Abstract
Staphylococcus aureus readily infects humans, causing infections from mild superficial skin infections to lethal bacteremia and endocarditis. Transporters produced by S. aureus allow the pathogen to adapt to a variety of settings, including survival at sites of infection and in the presence of antibiotics. The native functions of many transporters are unknown, but their potential dual contribution to fitness and antimicrobial resistance highlights their importance in staphylococcal infections. Here, we show that S. aureus NorD, a newly recognized efflux pump of the major facilitator superfamily, contributes to fitness in a murine subcutaneous abscess model. In community-associated methicillin-resistant S. aureus (CA-MRSA) strain MW2, norD was selectively upregulated 36-fold at the infection site relative to growth in vitro, and the norD mutant demonstrated significant fitness impairment in abscesses, with fitness 20- to 40-fold lower than that of the parent MW2 strain. Plasmid-encoded NorD could complement the fitness defect of the MW2 norD mutant. Chromosomal norD expression is polycistronic with the upstream oligopeptide permease genes (opp1ABCDF), which encode an ABC oligopeptide transporter. Both norD and opp1 were upregulated in abscesses and iron-restricted culture medium and negatively regulated by Fur, but only NorD contributed to fitness in the murine abscess model.
INTRODUCTION
Staphylococcus aureus is a versatile pathogen that causes a variety of diseases from food poisoning, skin infections, and abscesses to lethal bacteremia and endocarditis (21). Its adaptability is facilitated by a range of virulence factors and by factors that contribute to its survival and fitness in multiple environments within the host (2). Among the contributors to S. aureus fitness are a range of transmembrane transporters that can mediate uptake of nutrients or export of metabolites and toxic substances, including in some cases export of antimicrobial agents causing antimicrobial resistance (5, 9, 15, 16, 26, 28).
We found previously that several staphylococcal efflux pumps were selectively overexpressed within the milieu of experimental subcutaneous abscesses and that these pumps not only contributed to antimicrobial resistance when overexpressed in vitro but also contributed to bacterial survival within the abscess in the absence of antimicrobial agents (12). Both NorB and Tet38, members of the major facilitator superfamily (MFS) of secondary transporters encoded on the S. aureus chromosome, contributed selectively to fitness in the abscess environment and conferred low-level resistance to fluoroquinolones and tetracycline, respectively.
In order to identify other transporters that might also be important for bacterial survival in this environment, we evaluated the MFS transporter NorD. Like norB and tet38, norD expression was selectively upregulated in abscesses. Herein, we present data indicating that NorD contributes substantially to S. aureus fitness in an abscess infection model, and we identified a low-iron environment as a potential physiologic trigger for its upregulation in vivo. The selective fitness defect of a norD mutant is substantially greater than that reported for norB (12). Interestingly, expression of norD appears to be polycistronic with the upstream oligopeptide permease ABC family transporter opp1ABCDF, together forming the opp1-norD operon. Although opp1 alone appears to contribute little to S. aureus fitness in the abscess environment, all genes in the operon are coregulated in a low-iron medium in vitro and during infection. Intact fur (encoding the ferric uptake repressor) but not mgrA was necessary for norD induction under iron-limiting conditions.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All S. aureus strains, with the exception of RN4220 (r−) and RN6390, were derivatives of methicillin-resistant S. aureus (MRSA) strain MW2 (1, 2). S. aureus strains were cultivated in Trypticase soy broth (TSB) (Difco, Sparks, MD), and Escherichia coli strains were grown in Luria-Bertani broth (LB) (Difco) at 37°C, unless stated otherwise. The following antibiotics were obtained from Fisher (Sewanee, GA) or Sigma (St. Louis, MO) and used at the concentrations indicated: ampicillin, 100 μg/ml; chloramphenicol, 10 μg/ml; kanamycin, 50 μg/ml; tetracycline, 3 μg/ml; and anhydrotetracycline, 100 μg/ml. 5-Bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (Sigma) was used at 60 μg/ml in LB agar. To measure the induction of NorD in iron-restricted medium, strain MW2 was cultivated for 30 min in TSB supplemented with 400 μM dipyridyl (Sigma) with or without the addition of 50 μM ferrous sulfate (Sigma) (30).
