Abstract
Type IV secretion systems (T4SS) are multiprotein structures that direct the translocation of specific molecules across the bacterial cell envelope. As in other bacteria, pathogenicity of the genus Brucella essentially depends on the integrity of the T4SS-encoding virB operon, whose expression is regulated by multiple transcription factors belonging to different families. Previously, we identified IHF and HutC, two direct regulators of the virB genes that were isolated from total protein extracts of Brucella. Here, we report the identification of MdrA, a third regulatory element that was isolated using the same screening procedure. This transcription factor, which belongs to the MarR-family of transcriptional regulators, binds at two different sites of the virB promoter and regulates expression in a growth phase-dependent manner. Like other members of the MarR family, specific ligands were able to dissociate MdrA from DNA in vitro. Determination of the MdrA-binding sites by DNase I footprinting and analyses of protein-DNA complexes by electrophoresis mobility shift assays (EMSAs) showed that MdrA competes with IHF and HutC for the binding to the promoter because their target DNA sequences overlap. Unlike IHF, both MdrA and HutC bound to the promoter without inducing bending of DNA. Moreover, the two latter transcription factors activated virB expression to similar extents, and in doing so, they are functionally redundant. Taken together, our results show that MdrA is a regulatory element that directly modulates the activity of the virB promoter and is probably involved in coordinating gene expression in response to specific environmental signals.
INTRODUCTION
Type IV secretion systems (T4SSs) are multiprotein structures that direct the translocation of specific molecules across the bacterial cell envelope (3). During evolution, these machineries have specialized to secrete different kinds of substrates, which led to the emergence of the different biological functions that T4SSs can exert. Both the Escherichia coli Tra and the Agrobacterium tumefaciens VirB T4SSs are responsible for secreting protein-DNA complexes during plasmid conjugation and oncogenic transfer DNA (T-DNA) translocation, respectively (48, 54). Instead, other T4SSs, such as the Helicobacter pylori Cag system, the Legionella pneumophila Dot/Icm system, or the Bordetella pertussis Ptl system, have evolved to deliver protein substrates toward the eukaryotic host, where they subvert cellular processes and favor the development of the bacterial infection (8, 44). As in all these pathogenic bacteria, virulence of Brucella is also fully dependent on the integrity of a T4SS, since deletion of one or more T4SS-encoding virB genes completely abrogates the ability of this genus of facultative intracellular bacteria to colonize its eukaryotic host (30, 47).
Brucella comprises many species, which are the causative agents of a worldwide-distributed zoonotic disease known as brucellosis. The members of Brucella differ in their host preference, affecting mammals that range from rodents to cetaceans (52). Brucella abortus, the causal agent of bovine brucellosis, is one of the species that can be transmitted from infected animals to humans through inhalation, by contact with skin wounds or mucosa, or by consumption of contaminated dairy products. In the natural host, brucellosis produces sterility in males and abortion in pregnant females, whereas in humans it causes debilitating symptoms and complications that can lead to death (16).
The ability of Brucella to infect and persist within the mammalian host relies on different mechanisms that allow this bacterium to overcome the innate and adaptive immune host responses. Among all these strategies, bacterial survival and intracellular replication into the host professional phagocytes are key processes underlying brucellosis. After entering the host macrophages, the bacterium is located in the lumen of the so-called Brucella-containing vacuole (BCV) (32). During the first stages of the intracellular infection, this compartment undergoes sustained interactions with endoplasmic reticulum (ER) membranes. This process, which depends essentially on the activity of the VirB system, is necessary for the biogenesis of an ER-derived bacterial replication compartment, since abrogation of the early BCV-ER interactions prevents Brucella from multiplying intracellularly and leads to bacterial degradation in lysosomes (7). Recently, screening procedures based on different methods led to the identification of proteins that are translocated into the host cell cytoplasm in a VirB-dependent manner (10, 22). Until now, the function of these substrates in the establishment of the host cell infection has not been described except for RicA, a protein that interacts with and recruits the small GTPase Rab2 into BCVs during the intracellular trafficking of Brucella (9).
The first reported evidence of intracellular regulation of the VirB T4SS showed that expression of the virB genes is induced after internalization of the bacterium in macrophage cell lines (4). Such induction is dependent on the acidification of the BCV and reaches the maximal level of expression at 5 h postinfection. Subsequently, expression of the virB operon is rapidly turned off, which suggests that the system is controlled by regulatory mechanisms that respond to specific signals sensed by the bacterium within the host cell (46).
The main regulatory protein controlling expression of the Brucella virB genes is VjbR, a quorum sensing (QS)-related regulator that binds the virB promoter (PvirB) at position −94 relative to the transcription start site (2, 12). In addition, many other regulators were shown to directly or indirectly influence virB expression, including the stringent response mediator Rsh and transcription factors belonging to different families (13, 18, 37). In addition, although their binding sites in the virB promoter have not yet been reported, it was recently shown that both the two-component regulator BvrR and the second QS-related transcription factor BabR/BlxR participate in regulation of virB expression by directly interacting with PvirB, as demonstrated by pulldown experiments and electrophoresis mobility shift assays (EMSAs), respectively (6, 25).
In previous works from our group, we identified two transcriptional regulators that directly contribute to induction of virB expression: (i) IHF, a nucleoid-associated protein that binds to PvirB at position −162.5 and induces a DNA bend that is necessary for the promoter activation (46); and (ii) HutC, a GntR family regulator that binds at position −188 and links virB expression to histidine catabolism (45). Here, we report the identification of MdrA, a third regulatory element that was isolated using the same screening procedure. Our experiments showed that this transcription factor binds at two different sites of PvirB and regulates virB expression in a growth phase-dependent manner. Analysis of promoter activities under different mutant backgrounds revealed that MdrA exerts its regulatory role in redundancy with the function of HutC. Besides, consistently with both the architecture and localization of the MdrA-binding sites within the promoter region, we also observed that this transcription factor competes with IHF and HutC for the binding to PvirB. As is usually the case with MarR family members, our experiments showed that the presence of a specific ligand dissociates MdrA from a target DNA binding site. Such ligand-induced disruption of promoter binding indicates that this process may allow MdrA to receive and respond to environmental signals.
MATERIALS AND METHODS
Media and culture conditions.
Brucella strains were grown in tryptic soy broth (TSB) or in Gerhardt-Wilson minimal medium (15) or MM1 (45) at pH 4.5, 5.5, or 7.0. Bacteria were cultured at 37°C in a rotary shaker (250 rpm). Media were supplemented with kanamycin (50 μg ml−1) and/or nalidixic acid (5 μg ml−1) as needed. Sodium deoxycholate (DOC) (Sigma) was eventually added to the media at a final concentration of 100, 250, 500, 750, or 1,000 μM. Escherichia coli strains were cultured in Luria-Bertani (LB) broth at 37°C in a rotary shaker at 250 rpm. Media were supplemented with ampicillin (100 μg ml−1) or kanamycin (25 μg ml−1) as needed.
Isolation and identification of MdrA.
The protein detected by EMSA was isolated from exponential-phase cultures (optical density at 600 nm [OD600], ∼0.5) of the avirulent Brucella abortus Δpgm strain (51). Bacteria were harvested and disrupted as described previously (46). After ultracentrifugation and filtration, the total protein extract was fractionated by ammonium sulfate precipitation. The different fractions were analyzed by EMSA using probe vir-up or the control probe B10. The positive fraction (0 to 45% ammonium sulfate saturation) was suspended in 35 mM phosphate buffer (pH 6.8) containing 3 mM β-mercaptoethanol and dialyzed against the same buffer overnight at 4°C. The solution was loaded onto a Mono-S column and eluted with a linear gradient of 35 mM phosphate buffer (pH 6.8), 3 mM β-mercaptoethanol, and 1 M NaCl. The DNA-binding activity of the fractions was again analyzed by EMSA using the probes described above. Positive fractions were subjected to affinity chromatography: the biotinylated probe biot-vir-up, which corresponds to the −430 to −202 region of PvirB, was constructed by PCR using Taq, the 5′-biotinylated primer pvd228, the primer pirup, and genomic DNA of B. abortus 2308 as the template. The biotinylated control probe was constructed as described previously (45). The biotinylated probes were bound to streptavidin paramagnetic spheres (Promega), and a binding reaction was performed using binding buffer and the positive fractions of the Mono-S column. After two washes with binding buffer with 0.2 M NaCl, the DNA-bound proteins were eluted with 0.85 M NaCl and analyzed by 12.5% SDS-PAGE. The gel was silver stained with a mass spectrometry-compatible method for visualizing the protein bands. A band that was observed with the probe biot-vir-up but was absent in samples from the control probe was excised from the gel and analyzed by mass spectrometry by Vital Probes Inc. (Mayfield, PA).
