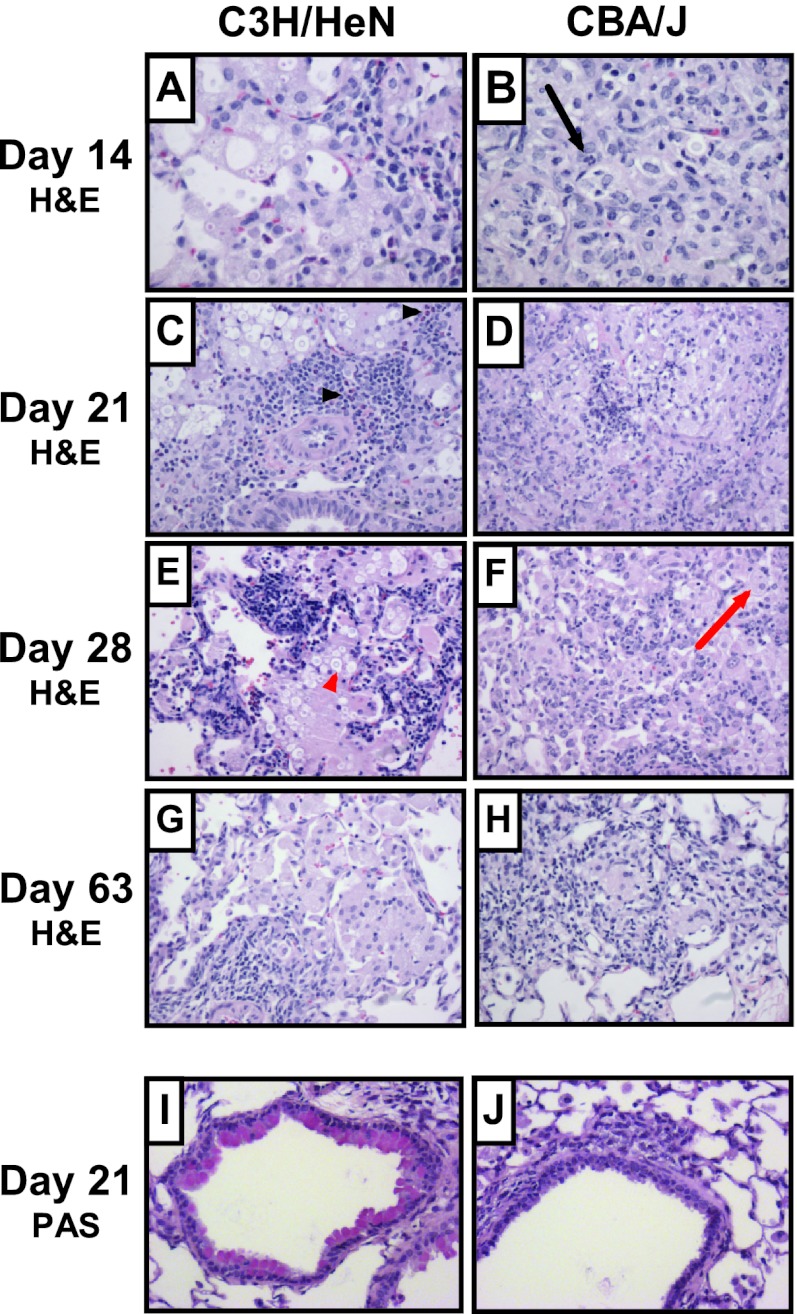
To investigate whether the C3H/HeN inbred strain presented an allergic bronchopulmonary response following cryptococcal infection as previously observed with susceptible C57BL/6J mice, we performed a comparative histological analysis of the pulmonary inflammatory response between C3H/HeN and CBA/J mice (Fig. 2). PAS or H&E staining of fixed lung sections did not demonstrate any significant differences between uninfected C3H/HeN and CBA/J mice (data not shown). At all time points and in both strains, inflammation was predominantly macrophage rich with fewer lymphocytes (Fig. 2A to H). The CBA/J mice showed a more extensive and nearly confluent infiltrate at day 14, while the C3H/HeN mice had moderate patchy involvement that peaked at day 21. Focal necrosis was present in CBA/J mice at days 14 and 21 but was never observed in the C3H/HeN mice. At days 14, 21, and 28, there was an increased presence of eosinophils in the C3H/HeN mice, while there were increased neutrophils in the CBA/J mice. H&E staining also demonstrated a greater presence of visible C. neoformans cells at 14, 21, and 28 days postinfection in C3H/HeN mouse lungs than CBA/J mouse lungs. At day 63, both mouse strains showed resolving inflammation that was characterized by a relatively mild patchy histiocytic infiltrate with lymphocytes and few granulocytes. Histiocytic aggregates and occasional giant cells were suggestive of ill-formed granulomas in the CBA/J mice. PAS staining demonstrated goblet cell metaplasia with airway epithelial cell mucus accumulation in C3H/HeN mouse airways at days 14, 21, and 28 after infection (Fig. 2I, representative of these time points). Mucus was not detectable in the C3H/HeN mouse airways at day 63 and was never detected in the CBA/J mouse airways after infection (Fig. 2J, representative of all time points).
Fig 2.
Lung histology following C. neoformans infection. Inbred 7-week-old female C3H/HeN and CBA/J mice were intratracheally infected with 104 CFU of C. neoformans ATCC 24067, and lungs were fixed, excised, paraffin embedded, and stained with H&E (A to H) or PAS (I and J) at the indicated time points. Photomicrographs of representative lung sections from 2 to 4 mice were taken at a magnification of ×200. Airway epithelial mucus stains red with PAS; highlighted cells include C. neoformans (red arrowhead), macrophage (red arrow), neutrophil (black arrow), and eosinophils (black arrowheads).
