Abstract
Arginine deiminase (ADI), carbamate kinase (CK), and ornithine transcarbamoylase (OTC) constitute the ADI system. In addition to metabolic functions, the ADI system has been implicated in the virulence of certain pathogens. The pathogenic intracellular bacterium Salmonella enterica serovar Typhimurium possesses the STM4467, STM4466, and STM4465 genes, which are predicted to encode ADI, CK, and OTC, respectively. Here we report that the STM4467 gene encodes an ADI and that ADI activity plays a role in the successful infection of a mammalian host by S. Typhimurium. An STM4467 deletion mutant was defective for replication inside murine macrophages and was attenuated for virulence in mice. We determined that a regulatory protein encoded by the STM4463 gene functions as an activator for STM4467 expression. The expression of the ADI pathway genes was enhanced inside macrophages in a process that required STM4463. Lack of STM4463 impaired the ability of S. Typhimurium to replicate within macrophages. A mutant defective in STM4467-encoded ADI displayed normal production of nitric oxide by macrophages.
INTRODUCTION
Salmonella enterica serovar Typhimurium is a facultatively intracellular bacterium that can cause a diverse spectrum of diseases. In the course of systemic infection of a mammalian host, S. Typhimurium can survive within the macrophage phagosome (13). For this purpose, S. Typhimurium expresses gene products to avoid killing by microbicides that are produced inside the phagosome (25, 39). Previous studies have also revealed that purine, pyrimidine, and amino acid auxotrophs of S. Typhimurium are unable to survive inside macrophages (18, 27). In addition, S. Typhimurium appears to activate an alternative metabolic pathway for the utilization of carbon sources during growth inside macrophages (15). These findings demonstrate that bacterial metabolism is a crucial determinant for the successful pathogenesis of S. Typhimurium (7, 32).
In general, three enzymes—arginine deiminase (ADI), ornithine transcarbamoylase (OTC), and carbamate kinase (CK)—constitute the ADI system, which catalyzes the conversion of l-arginine into ornithine, ammonia, and carbon dioxide with the formation of ATP (44). This enzymatic pathway is widely distributed in various bacterial species and is known to provide cellular energy, particularly under oxygen-limited conditions (4, 9, 20, 44). In addition to its metabolic function, the ADI system is also employed to protect some bacteria from stressful conditions. In oral streptococci and Streptococcus pyogenes, the ADI system helps the bacteria resist acidity by generating ammonia and thus neutralizing an acidic pH (4, 11). Moreover, the ADI system sometimes, though rarely, plays a role in bacterial pathogenesis. ADI is necessary for S. pyogenes to invade and survive inside epithelial cells (11). Additionally, a lack of ADI has been found to impair the survival of Listeria monocytogenes in the spleen during infection in a mouse model (41).
The STM4467, STM4466, and STM4465 genes of S. Typhimurium are predicted to encode ADI, CK, and OTC, respectively. The ADI pathway of S. Typhimurium appears to be functional, because the expression of the putative ADI cluster increased OTC activity (43). In the present study, we investigated the role of the ADI system in S. Typhimurium pathogenesis. We found that an S. Typhimurium strain lacking the STM4467-encoded ADI was defective in its ability to replicate inside macrophages and was attenuated for virulence in mice. We also revealed that the STM4463-encoded regulator contributes to S. Typhimurium virulence, at least in part, by upregulating the expression of the ADI gene cluster within macrophages.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. The S. Typhimurium strains were derived from strain 14028s. Phage P22-mediated transductions were performed as described previously (28). The bacteria were grown at 37°C in Luria-Bertani (LB) medium. Ampicillin, chloramphenicol, kanamycin, and isopropyl-β-d-thiogalactopyranoside (IPTG) were used at 50 μg/ml, 25 μg/ml, 50 μg/ml and 0.5 mM, respectively.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| S. enterica serovar Typhimurium strains | ||
| 14028s | Wild type | 18 |
| CH102 | ΔSTM4467 | This study |
| CH110 | PSTM4467::lacZY (Kmr) | This study |
| CH111 | PSTM4467::lacZY (Kmr) ΔSTM4463::Cmr | This study |
| CH201 | ΔSTM4463::Cmr | This study |
| Plasmids | ||
| pUHE21-2lacIq | reppMBI Apr lacIq | 42 |
| pACYC184 | repp15A Cmr Tetr | 5 |
| pKD13 | repR6Kγ Apr-FRT Kmr-FRT | 10 |
| pKD3 | repR6Kγ Apr-FRT Cmr-FRT | 10 |
| pKD46 | reppSC101(Ts) Apr ParaBAD γ β exo | 10 |
| pCP20 | reppSC101(Ts) Apr Cmr cI857 λ PRflp | 10 |
| pCE70 | repR6Kγ Apr-FRT lacZY+ | 35 |
| p4467 | pACYC184-STM4467 | This study |
| p4463 | pUHE21-2lacIq-STM4463 | This study |
| pPM4463 | pACYC184-STM4463 | This study |
Construction of strains.
S. Typhimurium strain CH102, in which the STM4467 gene is deleted, was constructed using the one-step gene inactivation method (10). The Kmr cassette from plasmid pKD13 (10) was amplified using primers STM67-lamb-F (5′-ACTCCTTCTTTATTCTTGTAATTATGTAAAAGGTATAATGTGTAGGCTGGAGCTGCTTCG-3′) and STM67-lamb-R (5′-CGCGACGACCAGTGTGCGTTTGTTTTCCATAACGTCTCCTATTCCGGGGATCCGTCGACC-3′). The resulting PCR product was integrated into the STM4467 region in strain 14028s, and the Kmr cassette was subsequently removed using plasmid pCP20 (10). In strain CH201, the STM4463 gene is deleted. For the construction of this strain, the Cmr cassette of pKD3 (10) was amplified using primers STM63-lamb-F (5′-CGTTGATATCAATAATAAAGATAAGGTGCATTTATGAAGGTGTAGGCTGGAGCTGCTTCG-3′) and STM63-lamb-R (5′-ATTAATGCATGATTTACTCATCGCAAACGGTTCTTATGAAATATGAATATCCTCCTTAGTTC-3′) and was integrated into the STM4463 region in strain 14028s. Deletion of the corresponding genes was verified by colony PCR. Strain CH110, which carries a transcriptional STM4467-lacZ fusion, was constructed as described previously (14). The lacZY genes were introduced into the FLP recombination target (FRT) site in strain CH102 by using plasmid pCE70 (35).