Construction of plasmids.
For construction of pK18mob-sacB-ΔmdrA, two PCRs were carried out using Pfx (Invitrogen), genomic DNA of B. abortus 2308 as the template, and primers SpeMarA (5′-GGACTAGTTGACGATATGTTTGCCGCAT-3′) and COMMarA (5′-CTGCAAGAAGGAGAGTATCGAATGCAGGCGCTCGACAAGC-3′) or SpeMarB (5′-GGACTAGTAACCGCTCTTGCGGCAATC-3′) and COMMarB (5′-TCGATACTCTCCTTCTTGCAGTTTATGGCGATGAACAAGTCG-3′). Both PCR products, corresponding to two 400-bp flanking regions of mdrA, were annealed and used as the templates for a PCR performed with primers SpeMarA and SpeMarB. The product was digested with SpeI and cloned into the kanamycin-resistant (Kanr) plasmid pK18mob-sacB (42).
For construction of the complementation plasmid pK18mob-sacB-Knock-in-mdrA, a PCR was carried out using Pfx, genomic DNA of B. abortus 2308 as the template, and primers SpeMarA (5′-GGACTAGTTGACGATATGTTTGCCGCAT-3′) and SpeMarB (5′-GGACTAGTAACCGCTCTTGCGGCAATC-3′). The PCR product, which contains the wild-type sequence of the mdrA gene and two 400-bp flanking regions, was digested with SpeI and cloned into the kanamycin-resistant plasmid pK18mob-sacB.
For construction of the expression vector pQE-31-mdrA, a PCR was performed using Pfx, genomic DNA of B. abortus 2308 as the template, and primers BamMarII (5′-CGGGATCCGACCAATACCCAGCGCAAGAT-3′) and HindMar (5′-GGTACCTCAGCCGCGAGATGGCGT-3′). The PCR products were digested with BamHI and HindIII and cloned into plasmid pQE-31 (Qiagen).
Construction of B. abortus ΔmdrA strain.
Plasmid pK18mob-sacB-ΔmdrA was transferred to B. abortus 2308 by biparental conjugation. Kanr colonies were selected as single-homologous recombinants. Selection with sucrose, excision of plasmids, and generation of deletion mutants were performed as described previously (46). PCR analyses of kanamycin-susceptible (Kans) colonies were carried out with primers SpeMarA and SpeMarB to identify clones that contained the deletion of mdrA.
Construction of the double mutant strain B. abortus ΔhutC ΔmdrA.
Plasmid pK18mob-sacB-ΔmdrA was transferred to B. abortus ΔhutC (45) by biparental conjugation. Kanr colonies were selected as single-homologous recombinants. After selection with sucrose and excision of the plasmid, a PCR analysis of Kans colonies was carried out with primers SpeMarA and SpeMarB to identify clones that contained the deletion of mdrA in the ΔhutC mutant background.
Construction of the knock-in complemented strain B. abortus ΔhutC ΔmdrA-KI.
Plasmid pK18mob-sacB-Knock-in-mdrA was transferred to the B. abortus ΔhutC ΔmdrA strain by biparental conjugation. Kanr colonies were selected as single-homologous recombinants. After selection with sucrose and excision of the plasmid, a PCR analysis of Kans colonies was carried out with primers SpeMarA and SpeMarB to identify clones that incorporated the wild-type mdrA gene.
Construction of strains containing a single PvirB-lacZ chromosomal transcriptional fusion.
Plasmid pK18mob-PvirB-lacZ was transferred by biparental conjugation into the wild-type strain B. abortus 2308 or into B. abortus ΔmdrA, B. abortus ΔhutC, B. abortus ΔhutC ΔmdrA, or B. abortus ΔhutC ΔmdrA-KI. Kanr colonies were selected as single-homologous recombinants.
Electrophoresis mobility shift assays.
32P labeling of probes, incubations, and running conditions were as described previously (46). The radiolabeled probe vir-up was constructed using Taq (Invitrogen), primers pvirup (5′-ATGACAGGCATATTTCAAC-3′) and pvd228 (5′-GTGATTTTCATATTTTTGCGG-3′), and genomic DNA of B. abortus 2308 as the template. The radiolabeled probes vir-down and vir-down-ihf− were constructed by PCR using primers pvd229 and pvirdownI, and genomic DNA of B. abortus 2308 or plasmid pBluescript-PvirB-IHF-sacB/R as the template, respectively, as described previously (46). The radiolabeled control probe B10 was constructed with primers B10Qu and B10d2, as described previously (45). The radiolabeled probe 286/59 was constructed using primers pvu144 (5′-CGGGATCCGATTCTTGGTCGGGTTAC-3′) and pvirdownIII (5′-TGTTTAAGCCGATATATGGAC-3′) and genomic DNA of B. abortus 2308 as the template.
The unlabeled competitors were constructed by PCR using Taq, genomic DNA of B. abortus 2308 as the template, and primers pvirup and pvd228 for competitor up, primers pvu77 (5′-CGGGATCCGATGCCGCCTAATGGAGC-3′) and pvd228 for competitor 77, primers pvu144 and pvd228 for competitor 144, and primers pvu229 and pvirdownI for competitor down. The unlabeled competitors A1 and B1 were constructed as described previously (46). Binding reactions were performed in binding buffer (15 mM Tris-HCl [pH 8.0], 0.5 mM EDTA, 10 μg ml−1 bovine serum albumin [BSA], 1 mM dithiothreitol [DTT], 30 mM KCl, 6% glycerol, 50 μg ml−1 salmon sperm DNA).
DNase I footprinting.
Labeling reactions, purification of probes, binding reactions, DNase I digestion, purification of digested fragments, and electrophoresis conditions were performed as described previously (46). Probe vir-upII was constructed using Taq, primer pvd228, the 32P-labeled primer pvirup, and genomic DNA of B. abortus 2308 as the template. Probe vir-downII was constructed using primer pvu144, the 32P-labeled primer pvirdownI, and genomic DNA of B. abortus 2308 as the template. Probe vir-upIII was constructed using primer pvirup, the 32P-labeled primer pvirdownIII, and genomic DNA of B. abortus 2308 as the template.
Expression and purification of recombinant proteins.
Recombinant IHF and HutC were prepared as described previously (45, 46). Recombinant MdrA was prepared as follows. Plasmid pQE-31-mdrA was transferred into E. coli M15 (pREP4) (Qiagen). Transformed bacteria were cultured in LB until the OD600 reached 0.6 and were induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). After 4 h, bacteria were harvested, suspended in lysis buffer (20 mM Tris-HCl [pH 7.6], 1 mM phenylmethylsulfonyl fluoride [PMSF]), and disrupted by sonication. After centrifugation, NaCl was added to a final concentration of 0.5 M, and the sample was loaded into a Hi-Trap nickel-chelating column (Amersham Biosciences). After washing with buffer B (20 mM Tris-HCl [pH 7.6], 0.5 M NaCl, 50 mM imidazole), the column was equilibrated with buffer A (20 mM Tris-HCl [pH 7.6], 0.5 M NaCl) and eluted with a linear gradient of buffer C (20 mM Tris-HCl [pH 7.6], 0.5 M NaCl, 1 M imidazole). Eluates were analyzed by 12.5% SDS-PAGE, and the fractions containing the recombinant protein MdrA (purity, ∼95%) were pooled and dialyzed against buffer D (20 mM Tris-HCl [pH 7.6], 0.5 M NaCl, 3 mM β-mercaptoethanol).
