Abstract
Salmonella enterica serovar Enteritidis is an important food-borne pathogen, and chickens are a primary reservoir of human infection. While most knowledge about Salmonella pathogenesis is based on research conducted on Salmonella enterica serovar Typhimurium, S. Enteritidis is known to have pathobiology specific to chickens that impacts epidemiology in humans. Therefore, more information is needed about S. Enteritidis pathobiology in comparison to that of S. Typhimurium. We used transposon mutagenesis to identify S. Enteritidis virulence genes by assay of invasiveness in human intestinal epithelial (Caco-2) cells and chicken liver (LMH) cells and survival within chicken (HD-11) macrophages as a surrogate marker for virulence. A total of 4,330 transposon insertion mutants of an invasive G1 Nalr strain were screened using Caco-2 cells. This led to the identification of attenuating mutations in a total of 33 different loci, many of which include genes previously known to contribute to enteric infection (e.g., Salmonella pathogenicity island 1 [SPI-1], SPI-4, SPI-5, CS54, fliH, fljB, csgB, spvR, and rfbMN) in S. Enteritidis and other Salmonella serovars. Several genes or genomic islands that have not been reported previously (e.g., SPI-14, ksgA, SEN0034, SEN2278, and SEN3503) or that are absent in S. Typhimurium or in most other Salmonella serovars (e.g., pegD, SEN1152, SEN1393, and SEN1966) were also identified. Most mutants with reduced Caco-2 cell invasiveness also showed significantly reduced invasiveness in chicken liver cells and impaired survival in chicken macrophages and in egg albumen. Consequently, these genes may play an important role during infection of the chicken host and also contribute to successful egg contamination by S. Enteritidis.
INTRODUCTION
Salmonella enterica serovar Enteritidis is a major cause of food-borne gastroenteritis. Poultry is the primary reservoir for S. Enteritidis, and epidemiological studies have implicated the contaminated egg as a primary vehicle for human infection (67). S. Enteritidis usually does not cause a disseminated systemic disease in humans but is clinically manifested as gastroenteritis and diarrhea (10). In chickens, S. Enteritidis is able to colonize the gastrointestinal epithelium. The pathogen can be shed in feces, leading to horizontal transmission within the flock, which can eventually result in contaminated meat during the slaughter and production process (51). An additional characteristic of S. Enteritidis infection of chickens, however, is its ability to produce persistent and asymptomatic systemic infection of the liver, spleen, and other internal organs, including reproductive tract tissues (28, 32). Persistent systemic infections of adult chickens with S. Enteritidis can intermittently disseminate to eggs, leading to production of internally contaminated eggs (28, 32). In addition to persistence within tissues, S. Enteritidis is more capable than other Salmonella serotypes of surviving the microbicidal properties of egg albumen by producing a capsular-like lipopolysaccharide (LPS) (66). Consequently, S. Enteritidis is the serotype that is primarily responsible for egg-borne outbreaks of salmonellosis throughout the world. It is thought that organ invasion must be prevented to impede the infection pathway that results in egg contamination. In addition, more knowledge is needed about how to interfere with the growth of S. Enteritidis on farms, especially because it has a robust metabolism and the ability to grow to high cell density (63).
Most knowledge of nontyphoidal Salmonella pathogenesis results from research conducted on Salmonella enterica serovar Typhimurium in both in vitro-cultured mammalian cells and in vivo murine infection models. While it is believed that S. Enteritidis and S. Typhimurium share virulence mechanisms for epithelial cell invasion and survival within macrophages, recent studies have identified several host-specific and serotype-specific bacterial factors that contribute to virulence of S. Enteritidis. For instance, Silva et al. (84) recently showed that systemic infection of mice by S. Enteritidis requires several genes or genomic islands that are absent from S. Typhimurium and most other Salmonella serotypes. Additionally, a genome-wide screen using in vivo expression technology (IVET) and transposon-mediated mutagenesis has identified several S. Enteritidis genes that contribute to S. Enteritidis tropism to the avian reproductive tract and survival in egg albumen (11, 29). Moreover, it has been reported that Salmonella requires different genes in different hosts, suggesting that Salmonella harbors both conserved and host-specific colonization factors (7, 9, 56, 64, 80, 84), reinforcing the need to study the genetic basis of the pathogenesis of S. Enteritidis, the Salmonella serotype responsible for the largest proportion of food-borne gastroenteritis.
Several studies using site-directed and random-transposon mutagenesis have been conducted to identify genes involved in S. Typhimurium pathogenesis in both in vitro and in vivo models. However, limited studies have been extended to study the genetic basis of S. Enteritidis pathogenesis. Consequently, our understanding of the mechanisms involved in S. Enteritidis pathogenicity in both humans and chickens is relatively poor. The objective of this study was to employ transposon mutagenesis to identify serotype-specific and common virulence genes of S. Enteritidis that are required for invasion in differentiated human intestinal epithelial (Caco-2) cells. The phenotypes of selected mutants with impaired Caco-2 cell invasiveness were confirmed by trans-complementation of disrupted genes in the mutant strains. In addition, we tested the Caco-2 cell invasion-attenuated mutants of S. Enteritidis for their invasion attenuation in chicken liver (LMH) cells and intracellular-survival defects in chicken macrophages, as well as testing for impaired survival in egg albumen.
MATERIALS AND METHODS
Bacterial strains and growth media.
The S. Enteritidis G1 Nalr strain used in this study is a phage type 4 strain and was chosen because it is invasive in cultured intestinal epithelial cells and virulent in orally inoculated chickens and mice (79, 83). A spontaneous nalidixic-acid-resistant mutant of 22079 (G1 Nalr) was derived according to the methods described previously (80, 81). A collection of 12 Escherichia coli S17-λ pir isolates containing uniquely tagged pUT mini-Tn5 plasmids were obtained from Roger Levesque and Francois Sanschagrin (Université Laval, Quebec, Canada). The random-transposon mutant library was generated, and randomness was assessed using Southern blotting as described previously (80). A total of 4,992 mutants were arrayed in a 96-well plate format and stored at −80°C in 15% (vol/vol) phosphate-buffered glycerol. Unless otherwise noted, S. Enteritidis and E. coli strains were grown in Luria-Bertani (LB) broth (Difco). When needed, the medium was supplemented with 1.6% (wt/vol) Bacto Agar, ampicillin (Am) (100 mg ml−1), nalidixic acid (Na) (30 mg ml−1), and kanamycin (Km) (50 mg ml−1).
Screening of S. Enteritidis transposon mutants for the ability to invade cultured Caco-2 cells.
Human colon adenocarcinoma (Caco-2) cells were routinely grown in Dulbecco's modified Eagle medium (DMEM) (Invitrogen) supplemented with 10% (vol/vol) fetal bovine serum (FBS) (Invitrogen), as described previously (83). Propagated cells were seeded at approximately 106 cm−2 into 75-cm2 flasks (BD Falcon) and incubated for 5 days in 95% air/5% CO2 at 37°C. Cells were harvested by trypsinization and suspended in DMEM with 10% FBS and gentamicin (10 μg/ml). A Caco-2 cell suspension (200 μl) was plated onto each well of a 96-well flat-bottom polystyrene tissue culture-treated plate (Nunc) and incubated. The cells reached confluence at 4 days, after which the medium was changed every second day until 13 days postincubation. At 13 days postincubation, well-differentiated monolayers with a cell density of approximately 5 × 105 cells/well were obtained. The state of cell differentiation was determined as described previously (83).
In preliminary screening, the ability of mutants to invade well-differentiated Caco-2 cells was assessed using a gentamicin protection assay in a 96-well plate format. Briefly, each 96-well plate containing the frozen transposon mutants was thawed on ice. An aliquot of each mutant (10 μl) was transferred to a fresh 96-well plate with 200 μl of LB containing Na and Km. The plates were incubated overnight (16 h) at 37°C with shaking at 150 rpm. On the day of the assay, the cell culture medium covering the Caco-2 cell monolayers was replaced with 100 μl fresh prewarmed DMEM with 10% heat-inactivated FBS without any antibiotics. The overnight cultures of mutants were resuspended by pipetting up and down using 96-well liquidator (Rainin), and 1 μl of bacterial suspension containing approximately 5 × 106 CFU was added to the 96-well plate containing Caco-2 cell monolayers to obtain a multiplicity of infection (MOI) of approximately 10:1 (106 CFU of S. Enteritidis and 105 Caco-2 cells). Given the large number of mutants screened in this study, it was not possible to optimize the CFU or optical density of the mutants prior to infection. Nevertheless, the number of CFU of each of the mutants was determined immediately after infection by plating serial 10-fold dilutions of overnight cultures onto LB agar containing Km and Na. Infected plates were centrifuged at 1,500 rpm for 3 min to facilitate contact between bacteria and Caco-2 cells, followed by incubation at 37°C for 2 h to allow bacterial invasion of the cells. The cell culture medium was then replaced with 100 μl of fresh DMEM containing gentamicin (200 μg/ml), and the cells were incubated for 2 h to kill extracellular bacteria. Following incubation, the medium was discarded, and the monolayers were washed twice with 100 μl phosphate-buffered saline (PBS) to remove residual gentamicin, followed by incubation at 37°C for 10 min in the presence of 100 μl PBS containing 0.5% Triton X-100 to lyse the cells. Serial 10-fold dilutions of the cell lysate were plated onto LB agar containing Na and Km to determine the numbers of CFU of mutants that survived gentamicin treatment and hence had invaded the Caco-2 cells. Each mutant was tested once during this preliminary screening and compared with the wild-type (WT) parent as a positive control. The percent invasion for each mutant and the WT parent strain was calculated using the following formula: intracellular CFU/inoculum CFU × 100. Mutants that showed ≥10-fold reduction in invasiveness compared with the WT parent were selected for secondary screening. Secondary screening was performed essentially similarly to the preliminary screening, with the exception that each mutant was tested in duplicate. Mutants that consistently showed a ≥10-fold reduction in invasiveness during secondary screening were selected for tertiary screening using standard gentamicin protection assays (confirmatory assays) in 12-well plates in which each mutant was tested in duplicate as described previously (83).
Cloning and sequencing.
Chromosomal DNA extracted from transposon mutants was digested with restriction enzymes (EcoRI, PstI, or KpnI) that cut on either end of the transposon, leaving the Km resistance gene intact. The digested fragments were ligated into the pUC19 vector (Invitrogen) and transformed into E. coli TOP10 (Invitrogen), and transformants were selected on LB plates containing Km. Plasmids containing both transposons and flanking regions were purified using a QIAprep Spin Miniprep Kit (Qiagen) and sequenced at the Washington State University (WSU) Genomic Core using the primers reading out from the left (5′-TGAACACTGGCAGAGCATTACGC-3′) or right (5′-TGTAACATCAGAGATTTTGAGACACAACG-3′) junction of the transposon. The DNA sequences of the disrupted genes flanking the transposon insertion were assembled and identified by searching the genome sequence of the recently sequenced S. Enteritidis PT4 strain P125109 in the NCBI GenBank database (GenBank accession no. AM933172) (93).
