Abstract
Clostridium perfringens type C strains are the only non-type-A isolates that cause human disease. They are responsible for enteritis necroticans, which was termed Darmbrand when occurring in post-World War II Germany. Darmbrand strains were initially classified as type F because of their exceptional heat resistance but later identified as type C strains. Since only limited information exists regarding Darmbrand strains, this study genetically and phenotypically characterized seven 1940s era Darmbrand-associated strains. Results obtained indicated the following. (i) Five of these Darmbrand isolates belong to type C, carry beta-toxin (cpb) and enterotoxin (cpe) genes on large plasmids, and express both beta-toxin and enterotoxin. The other two isolates are cpe-negative type A. (ii) All seven isolates produce highly heat-resistant spores with D100 values (the time that a culture must be kept at 100°C to reduce its viability by 90%) of 7 to 40 min. (iii) All of the isolates surveyed produce the same variant small acid-soluble protein 4 (Ssp4) made by type A food poisoning isolates with a chromosomal cpe gene that also produce extremely heat-resistant spores. (iv) The Darmbrand isolates share a genetic background with type A chromosomal-cpe-bearing isolates. Finally, it was shown that both the cpe and cpb genes can be mobilized in Darmbrand isolates. These results suggest that C. perfringens type A and C strains that cause human food-borne illness share a spore heat resistance mechanism that likely favors their survival in temperature-abused food. They also suggest possible evolutionary relationships between Darmbrand strains and type A strains carrying a chromosomal cpe gene.
INTRODUCTION
Clostridium perfringens, an anaerobic, Gram-positive bacterium, exists ubiquitously in soils, sewage, food, and the normal intestinal flora of humans and animals (22). It is also a very important pathogen because of its ability to produce at least 16 different toxins, although individual isolates never express this entire toxin arsenal. On the basis of the production of four typing toxins (alpha, beta, epsilon, and iota), C. perfringens is classified into five types (A, B, C, D, and E). Each type is associated with different diseases that affect humans or animals (22). In livestock species, type C isolates cause fatal necrotizing enteritis and enterotoxemia (22). Type C strains are also the only non-type-A C. perfringens strains that cause human disease (22), which is referred to as enteritis necroticans, also known as pigbel or Darmbrand. Enteritis necroticans is a fatal disease that involves vomiting, severe abdominal pain, intestinal necrosis, and bloody stool (13, 14). Acute cases can result in rapid death.
By definition, type C isolates must produce alpha-toxin (CPA) and beta-toxin (CPB). CPB, a 35-kDa pore-forming polypeptide encoded by the plasmid-borne cpb gene, is necessary for type C strains to cause either necrotizing enteritis or enterotoxemia in animals (20, 31, 35–37). CPA is a 43-kDa protein with phospholipase C activity and the ability to activate endogenous signaling pathways in host cells (34). Besides CPA and CPB, some type C isolates produce additional toxins, such as the enterotoxin (CPE), which is encoded by a plasmid-borne cpe gene in all of the cpe-positive type C strains examined to date (11, 18). CPE, a 35-kDa pore-forming polypeptide expressed during sporulation, contributes to several important human and veterinary enteric diseases caused by type A strains (21). About 75 to 80% of all cases of C. perfringens type A food poisoning, which is the second most common bacterial food-borne illness in the United States (6, 32), are caused by type A strains carrying a chromosomal cpe gene (8, 9, 21, 26). In contrast, cpe-positive type A strains that cause human non-food-borne gastrointestinal diseases or animal diseases invariably carry the cpe gene on a large plasmid (8, 9, 18, 25, 38).
C. perfringens type A food poisoning isolates with a chromosomal cpe gene typically produce spores that are extremely resistant to standard food hygiene approaches such as heat, cold, or chemical preservatives (16, 29). For example, it was shown that spores made by type A chromosomal cpe-positive isolates exhibit, on average, a 60-fold higher decimal reduction value at 100°C (i.e., the D100 value, which is the time that a culture must be kept at 100°C to reduce its viability by 90%) than the less resistant spores produced by either type A isolates carrying a plasmid-borne cpe gene or non-type-A strains (17, 29). The exceptional heat resistance properties of spores made by most type A chromosomal cpe-positive strains is thought to favor their survival in temperature-abused foods (16, 17, 21, 29).
Alpha/beta-type small acid-soluble proteins (SASPs) protect spores from heat, cold, or sodium nitrite by binding to spore DNA (19, 28, 33). A variant of small, acid-soluble protein 4 (Ssp4) was recently identified as an important contributor to the exceptional resistance properties of the spores made by most type A chromosomal-cpe-carrying isolates (17, 19). Those chromosomal-cpe-carrying strains that produce highly heat-resistant spores make an Ssp4 variant with an Asp substitution at residue 36, instead of the Gly residue present at that position in the Ssp4 variant made by C. perfringens strains that produce more heat-sensitive spores (17, 19).
Darmbrand was a severe human illness that occurred in the malnourished people of northern Germany from 1944 to 1949 (14, 39). The disease was often fatal because of necrotic inflammation of the small intestine, especially in the jejunum. Clostridium welchii, now named C. perfringens, was identified as the causative bacterium. Because C. welchii Darmbrand isolates produced unusually heat-resistant spores that could resist boiling for 1 to 4 h, they were initially classified as C. welchii type F (39). For example, one reported Darmbrand case occurred in a person who ate canned rabbit meat that had been boiled for 2 h (39). However, when the production of CPB by Darmbrand isolates was later detected using beta-antitoxin (14, 39), these type F isolates were reclassified as type C strains.
Darmbrand isolates have been little studied since the 1940s and 1950s, so the present investigation applied modern techniques to characterize several Darmbrand strains genotypically and phenotypically and compare them with other C. perfringens human enteric disease strains.
MATERIALS AND METHODS
Bacteria, media, and reagents.
The C. perfringens isolates examined in this study, including the 1940s Darmbrand strains, are listed and described in Table 1. Carriage of cpb, cpe, pfoA, and cpb2 genes by these isolates was evaluated by PCR as described below. All isolates were stored as cooked meat medium stock cultures at −20°C in our lab. Fluid thioglycolate (FTG) medium (Difco Laboratories) and TGY medium (3% tryptic soy broth [BD], 2% glucose [Fisher Scientific], 1% yeast extract [BD], 0.1% sodium thioglycolate [Sigma-Aldrich]) were used for broth cultures. Brain heart infusion (BHI; Difco Laboratories) agar plates were used for bacterial colony counting, and LB broth (10% tryptone [BD], 5% yeast extract [BD], 5% sodium chloride [Fisher Scientific]) was used to grow Escherichia coli. Modified Duncan-Strong (MDS) medium (7) was used to induce the sporulation of all C. perfringens isolates and CPE production by the cpe-positive strains.
Table 1.
Isolates used in this study
PCR analyses of carriage of cpe, cpb, pfoA, and cpb2 genes.
Template DNA for PCRs was extracted from overnight TGY medium cultures and then purified using the MasterPure Gram-positive DNA purification kit (Epicentre, Madison, WI). Each PCR mixture (20 μl) contained 2 μl of template DNA, 10 μl of Taq complete 2× Master Mix (New England BioLabs), and 1 μl of each primer pair (1 μM final concentration). The primers used in this investigation are listed in Table 2. The amplification conditions used for these PCR analyses were 1 cycle of 94°C for 2 min and 35 cycles of 94°C for 30 s, 55°C for 40 s, and 68°C for 40 s, followed by a final single extension of 8 min at 68°C. The PCR products were electrophoresed on a 1.5% agarose gel and stained with ethidium bromide for visualization.
Table 2.