β-Gal activity determinations.
β-Galactosidase (β-Gal) activity was determined in whole bacterial cells as described previously (46), and it was expressed in Miller units by the following formula: [A420/(volume × OD600)] ×100.
RESULTS
Isolation and identification of MdrA.
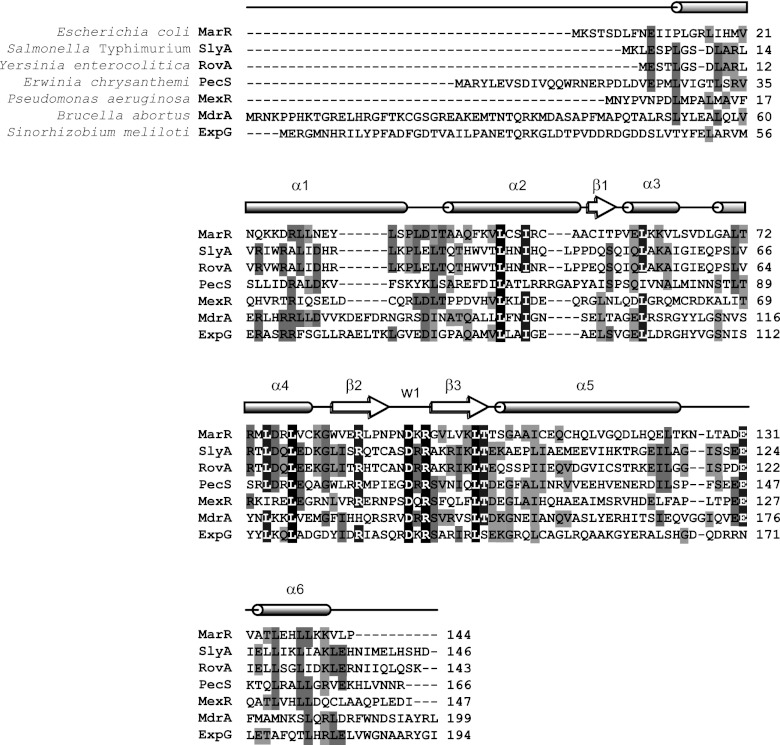
In previous studies, two transcription factors implicated in the control of expression of the virB operon were isolated by affinity chromatography from B. abortus total protein extracts (45, 46). Such analyses were performed by using DNA probes that contained sequences corresponding to positions −201 to +24 relative to the transcription start site of the virB promoter (PvirB). In order to identify possible additional regulatory proteins interacting with further upstream regions, we performed EMSAs using crude protein extracts and a radioactively labeled probe corresponding to positions −430 to −202 of PvirB. As shown in Fig. 1, a protein-DNA complex was detected using total protein extracts of B. abortus grown in rich medium (TSB) until exponential phase, whereas no signal was observed with stationary-phase extracts. To isolate the factor responsible for the observed complex, proteins were fractionated by ammonium sulfate precipitation and ion-exchange chromatography, followed by EMSA of each fraction during the purification process (see Materials and Methods). Subsequently, the partially purified protein fractions were subjected to affinity chromatography using a biotin-labeled probe, vir-up, or a nonrelated control probe bound to streptavidin-coated paramagnetic spheres, and the corresponding eluates were analyzed by SDS-PAGE. Using this procedure, we observed a specific band, which was excised from the gel and identified by mass spectrometry as YP_414216, a protein belonging to the MarR family of transcriptional regulators (14). Because of its biochemical properties and its role on regulation of virB expression (see below), we called this protein MdrA (for MarR-like sodium deoxycholate-responsive activator). As revealed by BLAST analyses, MdrA shares 29% identity and 44% similarity with the Escherichia coli prototypical member MarR (24), whereas it displayed slightly higher homology with other representative MarR members such as PecS, SlyA, and RovA (identity and similarity, 33% and 54%, 30% and 49%, and 31% and 50%, respectively) (21, 27, 34). As shown in Fig. 2, a multiple sequence alignment indicated that MdrA displays a high degree of amino acid conservation at positions corresponding to the winged helix-turn-helix DNA-binding domain (β1-α3-α4[recognition helix]-β2-W1[wing]-β3), which is characteristic of this family of transcriptional regulators (1).
Fig 1.
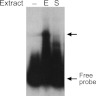
Identification of MdrA. EMSA performed with probe vir-up and total crude protein extracts from exponential (E) or stationary-phase (S) cultures of B. abortus. Arrows indicate the positions of the free probe and of a protein-DNA complex.
Fig 2.
Sequence alignment of MdrA and different representative members of the MarR family of transcriptional regulators. Relative positions of secondary-structure elements are shown as a schematic representation of α-helices, β-sheets, and the wing region (W1), according to the Escherichia coli MarR crystal structure described by Alekshun et al. (1). White letters identify residues identical in all (highlighted in black) or in six (highlighted in dark gray) of the seven MarR homologs considered. Black letters identify residues identical in five or four (highlighted in gray) or in three (highlighted in light gray) of the seven MarR homologs. The sequence alignment was performed using ClustalW (49).
MdrA interacts with PvirB at two different binding sites.
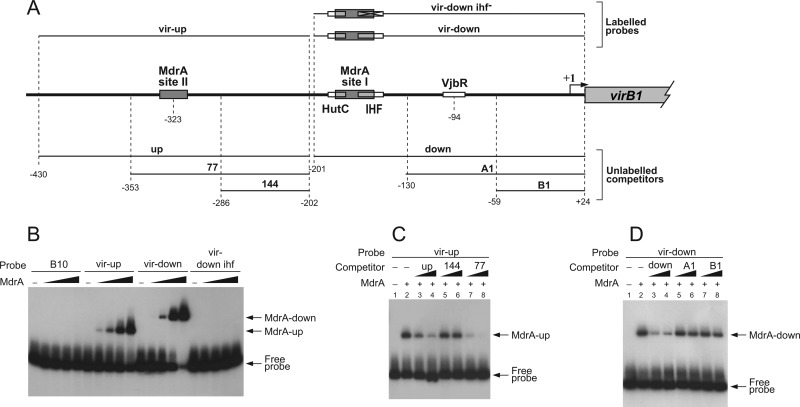
To study the MdrA-PvirB interaction, a His-tagged recombinant protein was expressed and assayed by EMSA using different radioactively labeled probes (Fig. 3A). As shown in Fig. 3B, recombinant MdrA interacted with probe vir-up, showing an electrophoretic mobility similar to that of the native nucleoprotein complex, whereas no signal was observed with the control probe B10. Incubation of MdrA with probe vir-down also resulted in the formation of a protein-DNA complex, which indicated that this regulator specifically recognizes two separated regions of the promoter (Fig. 3B). On the other hand, incubation of MdrA with a probe that lacks the IHF-binding site produced no signal by EMSA (Fig. 3B), suggesting that the MdrA-binding site overlaps that of IHF. Competition experiments performed in the presence of an excess of different unlabeled fragments indicated that the sequences recognized by MdrA in PvirB are located at two regions residing between positions −353 and −286 and between −201 and −130 (Fig. 3C and D). Here, we will refer to the downstream- and upstream-located elements recognized by this protein in PvirB as the MdrA-binding sites I and II, respectively.
Fig 3.