Construction of complementation plasmids.
Forward and reverse primer pairs were designed based on the S. Enteritidis P125109 sequence (Table 1) and used to amplify and clone full-length genes into the low-copy-number plasmid pACYC184 (New England BioLabs) at BamHI/SphI, BamHI/SalI, or SphI/SalI sites, thereby disrupting the tetracycline resistance gene of the plasmid. The forward primers contained Shine-Dalgarno sequences derived from the expression vector pET101 (Invitrogen), enabling heterologous genes to be controlled by the promoter of the tetracycline resistance gene. The electrocompetent cells of each mutant were prepared as follows: overnight cultures were diluted 1:200 using 50 ml of SOB medium (2% tryptone, 0.5% yeast extract, 0.05% NaCl, 2.5 mM KCl, 10 mM MgCl2) and grown to an optical density at 595 nm (OD595) of 0.45 to 0.6. The cells were chilled on ice for 30 min and spun at 7,800 rpm for 10 min at 4°C; the supernatant was aspirated, and the pellet was washed twice with equal volumes of ice-cold 10% glycerol, followed by resuspension in 500 μl of 10% glycerol. The plasmid constructs were electroporated at 2.2 kV using Micro Pulser (Bio-Rad).
Table 1.
Oligonucleotide primers used in this study
| Primera | Gene or ORF | Oligonucleotide sequence (5′–3′)b | Annealing temp (°C) | Amplicon size (bp) |
|---|---|---|---|---|
| FljB_F_RFB | fljB | ATGCggatccAGGAGATATACCATGGCACAAGTCATTAATACAAAC | 50.5 | 1,518 |
| FljB_R | TAGCgcatgcTTAACGCAGTAAAGAGAGGAC | 50.4 | ||
| RfbN_F_RFB | rfbN | ATGCggatccAGGAGATATACCATGAAAATAACATTAATTATTCCCAC | 48.5 | 945 |
| RfbN_R | TAGCgtcgacTTATTTTATCTCTTTTGACGAAG | 46.3 | ||
| SEN0034_F_RFB | SEN0034 | ATGCggatccAGGAGATATACCATGAAAAATAAGAAATATATTAATAG | 43.7 | 762 |
| SEN0034_R | TAGCgcatgcTTAAAACTCGAAGCCGATC | 46.7 | ||
| SEN1393_F_RFB | SEN1393 | ATGCggatccAGGAGATATACCATGTTAGGTATTTTCAGAAAGA | 45.5 | 396 |
| SEN1393_R | TAGCgcatgcTCACCACTGCACACAGCG | 52.6 | ||
| SEN1966_F_RFB | SEN1966 | ATGCggatccAGGAGATATACCATGGGTCGCAAACGTGCG | 52.6 | 990 |
| SEN1966_R | TAGCgcatgcTCAACGCTTCCTGTCCAGG | 53.2 |
F, forward primer; R, reverse primer.
The 5′ ATGC overhang in the forward primer and the 5′ TAGC overhang in the reverse primer are underlined. Restriction enzyme sites (BamHI, SphI, and SalI) in the forward and reverse primers are shown in lowercase. The italicized letters denote E. coli Shine-Delgarno sequences and spacers.
In vitro growth curves.
Each strain was grown overnight (16 h) in LB broth containing Km and Na. The overnight cultures were normalized to an OD595 of 2.0 (equivalent to 108 CFU/ml) in LB broth. The cultures were then diluted 10-fold in two different media (LB broth containing Na and Km and DMEM) to a final concentration of approximately 5 × 103 CFU/200 μl. Subsequently, 200 μl of this suspension was added to the wells of a honeycomb microplate (Growth Curves), followed by incubation at 37°C for 16 h (LB medium) or 4 h (DMEM) in continuous-shaking mode in a BioscreenC instrument (Growth Curves). Each strain was tested in triplicate in two independent experiments. The turbidity was measured every 30 min as the absorbance at 420 to 580 nm using the wide-band filter. The absorbance values of blanks (uninoculated media) were subtracted from the absorbance values of each sample. The area under the curve (AUC) for each strain was calculated to determine the differences in growth of the mutants.
Motility assays.
Bacterial motility was tested at 37°C as described previously, and the mutant strain was classified as motility impaired if the percent diameter of growth was <20% relative to the WT parent strain (82). The motility of each mutant strain was tested in triplicate in at least two independent experiments.
Determination of MIC.
For select mutants, the MICs of kasugamycin (Sigma) and polymyxin (Sigma) were determined by the broth dilution method as described previously (100). The concentrations of kasugamycin and polymyxin ranged from 25 μg to 1,000 μg and 0.25 μg to 4 μg/ml, respectively. Two independent experiments were conducted for each antibiotic.
LMH invasion assay.
LMH cells, a chicken hepatocellular carcinoma cell line (46), were cultured and maintained in Weymouth's medium (Sigma) with 10% FBS (Sigma) in T75 flasks pretreated with 0.1% gelatin for 15 min at room temperature. Prior to invasion assays, LMH cells were seeded in 12-well flasks to a cell density of 1 × 106 cells per well and incubated for 24 h at 37°C in a humidified 5% CO2 incubator. The cells were rinsed twice with prewarmed PBS supplemented with 5% heat-inactivated FBS (wash buffer). The bacteria were grown for 16 h at 37°C in 5 ml LB broth on a shaker platform, subcultured at 1:100 in fresh LB broth (3 ml), and grown to late log phase (3 h) at 37°C with shaking. The culture suspensions were centrifuged and resuspended in Weymouth's medium with 10% FBS, followed by storage at 4°C until they were used in the assay. The number of CFU/ml was determined by plating 10-fold dilution series of the suspensions on LB agar. Approximately 5 ×107 CFU of bacterial suspension in Weymouth's medium was added to each well to achieve an MOI of approximately 50. Each plate was then centrifuged at 1,000 × g for 5 min to promote bacterium-host cell contact and incubated at 42°C for 2 h. To quantitate cell invasion, the S. Enteritidis-inoculated cells were rinsed three times with PBS and treated for 30 min with medium containing gentamicin (200 μg/ml) to kill extracellular bacteria. The cells were washed three times in PBS and lysed with 1 ml 0.5% (vol/vol) Triton X-100 (Sigma) for 10 min at 42°C. The viable counts of the intracellular bacteria in the lysate were made on LB agar. Each mutant was tested in triplicate in two independent experiments. The percent invasion for each mutant was calculated as follows: intracellular CFU/inoculum CFU × 100.
HD-11 entry and survival assay.
Cells from the chicken macrophage cell line HD-11 (5) were seeded in T75 flasks in Isacove's minimal essential medium (IMDM) (Sigma) containing 10% FBS. Prior to the entry and survival assay, HD-11 cells were cultured in 12-well flasks to a density of 1 × 106 cells per well and incubated for 48 h at 42°C in a humidified 5% CO2 incubator. The bacterial culture and infection MOIs were similar to those described for LMH cells. After addition of infection inoculum, each plate was centrifuged as described above. Adhesion and subsequent entrance of bacteria in cells proceeded for 30 min at 42°C, at which point gentamicin (200 μg/ml) was added to kill extracellular bacteria. To confirm that all extracellular Salmonella bacteria were killed by gentamicin treatment, samples of the culture medium were plated at 30 min and 8 h post-gentamicin treatment. At 30 min (entry) and at 8 h (intramacrophage survival) postinfection, the cells were rinsed three times with wash buffer and lysed with 0.5% (vol/vol) Triton-X, and dilutions of the lysate were plated on LB agar for viable counts of the intracellular bacteria. Each mutant was tested in triplicate in two independent experiments. Entry was calculated as follows: intracellular CFU at 30 min/inoculum CFU × 100. Intramacrophage survival was calculated as follows: intracellular CFU at 8 h/intracellular CFU at 30 min × 100.
Growth in egg albumen.
The ability of S. Enteritidis strains to survive in egg albumen was quantified as described previously with minor modifications (79). Organic, unfertilized, antibiotic-free eggs (Chino Valley Ranchers, CA) were disinfected by immersion in 70% ethanol and aseptically broken to collect the egg albumen in a sterile container. Aliquots (1 ml) of egg albumen were inoculated with an overnight culture of each bacterial strain to a final concentration of ∼600 CFU/ml and incubated at 25 ± 2°C for 24 h after thorough mixing. Next, the bacterium-albumen mixture was diluted in maximal-recovery diluent (Oxoid) and plated on LB agar to enumerate the viable-bacteria counts. Uninoculated egg albumen served as a negative control. The percent growth in egg albumen was calculated using the following formula: CFU at 24 h/CFU at 0 h × 100. Each mutant strain was tested in duplicate in three independent experiments.
Data analysis.
The differences in the invasiveness of the mutant strains in Caco-2, LMH, and HD-11 cells; the growth index in egg albumen; and the growth curves of mutant strains were analyzed by analysis of variance (ANOVA), followed by Fisher's least significant difference (LSD) multiple-comparison test. The probability level for significance was taken as a P value of <0.05. All statistical analysis was performed using NCSS 2007 (NCSS, Kaysville, UT).
RESULTS AND DISCUSSION
Screening of the S. Enteritidis mutant library.