Primers used in this study
| Primer | Sequence | Reference |
|---|---|---|
| gyrB-F | 5′ ATTGTTGATAACAGTATTGATGAAGC | 10 |
| gyrB-R | 5′ ATTTCCTAATTTAGTTTTAGTTTGC | |
| sigK-F | 5′ CAATACTTATTAGAATTAGTTGGTAG | |
| sigK-R | 5′ CTAGATACATATGATCTTGATATACC | |
| sod-F | 5′ CAAAAAAAGTCCATTAATGTATCCAG | |
| sod-R | 5′ TTATCTATTGTTATAATATTCTTCAC | |
| groEL-F | 5′ TACAAGATTTATTACCATTACTTGAG | |
| groEL-R | 5′ CATTTCTTTTTCTGGAATATCTGC | |
| pgk-F | 5′ GACTTTAACGTTCCATTAAAAGATGG | |
| pgk-R | 5′ CTAATCCCATGAATCCTTCAGCGATG | |
| nadA-F | 5′ ATTAGCACATTATTATCAAATTCCTG | |
| nadA-R | 5′ TTATATGCCTTTAATCTTAAATCCTC | |
| colA-F | 5′ ATTAGAAAGTTTATGTACAATAGGTG | |
| colA-R2 | 5′ AAGACATTCTATTATTTCTATCGTAAGC | |
| plc-F | 5′ AGGAACTCATGCTATGATTGTAACTC | |
| plc-R | 5′ GGATCATTACCCTCTGATACATCGT | |
| Ssp4-F | 5′ ATGAGCAAGACACCATTAAAAAA | 17 |
| Ssp4-R | 5′ TTACTTTTCGTCAACGTGAGG | |
| cpbF | 5′ GCAGGATCCATGAAGAAAAAATTTAT | This study |
| cpbR | 5′ ATACTCGAGCTAAATAGCTGTTACTTT | |
| cpeF | 5′ AAAGGAGATGGTTGGATATTAGG | This study |
| cpeR | 5′ GTCCAAGGGTATGAGTTAGAAG | |
| pfoA-F | 5′ TTTATGAACTTAACAAATGAGGGG | This study |
| pfoA-R | 5′ CTACTCCAAGTGAGTTTTCAAGG | |
| cpb2-F | 5′ AGATTTTAAATATGATCCTAACC | |
| cpb2-R | 5′ CAATACCCTTCACCAAATACTC | |
| F1 | 5′ TCTAGTTACCCTAGAAAGCATTACT | 11 |
| R1 | 5′ GGAAGGTCCTCACTTATCAT | |
| F2 | 5′ ATGATAAGTGAGGACCTTCC | |
| R2 | 5′ GCTCTAAAAAAGAGCTTAAAAGCA | |
| F3 | 5′ CTGCTTTTAAGCTCTTTTTTAGAGC | |
| R3 | 5′ TGGTCATATTTCATGTATAACT | |
| F4 | 5′ AGTTATACATGAAATATGACCA | |
| R4 | 5′ CCTCCTTTTGTATATAGATGATCTG | |
| F5 | 5′ CAGATCATCTATATACAAAAGGAGG | |
| R5 | 5′ CCAGTTAACACCATTCCAATTAAGA | |
| TnF | 5′ ATACATTAACTAACTTAGAACGTAC | 30 |
| BetaR | 5′ GAAAGAAACTGTTATTATCTTAATTG | |
| dcmRseq | 5′ TCACCCAACAAGTAACTATAATG | 18 |
| cpeMR | 5′ TTAGAACAGTCCTTAGGTGATGGA |
PCR analysis and sequencing of the ssp4 gene.
PCR was performed to detect the presence of the ssp4 gene in Darmbrand isolates using purified DNA, prepared as described above, and specific primers listed in Table 2. The PCR conditions were 1 cycle of 94°C for 2 min and 35 cycles of 94°C for 30 s, 52°C for 30 s, and 68°C for 30 s, followed by a final single extension of 8 min at 68°C. The ssp4 PCR products were electrophoresed on a 1.5% agarose gel and stained with ethidium bromide for visualization. For sequencing analyses, the PCR products were cloned into the pCR2.1-TOPO vector (Invitrogen) and then transformed into E. coli DH5α competent cells (Invitrogen), which were then cultured overnight on BHI agar plates containing 50 μg/ml kanamycin at 37°C. Single colonies were subcultured in LB broth overnight at 37°C, and the plasmid was then extracted with a QIAprep Spin Miniprep kit (Qiagen). The PCR products in the plasmid were then sequenced at the University of Pittsburgh Core DNA Sequencing Facility.
Southern blot analyses.
Genomic C. perfringens DNA was purified using the MasterPure Gram-positive DNA purification kit (Epicentre, Madison, WI). Each purified DNA sample (2.5 μg) was digested overnight with XbaI at 37°C according to the manufacturer's (New England BioLabs) instructions. The digested DNA samples were electrophoresed on a 1% agarose gel, and the separated DNA digestion products were then transferred onto positively charged nylon membranes (Roche) for hybridization with either a cpe or a cpb probe. Digoxigenin (DIG)-labeled cpe or cpb probes were prepared using a PCR DIG probe synthesis kit (Roche); the primers used to make the cpb and cpe probes are listed in Table 2. After hybridization with these probes, the Southern blot was developed by using reagents from the DIG DNA labeling and detection kit (Roche) according to the manufacturer's instructions.
Pulsed-field gel electrophoresis (PFGE).
Individual C. perfringens isolates were grown overnight in 10 ml of FTG broth at 37°C. A 0.1-ml aliquot of each overnight culture was then inoculated into 10 ml of TGY medium. After overnight growth at 37°C, the TGY medium cultures were collected to prepare genomic DNA agarose plugs. Bacterial cells were harvested by centrifugation at 9,000 × g, washed three times with TES buffer (1 M Tris, 0.5 M EDTA, 5 M sucrose [pH 8.0]), resuspended in 200 μl of TE buffer, and then embedded in 200 μl of melted 2% certified low-melting-point agarose (Bio-Rad Laboratories). Plugs were prepared and cut into 2- to 3-mm slices. The agarose-embedded bacterial cells were lysed overnight at 37°C in lysis buffer (500 μM EDTA [pH 8.0], 0.5% Sarkosyl, 0.5% lysozyme [Sigma], 0.4% deoxycholic acid) with gentle shaking of the plugs, followed by a 2-day incubation at 55°C in 0.2% proteinase K (Gene Choice)–500 μM EDTA (pH 8.0) buffer. PFGE of these samples was then performed by using a 1% agarose gel in 0.5× TBE buffer at 14°C and the CHEF-DR II system (Bio-Rad Laboratories). The gel running parameters, as described previously (11, 18), were as follows: initial pulse, 1 s; final pulse, 25 s; voltage, 6 V/cm; time, 24 h.
Southern blot analyses of pulsed-field gels.
The DIG-labeled DNA probes used for pulsed-field Southern blot analyses were the same cpe and cpb probes described above for Southern blot assays of regular agarose gels. Southern hybridization of pulsed-field gels was performed as described previously (30). CSPD substrate (Roche) was used for detection of the Southern blot-hybridized bands according to the manufacturer's protocol.
Western blot analyses of CPB and CPE production by Darmbrand isolates.
Isolates were grown overnight in FTG medium at 37°C. An aliquot (100 μl) of each culture was then inoculated into 10 ml of TGY medium (for CPB production) or 10 ml of fresh MDS medium (for CPE production). After overnight incubation at 37°C, cultures were centrifuged at 8,000 × g for 5 min and the same volume of each culture supernatant was mixed with SDS-PAGE loading buffer and boiled for 5 min. The samples were electrophoresed on a 12% polyacrylamide gel containing SDS, and separated proteins were then transferred onto nitrocellulose membranes. The membranes were blocked with Tris-buffered saline–Tween 20 (0.05%, vol/vol) and nonfat dry milk (5%, wt/vol) for 1 h before each membrane was probed with either rabbit anti-CPE polyclonal antiserum (15) or mouse anti-CPB monoclonal antibody (20, 31). Bound antibody was then detected after incubation with a horseradish peroxidase-conjugated, species-specific antibody and addition of SuperSignal West Pico Chemiluminescent Substrate (Pierce).
Measurement of spore heat resistance.
The moist heat resistance of spores made by Darmbrand isolates was determined by calculating their D100 values (17, 29). For this purpose, an aliquot (0.1 ml) of an overnight FTG medium culture of each isolate was inoculated into 10 ml of fresh MDS medium (7) and incubated overnight at 37°C. The overnight culture was then heat shocked for 20 min at 70°C to kill the vegetative cells. After covering the cells and boiling them for 0 to 60 min, each heat-shocked culture was 10-fold serially diluted from 10−2 to 10−7 with sterile water and plated onto BHI agar plates for colony counting. The plates were incubated overnight under anaerobic conditions. Each experiment was separately repeated three times.
MLST analysis.