Construction of plasmids.
Plasmid p4467 expresses the STM4467 gene from its own promoter. For the construction of this plasmid, the STM4467 gene was amplified using PCR with primers STM4467-pACYC-F (5′-TTGTTTTTTGAAGCTTTCTGACCC-3′) and STM4467-pACYC-R (5′-ACGACCAGCATGCGTTTGTTTT-3′) and with chromosomal DNA from strain 14028s as a template. The product was introduced between the HindIII and SphI restriction sites of pACYC184 (5). To construct plasmid p4463, in which the STM4463 gene is expressed from the lac promoter, the STM4463 gene was amplified using primers STM4463-pUHE-F (5′-AAATGTGATGAATTCCGCCAGTCC-3′) and STM4463-pUHE-R (5′-TGAACCATGGATCCTCCCGGC-3′). The PCR product was introduced between the EcoRI and BamHI restriction sites of pUHE21-2lacIq (42). Plasmid pPM4463, which expresses the STM4463 gene from its own promoter, was also constructed. The STM4463 gene was amplified using primers STM4463-pACYC-F (5′-GAAAGTCTGAATTCCGGCCTCTC-3′) and STM4463-pACYC-R (5′-TTTACTCATCGCATGCGGTTCTTATG-3′). The PCR product was introduced between the HindIII and SphI restriction sites of pACYC184 (5). The sequences of the STM4467 and STM4463 coding regions on the recombinant plasmids were verified by nucleotide sequencing.
Determination of ADI activity.
The ADI activities of cell extracts were measured using a chemical colorimetric method based on the production of l-citrulline from l-arginine (3). A total of 50 ml of a bacterial culture grown in LB medium was harvested, and the cell pellet was suspended in 3 ml of lysis buffer (10 mM Tris [pH 8.0] containing 0.3 M NaCl) and was disrupted by sonication. After removal of the cellular debris by centrifugation, 0.4 ml of 10 mM l-arginine in 100 mM potassium phosphate buffer (pH 7.2) was added to 1 ml of cell extract. After a 60-min incubation at 37°C, 250 μl of a 1:3 (vol/vol) mixture of 95% sulfuric acid and 85% phosphoric acid and 250 μl of a 3% diacetyl monooxime solution were added, and the mixtures were boiled for 15 min. The development of an orange color was monitored at 490 nm.
β-Galactosidase assay.
β-Galactosidase assays were performed in duplicate, and the activity was determined as described previously (36).
Gentamicin protection assay.
The gentamicin protection assay was conducted as described previously (7). J774A.1 macrophage cells were grown in Dulbecco modified Eagle medium (DMEM) (Invitrogen, Grand Island, NY) supplemented with 10% fetal bovine serum (FBS), penicillin (50 U/ml), and streptomycin (50 U/ml). Prior to bacterial infection, a monolayer of 1 × 105 J774A.1 cells was prepared in a 24-well tissue culture plate and was incubated in DMEM–10% FBS without antibiotics at 37°C for 1 h under 5% CO2. A bacterial culture grown to stationary phase with aeration was applied to the cell monolayer at a multiplicity of infection (MOI) of 10. After 1 h of incubation, the wells were washed three times with prewarmed phosphate-buffered saline (PBS) to remove extracellular bacteria and were then incubated for 1 h with the prewarmed medium supplemented with 100 μg/ml of gentamicin to kill extracellular bacteria. Afterward, the wells were washed three times with PBS, lysed in 1% Triton X-100 for 30 min, and then diluted in PBS. A dilution of the suspension was plated on LB agar medium to enumerate the CFU.
Mouse virulence assay.
Bacterial cells grown overnight in LB medium were pelleted, washed, and resuspended in PBS. Eight-week-old C3H/HeN female mice were used to assess the virulence of S. Typhimurium strains. Approximately 104 bacterial cells in 200 μl of PBS were injected intraperitoneally into groups of mice (5 mice/group), and the survival of the mice was recorded over 3 weeks. To analyze bacterial colonization in organs, the mice were sacrificed at 5 days after infection, and the spleens and livers were removed aseptically. The organs were homogenized in 1 ml of ice-cold PBS and were serially diluted. Bacterial loads were determined by plating the diluents on LB agar plates.
Determination of nitrite concentration.
J774A.1 macrophages were infected with bacteria as described above in triplicate. The supernatants were harvested at 18 h after infection. The nitrite concentration was measured using the Griess assay (21). Briefly, 50 μl of culture supernatants was mixed with an equal volume of Griess reagent (Promega). The absorbance was measured after 10 min at 550 nm in an enzyme-linked immunosorbent assay (ELISA) microreader (Sunrise Basic; Tecan). NaNO2 was used to establish the standard nitrite concentration in the supernatants.
RNA isolation and qRT-PCR analysis.
For the extraction of RNA from S. Typhimurium growing inside J774A.1 macrophages, an infection experiment was conducted as described above except for the increased volume of macrophage cultures (50 ml in a 75-cm2 T-flask). At 1 h, 6 h, and 18 h after infection, the macrophage monolayers were washed, lysed in 1% Triton X-100, and centrifuged at 1,000 rpm for 5 min to pellet the lysed macrophages. From the supernatant that contained the intracellular bacteria, RNA was extracted using the RNeasy Mini Kit (Qiagen). The RNA samples were then treated with RNase-free DNase (Ambion), and cDNA was synthesized using Omniscript reverse transcription reagents (Qiagen) and random hexamers (Invitrogen). The cDNA was quantified by using the 2× iQ SYBR green Supermix (Bio-Rad), and real-time amplification of the PCR products was performed using the iCycler iQ real-time detection system (Bio-Rad). The calculated threshold cycle (CT) corresponding to a target gene was normalized to the CT of the control gene, rpoD. The sigma factor gene rpoD was chosen as a control because no significant variation in rpoD expression was observed inside macrophages (17). The sequences of the primers used in the quantitative reverse transcription-PCR (qRT-PCR) analysis are listed in Table 2.