Construction of norD, opp1, mgrA, and fur mutants.
We constructed an in-frame deletion, omitting all but the first 6 and the last 5 codons of norD (MW2386) in S. aureus MW2 (Fig. 1). The in-frame deletion fragment was generated using splicing by overlap extension PCR (11, 17, 19) with the primers norD-UpF with norD-UpR and norD-DnF with norD-DnR (Table 1). The insert was first cloned in pCR2.1, a TA cloning vector, in E. coli TOP10 (Invitrogen) and was PCR amplified using primers attB1-norD-F and attB2-norD-R to add attB1 and attB2 sites on either end of the amplicon. These two attB sites facilitated Gateway cloning (BP clonase II enzyme mix; Invitrogen) of the PCR products into shuttle vector pKOR1 (3) to generate pΔnorD. The correct construct was confirmed by DNA sequencing, and pΔnorD was electroporated into S. aureus RN4220, isolated, and then electroporated into S. aureus MW2. Allelic exchange using plasmid pΔnorD was performed as previously described (3). The extent of the in-frame deletion of 386 amino acids resulted in a ΔnorD mutant, which retained only 11 intact codons. The MW2 norD mutant was confirmed by sequence analysis of its PCR product. opp1 and mgrA in-frame deletions were also constructed in strain MW2 using the same strategy. Specifically, out of 1,657 codons in opp1, there are 32 codons retained in the MW2 opp1 strain, and norD is intact in this strain. The MW2 mgrA deletion mutant retains 20 of 148 mgrA codons. The PCR primers used for constructing the deletion mutants are listed in Table 1. The MW2 fur mutant was constructed by bacterial phage transduction. Briefly, an S. aureus phage 85 lysate was prepared from Newman fur::tet (kindly provided by Dominique Missiakas and Chuan He). A single MW2 transductant isolated on LB plates containing 5 μg/ml tetracycline was isolated and phenotypically characterized; the fur open reading frame (ORF) disruption was confirmed by PCR.
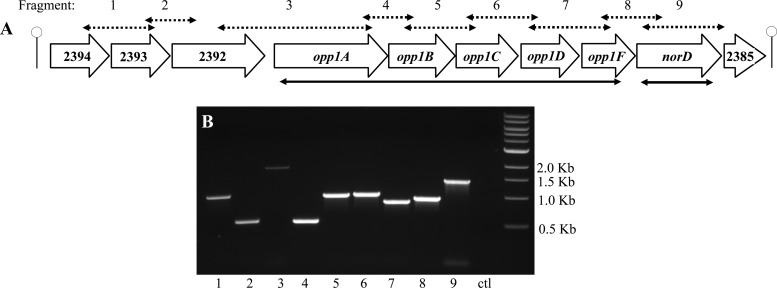
Fig 1.
Schematic depiction of the S. aureus strain MW2 opp1-norD chromosomal operon from MW2394 to MW2385. (A) Stemmed loops indicate the putative Rho-independent terminators. The extent of deletions in the opp1ABCDF and norD genes are indicated by the solid black double-headed arrows below the schematic map. Double-headed dashed arrows indicate cotranscription of genes demonstrated by RT-PCR of cDNA synthesized using a reverse primer to MW2385. (B) PCR products of the predicted sizes arrayed by agarose gel electrophoresis. The control lane (ctl) containing the RNA sample as the PCR template demonstrates no residual DNA after DNase I digestion.
Table 1.