To investigate the genetic relatedness of the Darmbrand isolates with other C. perfringens strains, multilocus sequencing typing (MLST) analysis of the Darmbrand strains was performed by sequencing eight housekeeping genes. As described previously for other C. perfringens strains (10), these genes included plc (encodes alpha-toxin), colA (encodes collagenase A), sodA (superoxide dismutase gene), groEL (heat shock protein gene), sigK (encodes sigma factor K), pgk and nadA (encode putative metabolism genes), and gyrB (DNA gyrase B gene). The primers used to amplify these eight housekeeping genes are listed in Table 2. Each PCR mixture (50 μl) contained 5 μl of template DNA, 25 μl of Taq complete 2× Master Mix (New England BioLabs), 2.5 μl of each primer pair (1 μM final concentration), and 15 μl of PCR grade water. PCRs for all eight housekeeping gene amplifications were performed under the same conditions: 1 cycle of 94°C for 2 min and 35 cycles of 94°C for 30 s, 55°C for 60 s, and 68°C for 60 s, followed by a final single extension of 10 min at 68°C. The PCR products were electrophoresed on a 1.5% agarose gel and stained with ethidium bromide for visualization. PCR products were purified with a QIAquick PCR purification kit (GIAGEN) and then sequenced at the University of Pittsburgh Core DNA Sequencing Facility. All sequence data were concatenated to produce an in-frame 5,274-bp nucleotide sequence according to the genome arrangement of each gene in strain 13. Sequence information for these eight housekeeping genes in three reference strains, whose whole genomes have been completely sequenced and assembled, was also used in this analysis. Concatenated sequence data were applied to phylogenetic analysis in the ClustalW format using Winxyz software.
Overlapping PCR and long-range PCR analyses to evaluate cpb locus organization in type C Darmbrand isolates.
Long-range and overlapping PCR analyses were performed to evaluate the diversity of the cpb locus among the five Darmbrand isolates identified as type C (see Results). These analyses used the primers listed in Table 2 and template DNA purified using the MasterPure Gram-positive DNA purification kit. Each PCR mixture contained 5 μl of template DNA, 25 μl of Taq 2× Master Mix (New England BioLabs), 1 μl of forward primer, 1 μl of reverse primer (1 μM final concentration), and 20 μl of distilled water. Long-range PCRs were performed with a Techne thermocycler using the following PCR conditions for the amplifications: 95°C for 2 min and 35 cycles of 94°C for 30 s, 54°C for 30 s, and 65°C for 3 min, followed by a final single extension of 10 min at 65°C. Overlapping PCRs were performed under the following amplification conditions: 35 cycles of 94°C for 30 s, 54°C for 30 s, and 68°C for 1 min, followed by a final single extension of 10 min at 68°C. PCR products were separated on a 1.5% agarose gel and visualized by ethidium bromide staining. PCR products were then purified with the QIAquick PCR purification kit (Qiagen) and sequenced at the University of Pittsburgh Core DNA Sequencing Facility.
To study sequences downstream of the cpb gene in Darmbrand strains, plasmid DNA was extracted from fresh overnight TGY medium cultures using the QIAprep Spin Miniprep kit (Qiagen). The extracted plasmids were then used as templates for sequencing downstream of the cpb gene.
PCR identification of possible circular transposition intermediates carrying the cpb gene.
As indicated above, genomic C. perfringens DNA was freshly extracted from BHI agar cultures grown anaerobically overnight. Each PCR mixture contained 5 μl of template DNA, 25 μl of Taq 2× Master Mix (New England BioLabs), 1 μl each of primers TnF and betaR (1 μM final concentration), and 20 μl of distilled water. PCR amplification was then performed with a Techne thermocycler under the following conditions: 95°C for 2 min and 35 cycles of 94°C for 30 s, 54°C for 30 s, and 68°C for 1.5 min, followed by a final single extension of 7 min at 68°C. PCR products were separated on a 1.5% agarose gel and visualized by ethidium bromide staining. PCR products were then excised from the gel using a Quantum Prep Freeze N Squeeze DNA gel extraction spin column (Bio-Rad) and sequenced at the University of Pittsburgh Core DNA Sequencing Facility.
PCR identification of potential cpe gene mobilization.
Each PCR mixture contained 5 μl of template DNA, which was freshly extracted from single colonies growing overnight on anaerobically cultured BHI agar plates, 25 μl of Taq 2× Master Mix (New England BioLabs), 1 μl each of primers dcmRseq and cpeMR (1 μM final concentration), and 20 μl of distilled water. PCR amplification was then performed with a Techne thermocycler under the following conditions: 95°C for 2 min and 35 cycles of 94°C for 30 s, 54°C for 30 s, and 68°C for 2 min, followed by a final single extension of 10 min at 68°C. PCR products were separated on 1.5% agarose gels and visualized by ethidium bromide staining. PCR products were then purified with the QIAquick PCR purification kit (Qiagen) and sequenced at the University of Pittsburgh Core DNA Sequencing Facility.
Statistical analyses.
Statistical analyses were performed using the Student t test.
Nucleotide sequence accession numbers.
The sequences determined in this investigation have been deposited in the GenBank database under accession numbers JX267142 to JX267169, JX276507 to JX276534, and JX308274 to JX308280.
RESULTS
PCR analysis of cpb, cpe, pfoA, and cpb2 gene carriage by Darmbrand isolates.
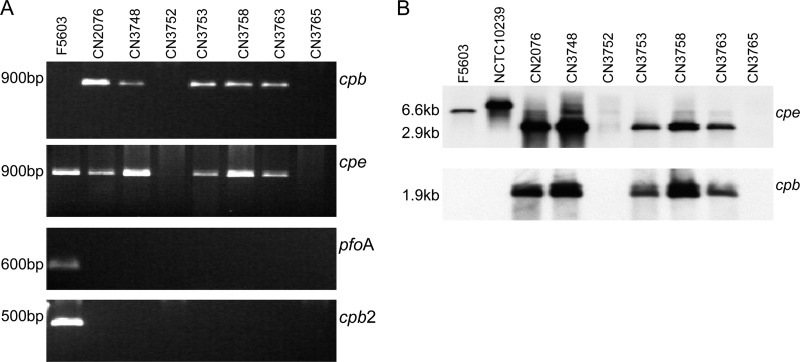
Using DNA prepared from Darmbrand strains CN2076, CN3748, CN3753, CN3758, and CN3763, PCR assays amplified products of the expected size indicating carriage of the cpb and cpe genes (Fig. 1A). However, DNA from Darmbrand strains CN3752 and CN3765 (Fig. 1A) did not support PCR amplification of either cpb or cpe products. In addition, these PCR assays revealed that none of these seven Darmbrand strains carry the pfoA or cpb2 gene (Fig. 1A). Consistent with their lack of the pfoA gene, the seven Darmbrand isolates did not produce the typical perfringolysin O-induced inner zone of beta hemolysis that develops when most C. perfringens strains are grown on sheep blood agar plates (data not shown).
Fig 1.
PCR assay and Southern blot analysis. (A) PCR analysis of cpb, cpe, pfoA, and cpb2 gene carriage in the seven Darmbrand isolates surveyed. Type A cpe-positive isolate F5603 was also included as a positive control for detection of the cpe, cpb2, and pfoA genes (10, 25). Expected sizes of PCR products are indicated on the left. (B) Southern blot hybridization. The genomic DNA of each isolate was digested with XbaI overnight at 37°C and then subjected to 1% agarose electrophoresis prior to Southern hybridization with a DIG-labeled cpe or cpb probe. Type A cpe-positive isolates F5603 and NCTC10239 were used as positive controls for the cpe gene. The sizes of detected bands are shown on the left side of the blots.
Southern blot assays using cpe and cpb probes confirmed the carriage of the cpe and cpb toxin genes by the five Darmbrand isolates that had tested positive by PCR for the possession of those two toxin genes (Fig. 1B). Thus, those five strains genotypically belong to type C, while Darmbrand strains CN3752 and CN3765 are classified genotypically as cpe-negative type A (Table 1).
Pulsed-field gel Southern blot analysis to investigate cpb and cpe gene locations in Darmbrand isolates.
PFGE conditions have been established that allow C. perfringens plasmid, but not chromosomal, DNA to enter a pulsed-field gel and migrate according to its molecular size (11, 18, 25, 30). Previous studies using Southern blot analyses of similar pulsed-field gels demonstrated that most type A food poisoning isolates carry a chromosomal cpe gene, while cpe-positive type A non-food-borne disease isolates and type C animal disease isolates carry the cpe gene on large plasmids (7–9, 18, 26).