Table 2.
Primers used in qRT-PCR analysis
| Primer | Target gene | Sequence (5′ to 3′) |
|---|---|---|
| STM4467-RT-F | STM4467 | CTGGCTACTGGATACGCAAA |
| STM4467-RT-R | STM4467 | GACGCCGTTATATATCCAGC |
| STM4466-RT-F | STM4466 | AACCGCTGGAGGCTGATATT |
| STM4466-RT-R | STM4466 | ATGATTCTTCAGCGCCTGTT |
| STM4465-RT-F | STM4465 | GGATGCGAAAAGCAAACACT |
| STM4465-RT-R | STM4465 | GGACGCGAGCAGTATCTTTC |
| STM4463-RT-F | STM4463 | TTGTCAGCGCCTGATTAGTG |
| STM4463-RT-R | STM4463 | ACCATTTCGGCTATTGAACG |
| ssaG-RT-F | ssaG | AGTGGATATGCTCTCCCACA |
| ssaG-RT-R | ssaG | AGGCAAATTGCGCTTTAATC |
| rpoD-RT-F | rpoD | GATGAAGATGCGGAAGAAGC |
| rpoD-RT-R | rpoD | GGTAATGGCTTCCGGGTATT |
Statistical analysis.
Statistical analyses were conducted using the GraphPad Prism program (version 5.0). Survival curves from animal experiments were analyzed by the log rank test, and all other results were analyzed by the unpaired t test. Data are presented as means ± standard deviations. A P value of <0.05 was considered statistically significant.
Ethics statement.
This study was carried out according to the recommended protocol for the care and use of laboratory animals from the Institute of Laboratory Animal Resources at Seoul National University, based on the Korean Animal Protection Law and Korea Food and Drug Administration regulations on laboratory animals. The protocol was approved by the Committee on the Ethics of Animal Experiments of Seoul National University (Institutional Animal Care and Use Committee permit number SNU-120616-1).
RESULTS AND DISCUSSION
The STM4467 gene encodes ADI in S. Typhimurium.
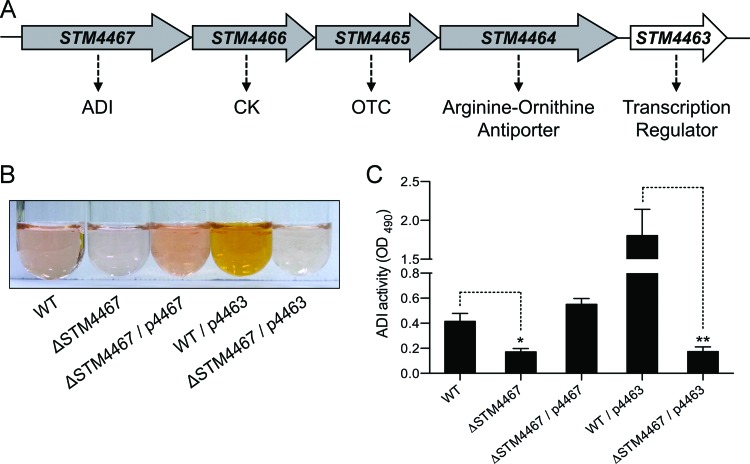
In the S. Typhimurium genome, the STM4467, STM4466, and STM4465 genes are clustered into an operon-like structure (Fig. 1A) and are predicted to encode enzymes of the ADI system: ADI, CK, and OTC, respectively (Fig. 1A). It has been reported that when Salmonella is grown in LB medium containing 0.4 M NaCl without agitation, the expression of this gene cluster is induced to promote OTC activity (43). Although this finding suggests that the ADI pathway might be functional in S. Typhimurium, the genes responsible for the enzymatic activities of the ADI pathway have remained unknown. Therefore, we compared the ADI activity of wild-type Salmonella with that of its isogenic STM4467 deletion mutant. When the ADI activity assay was conducted using a cell extract prepared from the wild-type strain, an orange color developed (Fig. 1B), which indicated the ADI-catalyzed production of citrulline from arginine (3, 41). However, in the STM4467 deletion mutant, the enzyme activity was poorly detected (Fig. 1B) and was only ∼40% of that present in the wild-type strain (Fig. 1C). Expression of the STM4467 gene from a plasmid enabled the STM4467 deletion mutant to produce citrulline at levels even higher than those of the wild-type strain (Fig. 1B and C). Thus, these results indicate that the STM4467 gene either encodes ADI or is required for full ADI activity.
Fig 1.
The STM4467 gene is required for ADI activity. (A) Schematic representation of the ADI pathway gene cluster in S. Typhimurium. (B) The ADI activities of the wild-type (WT) strain (14028s), the STM4467 deletion mutant (CH102), strain CH102 carrying plasmid p4467 (ΔSTM4467/p4467), the WT strain carrying plasmid p4463, and strain CH102 carrying plasmid p4463 (ΔSTM4467/p4463) were determined by using cell extracts grown in LB medium. The development of an orange color indicates the ADI-catalyzed production of l-citrulline from l-arginine. (C) Quantification of the ADI activity displayed by the S. Typhimurium strains described for panel B. Means and standard deviations from three independent experiments are shown (**, P < 0.01). OD490, optical density at 490 nm.
The STM4467 gene contributes to Salmonella virulence.
ADI is necessary for S. pyogenes to invade and survive inside epithelial cells (11). Wild-type L. monocytogenes survived longer in the spleen during a mouse infection than did a mutant lacking ADI (41). These findings suggested a role for ADI in bacterial pathogenesis.