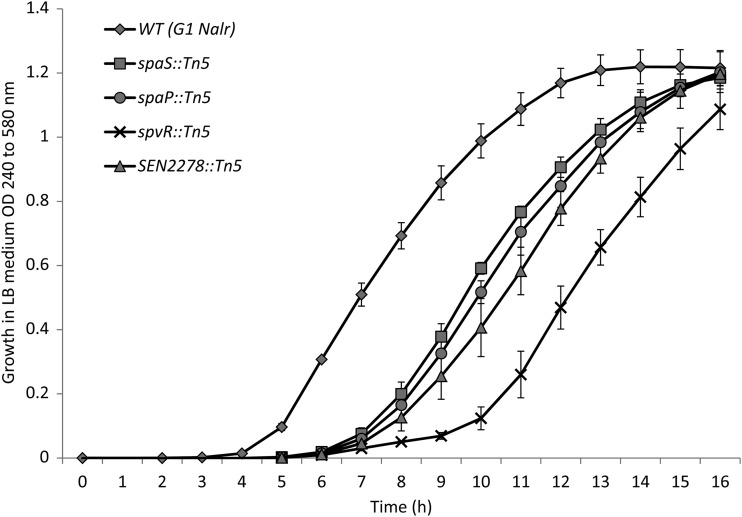
A total of 4,330 out of 4,992 mutants qualified for preliminary screening for their invasiveness in differentiated Caco-2 cells using a 96-well plate assay. The remaining mutants were excluded from the analysis because they were resistant to ampicillin, suggesting genomic integration of the backbone of the suicide vector, or they failed to grow in vitro in LB broth. The mean log10 intracellular CFU ± standard error (SE) (cell invasiveness) of the WT S. Enteritidis G1 Nalr strain was 5.71 ± 0.2, indicating that the strain is highly invasive. In the preliminary screening experiments, a total of 187 (4.3%) mutants showed 10-fold or lower invasiveness compared with the WT parent. These 187 mutants were subjected to secondary and tertiary screening to confirm the invasion attenuation, as described in Materials and Methods. This resulted in identification of a total of 33 mutants that consistently showed 10-fold to 100-fold lower invasiveness in differentiated Caco-2 cells than the WT parent (Table 2). Southern blot analysis confirmed that each of these mutants carried a single Tn5 transposon insertion (data not shown). Because mutants were routinely grown to stationary phase (16 h) in LB medium, we tested all 33 mutants for their growth kinetics in LB. The AUCs of four mutants (spaS::Tn5, spaP::Tn5, spvR::Tn5, and SEN2278::Tn5) were significantly lower than that of the WT parent (Fig. 1); however, all the strains reached similar densities at 16 h postincubation, suggesting that the impaired growth of mutants was primarily limited to the lag or early log phase. In addition, none of the above mutants showed growth defects in the cell culture medium (DMEM) routinely used for invasion assays with Caco-2 cells (data not shown). These data suggest that the reduced invasiveness of these mutants is not likely due to their in vitro growth defects.
Table 2.
Cell invasiveness and motility of S. Enteritidis mutants
| Functional category | Mutant | Gene/ORF | Function | Cell invasivenessa (mean log10 CFU ± SE) | % Motilityb |
|---|---|---|---|---|---|
| Virulence (pathogenicity islands) | 1F11 | orgA | SPI-1 TTSS structural protein | 3.57 ± 0.33 | 97 |
| 2F2 | prgH | SPI-1 TTSS structural protein | 3.37 ± 0.13 | 44 | |
| 1B12 | hilA | SPI-1 TTSS regulatory protein | 3.63 ± 0.24 | 76 | |
| 2B9 | sipC | SPI-1 TTSS effector protein | 3.81 ± 0.34 | 109 | |
| 1F2 | sicA | SPI-1 TTSS chaperone | 3.54 ± 0.35 | 99 | |
| 1C3 | spaS | SPI-1 TTSS structural protein | 3.46 ± 0.45 | 70 | |
| 2E8 | spaP | SPI-1 TTSS structural protein | 2.21 ± 0.48 | 52 | |
| 1E10 | spaO | SPI-1 TTSS structural protein | 3.08 ± 0.62 | 78 | |
| 2F6 | invI | SPI-1 TTSS structural protein | 2.93 ± 0.30 | 115 | |
| 1D3 | invB | SPI-1 TTSS chaperone | 3.84 ± 0.40 | 101 | |
| 1A5 | invG | SPI-1 TTSS structural protein | 3.69 ± 0.28 | 76 | |
| 2A2 | invF | SPI-1 TTSS regulatory protein | 3.56 ± 0.36 | 52 | |
| 1C9 | siiE | SPI-4 pathogenicity island protein | 3.67 ± 0.22 | 105 | |
| 2E3 | pipA | SPI-5 pathogenicity island protein | 2.82 ± 0.33 | 66 | |
| 2B4 | SEN0803 | SPI-14, putative acyl-CoA dehydrogenase | 3.82 ± 0.25 | 7 | |
| 1C5 | sinH | CS54 pathogenicity island protein (pseudogene) | 3.14 ± 0.16 | 90 | |
| Motility | 1F8 | fliH | Regulator of FliI (flagellar assembly protein) | 2.61 ± 0.73 | 7 |
| 1E9 | fljB | Flagellin | 2.97 ± 0.21 | 9 | |
| Fimbria | 1H3 | csgB | Curlin fimbria minor subunit | 3.33 ± 0.17 | 25 |
| 1E5 | pegD | Putative fimbrial-like adhesin protein | 3.39 ± 0.35 | 73 | |
| LPS biosynthesis | 2B5 | rfbM | Mannose-1-phosphate guanylyltransferase | 2.45 ± 0.49 | 56 |
| 1D10 | rfbN | Rhamnosyltransferase | 3.43 ± 0.50 | 6 | |
| DNA recombination, replication, and repair | 1F10 | SEN1152 | ϕSE12 bacteriophage tail fiber protein (pseudogene) | 2.91 ± 0.23 | 133 |
| 2F10 | SEN1393 | ϕSE14 bacteriophage protein | 3.35 ± 0.28 | 97 | |
| 1B3 | SEN1966 | ϕSE20 bacteriophage integrase protein (ϕST64B) | 3.63 ± 0.40 | 66 | |
| Translation | 2E7 | ksgA | Dimethyladenosine transferase | 4.12 ± 0.44 | 73 |
| Virulence plasmid | 1A9 | spvR | Plasmid virulence regulator protein | 3.64 ± 0.69 | 22 |
| Unknown | 2A5 | SEN0034 | Hypothetical protein | 3.21 ± 0.47 | 48 |
| 1E1 | SEN2278 | Hypothetical protein (ais protein) | 3.71 ± 0.30 | 45 | |
| 2D6 | SEN3503 | Hypothetical protein (putative sugar kinase) | 2.70 ± 0.46 | 56 | |
| Intergenic region | 2A6 | SEN1154-SopE | Unknown | 3.41 ± 0.64 | NT |
| 2B8 | phsA-sopA | Unknown | 3.51 ± 0.18 | NT | |
| 1F4 | proP-basS | Unknown | 2.83 ± 0.33 | NT |
Number of internalized bacteria per well of S. Enteritidis-inoculated Caco-2 cell monolayers treated with gentamicin for 2 h. The results represent the mean log10 CFU ± the standard error (SE) from at least five independent assays. The cell invasiveness (mean log10 CFU ± SE) of all the mutants was significantly lower (P <0.05) than that of the wild-type parent G1 Nalr (5.71 ± 0.2).
NT, not tested.
Fig 1.
Growth curve of S. Enteritidis mutants in LB medium 16 h postincubation at 37°C. The error bars indicate standard errors of the means.
Identification of the gene of insertion.
Rescue cloning and sequencing of the DNA flanking the transposon insertion site from each invasion-attenuated mutant identified 33 unique loci that were disrupted due to insertion of the transposon element (Table 2). Mapping of the genomic organization of insertion points in these 33 mutants with the S. Enteritidis P125109 genome (93) revealed that 16 mutants had transposon insertions in genes clustered in the Salmonella pathogenicity islands (SPIs), including SPI-1 (n = 12), SPI-4 (n = 1), SPI-5 (n = 1), SPI-14 (n = 1), and CS54 (n = 1). Additionally, three mutants had insertions in genes encoding bacteriophage proteins; six mutants had insertions in genes encoding flagellar (n = 2), fimbrial (n = 2), and LPS biosynthesis (n = 2) proteins; and one mutant each had insertions in genes that contribute to DNA methylation and regulation of plasmid virulence, respectively. In addition, three mutants had insertions in genes encoding hypothetical proteins. Interestingly, three invasion-attenuated mutants had insertions in the interspacer regions. While we report the invasion attenuation of these three mutants, we excluded the mutants from further characterization.
Pathogenicity island genes.
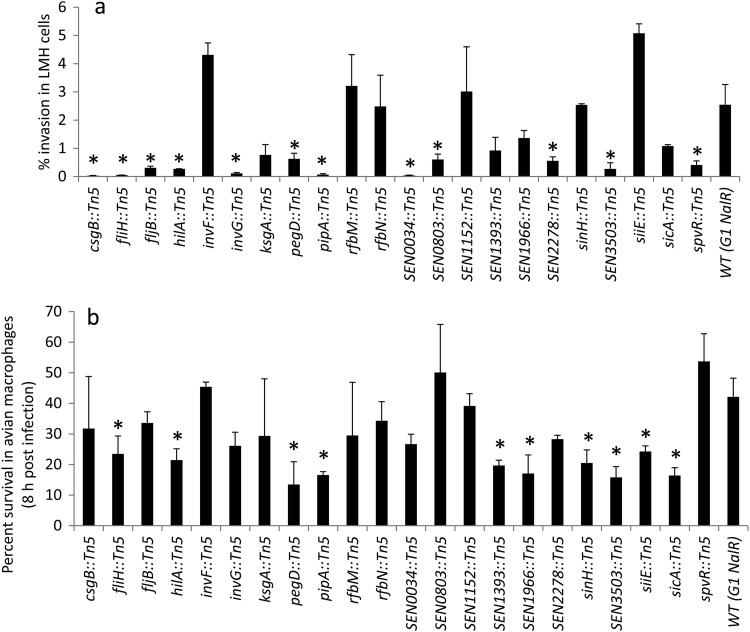
SPIs encode virulence factors and other proteins important for pathogenesis, persistence in the host, and host specificity (74). SPI-1 encodes a type 3 secretion system (T3SS) that is required for invasion in intestinal epithelial cells, and its functional expression is of central importance for translocation of the effector proteins and induction of intestinal responses typical for initiation of enteropathogenesis caused by Salmonella (27, 57, 58). Seven out of 12 SPI-1 mutants identified in this study had insertions in genes involved in the assembly of the SPI-1 T3SS needle complex (NC) that spans bacterial periplasmic space and inserts into the host epithelial cell membrane (49). The genes disrupted in these mutants encode components of the export apparatus that form the base (spaS, spaP, and spaO), inner ring (prgH), and outer ring (invG) of the NC (49, 55, 78, 99) and the invI and orgA genes that are essential for T3SS protein secretion or export (14, 42, 50, 54, 68, 90). Two mutants had insertions in hilA and invF genes that encode SPI-1 T3SS regulators. HilA is a central regulator of the SPI-1 T3SS and activates expression of the prg-org and inv-spa operons, whereas invF is the first gene in the inv-spa operon and is involved in induction of expression of effector proteins both inside and outside the SPI-1 T3SS (24). Two mutants had insertions in genes encoding SPI-1 T3SS chaperones, SicA and InvB, which are essential for efficient export of effector proteins encoded on the SPI-1 T3SS and elsewhere on the chromosome (8, 21, 22, 95). Finally, one transposon disrupted sipC, which encodes a multifunctional protein that is not only known to have T3SS effector translocation activities, but is also involved in actin nucleation and F-actin bundling (65). Because the role of SPI-1 T3SS in cell invasion is well established, the identification of several SPI-1 mutants in this study confirms that our screening assay sensitively detected meaningful phenotypes. Interestingly, it was recently reported that, in addition to intestinal colonization, the SPI-1 T3SS is also required for systemic colonization of S. Enteritidis in the spleen and liver in the chicken host (17, 20, 44). Therefore, we tested several SPI-1 mutants (hilA::Tn5, invF::Tn5, invG::Tn5, and sicA::Tn5) for the ability to invade chicken liver cells and survive within chicken macrophages (Fig. 2). The hilA::Tn5 mutant showed significantly reduced invasiveness in chicken liver cells and reduced survival within chicken macrophages (Fig. 2a). The invG::Tn5 mutant showed significantly reduced invasiveness in chicken liver cells, whereas the sicA::Tn5 mutant showed significantly reduced survival in chicken macrophages (Fig. 2). In contrast, the invF::Tn5 mutant was not attenuated in either model of chicken infection. The invasion attenuation of SPI-1 mutants, in particular the hilA::Tn5 mutant, which is a central regulator of SPI-1, suggests that the SPI-1 T3SS also contributes to systemic infection in chickens.