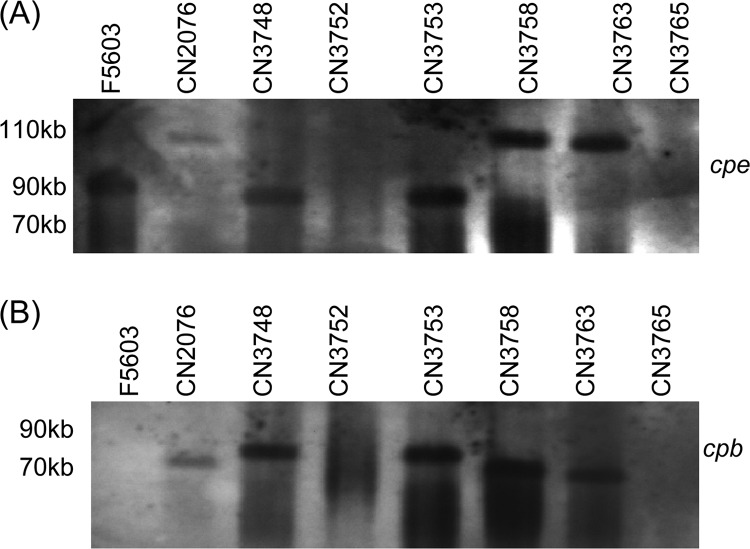
Therefore, the collection of Darmbrand isolates was subjected to a similar PFGE analysis, followed by sequential Southern hybridization with a cpe-specific probe. As shown in Fig. 2, these analyses showed that the five cpe-positive type C Darmbrand isolates surveyed carry the cpe gene on large plasmids, with a size of either ∼75 or ∼110 kb (Fig. 2A). It was notable that Darmbrand strain CN3758 was determined to carry an ∼110-kb cpe-bearing plasmid in this study since our laboratory previously reported (18) that the cpe-bearing plasmid in this strain is only ∼75 kb. However, when several stock cultures of this strain were rechecked during the present work, the cpe-bearing plasmid consistently ran with a size of ∼110 kb, suggesting possible mislabeling of the strain during the previous study (18).
Fig 2.
PFGE and Southern blot analysis of the seven Darmbrand isolates surveyed. Type A isolate F5603 was included as a positive control for a type A plasmid cpe-positive strain. (A) Genomic DNAs from all of the strains were subjected to PFGE prior to Southern blot hybridization with a DIG-labeled cpe-specific probe. (B) The same blot was stripped and then reprobed with a DIG-labeled cpb-specific probe. The migration of molecular size markers is indicated on the left side of the blots.
The same Southern blots were then stripped of the cpe probe and rehybridized with a cpb probe. Results from these analyses showed that, compared to their cpe-bearing plasmids, the cpb-bearing plasmids in these five type C Darmbrand strains are more diverse (Fig. 2B), with sizes ranging from ∼65 to 85 kb. Overlaying of the Fig. 2A and B blots confirmed (data not shown) that the cpb and cpe genes are present on two distinct plasmids in Darmbrand strains CN2076, CN3758, and CN3765. However, the cpb and cpe probes hybridized to the same location in the Southern blot assays using DNA from CN3748 and CN3753, which could indicate that the cpb and cpe genes in these two isolates are present on either (i) the same 85-kb plasmid or (ii) two different plasmids that comigrate on the pulsed-field gel because of their similarity in size.
Production of CPB and CPE by Darmbrand strains.
The expression of CPE and CPB by the Darmbrand isolates surveyed was evaluated by Western blot analysis. Using a CPE-specific antibody, CPE production was detected in sporulating cultures of isolates CN2076, CN3748, CN3753, CN3758, and CN3763 but not isolates CN3752 and CN3765 (Fig. 3A); those results are in agreement with the Fig. 1 PCR and Southern blot results indicating that the last two Darmbrand strains do not carry a cpe gene. Using a CPB-specific monoclonal antibody, CPB production was also detected in overnight TGY medium cultures of Darmbrand isolates CN2076, CN3748, CN3753, CN3758, and CN3763 but not in those of CN3752 and CN3765 (Fig. 3B), which also matches the Fig. 1 PCR and Southern blot results indicating that the last two Darmbrand strains do not carry a cpb gene.
Fig 3.
Western blot analysis of CPE and CPB production by Darmbrand isolates CN2076, CN3748, CN3752, CN3753, CN3758, CN3763, and CN3765. Type A isolate F5603 was used as a positive control for CPE production. (A) Detection of the production of CPE using a CPE-specific polyclonal antibody. The bacteria were cultured in MDS medium overnight at 37°C, and culture supernatants were then subjected to CPE Western blotting. The migration of purified CPE (35 kDa) is shown in the right lane of the blot. (B) Detection of CPB production. The bacteria were grown in TGY broth overnight at 37°C, and culture supernatants were then subjected to Western blotting using a CPB-specific monoclonal antibody. The migration of purified CPB (35 kDa) is shown in the right lane of the blot.
Darmbrand strain spore heat resistance.
The ability of the Darmbrand isolates surveyed to sporulate was first assessed by colony counting after MDS medium cultures had been heat shocked for 20 min at 70°C. Those analyses revealed approximately similar levels of sporulation by all seven Darmbrand isolates under these culture conditions (Fig. 4).
Fig 4.
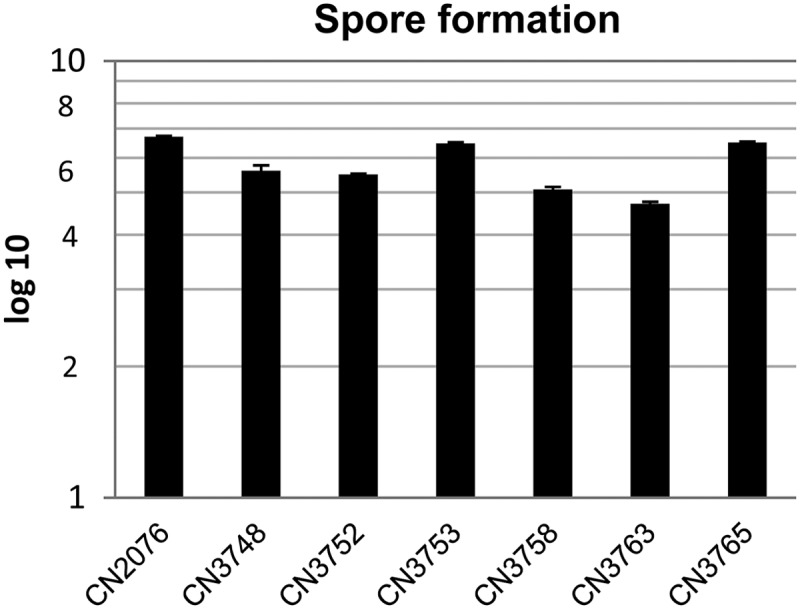
Spore formation by Darmbrand strains CN2076, CN3748, CN3752, CN3753, CN3758, CN3763, and CN3765. The bacteria were grown in MDS medium overnight at 37°C for sporulation, and the overnight cultures were then heat shocked for 20 min at 70°C. After 10-fold serial dilution with distilled water, the bacteria were plated onto BHI agar plates and grown overnight at 37°C for colony counting. Shown are mean results (log10 scale) from three independent repetitions. Error bars depict standard deviations.
Therefore, the heat resistance properties of spores of these Darmbrand isolates were assessed by boiling each culture for various times. Except for strain CN3765, which withstood boiling for up to 15 min, viable spores were recovered from all of the Darmbrand isolate cultures after boiling for 60 min (data not shown). When a D100 value (the time at 100°C needed to reduce spore viability by 1 log10) was calculated for spores of each Darmbrand isolate, the values determined ranged from 7 to 40 min (Table 3). For comparison, these values are significantly longer than the <1-min and ∼2.4-min D100 values calculated (Table 3), respectively, for C. perfringens strains F5603 and CN5388, which are (respectively) a type A strain carrying a cpe-bearing plasmid (2, 8) and a cpe-positive type C pigbel strain (19).
Table 3.
Spore heat (100°C) resistance
Sequencing of the ssp4 gene of Darmbrand isolates.