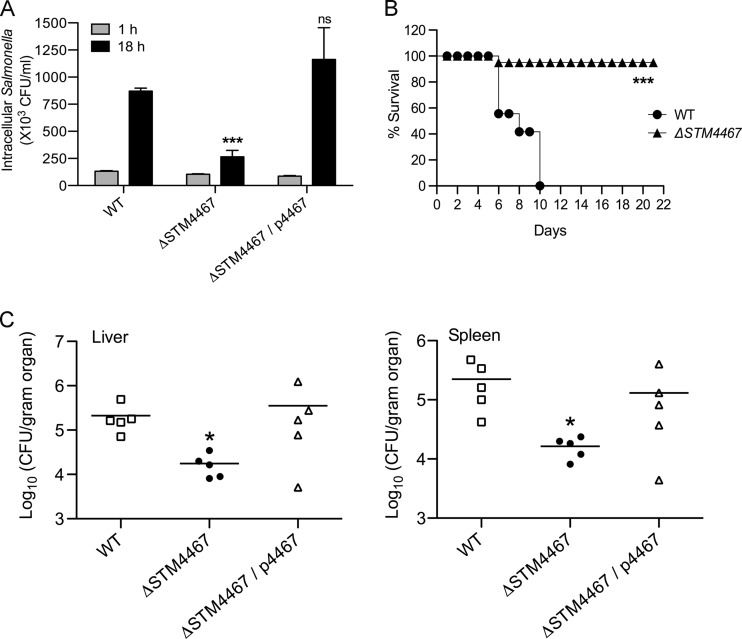
To explore whether the STM4467-encoded ADI contributes to Salmonella virulence, we initially compared the replication abilities of the wild-type and STM4467 deletion strains within murine macrophages. The gentamicin protection assay revealed that the CFU count of the STM4467 deletion mutant within the macrophages at 18 h postinfection was only ∼30% of that of the wild type (Fig. 2A). This result was not due to differences in the phagocytosis of the two strains, because the intracellular numbers of the two strains were similar at an earlier time point (i.e., 1 h) after infection (Fig. 2A). The phenotypic defect of the STM4467 deletion strain was due to the absence of STM4467 function, as evidenced by the fact that expression of the STM4467 gene from a plasmid enabled the STM4467 deletion mutant to replicate within the macrophages at a level similar to that of the wild-type strain (Fig. 2A).
Fig 2.
The STM4467 gene contributes to S. Typhimurium virulence. (A) J774A.1 macrophage cells were infected with the wild-type (WT) strain (14028s), the STM4467 deletion mutant (CH102), or strain CH102 harboring the p4467 plasmid (ΔSTM4467/p4467). The numbers of intracellular bacteria were determined at 1 and 18 h after infection by using the gentamicin protection assay. Means and standard deviations from at least three independent experiments are shown. Triple asterisks indicate that the numbers of bacteria were significantly different (P < 0.001) from those of the WT strain at 18 h postinfection; ns, not significantly different. (B) Groups of C3H/HeN mice (5 mice/group) were injected intraperitoneally with ∼104 cells of the WT or STM4467 deletion strain. The survival of the mice was monitored daily for 3 weeks. The results of one of two independent experiments (P < 0.001), which yielded similar results, are shown. The results of the other experiment are shown in Fig. S1 in the supplemental material. (C) Groups of C3H/HeN mice (5 mice/group) were infected with the WT strain, the STM4467 deletion mutant, or strain CH102 harboring plasmid p4467 as described for panel B. At 5 days after infection, the numbers of bacteria in the liver and spleen were determined. An asterisk indicates that the numbers of bacteria were significantly different (P < 0.05) from those of the WT strain.
To test whether the STM4467 deletion mutant might be attenuated for virulence in mice, we injected Salmonella intraperitoneally into groups of 5 mice. As shown in Fig. 2B, all of the mice inoculated with the wild-type strain died within 10 days, whereas 90% of the mice that received the STM4467 mutant survived over 3 weeks postinfection. We further verified the virulence phenotype of the STM4467 deletion mutant by determining the numbers of bacterial cells in organs of mice. In both the liver and the spleen, the numbers of STM4467 deletion mutant cells were ∼10-fold lower than those of wild-type cells at 5 days postinfection (Fig. 2C). This phenotypic difference was due to the absence of STM4467 function, as evidenced by the fact that the STM4467 deletion strain carrying the STM4467 expression plasmid was able to colonize the liver and spleen as efficiently as the wild-type strain (Fig. 2C). Taken together, these results suggest that in the absence of the STM4467-encoded ADI activity, S. Typhimurium cannot avoid killing by macrophages and thus is attenuated for virulence.
Expression of the ADI pathway genes is enhanced inside macrophages.
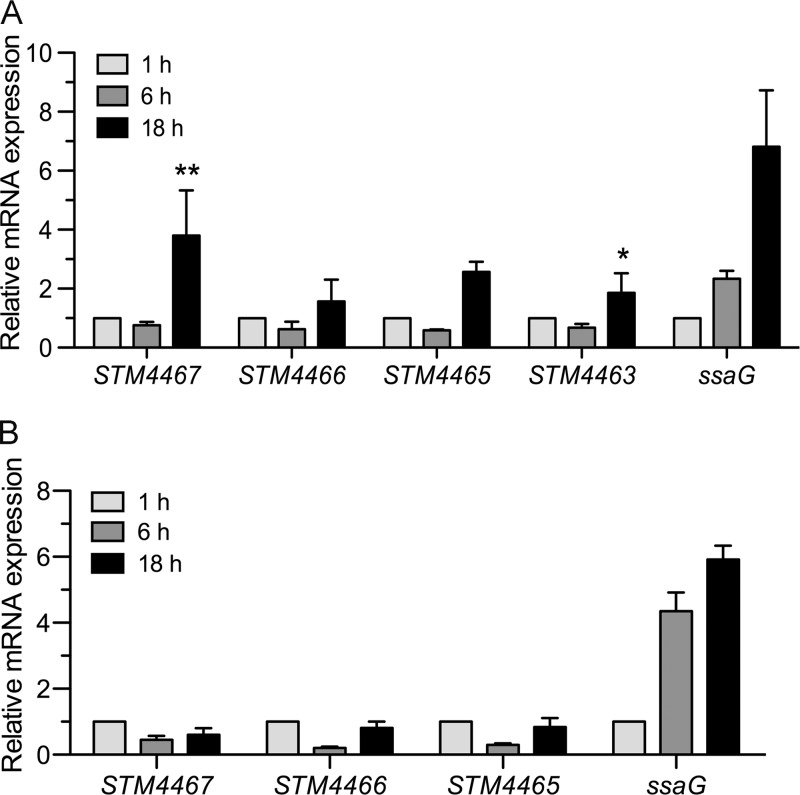
The attenuation of the virulence of the STM4467 mutant suggested that expression of the STM4467 gene might increase when Salmonella cells are inside macrophages. To test this hypothesis, we isolated RNA from wild-type Salmonella cells grown within macrophages and determined the transcription levels of genes via qRT-PCR. We found that STM4467 transcription was enhanced in Salmonella cells growing inside macrophages: the STM4467 mRNA levels increased ∼4-fold at 18 h over those at 1 h after bacterial entry into the macrophages (Fig. 3A). In agreement with the hypothesis that the ADI system genes constitute an operon that produces a polycistronic mRNA (2, 22), the transcript levels of the STM4466 and STM4465 genes, which putatively encode CK and OTC, respectively (Fig. 1A), were also elevated at 18 h after phagocytosis (Fig. 3A).