Analysis of the interaction between MdrA and the virB promoter. (A) Schematic representation of genomic sequences corresponding to the regulatory region of the virB operon, radiolabeled probes used for EMSA, or unlabeled competitors. Positions relative to the transcription start site are indicated. MdrA-, HutC-, IHF-, and VjbR-binding sites are indicated as boxes. The diagonally crossed box indicates the nonrelated sequences that replace the IHF-binding site in probe vir-down-ihf−. (B) EMSA performed with the indicated probes and 0, 10, 20, 40, or 60 nM MdrA. (C) EMSA performed with probe vir-up, MdrA, and the indicated unlabeled DNA fragments as competitors. Protein concentrations were as follows: line 1, no protein; lines 2 to 8, 30 nM. Concentrations of unlabeled DNA competitors were as follows: lanes 1 and 2, no competitor DNA; lanes 3, 5, and 7, 150 ng; lanes 4, 6, and 8, 360 ng. (D) EMSA performed with probe vir-down, MdrA, and the indicated unlabeled DNA fragments as competitors. The concentrations of protein and unlabeled DNA competitors were the same as for panel C.
Analysis of the interaction of MdrA, IHF, and HutC with the downstream region of PvirB.
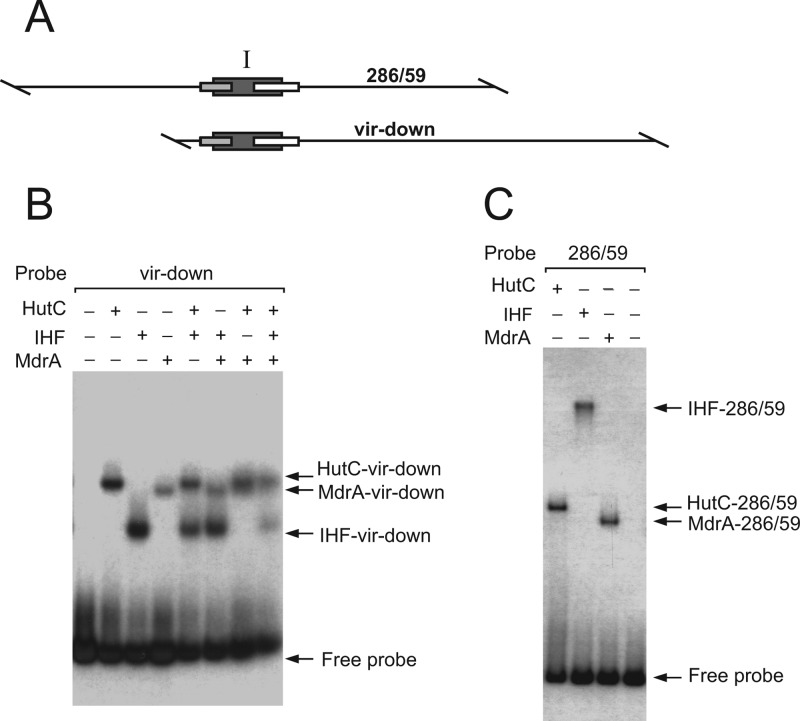
The above-described experiments showed that MdrA specifically binds to a 71-bp region of the promoter that also contains the binding sites for IHF and HutC. In previous works, we found that IHF and HutC compete with one another during binding to PvirB because their binding sites overlap (45, 46). Accordingly, we next asked if binding of MdrA to PvirB affects the interaction of IHF or HutC with the promoter. To address this question, we incubated probe vir-down with each of the three pairs of proteins or with all three proteins together. We then carried out EMSAs on the different protein combinations. Figure 4B shows that, separately, each of the regulators generated a signal corresponding to a binary protein-DNA complex, whereas no ternary complexes were observed with any of the combinations of two or more proteins. These results indicated that MdrA competes with IHF and HutC for the binding to the probe vir-down, suggesting that the MdrA-binding site I overlaps those of the two other regulators. The same results were obtained when EMSAs were carried out with probe 289/59, whose length is the same as that of probe vir-down but in which the location of the binding sites for MdrA, HutC, and IHF are close to the center rather than being near one end (data not shown). However, the electrophoretic mobility of the complex between IHF and probe 289/52 was substantially lower than that of the complex between IHF and probe vir-down (Fig. 4B and C). The reduced mobility of the IHF-289/52 complex is due to the IHF-induced bend of DNA, and the effect of bending on migration is more pronounced when the binding site is located near the center of the molecule (38). On the other hand, unlike IHF, the relative mobility of the MdrA- and HutC-nucleoprotein complexes remained constant regardless of the probe used, indicating that the last two transcriptional regulators do not induce bending of DNA upon binding (Fig. 4B and C).
Fig 4.
Analysis of the interaction between MdrA, HutC, and/or IHF with the downstream region of the virB promoter. (A) Schematic representation of probes vir-down and 286/59. Rectangles indicate the positions of the binding sites of HutC, IHF, and MdrA. (B) Coincubation experiment performed by EMSA with probe vir-down and different combinations of 20 nM MdrA, 20 nM HutC, or 40 nM IHF. (C) Coincubation experiment performed by EMSA with probe 286/59 and different combinations of MdrA, HutC, or IHF. Protein concentrations were as described for panel B.
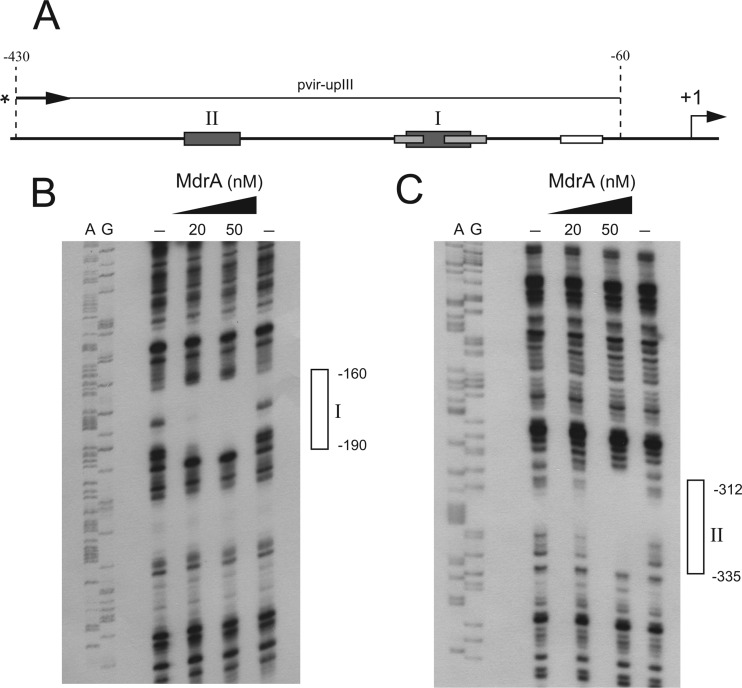
Determination of the MdrA-binding sites in PvirB.
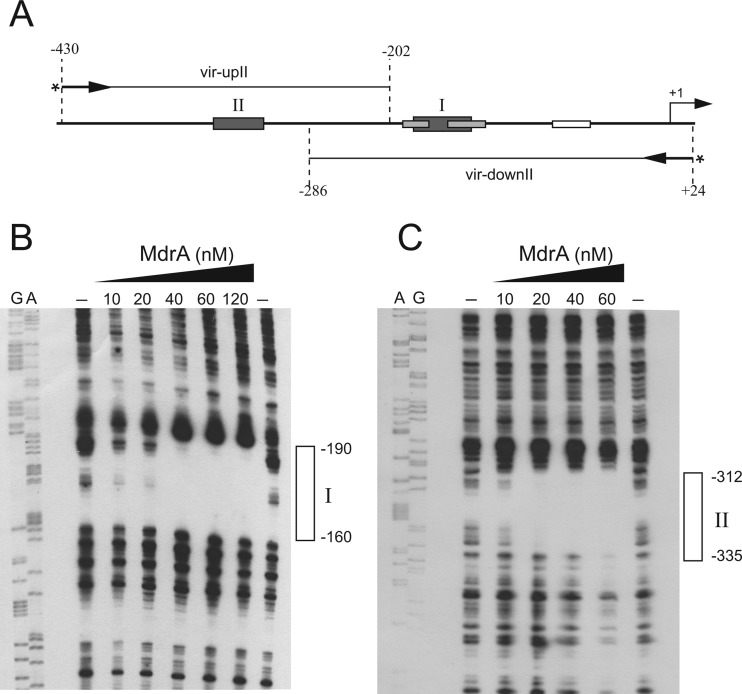
To identify both MdrA-binding sites I and II, we performed DNase I footprinting experiments using probes corresponding to different regions of PvirB: (i) a 228-bp fragment containing the upstream region of the promoter that was labeled at the 5′ end of the top strand and (ii) a 312-bp fragment containing the downstream region that was labeled at the 5′ end of the bottom strand (Fig. 5A). As shown in Fig. 5B, it was observed that MdrA bound to a 30-bp region extending from position −190 to −160, which corresponds to the MdrA-binding site I (Fig. 5B). DNase I footprinting experiments performed with the probe that contains sequences of the upstream region of PvirB showed that MdrA protected a 24-bp region that extends from position −335 to −312, which corresponds to the MdrA-binding site II (Fig. 5C). When a 370-bp fragment was end labeled at the top strand and used as a probe (Fig. 6A), it was observed that MdrA protected two regions corresponding to the MdrA-binding sites I and II (Fig. 6B and C), which indicated that this transcription factor is able to bind simultaneously to both operator sequences.