Sequences of primers used in this study
| Use and primer | Sequence |
|---|---|
| Construction of mutants | |
| mgrA-UpF | 5′-GTTTAAGATCGCAACAAACACA |
| mgrA-UpR | 5′-ATAAGCGATCAGACATTAAAGTTCTCCTCC |
| mgrA-DnF | 5′-GTCTGATCGCTTATTAGGTAAAGTCATTCATGC |
| mgrA-DnR | 5′-CTGCATCAACATGGATTTTAGA |
| attB1-mgrA-F | 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTGTTTAAGATCGCAACAAACACA |
| attB2-mgrA-R | 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCTGCATCAACATGGATTTTAGA |
| norD-UpF | 5′-TGTTGGAAGAAGTCGGTCTATC |
| norD-UpR | 5′-ACTACTCGCTCAAGCCATTGCACCTTTCAT |
| norD-DnF | 5′-GCAATGGCTTGAGCGAGTAGTCTTTAATGAAAA |
| norD-DnR | 5′-TAATACATGGTGGGAATTCATTCC |
| attB1-norD-F | 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTGTTGGAAGAAGTCGGTCTATC |
| attB2-norD-R | 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTAATACATGGTGGGAATTCATTCC |
| opp1-UpF | 5′-CTGAAACGCGAAACA |
| opp1-UpR | 5′-TTCTATTTTTCTCATTTGCTTTTCCTCTT |
| opp1-DnF | 5′-GCAAATGAGAAAAATAGAAGAACAAATTCCGACAAGC |
| opp1-DnR | 5′-AATACCACTGTTCAAC |
| attB1-opp1-F | 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCTGAAACGCGAAACA |
| attB2-opp1-R | 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTAATACCACTGTTCAAC |
| Real-time RT-PCR | |
| opp1A-RTf | 5′-ACGATCCACAAAGTACTATTGC |
| opp1A-RTr | 5′-TAAATGCGTCATCAATGCTGTT |
| opp1B-RTf | 5′-AATCCAGCTGTGACAATTTTAC |
| opp1B-RTr | 5′-GCATCGCTTCAAGTAACCAATTTT |
| opp1C-RTf | 5′-ATGTCTTTTTAGGATTAGCAGC |
| opp1C-RTr | 5′-GTCAGTACCTAGTAGATGTTGA |
| opp1D-RTf | 5′-TGGTAGCGGTAAATCAATCACT |
| opp1D-RTr | 5′-GCGATTCAGATAATGACAACATTGA |
| opp1F-RTf | 5′-GAAAGGTGTGTCATTTGAGTGT |
| opp1F-RTr | 5′-CCGGTTTCTCAATACCTAATAT |
| mw2385-RTf | 5′-TTTCGCAAAATTTATTTCGTCGTCC |
| mw2385-RTr | 5′-GCTCTACAAAGTTATATTGGCAGTA |
| mw2392-RTf | 5′-TTCGAAATCTTGCCAGATAT |
| mw2392-RTr | 5′-GCTTAAATGGTACAGCTGAAAA |
| mw2392-r | 5′-ATGCGCCTCGTTTTGTATT |
| mw2393-RTf | 5′-TGATATTGATCCACAAGCCG |
| mw2393-RTr | 5′-ACCTTTTGATCCGTAATTGT |
| mw2394-RTf | 5′-TGAAGTTCAAATGCCACAAG |
| mw2394-RTr | 5′-ACTTGTTTAACTGGAATCAC |
| norD-RTf | 5′-ATGAAAGGTGCAATGGCTTG |
| norD-RTr | 5′-GCTATGGCATTGATGATCAAAA |
| gmk-RTf | 5′-TATCAGGACCATCTGGAGTAGG |
| gmk-RTr | 5′-CATCAACTTCACCTTCACGC |
| Construction of norD complementary plasmid | |
| NdeI-PnorD-F | 5′-TGAACATATGAAAGGTGCAATGGCTTG |
| BamHI-PnorD-R | 5′-GTTTGGATCCTCATTAAAGACTACTCGCTGGGC |
Cloning of norD and construction of the norD complementation vector.