A variant small acid-soluble protein 4, which is encoded by the ssp4 gene, was recently demonstrated to be important for the exceptional resistance properties of spores made by most type A isolates carrying a chromosomal cpe gene (17). For that Ssp4 variant, the Asp located at residue 36 was shown to play a critical role when this protein mediates strong spore heat resistance; in contrast, C. perfringens strains that produce more heat-sensitive spores have a Gly residue present at Ssp4 position 36 (17).
To start assessing the basis for the relatively strong heat resistance of spores of Darmbrand strains shown in Table 3, the ssp4 open reading frames (ORFs) of the seven Darmbrand isolates surveyed were sequenced. Interestingly, the ssp4 gene of all seven Darmbrand isolates was found to encode an Asp at residue 36 of Ssp4 (Fig. 5).
Fig 5.
Deduced amino acid sequences encoded by the ssp4 ORF of Darmbrand strains CN2076, CN3748, CN3752, CN3753, CN3758, CN3763, and CN3765. Deduced Ssp4 sequences of type A strains F4969 and SM101, which produce (respectively) heat-sensitive spores and extremely heat-resistant spores (29) were used for comparison here. Instead of the Gly (G) residue present at position 36 in strain F4969, which forms heat-sensitive spores (19), Darmbrand strains encode an Ssp4 protein with an Asp (D) residue substitution at position 36, which was shown to increase the heat resistance of strain SM101 spores (17).
MLST analysis of the seven Darmbrand isolates.
Recent MLST studies indicated that chromosomal-cpe-bearing isolates share a genetic background and belong to a distinct cluster on the C. perfringens phylogenetic tree (10). Those MLST analyses also revealed that type A strains carrying a plasmid with cpe are somewhat related to one another but distinct from the type A chromosomal cpe-positive strains. Therefore, using the same eight representative housekeeping genes sequenced in that previous investigation, MLST analysis of the seven Darmbrand isolates surveyed was performed in order to find out whether those isolates are similar to one another and to determine if they resemble type A chromosomal cpe-positive strains or type A or C plasmid cpe-positive strains.
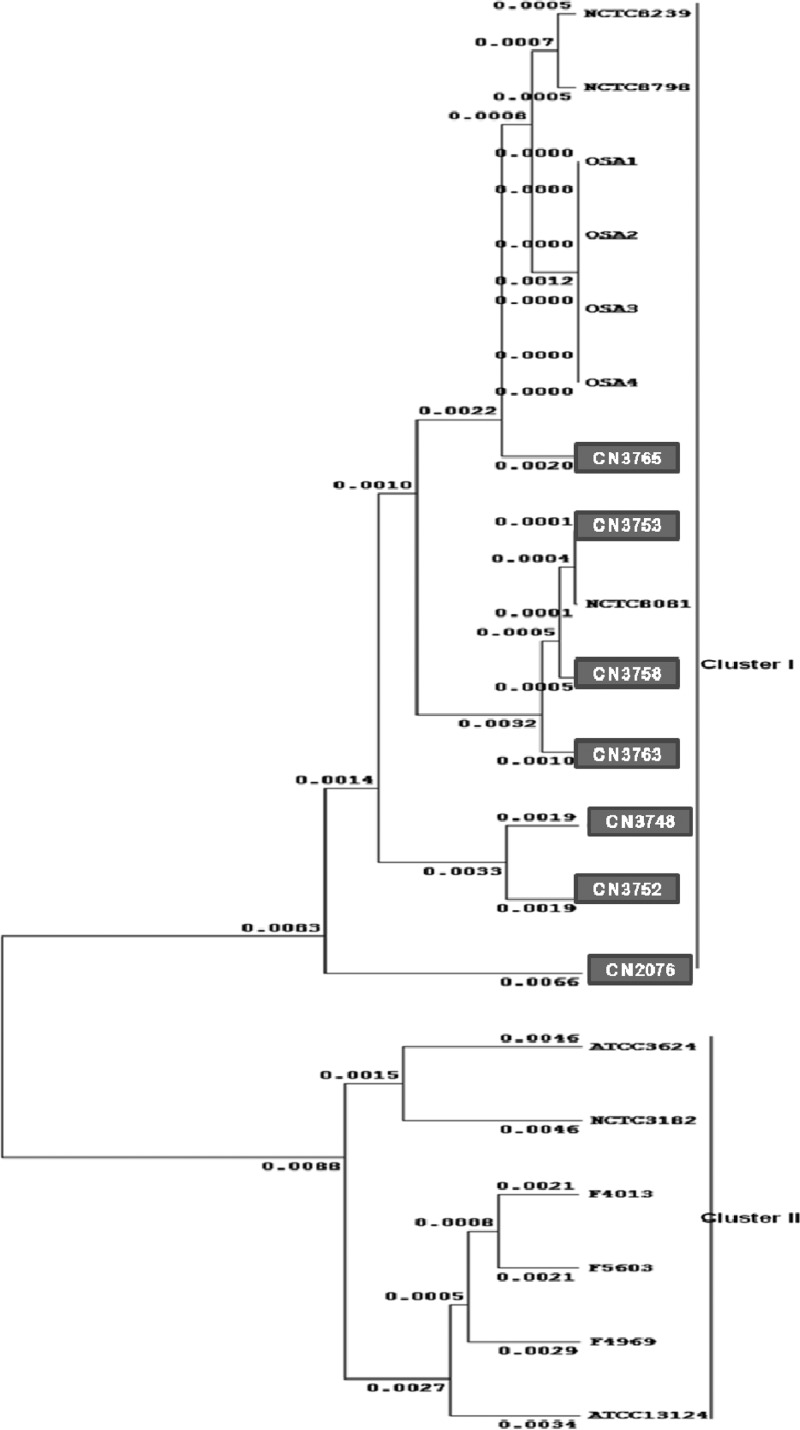
Results from the present MLST analyses indicated that these Darmbrand isolates share a genetic background with each other (Fig. 6). Furthermore, although these strains carry a plasmid-borne cpe gene, their genetic background resembles that of the previously characterized type A food poisoning chromosomal cpe-positive strains, as well as one type C Darmbrand strain (NCTC8081) not included in the present survey (10). All of the type A food poisoning chromosomal cpe-positive strains and the Darmbrand isolates, which produce heat-resistant spores, assembled into one definitive phylogenetic cluster (cluster I). However, all type A plasmid cpe-positive strains, type A cpe-negative isolates, and type C plasmid cpe-positive animal isolates assembled into a different cluster (cluster II).
Fig 6.
Phylogenetic relationships among 20 C. perfringens strains. Included are six previously genotyped (10) type A human food poisoning isolates carrying a chromosomal cpe gene (i.e., NCTC8239, NCTC8798, OSAKA1, OSAKA2, OSAKA3, and OSAKA4), the seven newly characterized Darmbrand isolates from this study (boxed), one previously examined (10) type C Darmbrand strain (NCTC8081), three previously genotyped (10) type A human sporadic-diarrhea isolates carrying a plasmid-borne cpe gene (F5603, F4969, and F4013), one cpe-negative type A strain ATCC 13124 (10), and one cpe-negative type C strain (NCTC3182) isolated from a diseased sheep (10). The phylogenetic tree was constructed by ClustalW format analysis using Winxyz software based on the concatenated 5,274-bp nucleotide sequence of eight housekeeping genes.
Overlapping PCR analyses of cpb locus organization in Darmbrand strains.