Fig 5.
Determination of the MdrA-binding sites in the virB promoter. (A) Schematic representation of probes vir-upII and vir-downII used for DNase I footprinting experiments. Positions relative to the transcription start site of PvirB are indicated. MdrA-, HutC-, IHF-, and VjbR-binding sites are indicated as boxes. An arrow and an asterisk indicate the primer used for construction of each 5′ radiolabeled probe. (B) DNase I footprinting experiment performed with probe vir-downII and MdrA at the indicated concentrations. Lanes A and G show sequence reactions performed by the Sanger method with primer vir-downI. The protected region of the MdrA-binding site I is indicated with an open rectangle. (C) DNase I footprinting experiment performed with probe vir-upII and MdrA at the indicated concentrations. Lanes A and G show sequence reactions performed by the Sanger method with primer vir-up. The protected region of the MdrA-binding site II is indicated as described for panel B.
Fig 6.
MdrA binds simultaneously to binding sites I and II. (A) Schematic representation of probe vir-upIII used for DNase I footprinting experiments shown in panels B and C. Positions relative to the transcription start site of PvirB are indicated. Rectangles indicate MdrA-, HutC-, IHF-, and VjbR-binding sites. The arrow and asterisk indicate the primer used for construction of the 5′ radiolabeled probe vir-upIII. (B and C) Different runs of a DNase I footprinting experiment performed with probe vir-upIII and MdrA at the indicated concentrations. Lanes A and G show sequence reactions performed by the Sanger method with primer pvirup. The protected regions of the MdrA-binding sites I and II are indicated with open rectangles.
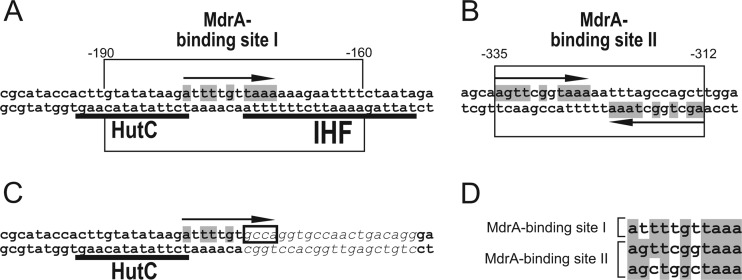
The DNase I footprinting experiments showed that the MdrA-binding site I overlaps those of both HutC and IHF (Fig. 5A), consistent with the observation that MdrA competed with these two transcriptional regulators for the binding to probe vir-down (Fig. 4B). The analysis of the protected regions revealed the presence of two partially conserved 11-bp motifs arranged as inverted repeats in the MdrA-binding site II (Fig. 7B). In contrast, the analysis of sequences corresponding to the MdrA-binding site I showed no obvious dyad symmetry. However, alignment of the 11-bp motifs together with the downstream protected region revealed the presence of a similar sequence located at the center of the MdrA-binding site I (Fig. 7A), suggesting that MdrA specifically recognizes this sequence. In agreement with this hypothesis, MdrA was unable to bind a probe lacking a sequence that is within the 11-bp motifs found in both MdrA-binding sites (Fig. 7C), which supports the notion that the sequence TAAA is part of the motif recognized by MdrA in the promoter.
Fig 7.
Sequence analysis of the MdrA-protected regions in the virB promoter. (A) Schematic representation of the MdrA-protected sequence in the binding site I. The rectangle indicates regions protected from DNase I cleavage in experiments performed with probe vir-upIII and vir-downII. Positions relative to the transcription start site are indicated. Sequences corresponding to the HutC- and IHF-binding sites are underlined. The arrow indicates the position of the 11-bp MdrA-binding consensus motif. Nucleotides that match the 11-bp MdrA-binding consensus motif are highlighted in gray. (B) Schematic representation of the MdrA-protected sequence in the binding site II. The open rectangle indicates the region protected from DNase I cleavage. Positions relative to the transcription start site are indicated. Arrows indicate the positions of the 11-bp MdrA-binding consensus motifs. Nucleotides that match the 11-bp MdrA-binding consensus motifs are highlighted in gray. Arrows indicate dyad symmetry. (C) Schematic representation of sequences corresponding to probe vir-down-ihf−. The HutC-binding site is indicated as described for panel A. Nucleotides that match the 11-bp MdrA-binding consensus motif are highlighted in gray. Nucleotides that replaced the IHF-binding site by a nonrelated sequence are indicated in italics. The position of nucleotides of the 11-bp MdrA-binding consensus motif that are not present in probe vir-down-ihf− is indicated by an open rectangle. (D) Alignment of sequences corresponding to the 11-bp MdrA-binding consensus motif found at the protected regions of the MdrA-binding sites I and II.
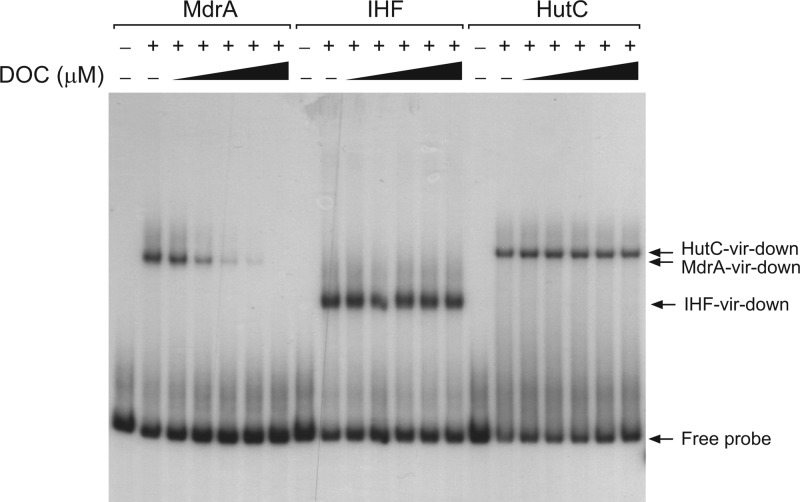
MdrA dissociates from DNA in the presence of sodium deoxycholate.
Several members of the MarR family are capable of directly binding specific ligands, which dissociate the regulator from DNA with a consequent modulation of gene expression (17). In Salmonella enterica serovar Typhimurium, it was recently found that sodium deoxycholate (DOC), a component of bile, interacts with MarR and interferes with its ability to bind the promoter of the marRAB operon (35). Accordingly, we investigated whether the DNA-binding activity of MdrA is susceptible to being affected by total bile salts or bile components. We observed by EMSA that formation of the MdrA-vir-down complex was impaired in the presence of total bile salts (data not shown). When individual components of bile were tested, we observed that binding of MdrA to PvirB was impaired by 250 μM DOC, whereas the binding of IHF or HutC was not affected by this bile salt at any of the assayed concentrations (Fig. 8). Therefore, these results demonstrated that, as is observed with other members of the MarR-family of transcriptional regulators, the interaction between MdrA and its operator sequences can be modulated by specific compounds, and such an ability may be involved in transduction of environmental stimuli to regulate MdrA-dependent gene expression.