The entire 1,194 bp of the norD coding sequence was amplified from S. aureus MW2 chromosomal DNA using primer pairs NdeI-PnorD-F and BamHI-PnorD-R. The amplicon was cloned into NdeI and BamHI sites of shuttle vector pOS1-Plgt to generate pNorD, in which norD transcription is under the control of the lipoprotein diacylglycerol transferase (lgt) promoter (37). These plasmids were electroporated into S. aureus RN4220 and then into MW2 norD and maintained by addition of 10 μg/ml chloramphenicol to the culture medium, unless otherwise stated.
Mouse subcutaneous abscess model.
Swiss Webster male mice aged 4 to 6 weeks were used for the subcutaneous abscess model, as previously described (6, 13). Briefly, exponential-phase S. aureus cultures were prepared by diluting overnight cultures 1:100 into TSB and incubating at 37°C with rotation until the culture medium reached an optical density at 600 nm (OD600) of 0.8. Cells of a single strain or a 1:1 mixture of mutant and parental strains were washed with and diluted 1:20 in phosphate-buffered saline (PBS). The cell suspension was then mixed with an equal volume of autoclaved Cytodex-1 beads (131 to 220 μm; Sigma) in PBS to generate the inoculum mentioned in the legend to Fig. 5. The suspension of cells and beads (0.2 ml) was injected subcutaneously in each shaved flank of a mouse anesthetized with ketamine and xylazine. After 48 h, the abscesses were excised and homogenized, and the bacterial burden was determined by enumerating CFU on LB agar plates. For in vitro experiments, 10 μl of the bacterial suspension was inoculated into 4 ml of TSB and grown for ∼24 h at 37°C with rotation. For fitness competition experiments, to distinguish the two strains, we used a streptomycin-resistant derivative of S. aureus MW2 designated MW2SM, which grows in the presence of 100 μg streptomycin/ml. MW2SM is a spontaneous mutant selected for its resistance to 500 μg streptomycin/ml, and its growth rate is the same as the parental strain MW2 (not shown). Their metabolic profiles are also identical as tested by API assays (bioMérieux) (J. Lee, unpublished data). The total CFU recovered from abscesses (or culture) of the two competing bacterial strains were evaluated using the competitive index (CI), which is defined as the output/input ratio. The ratio was calculated as follows: number of CFU of the competing strain/number of CFU of the MW2SM strain. For in vivo comparisons, a CI near one indicates similar numbers of CFU of the two competing strains recovered from abscesses, suggesting comparable fitness of the two strains. A CI of <1 indicates the fitness advantage of the parental strain MW2SM over the competitor. The difference between paired strains was analyzed using Wilcoxon signed rank test by comparing their CIs to one, a theoretical median. For comparison of two strains injected separately, the medians of recovered CFU were analyzed using the nonparametric Wilcoxon rank sum test. Each P value was estimated with two tails.
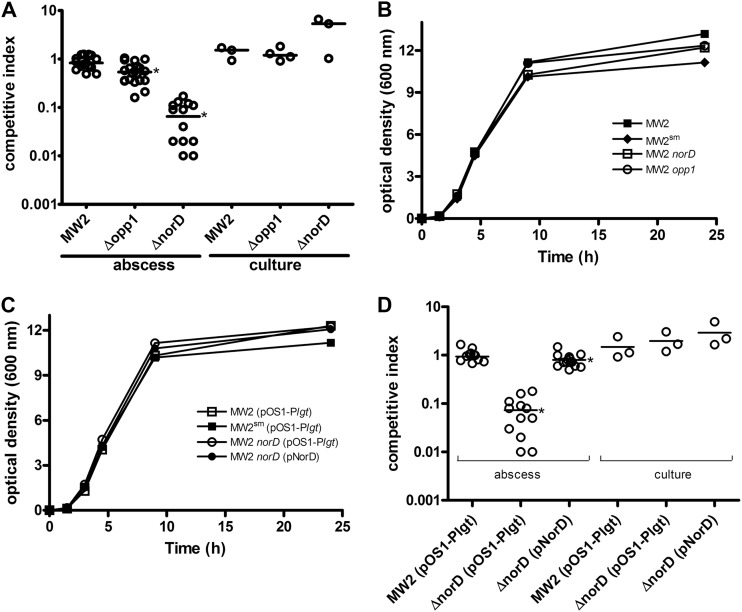
Fig 5.