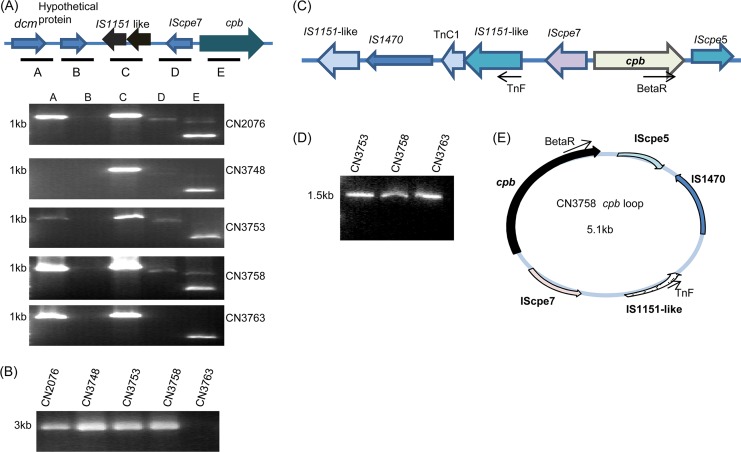
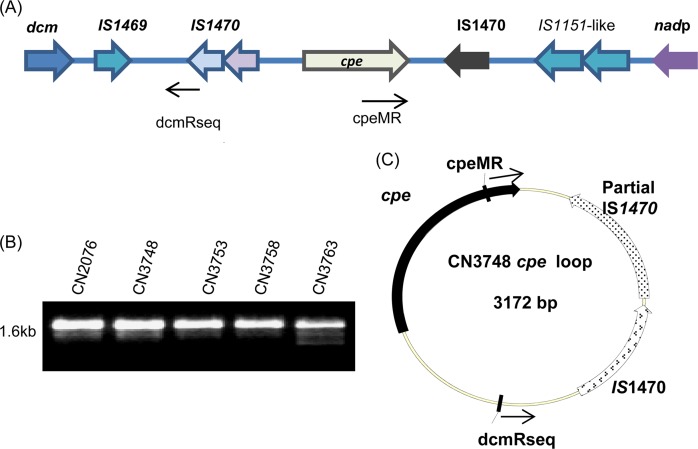
Previous studies in our lab had identified dcm, two IS1151-like sequences, and an IScpe7 sequence upstream of the cpb gene in type C strains JGS1495 and CN5388 (11), which do not have a Darmbrand origin. Therefore, overlapping PCR assays were performed to evaluate whether similar sequences are present upstream of the cpb gene in Darmbrand type C strains CN2076, CN3748, CN3753, CN3758, and CN3763. Five sets of primers (Table 2), amplifying PCR products A to E, showed the presence of IS1151-like sequence in all five isolates. The dcm gene was amplified from four Darmbrand type C isolates (CN2076, CN3753, CN3758, and CN3763) (Fig. 7A), but overlapping PCRs did not suggest the presence of these dcm genes near IS1151-like sequences or the cpb gene. Therefore, a long-range PCR analysis using primer sets F1/R5, F1/R3, and F3/R5 was performed. Long-range PCR with primer set F1/R3 and/or F1/R5 gave no PCR amplification (data not shown), which implied that the dcm genes are not adjacent to IS1151-like sequences or to the cpb gene in these isolates. PCR with F3/R5 suggested that IS1151-like sequences are located near the cpb gene in four of these isolates, i.e., CN2076, CN3748, CN37553, and CN3758, but not in CN3763 (Fig. 7B).
Fig 7.
Organization of the cpb locus and evidence of cpb gene mobilization. (A) Overlapping-PCR linkage of the cpb gene with IS1151 and the dcm gene in five type C Darmbrand isolates. The top panel shows a diagram of the cpb locus in C. perfringens type C isolate JGS1495 based upon the sequencing results released by the J. Craig Venter Institute;. Boxes A to E depict the overlapping PCRs of the panel below. The migration of a 1-kb size marker is shown at the left of the gels. (B) Long-range PCR analysis of the five type C Darmbrand isolates, i.e., CN2076, CN3748, CN3753, CN3758, and CN3763, using primers F3/R5. The size of the PCR product is shown at the left of the gel. (C) Arrangement of the cpb locus in strains CN2076, CN3748, CN3753, and CN3758 based upon our sequencing data. The reverse arrows show the orientation and binding of primers used for loop PCR. (D) PCR amplification of cpb-containing circular intermediates using primer pair TnF/BetaR with CN3753, CN3758, and CN3763 DNA extracted freshly from colonies grown overnight anaerobically on BHI agar plates. The migration of a 1-kb size marker is shown at the left lane of the gel. (E) Diagram of the circular intermediate derived from sequencing of the CN3758 loop PCR product in panel D that was amplified using primers TnF and BetaR.
Consistent with those PCR results, sequencing of the long-range PCR products generated with primers F3 and R5 showed that four Darmbrand strains carry a novel cpb locus that differs from the previously described cpb loci of type B strain ATCC 3626, type C veterinary strain JGS1495, or type C human pigbel strain CN5388 (11, 30). Specifically, this sequencing showed that, in those four Darmbrand isolates, two IS1151-like sequences, one IS1470 element, a TnC1 sequence, and IScpe7 are present upstream of the cpb gene, while an IScpe5 sequence is located downstream of the cpb gene (Fig. 7C).
PCR identification of possible circular transposition intermediates containing the cpb gene.
The results shown in Fig. 7C indicated that, when present in the Darmbrand strains surveyed, the cpb gene is associated with insertion sequences that might mobilize that toxin gene. Supporting this possibility is the fact that cpb-containing circular transposition intermediates have been detected in other C. perfringens strains (30).
Therefore, a reverse PCR was performed with primers in opposite orientations to evaluate whether similar cpb-containing circular intermediates might also be formed in the Darmbrand isolates surveyed. Primers TnF and BetaR amplified a 1.5-kb PCR product from isolates CN3753, CN3758, and CN3763 (Fig. 7D). Those PCR products were purified and sequenced, which confirmed the amplification of cpb, IS1470, IS1151, and IScpe5 sequences (Fig. 7E).
PCR identification of a possible circular transposition containing the cpe gene.
The sequence results shown in Fig. 8A indicated that the cpe gene is associated with insertion sequences that might mobilize the cpe gene. Supporting that possibility is the fact that potential cpe-containing circular transposition intermediates have been detected in some other C. perfringens strains (3, 18).
Fig 8.
Organization of the cpe gene locus and evidence of cpe gene mobilization. (A) Diagram of the cpe locus organization in isolate CN3748. The reverse arrows show the orientation and binding of primers dcmRseq and cpeMR used for loop PCR in panel B. (B) PCR amplification of cpe-containing circular intermediates using primer pair dcmRseq/cpeMR with CN2076, CN3748, CN3753, CN3758, and CN3763 DNA extracted freshly from colonies grown overnight anaerobically on BHI agar plates. The migration of a 1-kb size marker is shown to the left of the gel. (C) Diagram of the circular intermediate derived from sequencing of the CN3748 loop PCR product in panel B that was amplified using primer set dcmRseq/cpeMR.
Consequently, reverse PCRs were performed by using primers in opposite orientations to evaluate whether similar cpe-containing circular intermediates are formed in the cpe-positive Darmbrand isolates surveyed. Primers dcmRseq and cpeMR consistently amplified a 1.6-kb PCR product from the five cpe-positive Darmbrand isolates, i.e., CN2076, CN3748, CN3753, CN3758, and CN3763 (Fig. 8B). Those PCR products were purified and sequenced, which confirmed the amplification of a circular intermediate containing cpe sequences, an intact IS1470 sequence, and a partial IS1470 sequence (Fig. 8C), similar to results obtained previously with another Darmbrand isolate, i.e., CN2078 (18).
DISCUSSION
Food-borne illnesses, which remain a major public health problem, often involve foods that were cooked or stored at suboptimal temperatures. Therefore, it should be advantageous for C. perfringens strains that cause human food-borne illness to acquire an enhanced ability to survive in those temperature-abused foods (21). Consistent with this, ∼75 to 80% of C. perfringens type A food poisoning outbreaks involve chromosomal-cpe-carrying type A strains whose spores exhibit exceptional temperature resistance properties (21, 26, 29). Specifically, the spores made by most chromosomal cpe-positive type A strains possess D100 values ranging from 30 to 120 min, in contrast to the 0.5- to 2.7-min D100 values of spores made by other type A strains or most non-type-A strains (17, 29).
Many years ago, Darmbrand strains were also identified as possessing unusually strong heat resistance properties (39). Specifically, their spores were reported to survive boiling, which our results confirmed. However, to our knowledge, the D100 value of spores made by Darmbrand strains has not been determined, which precluded direct quantitative comparisons of the resistance properties of spores made by Darmbrand strains with those of spores from other C. perfringens strains. In response, the present investigation calculated that the D100 values of spores made by seven Darmbrand strains ranged from 7 to 40 min, which is near the range of D100 values determined for spores of most chromosomal cpe-positive type A strains and substantially greater than the D100 value ranges calculated for spores made by other C. perfringens strains. Therefore, these and previous data (17, 29) establish that two C. perfringens food-borne diseases of humans, i.e., C. perfringens type A food poisoning and Darmbrand, often involve strains that produce highly heat-resistant spores, which likely favors their survival in temperature-abused foods.