Fig 8.
Effect of DOC on the binding activity of MdrA. EMSA performed with probe vir-down and 30 nM MdrA, 50 nM IHF, or 30 nM HutC in the presence of 100, 250, 500, 750, or 1,000 μM DOC.
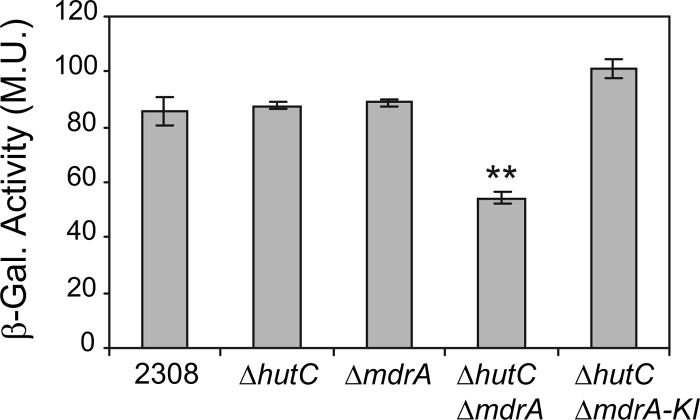
MdrA and HutC exert redundant roles on regulation of virB expression.
In order to determine the role of MdrA on transcriptional regulation of the virB genes, we introduced transcriptional fusions between PvirB and the lacZ reporter gene into B. abortus wild-type strain 2308 or into a ΔmdrA deletion mutant. Determination of β-galactosidase activities showed no differences between these strains after incubation of bacteria in rich medium (TSB) or in minimal media at different pH values in the presence of different carbon sources (data not shown). Based on these results, we hypothesized that the regulatory function of MdrA on PvirB is probably redundant with that of other transcription factors. Given that both MdrA and HutC bound to overlapping binding sites and that they appear to have similar structural roles, since neither of them induces DNA bending, we aimed to determine whether these two proteins exert redundant roles on virB expression. To this end, β-galactosidase activities of the PvirB-lacZ fusions were also assayed in both B. abortus ΔhutC and the B. abortus ΔhutC ΔmdrA double deletion mutant strain. Using all these genetic backgrounds, we observed no β-galactosidase activity differences between strains either when bacteria were grown in TSB until stationary phase (OD600, ≥3.5) or when they were grown in the above-mentioned minimal culture media (data not shown). However, in bacteria grown in TSB until exponential phase (OD600, ∼0.5 to 1.0), the ΔhutC ΔmdrA double deletion mutant showed a decrease of about 45% in promoter activity, whereas both the ΔhutC and the ΔmdrA single mutants showed the same levels of β-galactosidase activity as the wild-type strain (Fig. 9). Complementation of mdrA by a knock-in strategy in the double mutant restored the wild-type β-galactosidase activity levels (Fig. 9), thus confirming that MdrA exerts a growth-phase-dependent positive regulatory role on virB expression, functionally redundant with HutC. The analyses performed in the presence of DOC displayed the same pattern of β-galactosidase activity as that observed without this bile salt (data not shown), even at concentrations 3-fold higher than that which dissociated MdrA from the promoter. Thus, the sole presence of this bile salt was not a sufficient stimulus for modulating virB expression under standard culture conditions, which suggests that the capability of the MdrA-mediated regulation to respond to environmental signals may be restricted to specific environmental conditions.
Fig 9.
Role of MdrA in regulation of the virB promoter activity. B. abortus PvirB-lacZ (wild type) (strain 2308), B. abortus ΔhutC PvirB-lacZ, B. abortus ΔmdrA PvirB-lacZ, B. abortus ΔhutC ΔmdrA PvirB-lacZ, and B. abortus ΔhutC ΔmdrA-KI PvirB-lacZ were grown in rich medium (TSB) until exponential phase (OD600, 0.5 to 1). Subsequently, bacteria were harvested, and β-galactosidase activities were determined. Values are means ± standard deviations in Miller units (M.U.) of duplicate wells from a representative of three experiments. **, P < 0.01.
Taken together, these results revealed that MdrA controls expression of the virB genes in a growth-phase-dependent manner and exerts a regulatory role functionally redundant to that of a nonrelated factor.
DISCUSSION
Expression of the virB operon is under the control of transcriptional regulatory mechanisms that involve perception of environmental conditions related to the intracellular lifestyle of Brucella (4). Over the last years, several studies revealed that activity of the virB promoter is modulated by the direct action of many transcriptional regulators belonging to different families, including a nucleoid-associated protein (IHF), QS-related transcription factors, and a regulator of histidine catabolism (6, 12, 25, 45, 46). Here, using an approach that previously allowed us to isolate IHF and HutC, we identified MdrA, a protein belonging to the MarR family of transcription factors involved in regulation of virulence genes, aromatic catabolic pathways, or bacterial responses to environmental stress (53). The first evidence of interaction between MdrA and PvirB indicated that this regulator binds to the promoter at sequences located far upstream of the transcription start site (positions −430 to −202). Further EMSA analyses showed that MdrA was also able to interact with the downstream region, indicating that this regulator recognizes two distinct binding sites at PvirB (Fig. 3). These observations were confirmed by DNase I footprinting analyses that showed that MdrA is able to bind simultaneously to both regions, which are centered at positions −175 and −323.5 (MarR-binding sites I and II, respectively). It is remarkable that all positive regulators characterized so far in the MarR family act by means of displacing repressors (19, 40) or through prototypical activation mechanisms by interacting with RNA polymerase (RNAP) (28, 50). The latter mode of regulation may not account for the activity of MdrA on the virB promoter, since it activates transcription from regions located far upstream of position +1. We hypothesize that MdrA could interfere with the activity of an unknown repressor; a possible candidate may be BabR, which is a negative regulator that affects virB expression by 20% and whose binding site in the promoter has not yet been identified (6). Alternatively, contact between MdrA and RNAP may be mediated by additional elements that could introduce conformational changes in chromatin structure. Similar mechanisms were previously suggested for other positive regulators that bind far upstream from the transcription start site but whose mechanisms of activation have not yet been elucidated (e.g., EspR, RutR, TodT, and AlgR) (20, 26, 29, 39). It is worth mentioning that the possible involvement of elements that introduce modifications of the nucleoprotein structure of local sequences was also hypothesized for the LuxR-type regulator VjbR, since it binds at position −94 and consequently could not activate transcription of the virB promoter in a prototypical manner (2).
Our experiments revealed that the MdrA-binding site I overlaps those previously identified for IHF and HutC, which was consistent with the fact that these three transcription factors compete for binding to the promoter (Fig. 4). Similar structures have been reported for other promoters regulated by members of the MarR family (i.e., CbaR, MarR, and RovA), wherein the regulators bind to two or more operator sites (19, 24, 36). Furthermore, competition with nucleoid-associated proteins (i.e., H-NS and IHF) or with other global regulators (cyclic AMP receptor protein [CRP]) due to overlapping binding sites has also been reported to be part of the regulatory mechanisms involving MarR-related regulators (19, 40, 43). An additional feature shared by MdrA and most of the MarR homologs is the ligand-mediated modulation of its DNA-binding activity. However, it is important to highlight that MdrA is the first ligand-responsive positive regulator characterized in this family, since no ligand has yet been found for the other MarR-type transcriptional activators described so far (14). Curiously, MdrA is the second positive regulator of the virB promoter that dissociates from DNA in response to specific signals, since VjbR was previously shown to respond to acyl-homoserine lactones (2, 12). Our results indicated that either bile or micromolar concentrations of DOC specifically dissociated MdrA from DNA without affecting either IHF or HutC (Fig. 8). These observations have raised the hypothesis that after ligand recognition, MdrA could act as a signal transducer that modulates expression of the virB genes in response to exposure of Brucella to DOC within the host environment. Previous studies have described mechanisms in Brucella that confer resistance to DOC (11, 23, 33). Moreover, it was recently shown that Brucella is able to infect dendritic cells from murine Peyer's patches and to negatively modulate its activation through the action of the protein Btp1 (41). Although these lines of evidence support the notion that Brucella is able to overcome the action of bile salts and to invade antigen-presenting cells in the host's gut, we failed to observe any effect of DOC on virB expression in exponential-phase cultures under standard growth conditions. However, it cannot be excluded that, under some type of stress experienced by the bacterium within the host, this bile salt may permeate the bacterial membrane more easily and act as an environmental signal to induce an MdrA-mediated response. It is worth mentioning that it cannot be deduced that there exist any additional potential ligands for MdrA from the few MarR homologs reported so far in Rhizobiales, since their known ligands do not share structural similarity with DOC (5, 31). Therefore, further studies will be needed to determine whether this bile salt, or a structurally similar compound, can interact with MdrA in vivo and modulate virB expression.