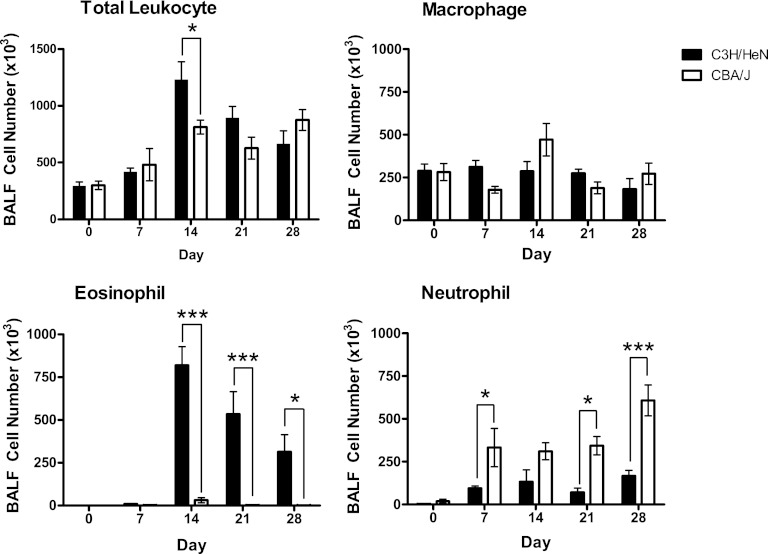
Cell recruitment to the airways and lung interstitium was assayed to characterize the inflammatory cell profiles associated with the differential pulmonary fungal burden and allergic response following experimental infection in C3H/HeN and CBA/J inbred mice (Fig. 3). The BALF revealed significantly greater total leukocyte recruitment to the airways of C3H/HeN mice than those of CBA/J mice at day 14 (1,230 × 103 ± 158 × 103 versus 814 × 103 ± 61 × 103; P < 0.05). Differential staining of leukocyte subsets revealed significantly greater eosinophilia in C3H/HeN mice at day 14 that continued through day 28 and accounted for the majority of the total airway cell population (day 14, 819 × 103 ± 110 × 103 for C3H/HeN mice versus 31 × 103 ± 15 × 103 for CBA/J mice [P < 0.001]; day 21, 534 × 103 ± 132 × 103 for C3H/HeN mice versus 3 × 103 ± 2 × 103 for CBA/J mice [P < 0.001]; day 28, 314 × 103 ± 100 × 103 for C3H/HeN mice versus 1 × 103 ± 1 × 103 for CBA/J mice [P < 0.05]). Notably, airway eosinophilia was almost completely absent in CBA/J mice throughout the course of the infection. In contrast, the CBA/J inbred strain presented a significantly greater neutrophilia at days 7, 21, and 28 following experimental C. neoformans infection (day 7, 333 × 103 ± 112 × 103 for CBA/J mice versus 95 × 103 ± 14 × 103 for C3H/HeN mice [P < 0.05]; day 21, 344 × 103 ± 54 × 103 for CBA/J mice versus 70 × 103 ± 25 × 103 for C3H/HeN mice [P < 0.05]; day 28, 108 × 103 ± 90 × 103 for CBA/J mice versus 166 × 103 ± 34 × 103 for C3H/HeN mice [P < 0.001]). No significant difference in macrophage recruitment to the airways was observed between C3H/HeN and CBA/J mice following cryptococcal infection.
Fig 3.
Airway leukocyte recruitment following C. neoformans infection. Airway leukocytes were isolated by BAL from inbred 7-week-old female C3H/HeN and CBA/J mice at days 0, 7, 14, 21, and 28 following intratracheal infection with 104 CFU of C. neoformans ATCC 24067. The total cell number in the BALF was determined by an automated cell counter. Leukocyte subsets were identified by Diff-Quik staining of BALF cell suspensions on cytospin slides. The absolute number of cells for each leukocyte subset was determined by multiplying the percentage of each cell type by the total number of leukocytes (n = 3 to 5 mice/strain/time point; *, P < 0.05; ***, P < 0.001).
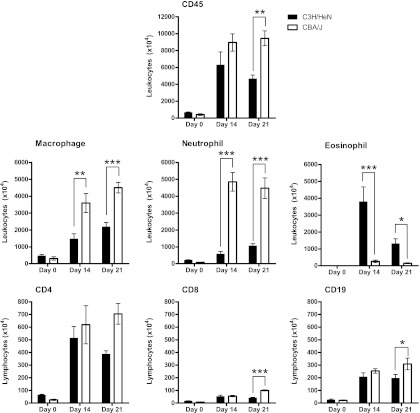
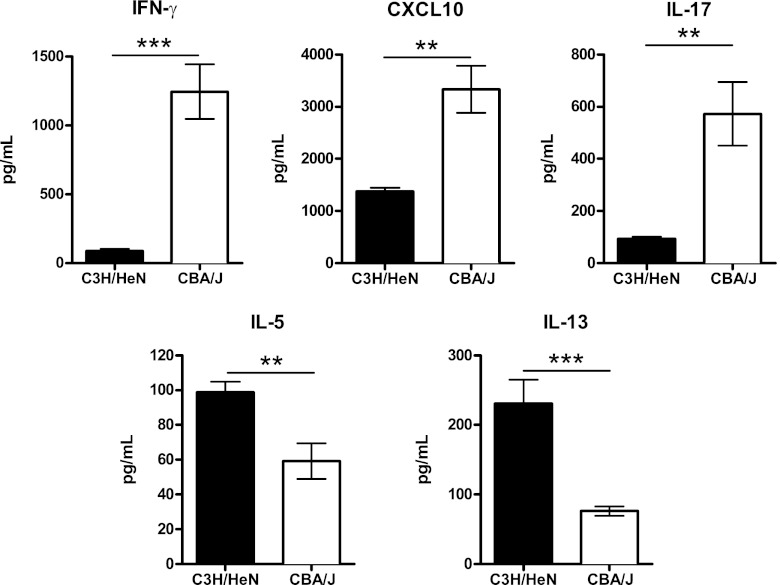
Leukocyte and lymphocyte recruitment to the interstitial compartment of the lungs was determined at day 14 and day 21, the time points after experimental infection that were associated with the most significant differences in airway cell recruitment between C3H/HeN and CBA/J mice. Enumeration of CD45+ cells selected from whole-lung digests using antibody-coated magnetic beads revealed that CBA/J mice recruited a significantly higher number than C3H/HeN mice at day 21 postinfection (9,448 × 104 ± 873 × 104 versus 4,650 × 104 ± 444 × 104; P < 0.01) (Fig. 4). Differential staining of CD45+ lung leukocytes confirmed the pattern of immune polarization between C3H/HeN and CBA/J mice that was observed by histology and bronchoalveolar lavage. Specifically, CBA/J mice presented significantly greater macrophage recruitment than C3H/HeN mice (day 14, 3,595 × 104 ± 561 × 104 versus 1,472 × 104 ± 309 × 104 [P < 0.01]; day 21, 4,508 × 104 ± 314 × 104 versus 2,193 × 104 ± 252 × 104 [P < 0.001]) and neutrophil recruitment (day 14, 4,853 × 104 ± 551 × 104 versus 575 × 104 ± 156 × 104 [P < 0.001]; day 21, 4,479 × 104 ± 611 × 104 versus 1,064 × 104 ± 130 × 104 [P < 0.001]). In contrast, susceptible C3H/HeN mice presented significantly greater tissue eosinophilia than CBA/J mice following cryptococcal infection (day 14, 3,800 × 104 ± 871 × 104 versus 266 × 104 ± 74 × 104 [P < 0.001]; day 21, 1,316 × 104 ± 288 × 104 versus 153 × 104 ± 26 × 104 [P < 0.05]). Flow cytometry analysis of CD4+, CD8+, and CD19+ lymphocyte populations was then performed on the isolated CD45+ lung cells. Following infection, CBA/J mice presented significantly higher CD8+ T-lymphocyte populations than C3H/HeN mice at day 21 (101 × 104 ± 5 × 104 versus 41 × 104 ± 5 × 104; P < 0.001). CBA/J mice also demonstrated a significant increase in CD19+ B lymphocytes at day 21 following infection (310 × 104 ± 46 × 104 versus 197 × 104 ± 29 × 104; P < 0.05) Cytokine expression was analyzed by protein ELISA to identify the pattern of immune polarization in the lungs of C3H/HeN and CBA/J mice following C. neoformans infection (Fig. 5). At day 14 postinfection, C3H/HeN mice presented significantly higher levels of the Th2-associated cytokines IL-5 (99 ± 6 pg/ml versus 59 ± 10 pg/ml; P < 0.01) and IL-13 (231 ± 34 pg/ml versus 76 ± 7 pg/ml; P < 0.001) than CBA/J mice. In contrast, the lungs of CBA/J mice demonstrated significantly higher expression of the Th1-associated cytokines CXCL10 (3,328 ± 450 pg/ml versus 1,380 ± 70 pg/ml; P < 0.001) and IFN-γ (1,245 ± 199 pg/ml versus 89 ± 16 pg/ml; P < 0.001) and the Th17-associated cytokine IL-17 (573 ± 122 pg/ml versus 93 ± 8 pg/ml; P < 0.01). In summary, following C. neoformans infection, C3H/HeN mice presented a Th2 pulmonary immune response characterized by airway and interstitial eosinophilia and higher expression of IL-5 and IL-13, while CBA/J mice developed airway and interstitial neutrophilia, greater interstitial macrophage infiltration, and higher expression of Th1- and Th17-associated cytokines.
Fig 4.
Whole-lung leukocyte and lymphocyte recruitment following C. neoformans infection. Whole-lung leukocytes were isolated using CD45+ antibody-conjugated magnetic beads from lung cell suspensions of inbred 7-week-old female C3H/HeN and CBA/J mice at day 14 and day 21 after intratracheal infection with 104 CFU of C. neoformans ATCC 24067. The total leukocyte number was determined by an automated cell counter, and leukocyte subsets were analyzed by Diff-Quik staining of cell suspensions on cytospin slides. CD45+ leukocytes were also stained with fluorochrome-labeled antibodies specific for CD4+ T-lymphocyte, CD8+ T-lymphocyte, and CD19+ B-lymphocyte subsets and analyzed by flow cytometry. The absolute number of cells was determined by multiplying the percentage of each cell type by the total number of leukocytes (n = 4 to 6 mice/strain/time point; *, P < 0.05; **, P < 0.01; ***, P < 0.001).
Fig 5.
Whole-lung cytokine expression pattern following C. neoformans infection. Whole-lung protein was collected at day 14 after intratracheal infection with 104 CFU of C. neoformans ATCC 24067. An individual sandwich ELISA was performed on the supernatant to determine the level of Th1 (CXCL10 and IFN-γ), Th17 (IL-17), and Th2 (IL-5, IL-13) cytokines (n = 3 to 5 mice for each strain; **, P < 0.01; ***, P < 0.001).
Complex genetic regulation of susceptibility to experimental C. neoformans infection.
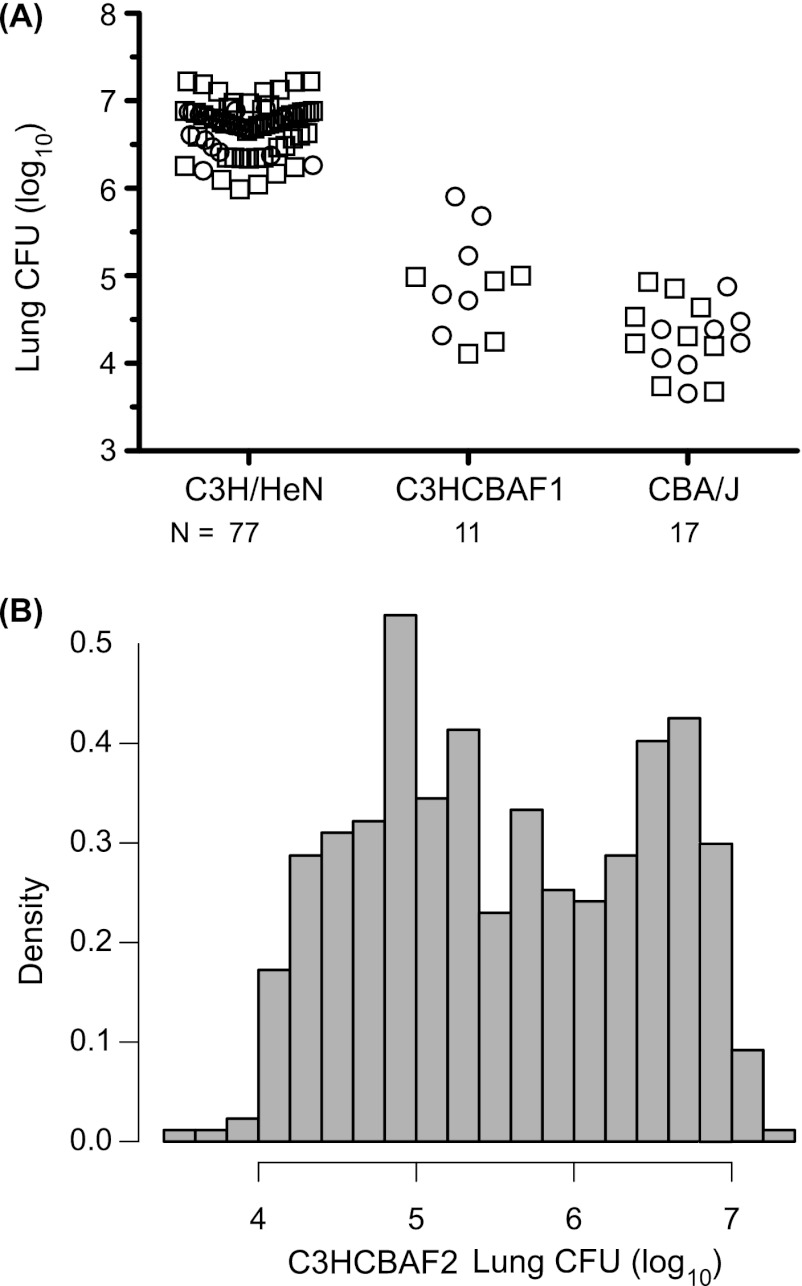
To elucidate the genetic regulation of differential susceptibility to experimental C. neoformans infection between C3H/HeN and CBA/J mice, genome-wide QTL analysis was performed using lung fungal burden as the phenotype. No significant difference in lung fungal burden was observed between male and female C3H/HeN, CBA/J, C3HCBAF1, or C3HCBAF2 mice. The mode of inheritance of susceptibility was first investigated in C3HCBAF1 mice using a standardized intratracheal infection with 104 CFU of C. neoformans ATCC 24067. C3HCBAF1 mice had a lung fungal burden at day 28 postinfection that was intermediate between the burdens of C3H/HeN and CBA/J mice (C3H/HeN versus C3HCBAF1 mice, P < 0.0001; CBA/J versus C3HCBAF1 mice, P < 0.01; Fig. 6A).