Fig 2.
Percent survival of S. Enteritidis mutants in LMH cells (a) and HD-11 cells (b). The asterisks indicate that the invasiveness of the mutant was significantly (P < 0.05) lower than that of the WT parent. The error bars indicate standard errors of the means.
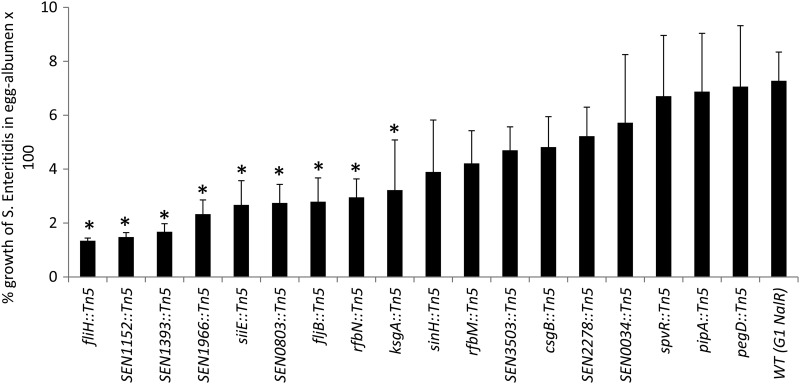
Our screen identified one SPI-4 mutant (siiE::Tn5) with significantly reduced invasiveness in differentiated Caco-2 cells compared with the WT parent. siiE is one of the six genes (siiABCDEF) encoding the SPI-4 type 1 secretion system (T1SS), where siiCDF forms a classical T1SS and siiE acts as a substrate, but the roles of siiAB are less well characterized. Interestingly, siiE encodes the largest protein (595 kDa) of the Salmonella proteome, with 53 immunoglobulin (Ig) domain repeats (30, 31). It has been previously reported that siiE functions as a nonfimbrial adhesin and is required for the binding and invasion of differentiated epithelial cells, but not in nondifferentiated cells (31). Additionally, insertion mutations in SPI-4 genes (siiCDE) in both S. Typhimurium and S. Enteritidis result in virulence attenuation for intraperitoneally inoculated mice (56, 84). Recently, systematic site-directed mutagenesis of SPI-4 genes in S. Typhimurium and S. Enteritidis revealed that all SPI-4 genes are also required for virulence in orally challenged mice (53). In contrast, studies investigating the contribution of SPI-4 in chickens are limited and have mostly observed intestinal colonization over a short time frame. For instance, S. Typhimurium mutants with disrupted SPI-4 genes (siiDEF) or S. Enteritidis mutants lacking the entire SPI-4 were not impaired for intestinal colonization in chickens (64, 73). In the current study, the siiE::Tn5 mutant showed significantly impaired survival in avian macrophages (Fig. 2b) and egg albumen (Fig. 3), suggesting that SPI-4 plays a role in the survival of S. Enteritidis within chicken macrophages and may also facilitate survival within egg albumen.
Fig 3.
Growth of S. Enteritidis mutants after incubation at 25 ± 2°C for 24 h in egg albumen. The asterisks indicate that the percent growth of the mutant was significantly (P < 0.05) lower than that of the WT parent. The error bars indicate standard errors of the means.
SPI-5 is known to play a role in the enteropathogensis of Salmonella (101). It is known that out of six SPI-5 genes (pipABCD and sopBE), sopB and pipBC are coregulated with SPI-1 and facilitate the cell invasion process (15, 71); however, the role of pipA in cell invasion has not been reported previously. In this study, one SPI-5 mutant (pipA::Tn5) was attenuated for invasiveness in differentiated Caco-2 cells and chicken LMH cells and showed impaired survival within chicken macrophages (Fig. 2). There is a great deal of variation in available data regarding the role of SPI-5 in the virulence of different Salmonella serotypes in different hosts. In S. Enteritidis, SPI-5 is required for systemic infection (84), but not for intestinal colonization in mice (45). In contrast, SPI-5 is not required for intestinal colonization in chickens (73), but several SPI-5 genes (pipBCD, orfX, and sopB) are significantly upregulated in the cecal contents of chickens orally inoculated with S. Enteritidis (18). In addition, the absence of SPI-5 also results in a slight reduction in heterophil infiltration in the chicken cecum at 5 days post-oral infection (73), suggesting that SPI-5 may also contribute to intestinal inflammation in chickens. The results of the current study suggest a previously unrecognized role of SPI-5 in systemic infection in chickens. A systematic study to test the functions of individual SPI-5 genes would be required to dissect the exact role of SPI-5 in the pathogenesis of S. Enteritidis infection in chickens.
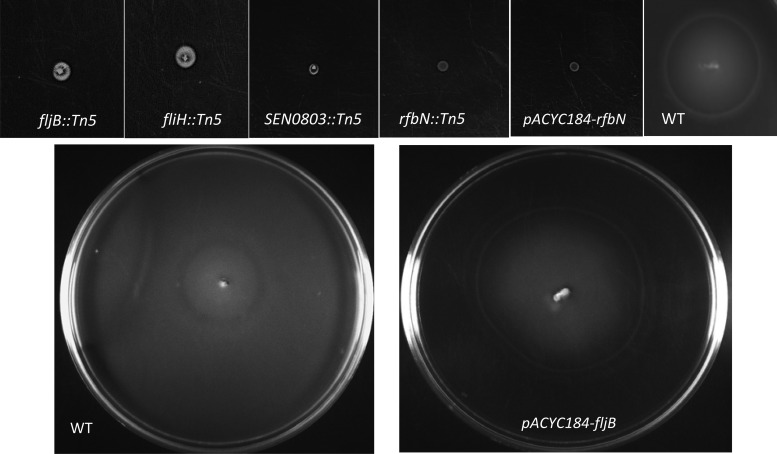
SPI-14 was first reported in Salmonella enterica serovar Gallinarum, a poultry-adapted serotype that is genetically and antigenically closely related to S. Enteritidis (80). Similar to S. Gallinarum, S. Enteritidis SPI-14 contains six open reading frames (ORFs) (SEN0800 to SEN0805). We identified one SPI-14 mutant with an insertion in SEN0803, which encodes a putative acyl-coenzyme A (Co-A) dehydrogenase that participates in alpha and beta dehydrogenation of acyl-CoA esters in both fatty acid and amino acid metabolism (92). In S. Typhimurium, a fatty acyl-CoA dehydrogenase is important for survival under conditions of carbon starvation (86), whereas in the plant pathogen Burkholderia cenocepacia, this enzyme is required for rough-colony morphology, biofilm production, and virulence (89). In our study, SEN0803::Tn5 showed significantly reduced invasiveness in differentiated Caco-2 cells (Table 2). In addition, SEN0803::Tn5 was impaired in motility (Table 1 and Fig. 4), suggesting that SEN0803 is not only involved in cell invasion, but is also associated with motility. Moreover, SEN0803::Tn5 was significantly impaired in its invasiveness in chicken liver cells (Fig. 2a), and its survival in egg albumen was significantly lower than that of the WT (Fig. 3); however, its survival within avian macrophages was not impaired (Fig. 2b). These results suggest that, similar to S. Gallinarum (80), SPI-14 may be required for S. Enteritidis systemic infection in chickens. This work raises the possibility that SPI-14 may also contribute to egg contamination. Systematic site-directed mutagenesis of each gene from SPI-14 may be required to further elucidate the role of SPI-14.
Fig 4.
Motility of S. Enteritidis mutants 7 h (top row) and 24 h (bottom row) postincubation at 37°C.
One of the mutants identified in our screen had a transposon insertion in sinH located on the CS54 island. Previous studies have shown that CS54 island genes, including sinH, are required for intestinal colonization and persistence of S. Typhimurium in mice (52). In this study, sinH::Tn5 showed significantly reduced invasiveness in Caco-2 cells (Table 2) and significantly reduced survival in chicken macrophages (Fig. 2b) but invaded chicken liver cells as well as the WT parent (Fig. 2a). Interestingly, sinH has been identified as a pseudogene due to a deletion mutation that results in a frameshift following codon 37 in a sequenced S. Enteritidis strain, P125109 (93). Therefore, we sequenced the entire ORF of sinH from our WT parent strain (G1 Nalr) and from strain P125109 (data not shown). Sequence analysis revealed that the sinH sequences from both strains were identical to the published sequence of P125109, consistent with the conclusion that sinH is a pseudogene. Currently, the mechanism underlying invasion attenuation in sinH::Tn5 is not understood, but it is possible that the transposon insertion in sinH (158 bp downstream of the frameshift mutation) may have led to polar effects on the other CS54 genes. Because pseudogenes are also known to be expressed in several strains of Salmonella (36, 69), further studies may be required to determine if the sinH gene is functionally expressed in S. Enteritidis.
Flagellar genes.