Fig 3.
Expression of the ADI system is enhanced inside macrophages in an STM4463-dependent manner. The transcription levels of the STM4467, STM4466, STM4465, STM4463, and ssaG genes in S. Typhimurium growing inside macrophages were determined via qRT-PCR. J774A.1 macrophages were infected with the wild-type strain (14028s) (A) or the STM4463 deletion strain (CH201) (B), and bacterial RNA was isolated at 1 h, 6 h, and 18 h after infection. To obtain the relative mRNA expression values on the y axis, the mRNA levels of each gene were divided by those of the rpoD gene, which were further normalized by the transcription levels displayed at 1 h after infection. Means and standard deviations from three independent experiments are shown. Asterisks indicate significant differences (**, P < 0.01; *, P < 0.05) in mRNA levels between the 1-h and 18-h samples.
In response to environmental cues inside the phagosome, Salmonella expresses a series of genes from Salmonella pathogenicity island 2 (SPI-2), which mediates its survival within macrophages (7, 24, 29, 31). Therefore, the induction of the SPI-2 gene ssaG under our experimental conditions (Fig. 3A) reflects the possibility that upregulation of the ADI pathway could occur in response to environmental cues inside the phagosome. Induction of the ADI pathway was observed only at 18 h after phagocytosis (Fig. 3A); this timing was slower than that of SPI-2 gene induction, which occurred at 6 h after bacterial entry into macrophages (Fig. 3A) (8). Thus, given that the environment within the Salmonella-containing phagosome is dynamically changing, the expression of the ADI system might be important at a stage of infection later than the onset of expression of the SPI-2 genes.
The STM4463 protein promotes the transcription of the STM4467 gene.
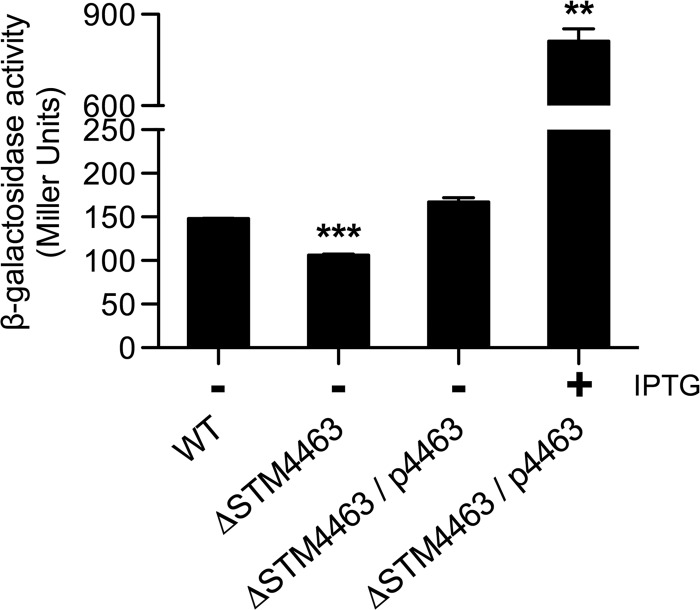
As in the structures of other known ADI operons (38, 44), the STM4463 gene encodes a transcriptional regulator downstream of the ADI gene cluster (Fig. 1A). This gene was formerly known as rosE, and the STM4463 protein was identified as a regulator that directly represses the expression of the std fimbrial operon (6). However, since homologs of STM4463 have been reported to regulate the expression of the ADI operon in other bacteria (12, 33, 34), we reasoned that the STM4463 protein might act as a regulator of Salmonella ADI expression. To examine STM4467 transcription, we constructed a strain that carried a transcriptional fusion of lacZ to the STM4467 gene. A β-galactosidase assay determined that in the STM4467-lacZ strain, the lack of STM4463 reduced STM4467 expression levels by ∼30% (Fig. 4). To further investigate the regulatory role of STM4463, we constructed plasmid p4463, in which expression of the STM4463 gene is under the control of the lac promoter. The β-galactosidase assay revealed that in the STM4467-lacZ strain lacking the STM4463 gene and harboring p4463, STM4467 transcription was restored to wild-type levels, which further increased ∼5-fold when the strain was grown with IPTG to induce STM4463 (Fig. 4). Salmonella ADI activity was also enhanced by STM4463 in a process that required STM4467: the overexpression of STM4463 increased ADI activity ∼5-fold in the wild-type strain but failed to do so for the STM4467 deletion mutant (Fig. 1B and C). Cumulatively, these results suggest that the STM4463 protein is a regulator that positively controls the ADI pathway of S. Typhimurium.
Fig 4.
The STM4463 protein promotes transcription of the STM4467 gene. Transcription levels of the STM4467 gene were determined using a β-galactosidase assay on S. Typhimurium strains carrying an STM4467-lacZ fusion. Strain CH110 (wild type [WT]), its isogenic STM4463 deletion mutant (CH111 ΔSTM4463), and strain CH111 harboring plasmid p4463 (ΔSTM4463/p4463) were grown in LB medium with (+) or without (−) 0.5 mM IPTG. Means and standard deviations from three independent experiments are shown. Asterisks indicate that the β-galactosidase levels of bacteria are significantly different (***, P < 0.001; **, P < 0.01) from those of the wild-type strain.