Recent studies determined that the exceptional heat resistance properties of spores typically made by type A strains with a chromosomal cpe gene is attributable, in large part, to their production of a variant Ssp4 protein (17). In this Ssp4 variant, an Asp (rather than a Gly) residue is present at position 36 (note that Ssp4 proteins can also vary at residue 72, but this second variation does not affect spore heat resistance properties [17]). Subsequent studies (19) showed that the Asp36 Ssp4 variant binds more tightly than the Gly36 Ssp4 variant to C. perfringens DNA, which presumably provides spore DNA with greater protection against stresses such as heat. Therefore, our present determination that Darmbrand isolates also carry an ssp4 gene that encodes the Ssp4 variant with the Asp36 residue strongly suggests a mechanistic basis for the strong heat resistance properties exhibited by spores made by Darmbrand strains. It is notable in this regard that, to date, the only identified C. perfringens isolates that encode the Ssp4 Asp36 variant are associated with food-borne human diseases, i.e., either C. perfringens type A food poisoning (17) or Darmbrand (this study). Interestingly, the one pigbel isolate (CN5388) examined in a previous investigation did not form highly heat-resistant spores (19), even though pigbel is a food-borne disease considered similar to Darmbrand. However, more pigbel isolates, which are very difficult to obtain, should be examined before any conclusions can be drawn regarding the relative heat resistance characteristics of the spores of pigbel strains.
Carriage of an ssp4 gene that encodes the Asp36 Ssp4 variant was not the only identified resemblance between Darmbrand strains and type A strains carrying a chromosomal cpe gene. While most C. perfringens strains carry a chromosomal pfoA gene that encodes perfringolysin O (27), both Darmbrand strains and type A strains carrying a chromosomal cpe gene are pfoA negative (10). Moreover, MLST analyses of eight housekeeping genes revealed that the genetic similarities between Darmbrand strains and type A food poisoning strains carrying a chromosomal cpe gene are even more pervasive than the simple absence of pfoA or the encoding of an Asp36 Ssp4 variant. By collating results from the present and previous (10) MLST studies, Darmbrand and type A chromosomal cpe-positive strains were found to group closely together on the C. perfringens phylogenetic tree; moreover, these strains have a genetic background distinctly different from that of plasmid cpe-positive type A to E veterinary strains. Therefore, Darmbrand and type A chromosomal cpe-positive strains represent a distinct subpopulation within the C. perfringens species. It should also be noted that the only other Darmbrand isolate (NCTC8081) that had been previously characterized by MLST (10) is also classified in this phylogenetic subgroup.
Darmbrand is usually attributed to type C strains of C. perfringens (13, 14). Therefore, it was notable that two of the seven Darmbrand isolates surveyed in this study were genotyped instead as type A. At least two explanations for these results are conceivable. First, these two type A Darmbrand strains may have been merely normal-flora C. perfringens strains that were isolated from Darmbrand patients, rather than the strains that actually caused the illness. This possibility seems less likely since the type A Darmbrand strains were genetically similar to type C Darmbrand strains with respect to their MLST results, the absence of a pfoA gene, the carriage of an ssp4 gene that encodes the Asp36 Ssp4 variant, and the production of highly heat-resistant spores. The alternative explanation is that these Darmbrand strains were originally type C but during the past 60+ years since their collection, they had lost their cpb plasmid. Similar “type degradation” has often been reported in the C. perfringens literature (23).
It was also notable that all of the type C Darmbrand strains surveyed produced CPE. To our knowledge, CPE production has not been previously associated with Darmbrand, which occurred before the identification of CPE in the late 1960s to the early 1970s (23). However, CPE has been proposed as a possible contributor to the pathogenesis of human pigbel (14), which is also an enteritis necroticans caused by type C strains. The relative importance of CPB versus CPE for the pathogenicity of Darmbrand or other CPE-positive type C strains is not clear at present, although both toxins are clearly active in the gastrointestinal tract and can also induce lethal enterotoxemia (5, 35).
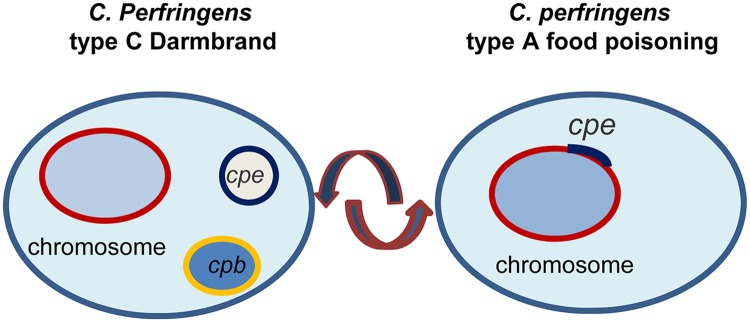
The cpe locus present in the five type C Darmbrand strains surveyed matched that reported previously for type C Darmbrand strain CN2078 (18). This plasmid-borne cpe locus of Darmbrand strains is also remarkably similar to the cpe locus present in type A chromosomal strains (3, 24, 26), except that, in Darmbrand strains, the IS1469 element is present upstream (rather than downstream) of the IS1470 element located upstream of the cpe gene. These findings, plus the genetic relatedness between Darmbrand strains and type A chromosomal cpe-positive strains as revealed by MLST, suggest evolutionary relationships between these strains. Several scenarios could be envisioned for this relationship. First, many toxin plasmids of C. perfringens are conjugative (1, 4, 12), so a C. perfringens strain with a housekeeping and spore resistance genetic background similar to that now found in Darmbrand and type A chromosomal cpe-positive strains may have conjugatively acquired a cpe-bearing plasmid like those now found in the Darmbrand strains, creating a Darmbrand precursor strain. Since this study found evidence of mobilization of the cpe gene by the adjacent IS sequences in Darmbrand strains, the cpe gene in one bacterium of that Darmbrand precursor strain might later have been inserted or recombined into the chromosome to give rise to type A chromosomal cpe-positive strains (Fig. 9). Another bacterium of the Darmbrand precursor strain may also have later conjugatively acquired a cpb plasmid, which converted it to a type C Darmbrand strain. The present and previous studies (11, 18) identified some Darmbrand strains that carry both their cpb and cpe genes on similar-sized plasmids, and a previous investigation provided (11) evidence that in at least two such Darmbrand strains (CN3753 and CN2078) those cpb and cpe genes are located on the same plasmid. Since this study found evidence of the mobilization of both the cpe and cpb genes in Darmbrand strains, it seems likely that, in some Darmbrand strains, a mobilized toxin gene was later inserted into a plasmid already carrying the other toxin gene; e.g., a mobilized cpb gene was inserted into a cpe-bearing plasmid or vice versa. An alternative scenario explaining the evolutionary relationship between Darmbrand strains and type A chromosomal cpe-positive strains is that the cpe gene on the chromosome of a type A strain, which can also be mobilized (3), may have moved onto a plasmid and then that strain later acquired a cpb-bearing plasmid by conjugation (Fig. 9).
Fig 9.
Possible evolutionary relationship between C. perfringens type A food poisoning isolates carrying a chromosomal cpe gene and type C Darmbrand isolates carrying a plasmid cpe gene.
Further research is under way to better understand the relationship between the cpe- and cpb-bearing plasmids found in various C. perfringens strains. This information will shed further light on the diversity and evolutionary relationships between these strains. Studies are also under way to better dissect the relative contributions of CPB and CPE to the pathogenicity of CPE-positive type C strains.
ACKNOWLEDGMENTS
This research was generously supported by grant R01 AI056177-09 from the National Institute of Allergy and Infectious Diseases.
We thank Paul Hauer for supplying a CPB-specific monoclonal antibody.