We identified two mutants with transposon insertions in flagellar genes. They included fljB, encoding a phase type 1/H1 flagellar filament protein flagellin, and fliH, a regulatory component of flagellar T3SS export assembly that prevents fliI (encoding an ATPase) from wasting ATP when fliI is not engaged in flagellar-protein export (3, 60). Both fljB::Tn5 and fliH::Tn5 mutants were strongly attenuated in invasiveness in differentiated Caco-2 cells compared with the WT parent (Table 2). As expected, both mutants also showed a nonmotile phenotype (Fig. 4). We were able to partially restore the invasion defects in fljB::Tn5 by trans-complementation of the WT fljB (Fig. 5); however, motility was completely restored after 24 h of incubation at 37°C (Fig. 4). These results are consistent with published reports showing that motility contributes to invasion in cultured epithelial cells (19, 23, 77, 83, 96). Two reports showed that the invasion defect in nonmotile Salmonella can be reversed by adding a gentle centrifugation step to facilitate intimate contact between bacteria and cells (25, 43). Note that fljB::Tn5 and fliH::Tn5 mutants exhibited invasion defects despite the fact that we applied a mild centrifugal force in all our experiments, and consequently, both fljB and fliH appear to be required for full invasion potential of S. Enteritidis. In addition to the invasion defects in Caco-2 cells, fljB::Tn5 and fliH::Tn5 mutants showed significantly reduced invasiveness in chicken liver cells (Fig. 2a). Moreover, both mutants showed significantly reduced survival in egg albumen (Fig. 3), whereas fliH::Tn5 also showed significantly reduced survival in chicken macrophages (Fig. 2b). These results corroborate our previous report that naturally occurring motility-impaired S. Enteritidis strains show reduced invasiveness in differentiated Caco-2 cells and impaired survival within avian macrophages and egg albumen. It has also been reported that motility-impaired S. Enteritidis is virulence attenuated in chickens orally inoculated with S. Enteritidis (2, 66, 79). The invasion defects of fljB::Tn5 and fliH::Tn5 mutants in chicken liver cells and impaired survival in chicken macrophages and egg albumen suggest that these genes not only contribute to systemic infection in chickens, but also may be involved in egg contamination.
Fig 5.
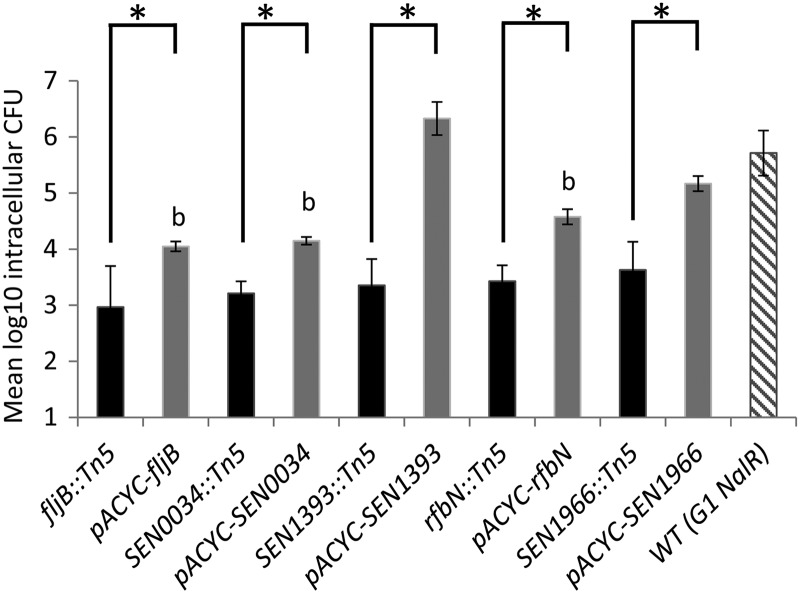
Invasion of differentiated Caco-2 cells by S. Enteritidis mutants. Functional complementation was achieved by in trans expression of full-length genes of pACYC184. The data are plotted as mean log10 intracellular CFU of viable bacteria surviving gentamicin treatment as described in Materials and Methods. The asterisks indicate that differences between the invasiveness of the mutant and that of complemented strains were statistically significant (P < 0.05). The letter b indicates that differences between the invasiveness of the complemented strains and that of the wild-type parent were statistically significant (P < 0.05). The results represent the means of at least four independent assays ± standard errors of the means.
LPS O-antigen biosynthesis genes.
The O-antigen component of the LPS encoded by the rfb gene cluster is considered an important virulence factor (102). Transposon mutagenesis studies have shown that mutations in O-antigen genes, including rfbN, attenuate the ability of S. Typhimurium to colonize chicken, calf, and pig intestine (9, 64). In addition, the rfbN mutant was negatively selected in a large-scale screening of S. Typhimurium genes required for long-term systemic infection in intraperitoneally inoculated mice (56). Similarly, an rfbM deletion in Salmonella enterica serovar Dublin leads to virulence attenuation in mice following oral inoculation (93). While these studies strongly suggest the role of O-antigen in virulence, its role in invasion of epithelial cells is controversial and appears to be dependent on the serotype. For instance, O-antigen mutants of Salmonella enterica serovars Typhi and Typhimurium are not defective for entry into epithelial cells (38, 41, 48, 59); however, mutants of Salmonella enterica serovars Choleraesuis and Enteritidis were defective for entry into epithelial cells (25, 88). We show that rfbN::Tn5 and rfbM::Tn5 were significantly attenuated in cell invasiveness in differentiated Caco-2 cells (Table 2) and that the invasion defect of the rfbN::Tn5 mutant could be recovered by trans-complementation of rfbN (Fig. 5). It has also been reported that O-antigen mutants of serovars Typhimurium and Dublin show impaired swarming motility (93, 94). The rfbN::Tn5 mutant was nonmotile (6% motility), whereas the motility of the rfbM::Tn5 mutant was substantially lower (56% motility) than that of the WT (Table 2). Trans-complementation of WT rfbN in the rfbN::Tn5 mutant partially rescued the invasion defects in Caco-2 cells (Fig. 5) but did not restore motility (Fig. 4). Nevertheless, we were able to rescue the motility defects in both the rfbN::Tn5 and rfbM::Tn5 mutants with the addition of surfactin (data not shown). This procedure is known to support swarming of the O-antigen mutants by lowering the surface tension and improving “wettability” (94). These data corroborate an earlier report that rfbN and rfbM genes contribute to swarming motility (94) and also reveal a previously unrecognized role of these genes in cell invasiveness. While O antigen is also considered a sensitive barometer of the ability of S. Enteritidis to survive environmental stress, invade organs, and efficiently contaminate eggs in chickens (33, 34, 40, 47, 61, 70), in serotypes S. Typhimurium and S. Dublin, the O antigen is not required for survival within murine macrophages (26, 93). Consistent with these reports, rfbN::Tn5 and rfbM::Tn5 mutants tested in this study did not show survival defects within chicken macrophages and were also not attenuated for invasion in chicken liver cells.
Fimbrial genes.
Fimbriae are proteinaceous, filamentous appendices on bacterial surfaces that enable the bacterial cell to make contact with eukaryotic or prokaryotic cells and inanimate surfaces. Adherence precedes colonization of surfaces and invasion of eukaryotic cells by the bacteria. Interestingly, a total of 13 fimbrial clusters have been identified in the S. Enteritidis genome (93). According to our screen, two mutants (csgB::Tn5 and pegD::Tn5) showed significantly reduced invasiveness in differentiated Caco-2 cells. The fimbrial operon csg contains at least six divergently transcribed genes (csgBA and csgDEFG) that are conserved in all Salmonella serovars and encode curli or thin aggregative fimbriae (4). For both S. Enteritidis and S. Typhimurium, the curli are needed for invasion of intestinal epithelial cells (19, 91). Deletion of csgB and csgD (a positive transcriptional regulator of csgBA) in S. Typhimurium results in virulence attenuation in mice (56, 98) and in a colonization defect in orally inoculated chickens, but not in cattle (64). The csgB::Tn5 mutant identified in this study did not exhibit survival defects in chicken macrophages but showed significantly reduced invasiveness in chicken liver cells (Fig. 2), suggesting that csgB may be needed for systemic infection in chickens. Curli also contribute to egg contamination by facilitating the attachment of S. Enteritidis to the vitelline membrane of the egg yolk (13). While the csgB::Tn5 mutant identified in this study showed reduced growth in egg albumen compared with the WT, the difference was not statistically significant.
In contrast to the csg operon, the peg fimbrial operon is restricted to S. enterica serovars Enteritidis, Gallinarum, and Paratyphi B (93). The peg operon consists of four genes (pegABCD), in which pegD encodes a putative export protein, pegA encodes a fimbrial subunit, and pegB and pegC encode chaperone and membrane usher proteins required for fimbrial biosynthesis (39). The invasion attenuation of the pegD::Tn5 mutant in differentiated Caco-2 cells reported here is the first in vitro role assigned to this gene. Earlier reports indicated that peg fimbriae contribute to cecal colonization in chickens and also facilitate oviduct colonization and egg contamination (12, 29). Moreover, Silva et al. (84) recently showed that pegABCD are required for the systemic phase of infection in mice, but the role of the peg operon in systemic infection in chickens has not been reported. We found that the pegD::Tn5 mutant was significantly reduced in invasiveness in chicken LMH cells (Fig. 2a). In addition, pegD::Tn5 showed significantly impaired ability to survive within chicken macrophages (Fig. 2b), and its survival in egg albumen was also lower than that of the WT parent (Fig. 3). Taken together, these data suggest that csgB and pegD not only contribute to invasion in differentiated Caco-2 cells, but may be needed for systemic infection in chickens and for the survival of S. Enteritidis within egg albumen. Further studies with defined, nonpolar mutants and trans-complemented strains will be required to confirm the roles of csg and peg operons in virulence in chickens.
Genes involved in DNA recombination, replication, and repair (phage-related genes).
Three mutants with insertions in genes SEN1152, SEN1393, and SEN1966 were significantly attenuated in invasiveness in differentiated Caco-2 cells. These genes are located in ϕ-related loci identified as “regions of difference (ROD)” between S. Enteritidis, S. Gallinarum, and S. Typhimurium (93). SEN1152 is located on ϕSE12, which represents a remnant of a gifsy-2 prophage in S. Enteritidis and carries the genes encoding the T3SS effector protein SopE and several other proteins (PipA, OmpX, and SodCI) previously implicated in Salmonella pathogenicity. Interestingly, according to the published genome sequence of S. Enteritidis strain P125109 (93), the SEN1152 coding DNA sequence has suffered a deletion in the N terminus and a frameshift mutation following codon 789, and therefore, it is considered a pseudogene. The sequence of SEN1152 in our WT parent (G1 Nalr) is identical to the published sequence, suggesting that SEN1152 is a pseudogene. Note that we also identified a second invasion-attenuated mutant (2A6) that had insertions in ϕSE12 located in the intergenic region (IR) between SEN1154 and sopE. The fact that insertion in the IR of ϕSE12 and an insertion in pseudogene SEN1152 (at 1,287 bp upstream of the frameshift mutation) impacted the invasiveness of S. Enteritidis suggests that ϕSE12 contributes to the virulence of S. Enteritidis. The second prophage gene, SEN1393, is located on the prophage ϕSE14 (SEN1378 to SEN1398), which also carries the S. Enteritidis-specific locus lyg and corresponds to an unstable genetic element that is not required for the systemic colonization of BALB/c mice (1, 76, 84). The third prophage-related gene, SEN1966, is located on SE20 (SEN1919A to SEN1966), which also carries fragments of sopE and orgA previously implicated in Salmonella pathogenicity. Interestingly, Silva et al. (84) recently reported that ϕSE20 also contributes to S. Enteritidis virulence in mice. In this study, we were able to restore the invasiveness of SEN1393::Tn5 and SEN1966::Tn5 by trans-complementation of SEN1393 and SEN1966, respectively, suggesting that these genes are required for invasion of S. Enteritidis in differentiated Caco-2 cells. Our results also show that SEN1393::Tn5 and SEN1966::Tn5 were significantly attenuated in survival within chicken macrophages (Fig. 2b). In addition, the invasiveness of these mutants in chicken liver cells was also slightly reduced (Fig. 2a). These results suggest that both SEN1393 and SEN1966 may be needed for systemic infection in chickens. Interestingly, all three mutants showed significantly reduced growth in egg albumen compared with the WT (Fig. 3). Collectively, these data provide the first evidence for the roles of SEN1152, SEN1393, and SEN1966 in Caco-2 cell invasion and also raise the possibility that these genes may contribute to egg contamination.