The S. aureus MW2 norD mutant exhibits a fitness defect in the murine subcutaneous abscesses. (A) Fitness competition between S. aureus MW2 and MW2 opp1, MW2 norD, and MW2SM mutants in the murine subcutaneous abscess model. Exponential-phase bacteria, together with Cytodex beads, were injected subcutaneously in both flanks of each mouse at a typical input ratio for each bacterial strain of 1:1. The inoculum was 1 × 106 to 5 × 106 CFU in 0.2 ml for each lesion. After 48 h, the bacterial load of each abscess was estimated by quantitative plate cultures. For in vitro TSB cultures, the numbers of bacteria were determined after 24 h. Competitor strains were distinguished by resistance to streptomycin in strain MW2SM. A competitive index (CI) was calculated as the output ratio of two strains divided by the input ratio. Each circle represents the CI for one abscess, and the horizontal bars represent medians for the group. (B and C) Growth curves of strains cultivated in TSB broth at 37°C. Representative results of repeated experiments are shown. (D) The fitness defect of the MW2 norD mutant in murine subcutaneous abscesses was complemented by plasmid-encoded NorD. All strains competed with MW2SM(pOS1-Plgt) as described above. The data shown in panels A and D are the results compiled from three independent experiments. An asterisk indicates that the median CI is significantly different from one.
Preparation of bacterial RNA and real-time RT-PCR.
Total S. aureus RNA was isolated using the RNeasy minikit (Qiagen, Valencia, CA). For exponential-phase bacterial RNA, cells were collected at an OD600 of 0.3 to 0.5, and the cells were lysed with lysostaphin (Sigma). For stationary-phase bacteria, RNA was extracted from overnight cultures (24 h) in TSB. Bacterial RNA expressed in vivo was isolated from abscesses as previously described (12). All RNA samples were digested with DNase I (Turbo DNA-free; Ambion, Austin, TX) to ensure the absence of detectable DNA by PCR. Real-time reverse transcription-PCR (RT-PCR) was performed in two steps. cDNA was synthesized using the Verso cDNA kit (ABgene, Epsom, Surrey, United Kingdom) with a gene-specific reverse primer. Quantitative PCR (qPCR) amplifications were conducted using SYBR green master mix (ABgene), as previously described (12). The primers used for reverse transcription and qPCR are listed in Table 1. Gene expression was estimated by the ΔΔCT method relative to the expression of the endogenous reference gene gmk (36). The difference of relative expression measured by RT-PCR was analyzed by Mann-Whitney U tests. Each P value was estimated with two tails.
RESULTS
norD, which encodes a highly conserved efflux pump among S. aureus strains, is upregulated during infection.
MW2386, a chromosomal gene of S. aureus MW2, encodes a protein with 397 amino acids. Sequence analysis predicts it to be an integral membrane protein with an N-terminal signal sequence and 12 membrane-spanning helices; it is a member of the MFS, as are the multidrug resistance transporters (NorA, NorB, and NorC) in S. aureus. Thus, we designated this gene norD. In addition to MW2, all of the 33 other S. aureus strains with available genome sequences (NCBI, 1 August 2012), harbor a chromosomal copy of norD, and BLAST results showed that the amino acid sequence is highly conserved (identity > 96%).