Footnotes
Published ahead of print 1 October 2012
REFERENCES
- 1. Bannam TL, et al. 2011. Necrotic enteritis-derived Clostridium perfringens strain with three closely related independently conjugative toxin and antibiotic plasmids. mBio 2:e00190–11 doi:10.1128/mBio.00190-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brett MM, Rodhouse JC, Donovan TJ, Tebbut GM, Hutchinson DN. 1992. Detection of Clostridium perfringens and its enterotoxin in cases of sporadic diarrhea. J. Clin. Pathol. 45:609–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brynestad S, Granum PE. 1999. Evidence that Tn5565, which includes the enterotoxin gene in Clostridium perfringens, can have a circular form which may be a transposition intermediate. FEMS Microbiol. Lett. 170:281–286 [DOI] [PubMed] [Google Scholar]
- 4. Brynestad S, Sarker MR, McClane BA, Granum PE, Rood JI. 2001. The enterotoxin plasmid from Clostridium perfringens is conjugative. Infect. Immun. 69:3483–3487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Caserta JA, et al. 2011. Development and application of a mouse intestinal loop model to study the in vivo action of Clostridium perfringens enterotoxin. Infect. Immun. 79:3020–3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. CDC 27 July 2011, posting date CDC estimates of foodborne illness in the United States: Clostridium perfringens. http://www.cdc.gov/foodborneburden/clostridium-perfringens.html CDC, Atlanta, GA [Google Scholar]
- 7. Collie RE, Kokai-Kun JF, McClane BA. 1998. Phenotypic characterization of enterotoxigenic Clostridium perfringens isolates from non-foodborne human gastrointestinal diseases. Anaerobe 4:69–79 [DOI] [PubMed] [Google Scholar]
- 8. Collie RE, McClane BA. 1998. Evidence that the enterotoxin gene can be episomal in Clostridium perfringens isolates associated with nonfoodborne human gastrointestinal diseases. J. Clin. Microbiol. 36:30–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cornillot E, et al. 1995. The enterotoxin gene (cpe) of Clostridium perfringens can be chromosomal or plasmid-borne. Mol. Microbiol. 15:639–647 [DOI] [PubMed] [Google Scholar]
- 10. Deguchi A, et al. 2009. Genetic characterization of type A enterotoxigenic Clostridium perfringens strains. PLoS One 4:e5598 doi:10.1371/journal.pone.0005598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gurjar A, Li J, McClane BA. 2010. Characterization of toxin plasmids in Clostridium perfringens type C isolates. Infect. Immun. 78:4860–4869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hughes ML, et al. 2007. Epsilon-toxin plasmids of Clostridium perfringens type D are conjugative. J. Bacteriol. 189:7531–7538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Johnson S, Gerding DN. 1997. Enterotoxemic infections, p 117–140 In Rood JI, McClane BA, Songer JG, Titball RW. (ed), The clostridia: molecular biology and pathogenesis. Academic Press, Ltd., London, United Kingdom [Google Scholar]
- 14. Lawrence GW. 1997. The pathogenesis of enteritis necroticans, p 198–207 In Rood JI, McClane BA, Songer JG, Titball RW. (ed), The clostridia: molecular biology and pathogenesis. Academic Press, Ltd., London, United Kingdom [Google Scholar]
- 15. Li J, Chen J, Vidal JE, McClane BA. 2011. The Agr-like quorum-sensing system regulates sporulation and production of enterotoxin and beta2 toxin by Clostridium perfringens type A non-food-borne human gastrointestinal disease strain F5603. Infect. Immun. 79:2451–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li J, McClane BA. 2006. Comparative effects of osmotic, sodium nitrite-induced, and pH-induced stress on growth and survival of Clostridium perfringens type A isolates carrying chromosomal or plasmid-borne enterotoxin genes. Appl. Environ. Microbiol. 72:7620–7625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li J, McClane BA. 2008. A novel small acid soluble protein variant is important for spore resistance of most Clostridium perfringens food poisoning isolates. PLoS Pathog. 4:e1000056 doi:10.1371/journal.ppat.1000056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li J, Miyamoto K, Sayeed S, McClane BA. 2010. Organization of the cpe locus in CPE-positive Clostridium perfringens type C and D isolates. PLoS One 5:e10932 doi:10.1371/journal.pone.0010932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li J, Paredes-Sabja D, Sarker MR, McClane BA. 2009. Further characterization of Clostridium perfringens small acid soluble protein-4 (Ssp4) properties and expression. PLoS One 4:e6249 doi:10.1371/journal.pone.0006249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ma M, Vidal J, Saputo J, McClane BA, Uzal F. 2011. The VirS/VirR two-component system regulates the anaerobic cytotoxicity, intestinal pathogenicity, and enterotoxemic lethality of Clostridium perfringens type C isolate CN3685. mBio 2:e00338–10 doi:10.1128/mBio.00338-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McClane BA, Robertson SL, Li J. 2013. Clostridium perfringens, p 465–489 In Doyle MP, Beuchat LR. (ed), Food microbiology: fundamentals and frontiers, 4th ed ASM Press, Washington, DC [Google Scholar]
- 22. McClane BA, Uzal FA, Miyakawa MF, Lyerly D, Wilkins T. 2006. The enterotoxic clostridia, p. 688–752 In Dworkin M, Falkow S, Rosenburg E, Schleifer KH, Stackebrandt E. (ed), The prokaryotes, 3rd ed Springer, New York, NY [Google Scholar]
- 23. McDonel JL. 1986. Toxins of Clostridium perfringens types A, B, C, D, and E, p 477–517 In Dorner F, Drews H. (ed), Pharmacology of bacterial toxins. Pergamon Press, Oxford, United Kingdom [Google Scholar]
- 24. Miyamoto K, Chakrabarti G, Morino Y, McClane BA. 2002. Organization of the plasmid cpe locus of Clostridium perfringens type A isolates. Infect. Immun. 70:4261–4272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Miyamoto K, et al. 2006. Complete sequencing and diversity analysis of the enterotoxin-encoding plasmids in Clostridium perfringens type A non-food-borne human gastrointestinal disease isolates. J. Bacteriol. 188:1585–1598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Miyamoto K, Wen Q, McClane BA. 2004. Multiplex PCR genotyping assay that distinguishes between isolates of Clostridium perfringens type A carrying a chromosomal enterotoxin gene (cpe) locus, a plasmid cpe locus with an IS1470-like sequence, or a plasmid cpe locus with an IS1151 sequence. J. Clin. Microbiol. 42:1552–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Myers GS, et al. 2006. Skewed genomic variability in strains of the toxigenic bacterial pathogen, Clostridium perfringens. Genome Res. 16:1031–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Raju D, Waters M, Setlow P, Sarker MR. 2006. Investigating the role of small, acid-soluble spore proteins (SASPs) in the resistance of Clostridium perfringens spores to heat. BMC Microbiol. 8:50 doi:10.1186/1471-2180-6-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sarker MR, Shivers RP, Sparks SG, Juneja VK, McClane BA. 2000. Comparative experiments to examine the effects of heating on vegetative cells and spores of Clostridium perfringens isolates carrying plasmid versus chromosomal enterotoxin genes. Appl. Environ. Microbiol. 66:3234–3240 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30. Sayeed S, Li J, McClane BA. 2010. Characterization of virulence plasmid diversity among Clostridium perfringens type B isolates. Infect. Immun. 78:495–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sayeed S, et al. 2008. Beta toxin is essential for the intestinal virulence of Clostridium perfringens type C disease isolate CN3685 in a rabbit ileal loop model. Mol. Microbiol. 67:15–30 [DOI] [PubMed] [Google Scholar]
- 32. Scallan E, et al. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg. Infect. Dis. 17:7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Setlow P. 2007. I will survive: DNA protection in bacterial spores. Trends Microbiol. 15:172–180 [DOI] [PubMed] [Google Scholar]
- 34. Titball RW, Naylor CE, Basak AK. 1999. The Clostridium perfringens alpha-toxin. Anaerobe 5:51–64 [DOI] [PubMed] [Google Scholar]
- 35. Uzal FA, et al. 2009. Development and application of new mouse models to study the pathogenesis of Clostridium perfringens type C enterotoxemias. Infect. Immun. 77:5291–5299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vidal JE, et al. 2012. Evidence that the Agr-like quorum sensing system regulates the toxin production, cytotoxicity and pathogenicity of Clostridium perfringens type C isolate CN3685. Mol. Microbiol. 83:179–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vidal JE, McClane BA, Saputo J, Parker J, Uzal FA. 2008. Effects of Clostridium perfringens beta-toxin on the rabbit small intestine and colon. Infect. Immun. 76:4396–4404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wen Q, Miyamoto K, McClane BA. 2003. Development of a duplex PCR genotyping assay for distinguishing Clostridium perfringens type A isolates carrying chromosomal enterotoxin (cpe) genes from those carrying plasmid-borne enterotoxin (cpe) genes. J. Clin. Microbiol. 41:1494–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zeissler J, Rassfeld-Sternberg L. 1949. Enteritis necroticans due to Clostridium welchii type F. Br. Med. J. 1(4597):267–269 [DOI] [PMC free article] [PubMed] [Google Scholar]