Fig 6.
Distribution of fungal burden in C3H/HeN and CBA/J mice and their F1 and F2 progeny following C. neoformans infection. Seven-week-old mice were intratracheally infected with 104 CFU of C. neoformans ATCC 24067. Lungs were harvested at 28 days postinfection, and the numbers of CFU were counted. (A) Dot plot of the numbers of CFU in the lungs of C3H/HeN, C3HCBAF1, and CBA/J mice. Symbols: squares, male mice; circles, female mice. (B) Histogram of the numbers of CFU in the lungs of 435 C3HCBAF2 mice.
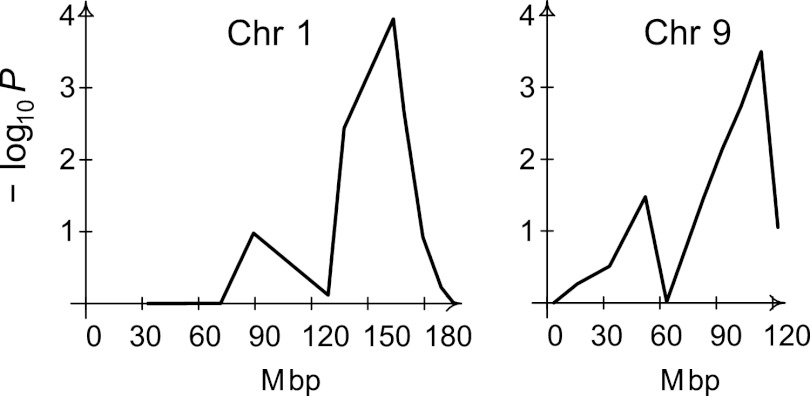
To dissect the genetic regulation of susceptibility to experimental cryptococcal infection, 435 C3HCBAF2 mice were intratracheally infected with 104 CFU of C. neoformans ATCC 24067 and the lung fungal burden was measured at day 28. C3HCBAF2 mice presented a continuous distribution of lung fungal CFU that was consistent with a complex genetic trait (Fig. 6B). The mean fungal burden among 217 female C3HCBAF2 mice was 5.62 ± 0.07 log10 CFU, while the mean fungal burden among 218 male C3HCBAF2 mice was 5.50 ± 0.05 log10 CFU. A high-density mouse genotyping microarray was used to identify 33,610 informative SNPs between C3H/HeN and CBA/J DNA mice; however, 11 large genomic regions (>40 Mb) did not possess any informative markers. A total of 94 SNPs with an average spacing of 24.1 Mb were genotyped in the entire C3HCBAF2 population. QTL analysis was performed using the entire F2 population, as sex was not shown to be a significant covariate of lung fungal burden by linear regression analysis. A non-Mendelian pattern of inheritance of SNP alleles on chromosomes 3 and 19 was observed. Marker regression, a method that does not rely on inference of the Mendelian inheritance of parental alleles in the F2 population, was used for the single-marker analysis. Genome-wide analysis of the 435 C3HCBAF2 mice revealed one significant QTL on chromosome 1 at SNP rs30599866 (163.55 Mb) and one significant QTL on chromosome 9 at SNP rs30136669 (113.82 Mb), based on 10,000 permutations of the data set (Fig. 7). For both of these loci, the additive model gave the highest probability of linkage (P = 0.0001 and P = 0.003 for rs30599866 and rs30136669, respectively; for the recessive model, P = 0.0004 and P = 0.0006, respectively; under the dominant model, all P values were >0.07). One other SNP on chromosome 16 reached the empirical threshold for suggestive QTL at a P value of 0.10. There were no significant pairwise interactions.
Fig 7.
QTL analysis for susceptibility to cryptococcal pneumonia. QTL mapping by single-marker regression was performed on the 435 C3HCBAF2 mice using lung fungal burden as the quantitative trait. Genome-wide −log10 P values based on 10,000 resamples are plotted against the physical position (Mbp) on chromosomes 1 and 9. Plots for the remaining chromosomes are not shown since the −log10 P values were greater than 0.05 for all markers on the chromosome. A P value of <0.01, which corresponds to a −log10 value of 2, was considered to be statistically significant evidence of linkage.
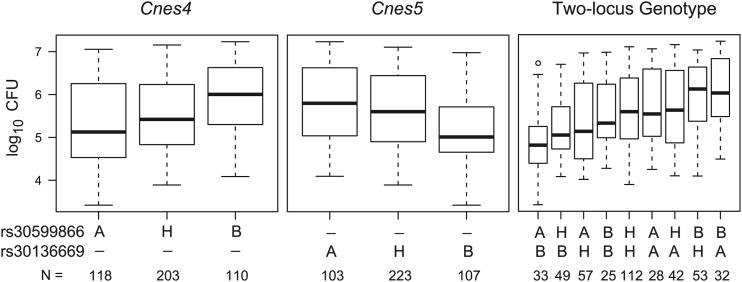
The QTL on chromosome 1, designated C. neoformans susceptibility locus 4 (Cnes4), showed a peak logarithm of the odds (LOD) score of 5.79 (P = 0.0001) with a 2-LOD confidence interval of 128.85 to 179.27 Mb and explained 6.0% of the phenotypic variance. The QTL on chromosome 9, designated Cnes5, showed a peak LOD score of 5.47 (P = 0.0002) with a 2-LOD confidence interval of 83.13 to 122.75 Mb and explained 5.6% of the phenotypic variance. Analysis of the effect of each parental allele at Cnes4 and Cnes5 on lung fungal burden revealed that C3H/HeN mice harbor a resistance allele at Cnes4 and a susceptibility allele at Cnes5 (Fig. 8). The combined effect of the alleles at both loci is also shown for the nine two-locus genotype groups. For an additive model, one would expect F2 mice homozygous for the C3H/HeN allele at Cnes4 and homozygous for the CBA/J allele at Cnes5 to be in the lower tail of the distribution in Fig. 6B, and conversely, one would expect F2 mice homozygous for the CBA/J allele at Cnes4 and for the C3H/HeN allele at Cnes5 to be in the upper tail of the distribution. Thus, the data are consistent with two strong additive loci, although other genetic and environmental factors as well as experimental variation also contribute to the observed variation in lung CFU.