The STM4463 regulator is necessary for Salmonella to express the ADI gene cluster and replicate inside macrophages.
Since the STM4463 protein appeared to enhance ADI expression (Fig. 4), we reasoned that the STM4463 regulator might be responsible for intramacrophage induction of the ADI system. To test this possibility, we examined the transcription of the STM4467, STM4466, and STM4465 genes within macrophages. qRT-PCR analysis revealed that in contrast to the wild-type strain, the STM4463 deletion mutant failed to induce the expression of these three genes at 18 h after phagocytosis (Fig. 3B). The failure of the STM4463 mutant to induce the ADI gene cluster is not due to a general expression defect, because the intraphagosomal induction of the SPI-2 gene ssaG was unaffected by the STM4463 deletion (Fig. 3B).
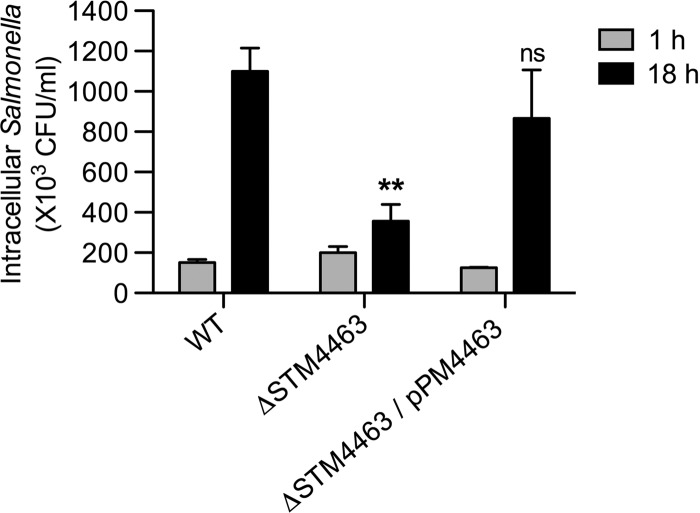
We then hypothesized that the STM4463 regulator could contribute to Salmonella virulence by activating ADI expression within macrophages. Indeed, a lack of STM4463 impaired the ability of Salmonella bacteria to replicate inside macrophages. The numbers of intracellular Salmonella bacteria with the STM4463 deletion were ∼3-fold lower than those of wild-type bacteria at 18 h after phagocytosis (Fig. 5). This defective phenotype was due to the function of STM4463, as evidenced by the fact that the replication ability of the STM4463 deletion mutant was recovered by expression of the STM4463 gene from a plasmid (Fig. 5). The numbers of bacteria of the wild-type strain and the STM4463 deletion strain inside the macrophages were similar at 1 h after infection, indicating that the STM4463 regulator did not interfere with phagocytosis.
Fig 5.
The STM4463 regulator is necessary for S. Typhimurium replication inside macrophages. J774A.1 macrophages were infected with the wild-type (WT) strain (14028s), an STM4463 mutant (CH201), or strain CH201 carrying plasmid pPM4463 (ΔSTM4463/pPM4463). The numbers of intracellular bacteria were determined at 1 h and 18 h after infection. Means and standard deviations from at least three independent experiments are shown. Asterisks indicate that the numbers of bacteria are significantly different (**, P < 0.01) from those of the WT strain at 18 h postinfection; ns, not significantly different.
In the wild-type strain, transcription levels of the STM4463 gene increased ∼2-fold at 18 h after phagocytosis (Fig. 3A), concurrently with ADI pathway induction (Fig. 3A). In addition, induction of the STM4463 regulator greatly increased the expression of the STM4467 gene (Fig. 4). Therefore, although the precise mechanism of control of STM4463 expression within macrophages is presently unclear, we propose that levels of the STM4463 regulator are enhanced by unknown signals present inside the phagosome and, in turn, activate expression of the ADI system.
It is possible that the failure of expression of the ADI gene cluster partially contributes to the defective survival of the STM4463 mutant within macrophages. The STM4463 regulator repressed the expression of the fimbrial operon std, which appeared necessary for full Salmonella virulence in mice (6). However, because the effects of STM4463 and std mutations on Salmonella virulence were assessed by bacterial colonization of organs of mice that were infected via an oral route (6), it remains unknown whether STM4463 regulation of the std operon could affect the survival of Salmonella inside macrophages.
STM4467 does not affect the levels of nitric oxide production inside macrophages.
After phagocytosis, bacterial cells are killed inside the phagosome by the actions of microbicidal products (19, 23). Of these antimicrobials, nitric oxide (NO) is synthesized from arginine by NO synthase (NOS), and the availability of arginine is one of the rate-limiting factors in cellular NO production (1, 16, 37). S. Typhimurium appears to have a means to control host arginine metabolism by which it can avoid NO toxicity. A recent study demonstrated that in Salmonella-infected macrophages, the upregulation of arginase II quenches arginine and reduces NO production (30). On the basis of these observations, we hypothesized that the enhanced ADI activity within the phagosome might reduce NO levels by consuming arginine, thus helping Salmonella to avoid NO-mediated killing by macrophages. To test this hypothesis, we measured NO levels within macrophages with or without Salmonella infection. The results indicated that infection by wild-type Salmonella dramatically increased the NO levels in macrophages (Fig. 6), in accordance with the observation that NOS activity is inducible upon bacterial infection (26). We found that the levels of NO production stimulated by Salmonella bacteria with the STM4467 deletion were comparable to those occurring upon infection with the wild-type strain (Fig. 6). Therefore, this result suggests that the STM4467-encoded ADI activity contributes to the intramacrophage survival of Salmonella via an alternative mechanism that is unrelated to NO production.
Fig 6.