The results presented here showed that MdrA and HutC, two independently isolated proteins belonging to different families of transcription factors, bind to PvirB at overlapping target DNA sequences and positively modulate expression of the virB genes to similar extents. Moreover, both regulators have been shown to play redundant roles since the regulatory activity of MdrA could not be determined unless a ΔmdrA ΔhutC double deletion mutant background was used to measure virB promoter activity (Fig. 9). In addition, our EMSAs indicated that, unlike IHF, neither MdrA nor HutC induced DNA bending upon binding to PvirB (Fig. 4), suggesting that these two transcriptional regulators exert similar structural roles in agreement with their redundant activity on modulation of virB expression.
The data obtained from the present work showed that the function of MdrA leads to a positive regulatory effect on virB expression, which does not take place in an all-or-nothing manner. Deletion of mdrA affected virB expression by 45%, which was a magnitude of difference detectable by quantitative measurements of β-Gal activity (Fig. 9) but not by Western blot experiments performed with an anti-VirB7 antibody (data not shown). Our results indicated that, similarly to what was previously observed with HutC, MdrA also acts as an activator that enhances virB promoter activity under defined conditions and appears to play an accessory regulatory role, probably acting to synchronize maximal virB expression with certain metabolic and/or environmental signals. It was previously observed that HutC activates intracellular expression of the virB genes within J774 macrophages (45). In contrast, MdrA does not appear to play any role in this experimental model, since deletion of mdrA did not produce any detectable intracellular effect and the ΔmdrA ΔhutC double mutant affected intracellular virB expression to the same extent as in the single ΔhutC deletion background (see Fig. S1 in the supplemental material). On the other hand, MdrA was necessary for virB expression in cultured bacteria at the exponential phase of the growth in rich medium, whereas in stationary phase we did not observe any MdrA-dependent effect (Fig. 9). In addition, it is worth noting that the growth phase wherein MdrA exerts its regulatory activity is coincident with the only condition under which it could be isolated (Fig. 1), indicating that this regulator is probably under the control of mechanisms that modulate its expression and/or its DNA-binding activity. Taken together, our observations suggest that the MdrA-mediated regulation of the virB genes may be a result of an adaptation to activate virB expression at stages of the infection process during which HutC is not acting. However, although functionality of MdrA was demonstrated, it still remains to be determined which are the conditions wherein MdrA-dependent expression is achieved in vivo. Besides, it will also be important to assess whether this MarR-related transcriptional regulator is able to perceive environmental signals in specific tissues/organs of the mammalian host during the course of the disease.
Supplementary Material
ACKNOWLEDGMENTS
We thank Joseph Connolly for help with mass spectrometry.
This work was supported by grant PICT05-38207 to R.S. and grant PICT06-651 to R.A.U. from the Agencia Nacional de Promoción Científica y Tecnológica, Buenos Aires, Argentina; by grant PIP2011-00336 to R.S. from the Argentinian Council of Research (CONICET); and by grant UBACyT X240 to A.Z. from the University of Buenos Aires, Argentina.
Footnotes
Published ahead of print 21 September 2012
This work is dedicated to the memory of Rodolfo A. Ugalde.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Alekshun MN, Levy SB, Mealy TR, Seaton BA, Head JF. 2001. The crystal structure of MarR, a regulator of multiple antibiotic resistance, at 2.3 A resolution. Nat. Struct. Biol. 8:710–714 [DOI] [PubMed] [Google Scholar]
- 2. Arocena GM, Sieira R, Comerci DJ, Ugalde RA. 2010. Identification of the quorum sensing target DNA-sequence and N-acyl homoserine lactone responsiveness in the Brucella abortus virB promoter. J. Bacteriol. 192:3434–3440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Backert S, Meyer TF. 2006. Type IV secretion systems and their effectors in bacterial pathogenesis. Curr. Opin. Microbiol. 9:207–217 [DOI] [PubMed] [Google Scholar]
- 4. Boschiroli ML, et al. 2002. The Brucella suis virB operon is induced intracellularly in macrophages. Proc. Natl. Acad. Sci. U. S. A. 99:1544–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Caswell CC, Baumgartner JE, Martin DW, Roop RM., II 2012. Characterization of the organic hydroperoxide resistance system of Brucella abortus 2308. J. Bacteriol. 194:5065–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Caswell CC, Gaines JM, Roop RM., II 2012. The RNA chaperone Hfq independently coordinates expression of the VirB type IV secretion system and the LuxR-type regulator BabR in Brucella abortus 2308. J. Bacteriol. 194:3–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Celli J, et al. 2003. Brucella evades macrophage killing via VirB-dependent sustained interactions with the endoplasmic reticulum. J. Exp. Med. 198:545–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Covacci A, Telford JL, Del Giudice G, Parsonnet J, Rappuoli R. 1999. Helicobacter pylori virulence and genetic geography. Science 284:1328–1333 [DOI] [PubMed] [Google Scholar]
- 9. de Barsy M, et al. 2011. Identification of a Brucella spp. secreted effector specifically interacting with human small GTPase Rab2. Cell. Microbiol. 13:1044–1058 [DOI] [PubMed] [Google Scholar]
- 10. de Jong MF, Sun YH, den Hartigh AB, van Dijl JM, Tsolis RM. 2008. Identification of VceA and VceC, two members of the VjbR regulon that are translocated into macrophages by the Brucella type IV secretion system. Mol. Microbiol. 70:1378–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Delpino MV, et al. 2007. A bile salt hydrolase of Brucella abortus contributes to the establishment of a successful infection through the oral route in mice. Infect. Immun. 75:299–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Delrue RM, et al. 2005. A quorum-sensing regulator controls expression of both the type IV secretion system and the flagellar apparatus of Brucella melitensis. Cell. Microbiol. 7:1151–1161 [DOI] [PubMed] [Google Scholar]
- 13. Dozot M, et al. 2006. The stringent response mediator Rsh is required for Brucella melitensis and Brucella suis virulence, and for expression of the type IV secretion system virB. Cell. Microbiol. 8:1791–1802 [DOI] [PubMed] [Google Scholar]
- 14. Ellison DW, Miller VL. 2006. Regulation of virulence by members of the MarR/SlyA family. Curr. Opin. Microbiol. 9:153–159 [DOI] [PubMed] [Google Scholar]
- 15. Gerhardt P. 1958. The nutrition of brucellae. Bacteriol. Rev. 22:81–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Godfroid J, et al. 2011. Brucellosis at the animal/ecosystem/human interface at the beginning of the 21st century. Prev. Vet. Med. 102:118–131 [DOI] [PubMed] [Google Scholar]
- 17. Grkovic S, Brown MH, Skurray RA. 2002. Regulation of bacterial drug export systems. Microbiol. Mol. Biol. Rev. 66:671–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Haine V, et al. 2005. Systematic targeted mutagenesis of Brucella melitensis 16M reveals a major role for GntR regulators in the control of virulence. Infect. Immun. 73:5578–5586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Heroven AK, Nagel G, Tran HJ, Parr S, Dersch P. 2004. RovA is autoregulated and antagonizes H-NS-mediated silencing of invasin and rovA expression in Yersinia pseudotuberculosis. Mol. Microbiol. 53:871–888 [DOI] [PubMed] [Google Scholar]