Hypothetical proteins.
We found three mutants (SEN0034::Tn5, SEN2278::Tn5, and SEN3503::Tn5) with insertions in genes encoding proteins with unknown functions. In addition to the invasion defects in differentiated Caco-2 cells, these mutants showed significantly lower invasiveness in chicken liver cells (Fig. 2a). SEN3503::Tn5 also showed reduced survival within chicken macrophages (Fig. 2b). In addition, all three mutants showed reduced growth in egg albumen compared with the WT, but these differences were significant only in the case of SEN0034::Tn5 (Fig. 3). SEN0034 encodes a putative hypothetical protein belonging to a family of nucleoside-specific channel-forming outer membrane proteins (Tsx) that are known to facilitate uptake of nucleosides. In addition, the gene upstream of SEN0034 encodes a putative secreted 5′-nucleotidase that is required for hydrolysis of extracellular nucleoside 5′-monophosphates (75), suggesting the possibility that SEN0034 is involved in nucleotide metabolism. While the underlying mechanism of SEN0034-mediated invasion of S. Enteritidis into cultured epithelial cells is not understood, we were able to partially restore the invasion defect of SEN0034::Tn5 by trans-complementation of SEN0034 (Fig. 5), indicating that SEN0034 indeed plays a role in cell invasion. SEN2278 is located at the 5′ end of the pmrF locus in S. Enteritidis and is homologous to ais (Al-inducible gene) in E. coli and pmrG (previously known as pagH) in S. Typhimurium. It has been shown that ais-pmrG expression is controlled by a PmrA-PmrB two-component regulatory system (35) whose products modify LPS and therefore are essential for resistance to polymixin B and other antimicrobial peptides (35, 85). The MIC of polymyxin B against SEN2278::Tn5 was lower (2 μg/ml) than that of the WT parent (4 μg/ml), consistent with SEN2278 contributing to resistance against polymyxin. Finally, the SEN3503 gene encodes a putative transcriptional regulator belonging to the FGGY kinases of proteins that are known to play an important role in the evolutionary diversity and adaptability of bacterial carbohydrate utilization machinery (103). While E. coli and B. subtilis genomes each contain six FGGY kinases with distinct specificities involved in catabolic pathways of different carbohydrates, the biological functions and biochemical substrate preferences of SEN3503 have not been experimentally characterized for S. Enteritidis. Overall, the above data suggest that SEN0034, SEN2278, and SEN3503 are required for invasion of cultured human intestinal epithelial cells, but the invasion defects observed in chicken liver cells, along with reduced survival in chicken macrophages and egg albumen, raise the possibility that these genes may also contribute to systemic infection in chickens and that they are possibly involved in egg contamination.
Miscellaneous genes.
One mutant with significantly reduced invasiveness in Caco-2 cells had a transposon insertion in the ksgA gene, which encodes a ribosomal protein methyltransferase. The ksgA::Tn5 mutant showed slightly reduced invasiveness in chicken liver cells, but its survival within chicken macrophages was similar to that of the WT parent (Fig. 2). Interestingly, the ksgA::Tn5 mutant showed significantly reduced survival in egg albumen (Fig. 3). Previous studies have shown that the deficiency of methyltransferase due to mutation in ksgA results in increased resistance to the antibiotic kasugamycin (6, 16) and also increases translational errors during both the initiation and elongation phases of protein synthesis (97). In this study, the MIC of kasugamycin for the ksgA::Tn5 mutant (500 μg/ml) was significantly higher than the MIC for the WT parent (150 μg/ml), suggesting that the function of ksgA was impacted due to the transposon insertion. While one study has shown that mutation in ksgA results in virulence attenuation of Yersinia pseudotuberculosis in mice (62), our study provides the first evidence for the role of ksgA in intestinal cell invasion and survival in egg albumen. Our screen also identified one mutant with a transposon insertion in a large virulence plasmid-borne spvR gene, a positive transcriptional regulator of the spv operon (spvABCD). The spvR::Tn5 mutant showed significantly reduced invasiveness in human intestinal cells and in chicken liver cells, but its survival within chicken macrophages and egg albumen was not affected. It is now well established that spvR is required for the full expression of virulence of Salmonella in both mice and chickens (72). The identification of spvR further confirms that our screen sensitively identified mutants with attenuated invasiveness.
Finally, three mutants with insertions in IRs showed significantly reduced invasiveness in Caco-2 cells. Southern blot analysis revealed that these mutants did not carry additional insertions elsewhere in the genome (data not shown). The transposon insertion in one mutant (2B8) was located 447 bp downstream of phsA (encoding thiosulfate reductase), a gene that contributes to anaerobic energy metabolism (37), and 174 bp upstream of sopA, which encodes a known SPI-1 T3SS-secreted effector protein essential for bacterial invasion of epithelial cells (87). In the second mutant, the transposon insertion was located 41 bp downstream of proP, encoding a proline/glycine betain transporter, and 125 bp downstream of basSR, encoding the two-component regulatory system. The third mutant had insertions in the IR between SEN1154 (pseudogene) and sopE, enciding a SPI-1 T3SS effector protein located on ϕSE12 that represents a remnant of a gifsy-2 prophage. In this case, the insertion was located 106 bp downstream of sopE and 96 bp downstream of SEN1154 (a pseudogene). Because the insertions in these mutants occurred in the downstream regions of the flanking genes, invasion defects are not likely due to the interruption within the promoter regions. In addition, we searched for possible noncoding RNA (ncRNA) in the Rfam database (http://rfam.sanger.ac.uk/search), but none of the IRs matched the existing ncRNAs in the database. While the mechanism underlying the observed invasion-defective phenotypes in these mutants currently remains unclear, it is likely that the transposon insertion in the IR may cause pleiotropic effects on the genes upstream or downstream, resulting in invasion attenuation. A global transcriptomic or proteomic analysis of these mutants may be required to unravel the underlying molecular mechanisms for their invasion attenuation.
Conclusions.
The design of our study was solely based on in vitro screening of S. Enteritidis transposon insertion mutants for defects in cell invasion using differentiated Caco-2 cells as a surrogate marker for virulence. In the first large-scale screen of this type, we identified several genes or genomic islands that have not been reported previously (e.g., SPI-14, ksgA, SEN0034, SEN2278, and SEN3503) or that are absent in S. Typhimurium or in most other Salmonella serovars (e.g., pegD, SEN1152, SEN1393, and SEN1966). Numerous mutants showing invasion defects in Caco-2 cells also presented invasion defects in LMH chicken liver cells, intracellular survival defects in HD-11 chicken macrophages, and/or impaired survival in egg albumen. The identification of these novel genetic factors will advance our current understanding of the genetic basis of pathogenesis of S. Enteritidis infection in both humans and chickens. Several genes identified in this study are parts of large operons. Thus, it is possible that in some insertional mutants a gene in which insertion occurred was not directly responsible for attenuation, for example, the relevant gene was the next one in the operon. In such cases, further studies involving allelic replacement without polar effect may confirm the gene responsible for attenuation. In addition, in vivo studies involving inoculation of mutants by various parenteral routes and determination of their growth curves in the reticuloendothelial system of chickens at various stages of infection will be required to confirm the specific roles of the S. Enteritidis genes in poultry infection. It would also be important to study the roles of these genes in egg contamination in a laying hen model. These studies may have profound implications for the design of either live attenuated or subunit vaccines for use in chickens.
ACKNOWLEDGMENTS
We are grateful to Roger Levesque and Francois Sanschagrin for their generous gift of the E. coli strains. We gratefully acknowledge the technical assistance of Carol Casavant, Lisa Orfe, Gail Deckert, Salma Al-Adwani, and Tarek Addwebi.
This project was funded in part with federal funds from the NIAID, National Institutes of Health, under contract no. N01-A1-30055 and by the Agricultural Animal Health Program, College of Veterinary Medicine, Washington State University. Hye-Young Kim was supported in part by National Institutes of Health grant RR07049.