Fig 8.
Effect of Cnes4 and Cnes5 alleles on lung fungal burden following C. neoformans infection. Box plots of the numbers of CFU for the C3HCBAF2 mice are shown for the single-locus genotypes at the linkage peak of rs30599866 (Cnes4) and rs30136669 (Cnes5) and for the two-locus genotypes. The solid bar indicates the median; the bottom and top of each box represent the first and third quartiles, respectively; the whiskers extend 1.5 times the interquartile range; the open circle represents a single value beyond the whiskers. All pairwise comparisons of the single-locus genotypes indicate that mice homozygous for the CBA/J allele (B) are significantly different from mice homozygous for the C3H/HeN allele (A) and from heterozygous mice (H). Among all pairwise comparisons of the two-locus genotype groups, the three most resistant groups (for Cnes4-Cnes5, A-B, H-B, and A-H) are significantly different from the two most susceptible groups (B-H and B-A). In addition, the A-B genotype group is significantly different from the H-H, A-A, and H-A genotype groups (P value range, <0.05 to <0.001).
DISCUSSION
The molecular mechanisms of robust immunity against C. neoformans are poorly characterized. To investigate genetic regulation of the host response against C. neoformans, we have used a well-established infection model that mimics clinical disease to characterize a panel of inbred mouse strains and perform complex trait analysis. The major findings of this report are as follows: (i) C3H/HeN inbred mice are more highly susceptible to intratracheal C. neoformans infection than the closely related CBA/J inbred strain; (ii) C3H/HeN inbred mice mount a Th2 polarized lung immune response to cryptococcal infection, while CBA/J inbred mice generate a Th1 and Th17 response; (iii) two QTLs, Cnes4 on chromosome 1 and Cnes5 on chromosome 9, regulate the development of progressive cryptococcal pneumonia in an additive manner; and (iv) C3H/HeN mice carry a resistance allele at Cnes4 and a susceptibility allele at Cnes5.
Laboratory mice represent a natural source of allelic diversity and are widely recognized as relevant hosts for the study of C. neoformans infection (9). A multitude of infection models exists, and the predominant outcome (pneumonia, meningitis, or disseminated disease) varies according to the fungal serotype and virulence, dose and route of infection, and host genetic background (8). Although no single model can perfectly recapitulate all clinical manifestations, low-dose intratracheal infection with C. neoformans ATCC 24067 closely mimics the course of human disease (pneumonia with a potential for meningitis) and is widely used for studies of pulmonary inflammation and immunity (22). Nonetheless, identification of genes that regulate host susceptibility to cryptococcal pneumonia using this method has been limited by the fact that natural variation in lung fungal burden and inflammatory responsiveness has been analyzed in only a few inbred strains (12, 20, 21).
In the current study, we used lung fungal burden at 28 days postinfection to confirm the relative susceptibility of four previously characterized inbred strains (C57BL/6J, BALB/c, CBA/J, SJL/J) and established that complement C5 deficiency is associated with susceptibility on multiple genetic backgrounds (FVB/N, SWR/J, DBA/2J, A/J). The most notable finding from the strain survey was that inbred C3H/HeN mice are highly susceptible to C. neoformans pneumonia, despite the presence of a functional C5 allele and the absence of other known immune defects. Subsequent analysis showed that C3H/HeN mice had a significantly higher lung fungal burden than CBA/J mice from day 21 to day 56 postinfection. The lung fungal burden of C3H/HeN mice reached a plateau from day 21 to 28 and subsequently decreased, while CBA/J mice showed a decreasing lung fungal burden after day 21. The late timing of these differences suggests that genetic susceptibility factors carried by the C3H/HeN strain create a relatively permissive environment for cryptococcal replication during the adaptive phase of immunity. At day 28, C3H/HeN mice also had significantly higher dissemination to the spleen than CBA/J mice; nevertheless, the lack of deaths or detectable brain dissemination in either strain at day 56 suggests that the differential susceptibility of C3H/HeN mice is most likely confined to the lungs.
Both cell-mediated and humoral immunity have been shown to play a protective role in experimental C. neoformans infection (23, 44). In mice, clearance of C. neoformans requires CD4+ and CD8+ T-lymphocyte subsets (24, 34), while antibody-mediated protective efficacy depends on several factors, including the host genetic background (44). Exudate macrophages that are derived from Ly6c-positive (Ly6c+) monocytes and recruited to the lung via a CCR2-dependent mechanism were also recently shown to mediate cryptococcal clearance (40). Notably, the susceptible C3H/HeN strain had significantly fewer CD8+ T lymphocytes and CD19+ B cells at day 21 postinfection and smaller numbers of macrophages in the lungs at day 14 and day 21 than resistant CBA/J mice. On the basis of these observations, it is possible that diminished recruitment and/or accumulation of one or more of these lymphoid or myeloid subpopulations mediates host susceptibility in C3H/HeN mice. At day 14 after C. neoformans infection, C3H/HeN mice also developed significantly higher airway and lung tissue eosinophilia, while CBA/J mice mounted a neutrophilic response. Pulmonary eosinophilia is a marker of a Th2 response and has been associated with chronic or progressive cryptococcal infection (21), while airway and lung tissue neutrophilia is characteristic of resistant inbred strains such as SJL/J and CBA/J (17). Despite these associations, the role of these inflammatory cell subsets in host defense against C. neoformans is complex. For example, anti-IL-5-mediated reduction of pulmonary eosinophilia in C57BL/6J mice did not significantly influence lung cryptococcal clearance (21). Similarly, neutrophil depletion with a monoclonal anti-granulocyte receptor-1 (Ly6g/c) Ab prior to cryptococcal infection increased survival of BALB/c mice but did not alter the lung fungal burden at day 7 postinfection; however, it is likely that a nonspecific effect on Ly6c+ myeloid cell subsets may have altered host inflammation without affecting pathogen replication (13, 33). We speculate that neutrophils are a crucial mediator of enhanced host defense in CBA/J mice, although confirmation of this hypothesis will require additional studies.