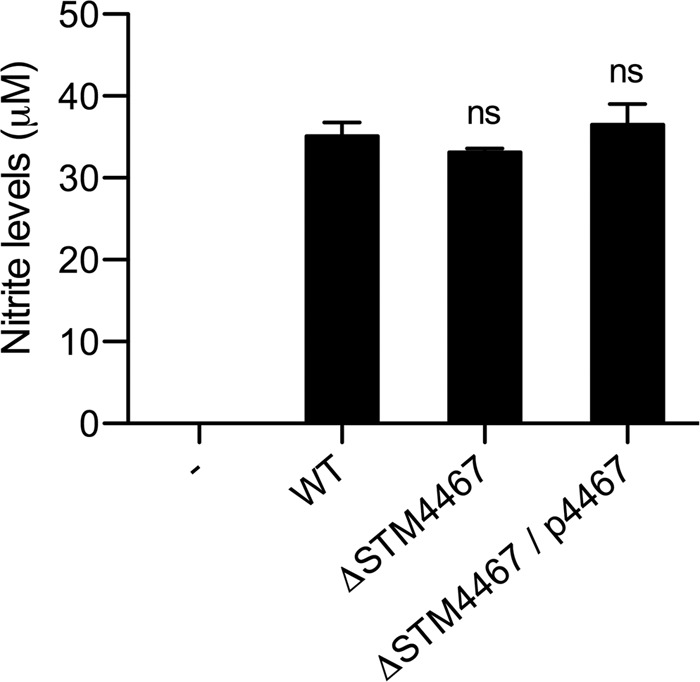
The STM4467-encoded ADI has no effect on NO2− generation by macrophages. J774A.1 macrophage cells were infected with the wild-type (WT) strain (14028s), an STM4467 deletion mutant (CH102 ΔSTM4467), or strain CH102 harboring plasmid p4467 (ΔSTM4467/p4467). At 18 h after infection, levels of NO2− production in the supernatants of macrophages were determined by using the Griess reaction. Note that NO2− was nearly absent from macrophages without (−) S. Typhimurium infection. Means and standard deviations from three independent experiments are shown. “ns” indicates that the nitrite levels of bacteria are not significantly different from those of the WT strain.
Concluding remarks.
In the present study, we established that the ADI pathway contributes to Salmonella pathogenesis. The ADI gene cluster was induced inside macrophages in a process that required the STM4463 regulator, the lack of which impaired the ability of Salmonella to replicate inside macrophages. To our knowledge, this is the first report to demonstrate the regulation of the ADI genes and their role in a pathogen growing inside host cells. Our findings raise important questions. First, how do levels of the STM4463 regulator increase within macrophages? Since the STM4463 gene appears to have its own promoter, an unknown trans-acting factor might act on the STM4463 regulatory region in response to environmental cues within the phagosome. Second, how does the ADI pathway contribute to Salmonella virulence? We were able to exclude the possibility that the ADI pathway reduced NO toxicity. In L. monocytogenes and S. pyogenes, ADI enzymes appeared to protect bacteria from acidic stress (4, 11, 43). Since Salmonella experiences an acidic pH within the phagosome (13, 40), we tested whether the ADI pathway is also implicated in the survival of Salmonella at acidic pHs. However, we found that the STM4467 deletion did not inhibit the growth of Salmonella at pH 5.5 (data not shown). Therefore, as proposed in a recent study (43), the ADI system might provide energy for intracellular Salmonella bacteria by producing ATP or might protect it from oxidative stress within the phagosome via production of the polyamine putrescine.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education, Science and Technology (2009-0069104; to D. Shin), and by the NRF through the World Class University program (R32-2008-000-10183-0; to S. Ryu).
Footnotes
Published ahead of print 24 September 2012
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1. Alderton WK, Cooper CE, Knowles RG. 2001. Nitric oxide synthases: structure, function and inhibition. Biochem. J. 357:593–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baur H, Luethi E, Stalon V, Mercenier A, Haas D. 1989. Sequence analysis and expression of the arginine-deiminase and carbamate-kinase genes of Pseudomonas aeruginosa. Eur. J. Biochem. 179:53–60 [DOI] [PubMed] [Google Scholar]
- 3. Boyde TR, Rahmatullah M. 1980. Optimization of conditions for the colorimetric determination of citrulline, using diacetyl monoxime. Anal. Biochem. 107:424–431 [DOI] [PubMed] [Google Scholar]
- 4. Casiano-Colon A, Marquis RE. 1988. Role of the arginine deiminase system in protecting oral bacteria and an enzymatic basis for acid tolerance. Appl. Environ. Microbiol. 54:1318–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chang AC, Cohen SN. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chessa D, Winter MG, Nuccio SP, Tukel C, Baumler AJ. 2008. RosE represses Std fimbrial expression in Salmonella enterica serotype Typhimurium. Mol. Microbiol. 68:573–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Choi J, et al. 2010. Salmonella pathogenicity island 2 expression negatively controlled by EIIANtr-SsrB interaction is required for Salmonella virulence. Proc. Natl. Acad. Sci. U. S. A. 107:20506–20511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cirillo DM, Valdivia RH, Monack DM, Falkow S. 1998. Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol. Microbiol. 30:175–188 [DOI] [PubMed] [Google Scholar]
- 9. Cunin R, Glansdorff N, Pierard A, Stalon V. 1986. Biosynthesis and metabolism of arginine in bacteria. Microbiol. Rev. 50:314–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Degnan BA, et al. 2000. Characterization of an isogenic mutant of Streptococcus pyogenes Manfredo lacking the ability to make streptococcal acid glycoprotein. Infect. Immun. 68:2441–2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dimova D, et al. 2000. Thermostability, oligomerization and DNA-binding properties of the regulatory protein ArgR from the hyperthermophilic bacterium Thermotoga neapolitana. Mol. Gen. Genet. 263:119–130 [DOI] [PubMed] [Google Scholar]
- 13. Drecktrah D, Knodler LA, Ireland R, Steele-Mortimer O. 2006. The mechanism of Salmonella entry determines the vacuolar environment and intracellular gene expression. Traffic 7:39–51 [DOI] [PubMed] [Google Scholar]
- 14. Ellermeier CD, Janakiraman A, Slauch JM. 2002. Construction of targeted single copy lac fusions using lambda Red and FLP-mediated site-specific recombination in bacteria. Gene 290:153–161 [DOI] [PubMed] [Google Scholar]
- 15. Eriksson S, Lucchini S, Thompson A, Rhen M, Hinton JC. 2003. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol. Microbiol. 47:103–118 [DOI] [PubMed] [Google Scholar]
- 16. Fang FC. 2004. Antimicrobial reactive oxygen and nitrogen species: concepts and controversies. Nat. Rev. Microbiol. 2:820–832 [DOI] [PubMed] [Google Scholar]