Footnotes
Published ahead of print 17 September 2012
REFERENCES
- 1. Agron PG, et al. 2001. Identification by subtractive hybridization of sequences specific for Salmonella enterica serovar enteritidis. Appl. Environ. Microbiol. 67:4984–4991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Allen-Vercoe E, Sayers AR, Woodward MJ. 1999. Virulence of Salmonella enterica serotype Enteritidis aflagellate and afimbriate mutants in a day-old chick model. Epidemiol. Infect. 122:395–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Apel D, Surette MG. 2008. Bringing order to a complex molecular machine: the assembly of the bacterial flagella. Biochim. Biophys. Acta 1778:1851–1858 [DOI] [PubMed] [Google Scholar]
- 4. Barnhart MM, Chapman MR. 2006. Curli biogenesis and function. Annu. Rev. Microbiol. 60:131–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Beug H, von Kirchbach A, Doderlein G, Conscience JF, Graf T. 1979. Chicken hematopoietic cells transformed by seven strains of defective avian leukemia viruses display three distinct phenotypes of differentiation. Cell 18:375–390 [DOI] [PubMed] [Google Scholar]
- 6. Binet R, Maurelli AT. 2009. The chlamydial functional homolog of KsgA confers kasugamycin sensitivity to Chlamydia trachomatis and impacts bacterial fitness. BMC Microbiol. 9:279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bispham J, Tripathi BN, Watson PR, Wallis TS. 2001. Salmonella pathogenicity island 2 influences both systemic salmonellosis and Salmonella-induced enteritis in calves. Infect. Immun. 69:367–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bronstein PA, Miao EA, Miller SI. 2000. InvB is a type III secretion chaperone specific for SspA. J. Bacteriol. 182:6638–6644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Carnell SC, et al. 2007. Role in virulence and protective efficacy in pigs of Salmonella enterica serovar Typhimurium secreted components identified by signature-tagged mutagenesis. Microbiology 153:1940–1952 [DOI] [PubMed] [Google Scholar]
- 10. CDC 2007. Salmonella serotype Enteritidis infections among workers producing poultry vaccine, Maine, November-December 2006. Morb. Mortal Wkly. Rep. 56:877–879 [PubMed] [Google Scholar]
- 11. Clavijo RI, Loui C, Andersen GL, Riley LW, Lu S. 2006. Identification of genes associated with survival of Salmonella enterica serovar Enteritidis in chicken egg albumen. Appl. Environ. Microbiol. 72:1055–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Clayton DJ, et al. 2008. Analysis of the role of 13 major fimbrial subunits in colonisation of the chicken intestines by Salmonella enterica serovar Enteritidis reveals a role for a novel locus. BMC Microbiol. 8:228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cogan TA, et al. 2004. Flagella and curli fimbriae are important for the growth of Salmonella enterica serovars in hen eggs. Microbiology 150:1063–1071 [DOI] [PubMed] [Google Scholar]
- 14. Collazo CM, Zierler MK, Galan JE. 1995. Functional analysis of the Salmonella typhimurium invasion genes invl and invJ and identification of a target of the protein secretion apparatus encoded in the inv locus. Mol. Microbiol. 15:25–38 [DOI] [PubMed] [Google Scholar]
- 15. Darwin KH, Robinson LS, Miller VL. 2001. SigE is a chaperone for the Salmonella enterica serovar Typhimurium invasion protein SigD. J. Bacteriol. 183:1452–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Demirci H, et al. 2010. Modification of 16S ribosomal RNA by the KsgA methyltransferase restructures the 30S subunit to optimize ribosome function. RNA 16:2319–2324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Desin TS, et al. 2009. Salmonella enterica serovar Enteritidis pathogenicity island 1 is not essential for but facilitates rapid systemic spread in chickens. Infect. Immun. 77:2866–2875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dhawi AA, et al. 2011. Adaptation to the chicken intestine in Salmonella Enteritidis PT4 studied by transcriptional analysis. Vet. Microbiol. 153:198–204 [DOI] [PubMed] [Google Scholar]
- 19. Dibb-Fuller MP, Allen-Vercoe E, Thorns CJ, Woodward MJ. 1999. Fimbriae- and flagella-mediated association with and invasion of cultured epithelial cells by Salmonella enteritidis. Microbiology 145:1023–1031 [DOI] [PubMed] [Google Scholar]
- 20. Dieye Y, Ameiss K, Mellata M, Curtiss R., III 2009. The Salmonella Pathogenicity Island (SPI) 1 contributes more than SPI2 to the colonization of the chicken by Salmonella enterica serovar Typhimurium. BMC Microbiol. 9:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dieye Y, Dyszel JL, Kader R, Ahmer BM. 2007. Systematic analysis of the regulation of type three secreted effectors in Salmonella enterica serovar Typhimurium. BMC Microbiol. 7:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ehrbar K, Friebel A, Miller SI, Hardt WD. 2003. Role of the Salmonella pathogenicity island 1 (SPI-1) protein InvB in type III secretion of SopE and SopE2, two Salmonella effector proteins encoded outside of SPI-1. J. Bacteriol. 185:6950–6967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Eichelberg K, Galan JE. 2000. The flagellar sigma factor FliA (sigma(28)) regulates the expression of Salmonella genes associated with the centisome 63 type III secretion system. Infect. Immun. 68:2735–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ellermeier JR, Slauch JM. 2007. Adaptation to the host environment: regulation of the SPI1 type III secretion system in Salmonella enterica serovar Typhimurium. Curr. Opin. Microbiol. 10:24–29 [DOI] [PubMed] [Google Scholar]
- 25. Finlay BB, Falkow S. 1989. Salmonella as an intracellular parasite. Mol. Microbiol. 3:1833–1841 [DOI] [PubMed] [Google Scholar]
- 26. Forbes SJ, Eschmann M, Mantis NJ. 2008. Inhibition of Salmonella enterica serovar Typhimurium motility and entry into epithelial cells by a protective antilipopolysaccharide monoclonal immunoglobulin A antibody. Infect. Immun. 76:4137–4144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Galan JE. 1996. Molecular genetic bases of Salmonella entry into host cells. Mol. Microbiol. 20:263–271 [DOI] [PubMed] [Google Scholar]
- 28. Gantois I, et al. 2009. Mechanisms of egg contamination by Salmonella enteritidis. FEMS Microbiol. Rev. 33:718–738 [DOI] [PubMed] [Google Scholar]
- 29. Gantois I, Ducatelle R, Pasmans F, Haesebrouck F, Van Immerseel F. 2008. Salmonella enterica serovar Enteritidis genes induced during oviduct colonization and egg contamination in laying hens. Appl. Environ. Microbiol. 74:6616–6622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gerlach RG, Jackel D, Geymeier N, Hensel M. 2007. Salmonella pathogenicity island 4-mediated adhesion is coregulated with invasion genes in Salmonella enterica. Infect. Immun. 75:4697–4709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gerlach RG, et al. 2007. Salmonella Pathogenicity Island 4 encodes a giant non-fimbrial adhesin and the cognate type 1 secretion system. Cell Microbiol. 9:1834–1850 [DOI] [PubMed] [Google Scholar]
- 32. Guard-Petter J. 2001. The chicken, the egg and Salmonella enteritidis. Environ. Microbiol. 3:421–430 [DOI] [PubMed] [Google Scholar]
- 33. Guard-Petter J, Keller LH, Rahman MM, Carlson RW, Silvers S. 1996. A novel relationship between O-antigen variation, matrix formation, and invasiveness of Salmonella enteritidis. Epidemiol. Infect. 117:219–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Guard-Petter J, Lakshmi B, Carlson R, Ingram K. 1995. Characterization of lipopolysaccharide heterogeneity in Salmonella enteritidis by an improved gel electrophoresis method. Appl. Environ. Microbiol. 61:2845–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gunn JS, et al. 1998. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol. Microbiol. 27:1171–1182 [DOI] [PubMed] [Google Scholar]
- 36. Hautefort I, et al. 2008. During infection of epithelial cells Salmonella enterica serovar Typhimurium undergoes a time-dependent transcriptional adaptation that results in simultaneous expression of three type 3 secretion systems. Cell Microbiol. 10:958–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Heinzinger NK, Fujimoto SY, Clark MA, Moreno MS, Barrett EL. 1995. Sequence analysis of the phs operon in Salmonella typhimurium and the contribution of thiosulfate reduction to anaerobic energy metabolism. J. Bacteriol. 177:2813–2820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hoare A, et al. 2006. The outer core lipopolysaccharide of Salmonella enterica serovar Typhi is required for bacterial entry into epithelial cells. Infect. Immun. 74:1555–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hultgren SJ, Normark S. 1991. Biogenesis of the bacterial pilus. Curr. Opin. Genet. Dev. 1:313–318 [DOI] [PubMed] [Google Scholar]
- 40. Humphrey TJ, et al. 1996. Isolates of Salmonella enterica Enteritidis PT4 with enhanced heat and acid tolerance are more virulent in mice and more invasive in chickens. Epidemiol. Infect. 117:79–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ilg K, et al. 2009. O-antigen-negative Salmonella enterica serovar Typhimurium is attenuated in intestinal colonization but elicits colitis in streptomycin-treated mice. Infect. Immun. 77:2568–2575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jones BD, Falkow S. 1994. Identification and characterization of a Salmonella typhimurium oxygen-regulated gene required for bacterial internalization. Infect. Immun. 62:3745–3752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jones GW, Richardson LA, Uhlman D. 1981. The invasion of HeLa cells by Salmonella typhimurium: reversible and irreversible bacterial attachment and the role of bacterial motility. J. Gen. Microbiol. 127:351–360 [DOI] [PubMed] [Google Scholar]
- 44. Jones MA, Hulme SD, Barrow PA, Wigley P. 2007. The Salmonella pathogenicity island 1 and Salmonella pathogenicity island 2 type III secretion systems play a major role in pathogenesis of systemic disease and gastrointestinal tract colonization of Salmonella enterica serovar Typhimurium in the chicken. Avian Pathol. 36:199–203 [DOI] [PubMed] [Google Scholar]
- 45. Karasova D, et al. 2010. Influence of 5 major Salmonella pathogenicity islands on NK cell depletion in mice infected with Salmonella enterica serovar Enteritidis. BMC Microbiol. 10:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kawaguchi T, Nomura K, Hirayama Y, Kitagawa T. 1987. Establishment and characterization of a chicken hepatocellular carcinoma cell line, LMH. Cancer Res. 47:4460–4464 [PubMed] [Google Scholar]
- 47. Keller LH, Benson CE, Krotec K, Eckroade RJ. 1995. Salmonella enteritidis colonization of the reproductive tract and forming and freshly laid eggs of chickens. Infect. Immun. 63:2443–2449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kihlstrom E, Edebo L. 1976. Association of viable and inactivated Salmonella typhimurium 395 MS and MR 10 with HeLa cells. Infect. Immun. 14:851–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kimbrough TG, Miller SI. 2002. Assembly of the type III secretion needle complex of Salmonella typhimurium. Microbes Infect. 4:75–82 [DOI] [PubMed] [Google Scholar]
- 50. Kimbrough TG, Miller SI. 2000. Contribution of Salmonella typhimurium type III secretion components to needle complex formation. Proc. Natl. Acad. Sci. U. S. A. 97:11008–11013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kimura AC, et al. 2004. Chicken consumption is a newly identified risk factor for sporadic Salmonella enterica serotype Enteritidis infections in the United States: a case-control study in FoodNet sites. Clin. Infect. Dis. 38(Suppl. 3):S244–S252 [DOI] [PubMed] [Google Scholar]