- 20. Lacal J, Busch A, Guazzaroni ME, Krell T, Ramos JL. 2006. The TodS-TodT two-component regulatory system recognizes a wide range of effectors and works with DNA-bending proteins. Proc. Natl. Acad. Sci. U. S. A. 103:8191–8196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ludwig A, et al. 1995. SlyA, a regulatory protein from Salmonella typhimurium, induces a haemolytic and pore-forming protein in Escherichia coli. Mol. Gen. Genet. 249:474–486 [DOI] [PubMed] [Google Scholar]
- 22. Marchesini MI, Herrmann CK, Salcedo SP, Gorvel JP, Comerci DJ. 2011. In search of Brucella abortus type IV secretion substrates: screening and identification of four proteins translocated into host cells through VirB system. Cell. Microbiol. 13:1261–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Martin FA, et al. 2009. Interplay between two RND systems mediating antimicrobial resistance in Brucella suis. J. Bacteriol. 191:2530–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Martin RG, Rosner JL. 1995. Binding of purified multiple antibiotic-resistance repressor protein (MarR) to mar operator sequences. Proc. Natl. Acad. Sci. U. S. A. 92:5456–5460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Martinez-Nunez C, et al. 2010. The two-component system BvrR/BvrS regulates the expression of the type IV secretion system VirB in Brucella abortus. J. Bacteriol. 192:5603–5608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mohr CD, Hibler NS, Deretic V. 1991. AlgR, a response regulator controlling mucoidy in Pseudomonas aeruginosa, binds to the FUS sites of the algD promoter located unusually far upstream from the mRNA start site. J. Bacteriol. 173:5136–5143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nagel G, Lahrz A, Dersch P. 2001. Environmental control of invasin expression in Yersinia pseudotuberculosis is mediated by regulation of RovA, a transcriptional activator of the SlyA/Hor family. Mol. Microbiol. 41:1249–1269 [DOI] [PubMed] [Google Scholar]
- 28. Navarre WW, et al. 2005. Co-regulation of Salmonella enterica genes required for virulence and resistance to antimicrobial peptides by SlyA and PhoP/PhoQ. Mol. Microbiol. 56:492–508 [DOI] [PubMed] [Google Scholar]
- 29. Nguyen Ple M, Bervoets I, Maes D, Charlier D. 2010. The protein-DNA contacts in RutR*carAB operator complexes. Nucleic Acids Res. 38:6286–6300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. O'Callaghan D, et al. 1999. A homologue of the Agrobacterium tumefaciens VirB and Bordetella pertussis Ptl type IV secretion systems is essential for intracellular survival of Brucella suis. Mol. Microbiol. 33:1210–1220 [DOI] [PubMed] [Google Scholar]
- 31. Perera IC, Grove A. 2010. Urate is a ligand for the transcriptional regulator PecS. J. Mol. Biol. 402:539–551 [DOI] [PubMed] [Google Scholar]
- 32. Pizarro-Cerda J, et al. 1998. Brucella abortus transits through the autophagic pathway and replicates in the endoplasmic reticulum of nonprofessional phagocytes. Infect. Immun. 66:5711–5724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Posadas DM, et al. 2007. The TolC homologue of Brucella suis is involved in resistance to antimicrobial compounds and virulence. Infect. Immun. 75:379–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Praillet T, Nasser W, Robert-Baudouy J, Reverchon S. 1996. Purification and functional characterization of PecS, a regulator of virulence-factor synthesis in Erwinia chrysanthemi. Mol. Microbiol. 20:391–402 [DOI] [PubMed] [Google Scholar]
- 35. Prouty AM, et al. 2004. Transcriptional regulation of Salmonella enterica serovar Typhimurium genes by bile. FEMS Immunol. Med. Microbiol. 41:177–185 [DOI] [PubMed] [Google Scholar]
- 36. Providenti MA, Wyndham RC. 2001. Identification and functional characterization of CbaR, a MarR-like modulator of the cbaABC-encoded chlorobenzoate catabolism pathway. Appl. Environ. Microbiol. 67:3530–3541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rambow-Larsen AA, Rajashekara G, Petersen E, Splitter G. 2008. Putative quorum-sensing regulator BlxR of Brucella melitensis regulates virulence factors including the type IV secretion system and flagella. J. Bacteriol. 190:3274–3282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Robertson CA, Nash HA. 1988. Bending of the bacteriophage lambda attachment site by Escherichia coli integration host factor. J. Biol. Chem. 263:3554–3557 [PubMed] [Google Scholar]
- 39. Rosenberg OS, et al. 2011. EspR, a key regulator of Mycobacterium tuberculosis virulence, adopts a unique dimeric structure among helix-turn-helix proteins. Proc. Natl. Acad. Sci. U. S. A. 108:13450–13455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rouanet C, Nomura K, Tsuyumu S, Nasser W. 1999. Regulation of pelD and pelE, encoding major alkaline pectate lyases in Erwinia chrysanthemi: involvement of the main transcriptional factors. J. Bacteriol. 181:5948–5957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Salcedo SP, et al. 2008. Brucella control of dendritic cell maturation is dependent on the TIR-containing protein Btp1. PLoS Pathog. 4:e21 doi:10.1371/journal.ppat.0040021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schafer A, et al. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73 [DOI] [PubMed] [Google Scholar]
- 43. Schielke S, et al. 2009. Expression of the meningococcal adhesin NadA is controlled by a transcriptional regulator of the MarR family. Mol. Microbiol. 72:1054–1067 [DOI] [PubMed] [Google Scholar]
- 44. Segal G, Purcell M, Shuman HA. 1998. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc. Natl. Acad. Sci. U. S. A. 95:1669–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sieira R, Arocena GM, Bukata L, Comerci DJ, Ugalde RA. 2010. Metabolic control of virulence genes in Brucella abortus: HutC coordinates virB expression and the histidine utilization pathway by direct binding to both promoters. J. Bacteriol. 192:217–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sieira R, Comerci DJ, Pietrasanta LI, Ugalde RA. 2004. Integration host factor is involved in transcriptional regulation of the Brucella abortus virB operon. Mol. Microbiol. 54:808–822 [DOI] [PubMed] [Google Scholar]
- 47. Sieira R, Comerci DJ, Sanchez DO, Ugalde RA. 2000. A homologue of an operon required for DNA transfer in Agrobacterium is required in Brucella abortus for virulence and intracellular multiplication. J. Bacteriol. 182:4849–4855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Stachel SE, Nester EW. 1986. The genetic and transcriptional organization of the vir region of the A6 Ti plasmid of Agrobacterium tumefaciens. EMBO J. 5:1445–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tran HJ, et al. 2005. Analysis of RovA, a transcriptional regulator of Yersinia pseudotuberculosis virulence that acts through antirepression and direct transcriptional activation. J. Biol. Chem. 280:42423–42432 [DOI] [PubMed] [Google Scholar]
- 51. Ugalde JE, Czibener C, Feldman MF, Ugalde RA. 2000. Identification and characterization of the Brucella abortus phosphoglucomutase gene: role of lipopolysaccharide in virulence and intracellular multiplication. Infect. Immun. 68:5716–5723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Whatmore AM. 2009. Current understanding of the genetic diversity of Brucella, an expanding genus of zoonotic pathogens. Infect. Genet. Evol. 9:1168–1184 [DOI] [PubMed] [Google Scholar]
- 53. Wilkinson SP, Grove A. 2006. Ligand-responsive transcriptional regulation by members of the MarR family of winged helix proteins. Curr. Issues Mol. Biol. 8:51–62 [PubMed] [Google Scholar]
- 54. Winans SC, Walker GC. 1985. Conjugal transfer system of the IncN plasmid pKM101. J. Bacteriol. 161:402–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.