The lung cytokine milieu and, specifically, the balance between Th1, Th17, and Th2 cytokines regulate the outcome of C. neoformans infection (1). Using various strategies, Th1 (IFN-γ, tumor necrosis factor alpha, IL-12, IL-18) and Th17 (IL-17a, IL-23) cytokines have been shown to be protective against C. neoformans, and Th2 (IL-4, IL-10, IL-13) mediators have been associated with chronic or progressive pulmonary infection (18, 19, 21, 26, 27, 35). In the present study, CBA/J mice intratracheally infected with C. neoformans expressed significantly higher levels of the Th1-associated cytokines CXCL10 and IFN-γ as well as the Th17-associated cytokine IL-17 in association with a significantly lower fungal burden and heightened neutrophilia in the lung. In contrast, C3H/HeN mice presented a Th2 lung immune response characterized by significantly higher expression of IL-5 and IL-13 that correlated with eosinophilia, goblet cell metaplasia, and a higher fungal burden (12, 21, 35). The heightened expression of Th2-associated cytokines has been shown to negatively regulate the Th17 pulmonary immune response in a Th1-independent manner and may explain the significantly reduced IL-17 expression observed in C3H/HeN mice following infection (35, 48).
The C3H and CBA mouse lines were created during the 1920s after a differential incidence of spontaneous tumor formation was observed among the offspring of a single progenitor pair (42, 54). Through phenotypic selection and serial brother-sister mating, two independent lines with a high (C3H) and a low (CBA) incidence of tumor formation were created and subsequently disseminated to other investigators (54). Both lines encode the H-2k haplotype at the major histocompatibility complex, and over 20 inbred substrains with unique phenotypic characteristics have been derived from the original stocks through segregation of residual heterozygosity. Notably, the C3H/HeJ substrain was relatively resistant to high-dose intravenous infection with C. neoformans (43), despite the fact that it carries a spontaneous loss-of-function point mutation in Toll-like receptor 4 that has been shown to recognize bacterial lipopolysaccharide and cryptococcal glucuronoxylomannan (47).
The C3H/HeN and CBA/J substrains arising from the original lines are divergent for several infectious disease-related traits, including pneumococcal pneumonia, cutaneous anthrax, and vaginal candidiasis, but due to their recent common origin, the genetic factors underlying these phenotypic variations have been difficult to map due to a paucity of informative markers (7, 37, 38, 42, 49, 51, 54). In the current study, a high-density mouse genotyping microarray that assayed 623,124 SNPs in the C3H/HeN and CBA/J inbred strains was used to identify 94 markers for genome-wide QTL analysis of C. neoformans susceptibility (56). Despite the comprehensive nature and depth of the array, several large genomic intervals were devoid of informative markers and most likely signify areas that are identical by descent between C3H/HeN and CBA/J inbred strains. Accordingly, QTL analysis was conducted using marker regression, a method that is accurate in the presence of noninformative genomic regions or in cases where deviation from Mendelian inheritance of parental alleles is encountered. Two major QTLs that regulate host resistance to C. neoformans, Cnes4 on chromosome 1 and Cnes5 on chromosome 9, were identified. Interestingly, Cnes4 (chromosome 1, 128.85 to 179.27 Mb) colocalizes with the peak position of a susceptibility locus for the fungus Histoplasma capsulatum (32) and the parasite Leishmania major (2). Furthermore, Cnes5 (chromosome 9, 83.13 to 122.75 Mb) encompasses loci associated with susceptibility to the intracellular eukaryotic pathogens Plasmodium chabaudi (15) and Plasmodium yoelii (39). It is tempting to speculate that the same genes on chromosome 1 and chromosome 9 regulate host resistance to diverse eukaryotic microbial pathogens; however, an equally plausible explanation is that more than one QTL is present within either or both of these large linkage intervals (31). The precise nature of the genetic variation that underlies Cnes4 and Cnes5 remains to be established in future studies.
The full complexity of a genetically regulated trait may be elucidated through a multiple-cross-mapping approach that takes advantage of a spectrum of polymorphisms among inbred strains (6, 29). In a previous report, three QTLs (Cnes1 to Cnes3) that regulate host resistance against C. neoformans pneumonia were identified among F2 mice of a single cross derived from susceptible C57BL/6J and resistant CBA/J strains (10). In the current study, QTL analysis of resistance against the same strain of C. neoformans among F2 mice derived from C3H/HeN and CBA/J strains revealed two distinct loci that have been named Cnes4 and Cnes5. The clear distinction between the QTLs identified in the previous and current studies suggests that the genetic regulation of host resistance to C. neoformans differs between C57BL/6J and C3H/HeN strains, despite their similar Th2 inflammatory response and high lung fungal burden. Analysis of allelic inheritance at the peak of each QTL interval revealed that the susceptible C3H/HeN strain contributed an allele associated with low fungal burden at Cnes4 and a high fungal burden at Cnes5. A comparison of all allelic combinations at Cnes4 and Cnes5 showed that these two loci have an additive effect such that F2 mice that inherit a C3H/HeN allele at Cnes4 and a CBA/J allele at Cnes5 are most resistant to cryptococcal pneumonia, while those with the opposite inheritance pattern are the most susceptible. The presence of resistance alleles within a susceptible inbred strain has been previously observed in experimental models of infection, such as a model of Salmonella enterica serovar Typhimurium infection (46), and further exemplifies how forward genetic approaches are able to delineate unforeseen levels of genetic complexity (45).
In conclusion, despite the clear heritable variation in host resistance among inbred mouse strains, the pathogenesis of C. neoformans disease and the genetic control of immune responsiveness against this fungal pathogen are poorly understood. The newly described Cnes4 and Cnes5 QTLs identified in the C3H/HeN inbred strain extend our knowledge of the underlying genetic architecture that regulates host susceptibility to progressive cryptococcal pneumonia. Validation and fine mapping of Cnes4 and Cnes5 will facilitate the identification of the candidate genes that these QTLs encode, and precise characterization of the underlying allelic variation will reveal molecular mechanisms of host defense and provide a better understanding of immune regulation.
ACKNOWLEDGMENTS
This work was supported by a grant from the Fonds de la Recherche en Santé du Québec (FRSQ) to the Research Institute of the McGill University Health Centre, the Canadian Institutes of Health Research (to S.T.Q.), a Canada Research Chair (to S.T.Q.), the Career Awards in the Biomedical Sciences program of the Burroughs Wellcome Fund (to S.T.Q.), the Quebec Respiratory Health Training Program (to S.T.Q. and E.I.L.), the Mathematics of Information Technology and Complex Systems (MITACS) through the Canadian Network of Centres of Excellence Program (to J.C.L.-O. and K.M.), and the National Sciences and Engineering Research Council of Canada (to J.C.L.-O.).