- 52. Kingsley RA, et al. 2003. Molecular and phenotypic analysis of the CS54 island of Salmonella enterica serotype Typhimurium: identification of intestinal colonization and persistence determinants. Infect. Immun. 71:629–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kiss T, Morgan E, Nagy G. 2007. Contribution of SPI-4 genes to the virulence of Salmonella enterica. FEMS Microbiol. Lett. 275:153–159 [DOI] [PubMed] [Google Scholar]
- 54. Klein JR, Fahlen TF, Jones BD. 2000. Transcriptional organization and function of invasion genes within Salmonella enterica serovar Typhimurium pathogenicity island 1, including the prgH, prgI, prgJ, prgK, orgA, orgB, and orgC genes. Infect. Immun. 68:3368–3376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kubori T, et al. 1998. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science 280:602–605 [DOI] [PubMed] [Google Scholar]
- 56. Lawley TD, et al. 2006. Genome-wide screen for Salmonella genes required for long-term systemic infection of the mouse. PLoS Pathog. 2:e11 doi:10.1371/journal.ppat.0020011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lostroh CP, Lee CA. 2001. The Salmonella pathogenicity island-1 type III secretion system. Microbes Infect. 3:1281–1291 [DOI] [PubMed] [Google Scholar]
- 58. Ly KT, Casanova JE. 2007. Mechanisms of Salmonella entry into host cells. Cell Microbiol. 9:2103–2111 [DOI] [PubMed] [Google Scholar]
- 59. Lyczak JB, et al. 2001. Epithelial cell contact-induced alterations in Salmonella enterica serovar Typhi lipopolysaccharide are critical for bacterial internalization. Cell Microbiol. 3:763–772 [DOI] [PubMed] [Google Scholar]
- 60. Macnab RM. 2004. Type III flagellar protein export and flagellar assembly. Biochim. Biophys. Acta 1694:207–217 [DOI] [PubMed] [Google Scholar]
- 61. Martin GD, et al. 2000. Invasiveness of Salmonella serotypes Typhimurium and Enteritidis of human gastro-enteritic origin for rabbit ileum: role of LPS, plasmids and host factors. J. Med. Microbiol. 49:1011–1021 [DOI] [PubMed] [Google Scholar]
- 62. Mecsas J, Bilis I, Falkow S. 2001. Identification of attenuated Yersinia pseudotuberculosis strains and characterization of an orogastric infection in BALB/c mice on day 5 postinfection by signature-tagged mutagenesis. Infect. Immun. 69:2779–2787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Morales CA, et al. 2005. Correlation of phenotype with the genotype of egg-contaminating Salmonella enterica serovar Enteritidis. Appl. Environ. Microbiol. 71:4388–4399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Morgan E, et al. 2004. Identification of host-specific colonization factors of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 54:994–1010 [DOI] [PubMed] [Google Scholar]
- 65. Myeni SK, Zhou D. 2010. The C terminus of SipC binds and bundles F-actin to promote Salmonella invasion. J. Biol. Chem. 285:13357–13363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Parker CT, Guard-Petter J. 2001. Contribution of flagella and invasion proteins to pathogenesis of Salmonella enterica serovar enteritidis in chicks. FEMS Microbiol. Lett. 204:287–291 [DOI] [PubMed] [Google Scholar]
- 67. Patrick ME, et al. 2004. Salmonella enteritidis infections, United States, 1985–1999. Emerg. Infect. Dis. 10:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Pegues DA, Hantman MJ, Behlau I, Miller SI. 1995. PhoP/PhoQ transcriptional repression of Salmonella typhimurium invasion genes: evidence for a role in protein secretion. Mol. Microbiol. 17:169–181 [DOI] [PubMed] [Google Scholar]
- 69. Perkins TT, et al. 2009. A strand-specific RNA-Seq analysis of the transcriptome of the typhoid bacillus Salmonella typhi. PLoS Genet. 5:e1000569 doi:10.1371/journal.pgen.1000569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Petter JG. 1993. Detection of two smooth colony phenotypes in a Salmonella enteritidis isolate which vary in their ability to contaminate eggs. Appl. Environ. Microbiol. 59:2884–2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Rodriguez-Escudero I, Ferrer NL, Rotger R, Cid VJ, Molina M. 2011. Interaction of the Salmonella Typhimurium effector protein SopB with host cell Cdc42 is involved in intracellular replication. Mol. Microbiol. 80:1220–1240 [DOI] [PubMed] [Google Scholar]
- 72. Rychlik I, Gregorova D, Hradecka H. 2006. Distribution and function of plasmids in Salmonella enterica. Vet. Microbiol. 112:1–10 [DOI] [PubMed] [Google Scholar]
- 73. Rychlik I, et al. 2009. Virulence potential of five major pathogenicity islands (SPI-1 to SPI-5) of Salmonella enterica serovar Enteritidis for chickens. BMC Microbiol. 9:268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Sabbagh SC, Forest CG, Lepage C, Leclerc JM, Daigle F. 2010. So similar, yet so different: uncovering distinctive features in the genomes of Salmonella enterica serovars Typhimurium and Typhi. FEMS Microbiol. Lett. 305:1–13 [DOI] [PubMed] [Google Scholar]
- 75. Sakai Y, et al. 1987. Properties of the membrane-bound 5′-nucleotidase and utilization of extracellular ATP in Vibrio parahaemolyticus. J. Gen. Microbiol. 133:2751–2757 [DOI] [PubMed] [Google Scholar]
- 76. Santiviago CA, et al. 2010. Spontaneous excision of the Salmonella enterica serovar Enteritidis-specific defective prophage-like element phiSE14. J. Bacteriol. 192:2246–2254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Schmitt CK, et al. 2001. Absence of all components of the flagellar export and synthesis machinery differentially alters virulence of Salmonella enterica serovar Typhimurium in models of typhoid fever, survival in macrophages, tissue culture invasiveness, and calf enterocolitis. Infect. Immun. 69:5619–5625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Schraidt O, et al. 2010. Topology and organization of the Salmonella typhimurium type III secretion needle complex components. PLoS Pathog. 6:e1000824 doi:10.1371/journal.ppat.1000824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Shah DH, et al. 2012. Salmonella Enteritidis strains from poultry exhibit differential responses to acid stress, oxidative stress, and survival in the egg albumen. Foodborne Pathog. Dis. 9:258–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Shah DH, et al. 2005. Identification of Salmonella gallinarum virulence genes in a chicken infection model using PCR-based signature-tagged mutagenesis. Microbiology 151:3957–3968 [DOI] [PubMed] [Google Scholar]
- 81. Shah DH, et al. 2005. Control of fowl typhoid using tissue culture medium waste after harvest of Korean wild ginseng (Panax ginseng). J. Appl. Poultry Res. 14:455–462 [Google Scholar]
- 82. Shah DH, Zhou X, Addwebi T, Davis MA, Call DR. 2011. In vitro and in vivo pathogenicity of Salmonella enteritidis clinical strains isolated from North America. Arch. Microbiol. 193:811–821 [DOI] [PubMed] [Google Scholar]
- 83. Shah DH, et al. 2011. Cell invasion of poultry-associated Salmonella enterica serovar Enteritidis isolates is associated with pathogenicity, motility and proteins secreted by the type III secretion system. Microbiology 157:1428–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Silva CA, et al. 2012. Infection of mice by Salmonella enterica serovar Enteritidis involves additional genes that are absent in the genome of serovar Typhimurium. Infect. Immun. 80:839–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Soncini FC, Groisman EA. 1996. Two-component regulatory systems can interact to process multiple environmental signals. J. Bacteriol. 178:6796–6801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Spector MP, et al. 1999. The medium-/long-chain fatty acyl-CoA dehydrogenase (fadF) gene of Salmonella typhimurium is a phase 1 starvation-stress response (SSR) locus. Microbiology 145:15–31 [DOI] [PubMed] [Google Scholar]
- 87. Steele-Mortimer O, et al. 2002. The invasion-associated type III secretion system of Salmonella enterica serovar Typhimurium is necessary for intracellular proliferation and vacuole biogenesis in epithelial cells. Cell Microbiol. 4:43–54 [DOI] [PubMed] [Google Scholar]
- 88. Stone BJ, et al. 1992. Identification of novel loci affecting entry of Salmonella enteritidis into eukaryotic cells. J. Bacteriol. 174:3945–3952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Subramoni S, Nguyen DT, Sokol PA. 2011. Burkholderia cenocepacia ShvR-regulated genes that influence colony morphology, biofilm formation, and virulence. Infect. Immun. 79:2984–2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Sukhan A, Kubori T, Wilson J, Galan JE. 2001. Genetic analysis of assembly of the Salmonella enterica serovar Typhimurium type III secretion-associated needle complex. J. Bacteriol. 183:1159–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Sukupolvi S, et al. 1997. Expression of thin aggregative fimbriae promotes interaction of Salmonella typhimurium SR-11 with mouse small intestinal epithelial cells. Infect. Immun. 65:5320–5325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Swigonova Z, Mohsen AW, Vockley J. 2009. Acyl-CoA dehydrogenases: dynamic history of protein family evolution. J. Mol. Evol. 69:176–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Thomson NR, et al. 2008. Comparative genome analysis of Salmonella Enteritidis PT4 and Salmonella Gallinarum 287/91 provides insights into evolutionary and host adaptation pathways. Genome Res. 18:1624–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Toguchi A, Siano M, Burkart M, Harshey RM. 2000. Genetics of swarming motility in Salmonella enterica serovar Typhimurium: critical role for lipopolysaccharide. J. Bacteriol. 182:6308–6321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Tucker SC, Galan JE. 2000. Complex function for SicA, a Salmonella enterica serovar Typhimurium type III secretion-associated chaperone. J. Bacteriol. 182:2262–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Van Asten FJ, Hendriks HG, Koninkx JF, Van der Zeijst BA, Gaastra W. 2000. Inactivation of the flagellin gene of Salmonella enterica serotype enteritidis strongly reduces invasion into differentiated Caco-2 cells. FEMS Microbiol. Lett. 185:175–179 [DOI] [PubMed] [Google Scholar]
- 97. van Buul CP, Visser W, van Knippenberg PH. 1984. Increased translational fidelity caused by the antibiotic kasugamycin and ribosomal ambiguity in mutants harbouring the ksgA gene. FEBS Lett. 177:119–124 [DOI] [PubMed] [Google Scholar]
- 98. van der Velden AW, Baumler AJ, Tsolis RM, Heffron F. 1998. Multiple fimbrial adhesins are required for full virulence of Salmonella typhimurium in mice. Infect. Immun. 66:2803–2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Wagner S, et al. 2010. Organization and coordinated assembly of the type III secretion export apparatus. Proc. Natl. Acad. Sci. U. S. A. 107:17745–17750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Wiegand I, Hilpert K, Hancock RE. 2008. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 3:163–175 [DOI] [PubMed] [Google Scholar]
- 101. Wood MW, et al. 1998. Identification of a pathogenicity island required for Salmonella enteropathogenicity. Mol. Microbiol. 29:883–891 [DOI] [PubMed] [Google Scholar]
- 102. Xiang SH, Haase AM, Reeves PR. 1993. Variation of the rfb gene clusters in Salmonella enterica. J. Bacteriol. 175:4877–4884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Zhang Y, Zagnitko O, Rodionova I, Osterman A, Godzik A. 2011. The FGGY carbohydrate kinase family: insights into the evolution of functional specificities. PLoS Comput. Biol. 7:e1002318 doi:10.1371/journal.pcbi.1002318 [DOI] [PMC free article] [PubMed] [Google Scholar]