- 17. Faucher SP, Porwollik S, Dozois CM, McClelland M, Daigle F. 2006. Transcriptome of Salmonella enterica serovar Typhi within macrophages revealed through the selective capture of transcribed sequences. Proc. Natl. Acad. Sci. U. S. A. 103:1906–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fields PI, Swanson RV, Haidaris CG, Heffron F. 1986. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc. Natl. Acad. Sci. U. S. A. 83:5189–5193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Flannagan RS, Cosio G, Grinstein S. 2009. Antimicrobial mechanisms of phagocytes and bacterial evasion strategies. Nat. Rev. Microbiol. 7:355–366 [DOI] [PubMed] [Google Scholar]
- 20. Gamper M, Zimmermann A, Haas D. 1991. Anaerobic regulation of transcription initiation in the arcDABC operon of Pseudomonas aeruginosa. J. Bacteriol. 173:4742–4750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Griess P. 1879. Bemerkungen zu der abhandlung der H. H. Weselsky und Benedikt “Ueber einige azoverbindungen.” Chem. Ber. 12:426–428 [Google Scholar]
- 22. Griswold A, Chen YY, Snyder JA, Burne RA. 2004. Characterization of the arginine deiminase operon of Streptococcus rattus FA-1. Appl. Environ. Microbiol. 70:1321–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hampton MB, Kettle AJ, Winterbourn CC. 1998. Inside the neutrophil phagosome: oxidants, myeloperoxidase, and bacterial killing. Blood 92:3007–3017 [PubMed] [Google Scholar]
- 24. Hansen-Wester I, Hensel M. 2001. Salmonella pathogenicity islands encoding type III secretion systems. Microbes Infect. 3:549–559 [DOI] [PubMed] [Google Scholar]
- 25. Haraga A, Ohlson MB, Miller SI. 2008. Salmonellae interplay with host cells. Nat. Rev. Microbiol. 6:53–66 [DOI] [PubMed] [Google Scholar]
- 26. Henard CA, Vazquez-Torres A. 2011. Nitric oxide and Salmonella pathogenesis. Front. Microbiol. 2:84 doi:10.3389/fmicb.2011.00084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hoiseth SK, Stocker BAD. 1981. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291:238–239 [DOI] [PubMed] [Google Scholar]
- 28. Hughes KT, Maloy SR. 2007. Advanced bacterial genetics: use of transposons and phage for genomic engineering. Academic Press, San Diego, CA [Google Scholar]
- 29. Kuhle V, Hensel M. 2004. Cellular microbiology of intracellular Salmonella enterica: functions of the type III secretion system encoded by Salmonella pathogenicity island 2. Cell. Mol. Life Sci. 61:2812–2826 [DOI] [PubMed] [Google Scholar]
- 30. Lahiri A, Das P, Chakravortty D. 2008. Arginase modulates Salmonella induced nitric oxide production in RAW264.7 macrophages and is required for Salmonella pathogenesis in mice model of infection. Microbes Infect. 10:1166–1174 [DOI] [PubMed] [Google Scholar]
- 31. Lim S, Kim B, Choi HS, Lee Y, Ryu S. 2006. Fis is required for proper regulation of ssaG expression in Salmonella enterica serovar Typhimurium. Microb. Pathog. 41:33–42 [DOI] [PubMed] [Google Scholar]
- 32. Lim S, et al. 2007. Mlc regulation of Salmonella pathogenicity island I gene expression via hilE repression. Nucleic Acids Res. 35:1822–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lu CD, Winteler H, Abdelal A, Haas D. 1999. The ArgR regulatory protein, a helper to the anaerobic regulator ANR during transcriptional activation of the arcD promoter in Pseudomonas aeruginosa. J. Bacteriol. 181:2459–2464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Maghnouj A, de Sousa Cabral TF, Stalon V, Vander Wauven C. 1998. The arcABDC gene cluster, encoding the arginine deiminase pathway of Bacillus licheniformis, and its activation by the arginine repressor ArgR. J. Bacteriol. 180:6468–6475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Merighi M, Ellermeier CD, Slauch JM, Gunn JS. 2005. Resolvase-in vivo expression technology analysis of the Salmonella enterica serovar Typhimurium PhoP and PmrA regulons in BALB/c mice. J. Bacteriol. 187:7407–7416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 37. Mori M, Gotoh T. 2004. Arginine metabolic enzymes, nitric oxide and infection. J. Nutr. 134:2820S–2825S (Discussion, 2853S.) [DOI] [PubMed] [Google Scholar]
- 38. Nishijyo T, Park SM, Lu CD, Itoh Y, Abdelal AT. 1998. Molecular characterization and regulation of an operon encoding a system for transport of arginine and ornithine and the ArgR regulatory protein in Pseudomonas aeruginosa. J. Bacteriol. 180:5559–5566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ohl ME, Miller SI. 2001. Salmonella: a model for bacterial pathogenesis. Annu. Rev. Med. 52:259–274 [DOI] [PubMed] [Google Scholar]
- 40. Rathman M, Sjaastad MD, Falkow S. 1996. Acidification of phagosomes containing Salmonella typhimurium in murine macrophages. Infect. Immun. 64:2765–2773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ryan S, Begley M, Gahan CG, Hill C. 2009. Molecular characterization of the arginine deiminase system in Listeria monocytogenes: regulation and role in acid tolerance. Environ. Microbiol. 11:432–445 [DOI] [PubMed] [Google Scholar]
- 42. Soncini FC, Vescovi EG, Groisman EA. 1995. Transcriptional autoregulation of the Salmonella typhimurium phoPQ operon. J. Bacteriol. 177:4364–4371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sonck KA, et al. 2009. The proteome of Salmonella Typhimurium grown under in vivo-mimicking conditions. Proteomics 9:565–579 [DOI] [PubMed] [Google Scholar]
- 44. Zuniga M, Perez G, Gonzalez-Candelas F. 2002. Evolution of arginine deiminase (ADI) pathway genes. Mol. Phylogenet. Evol. 25:429–444 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.