We thank Genevieve Houde and Isabelle Angers for technical assistance. We also acknowledge Markus Schnare for critically reviewing the manuscript, Laurel Holmes for assistance with figure preparation, and Qutayba Hamid for providing us with microscope facilities.
Footnotes
Published ahead of print 17 September 2012
REFERENCES
- 1. Arora S, Huffnagle GB. 2005. Immune regulation during allergic bronchopulmonary mycosis: lessons taught by two fungi. Immunol. Res. 33:53–68 [DOI] [PubMed] [Google Scholar]
- 2. Badalova J, et al. 2002. Separation and mapping of multiple genes that control IgE level in Leishmania major infected mice. Genes Immun. 3:187–195 [DOI] [PubMed] [Google Scholar]
- 3. Baddley JW, et al. 2008. Pulmonary cryptococcosis in patients without HIV infection: factors associated with disseminated disease. Eur. J. Clin. Microbiol. Infect. Dis. 27:937–943 [DOI] [PubMed] [Google Scholar]
- 4. Beck JA, et al. 2000. Genealogies of mouse inbred strains. Nat. Genet. 24:23–25 [DOI] [PubMed] [Google Scholar]
- 5. Broman KW, Wu H, Sen S, Churchill GA. 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19:889–890 [DOI] [PubMed] [Google Scholar]
- 6. Burgess-Herbert SL, Cox A, Tsaih SW, Paigen B. 2008. Practical applications of the bioinformatics toolbox for narrowing quantitative trait loci. Genetics 180:2227–2235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Calderon L, Williams R, Martinez M, Clemons KV, Stevens DA. 2003. Genetic susceptibility to vaginal candidiasis. Med. Mycol. 41:143–147 [DOI] [PubMed] [Google Scholar]
- 8. Capilla J, Clemons KV, Stevens DA. 2007. Animal models: an important tool in mycology. Med. Mycol. 45:657–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Carroll SF, Guillot L, Qureshi ST. 2007. Mammalian model hosts of cryptococcal infection. Comp. Med. 57:9–17 [PubMed] [Google Scholar]
- 10. Carroll SF, Loredo Osti JC, Guillot L, Morgan K, Qureshi ST. 2008. Sex differences in the genetic architecture of susceptibility to Cryptococcus neoformans pulmonary infection. Genes Immun. 9:536–545 [DOI] [PubMed] [Google Scholar]
- 11. Chayakulkeeree M, Perfect JR. 2006. Cryptococcosis. Infect. Dis. Clin. North Am. 20:507–544, v–vi [DOI] [PubMed] [Google Scholar]
- 12. Chen GH, et al. 2008. Inheritance of immune polarization patterns is linked to resistance versus susceptibility to Cryptococcus neoformans in a mouse model. Infect. Immun. 76:2379–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Daley JM, Thomay AA, Connolly MD, Reichner JS, Albina JE. 2008. Use of Ly6G-specific monoclonal antibody to deplete neutrophils in mice. J. Leukoc. Biol. 83:64–70 [DOI] [PubMed] [Google Scholar]
- 14. Deshaw M, Pirofski LA. 1995. Antibodies to the Cryptococcus neoformans capsular glucuronoxylomannan are ubiquitous in serum from HIV+ and HIV− individuals. Clin. Exp. Immunol. 99:425–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Foote SJ, et al. 1997. Mouse loci for malaria-induced mortality and the control of parasitaemia. Nat. Genet. 17:380–381 [DOI] [PubMed] [Google Scholar]
- 16. Garcia-Hermoso D, Janbon G, Dromer F. 1999. Epidemiological evidence for dormant Cryptococcus neoformans infection. J. Clin. Microbiol. 37:3204–3209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guillot L, Carroll SF, Homer R, Qureshi ST. 2008. Enhanced innate immune responsiveness to pulmonary Cryptococcus neoformans infection is associated with resistance to progressive infection. Infect. Immun. 76:4745–4756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hernandez Y, et al. 2005. Distinct roles for IL-4 and IL-10 in regulating T2 immunity during allergic bronchopulmonary mycosis. J. Immunol. 174:1027–1036 [DOI] [PubMed] [Google Scholar]
- 19. Herring AC, Lee J, McDonald RA, Toews GB, Huffnagle GB. 2002. Induction of interleukin-12 and gamma interferon requires tumor necrosis factor alpha for protective T1-cell-mediated immunity to pulmonary Cryptococcus neoformans infection. Infect. Immun. 70:2959–2964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hoag KA, Street NE, Huffnagle GB, Lipscomb MF. 1995. Early cytokine production in pulmonary Cryptococcus neoformans infections distinguishes susceptible and resistant mice. Am. J. Respir. Cell Mol. Biol. 13:487–495 [DOI] [PubMed] [Google Scholar]
- 21. Huffnagle GB, Boyd MB, Street NE, Lipscomb MF. 1998. IL-5 is required for eosinophil recruitment, crystal deposition, and mononuclear cell recruitment during a pulmonary Cryptococcus neoformans infection in genetically susceptible mice (C57BL/6). J. Immunol. 160:2393–2400 [PubMed] [Google Scholar]
- 22. Huffnagle GB, Lipscomb MF. 1992. Pulmonary cryptococcosis. Am. J. Pathol. 141:1517–1520 [PMC free article] [PubMed] [Google Scholar]
- 23. Huffnagle GB, Lipscomb MF, Lovchik JA, Hoag KA, Street NE. 1994. The role of CD4+ and CD8+ T cells in the protective inflammatory response to a pulmonary cryptococcal infection. J. Leukoc. Biol. 55:35–42 [DOI] [PubMed] [Google Scholar]
- 24. Huffnagle GB, Yates JL, Lipscomb MF. 1991. Immunity to a pulmonary Cryptococcus neoformans infection requires both CD4+ and CD8+ T cells. J. Exp. Med. 173:793–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Keane TM, et al. 2011. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477:289–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kleinschek MA, et al. 2006. IL-23 enhances the inflammatory cell response in Cryptococcus neoformans infection and induces a cytokine pattern distinct from IL-12. J. Immunol. 176:1098–1106 [DOI] [PubMed] [Google Scholar]
- 27. Kleinschek MA, et al. 2010. Administration of IL-23 engages innate and adaptive immune mechanisms during fungal infection. Int. Immunol. 22:81–90 [DOI] [PubMed] [Google Scholar]
- 28. Kronstad JW, et al. 2011. Expanding fungal pathogenesis: Cryptococcus breaks out of the opportunistic box. Nat. Rev. Microbiol. 9:193–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li R, Lyons MA, Wittenburg H, Paigen B, Churchill GA. 2005. Combining data from multiple inbred line crosses improves the power and resolution of quantitative trait loci mapping. Genetics 169:1699–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lui G, et al. 2006. Cryptococcosis in apparently immunocompetent patients. QJM 99:143–151 [DOI] [PubMed] [Google Scholar]
- 31. Mackay TF, Stone EA, Ayroles JF. 2009. The genetics of quantitative traits: challenges and prospects. Nat. Rev. Genet. 10:565–577 [DOI] [PubMed] [Google Scholar]
- 32. Mayfield JA, Rine J. 2007. The genetic basis of variation in susceptibility to infection with Histoplasma capsulatum in the mouse. Genes Immun. 8:468–474 [DOI] [PubMed] [Google Scholar]
- 33. Mednick AJ, Feldmesser M, Rivera J, Casadevall A. 2003. Neutropenia alters lung cytokine production in mice and reduces their susceptibility to pulmonary cryptococcosis. Eur. J. Immunol. 33:1744–1753 [DOI] [PubMed] [Google Scholar]
- 34. Mody CH, Lipscomb MF, Street NE, Toews GB. 1990. Depletion of CD4+ (L3T4+) lymphocytes in vivo impairs murine host defense to Cryptococcus neoformans. J. Immunol. 144:1472–1477 [PubMed] [Google Scholar]
- 35. Muller U, et al. 2007. IL-13 induces disease-promoting type 2 cytokines, alternatively activated macrophages and allergic inflammation during pulmonary infection of mice with Cryptococcus neoformans. J. Immunol. 179:5367–5377 [DOI] [PubMed] [Google Scholar]
- 36. Nadrous HF, Antonios VS, Terrell CL, Ryu JH. 2003. Pulmonary cryptococcosis in nonimmunocompromised patients. Chest 124:2143–2147 [DOI] [PubMed] [Google Scholar]
- 37. Oharaseki T, et al. 2005. Susceptibility loci to coronary arteritis in animal model of Kawasaki disease induced with Candida albicans-derived substances. Microbiol. Immunol. 49:181–189 [DOI] [PubMed] [Google Scholar]
- 38. Ohno N. 2004. Murine model of Kawasaki disease induced by mannoprotein–beta-glucan complex, CAWS, obtained from Candida albicans. Jpn. J. Infect. Dis. 57:S9–S10 [PubMed] [Google Scholar]
- 39. Ohno T, et al. 2001. Chromosomal mapping of the host resistance locus to rodent malaria (Plasmodium yoelii) infection in mice. Immunogenetics 53:736–740 [DOI] [PubMed] [Google Scholar]
- 40. Osterholzer JJ, et al. 2011. Chemokine receptor 2-mediated accumulation of fungicidal exudate macrophages in mice that clear cryptococcal lung infection. Am. J. Pathol. 178:198–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Paigen K, Eppig JT. 2000. A mouse phenome project. Mamm. Genome 11:715–717 [DOI] [PubMed] [Google Scholar]
- 42. Petkov PM, et al. 2004. An efficient SNP system for mouse genome scanning and elucidating strain relationships. Genome Res. 14:1806–1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rhodes JC, Wicker LS, Urba WJ. 1980. Genetic control of susceptibility to Cryptococcus neoformans in mice. Infect. Immun. 29:494–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rivera J, Zaragoza O, Casadevall A. 2005. Antibody-mediated protection against Cryptococcus neoformans pulmonary infection is dependent on B cells. Infect. Immun. 73:1141–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Roy MF, Riendeau N, Loredo-Osti JC, Malo D. 2006. Complexity in the host response to Salmonella Typhimurium infection in AcB and BcA recombinant congenic strains. Genes Immun. 7:655–666 [DOI] [PubMed] [Google Scholar]
- 46. Sebastiani G, et al. 1998. Mapping of genetic modulators of natural resistance to infection with Salmonella typhimurium in wild-derived mice. Genomics 47:180–186 [DOI] [PubMed] [Google Scholar]
- 47. Shoham S, Huang C, Chen JM, Golenbock DT, Levitz SM. 2001. Toll-like receptor 4 mediates intracellular signaling without TNF-alpha release in response to Cryptococcus neoformans polysaccharide capsule. J. Immunol. 166:4620–4626 [DOI] [PubMed] [Google Scholar]
- 48. Song C, et al. 2008. IL-17-producing alveolar macrophages mediate allergic lung inflammation related to asthma. J. Immunol. 181:6117–6124 [DOI] [PubMed] [Google Scholar]
- 49. Takashima K, et al. 1996. Establishment of a model of penicillin-resistant Streptococcus pneumoniae pneumonia in healthy CBA/J. mice. J. Med. Microbiol. 45:319–322 [DOI] [PubMed] [Google Scholar]
- 50. Voelz K, May RC. 2010. Cryptococcal interactions with the host immune system. Eukaryot. Cell 9:835–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Welkos SL, Keener TJ, Gibbs PH. 1986. Differences in susceptibility of inbred mice to Bacillus anthracis. Infect. Immun. 51:795–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wetsel RA, Fleischer DT, Haviland DL. 1990. Deficiency of the murine fifth complement component (C5). A 2-base pair gene deletion in a 5′-exon. J. Biol. Chem. 265:2435–2440 [PubMed] [Google Scholar]
- 53. Wheat WH, Wetsel R, Falus A, Tack BF, Strunk RC. 1987. The fifth component of complement (C5) in the mouse. Analysis of the molecular basis for deficiency. J. Exp. Med. 165:1442–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Whitmore AC, Whitmore SP. 1985. Subline divergence within L.C. Strong's C3H and CBA inbred mouse strains. A review. Immunogenetics 21:407–428 [DOI] [PubMed] [Google Scholar]
- 55. Wozniak KL, Hardison S, Olszewski M, Wormley FL., Jr 2012. Induction of protective immunity against cryptococcosis. Mycopathologia 173:387–394 [DOI] [PubMed] [Google Scholar]
- 56. Yang H, et al. 2009. A customized and versatile high-density genotyping array for the mouse. Nat. Methods 6:663–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zaragoza O, Alvarez M, Telzak A, Rivera J, Casadevall A. 2007. The relative susceptibility of mouse strains to pulmonary Cryptococcus neoformans infection is associated with pleiotropic differences in the immune response. Infect. Immun. 75:2729–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]