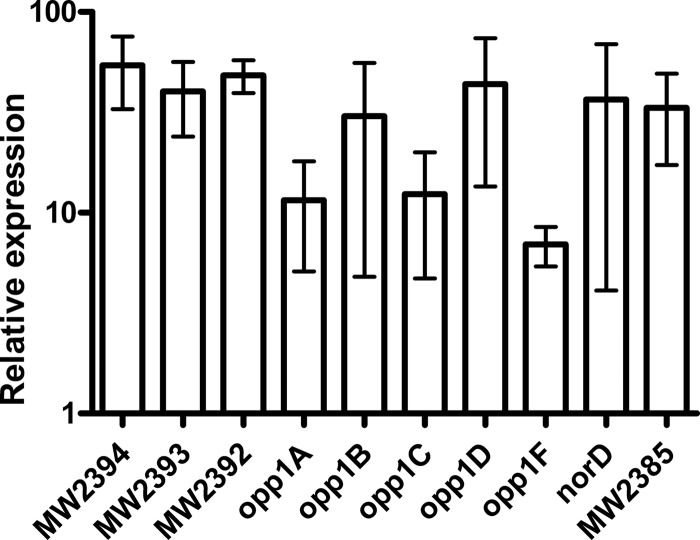
The similarity of the amino acid sequence and molecular structure of NorD to other efflux pumps suggests that NorD plays a role similar to that of other efflux pumps. We previously reported that efflux pump NorB in S. aureus MW2 confers fitness advantages at the site of infection in the murine subcutaneous abscess model (12). Thus, we infected mice with strain MW2 and assessed the relative expression of norD in total bacterial RNA recovered from an abscess versus in vitro cultures using qRT-PCR. Compared to its expression in vitro at stationary phase (24 h) in TSB, norD was upregulated about 36-fold in abscesses (Fig. 2). The effect was even greater compared to expression in exponential phase in vitro (90-fold [data not shown]). This result is similar to that for norB, which is also selectively upregulated (171-fold) in the mouse subcutaneous abscess model (12).
Fig 2.
Expression of the opp1-norD operon in S. aureus MW2 recovered from murine subcutaneous abscesses relative to in vitro gene expression. Relative expression was determined by real-time RT-PCR using specific primers for each gene to synthesize cDNA. Both the medians and the ranges of relative expression from at least two independent experiments are shown.
norD is linked to the opp1 operon, which is also selectively overexpressed in abscesses.
In strain MW2, norD is 11 bp downstream of opp1F, which encodes a subunit of the ABC type transporter Opp1, a putative oligopeptide permease (Fig. 1). Typically, five subunits constitute an Opp permease (15), including a substrate-binding protein (OppA), two permease proteins (OppB and OppC), and two ATP-binding proteins (OppD and OppF). The genome of S. aureus contains four opp loci, the constituent genes of which are expressed as an operon, including opp1ABCDF upstream of norD. Previous studies indicated that Opp1 does not transport nitrogen in S. aureus (5, 15), and its substrates for transport are unknown. Instead, disrupted opp1 has been reported to attenuate S. aureus in murine infection models (9, 26). Thus, we examined whether opp1 was also selectively upregulated in the subcutaneous abscess environment. The amount of transcripts of all opp1 genes, by means of qRT-PCR, increased from 7- to 54-fold in abscesses compared to their expression in vitro (Fig. 2).
opp1 and norD are regulated by environmental iron.
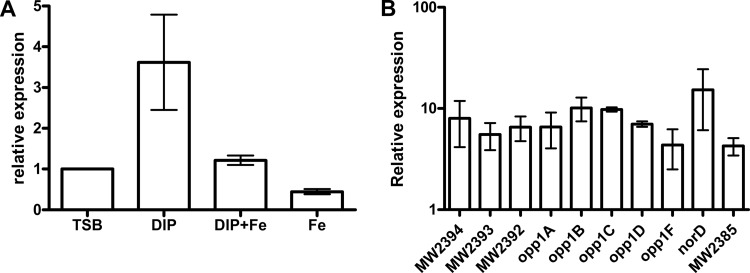
Upregulated expression of norD during infection implies that certain environmental signals that exist in vivo trigger changes in gene expression. A variety of such signals mimicking the in vivo settings, such as starvation (saline), oxidative stress (H2O2), and acid stress (pH 5.5), were tested in vitro for effects on norD expression. None of these signals induced upregulation of norD transcription (data not shown). In contrast, a shift to an acidic environment (pH 5.5) caused a 3-fold decrease in norD expression (data not shown). However, free iron restriction, which occurs at many sites of infection (7, 30), upregulated norD expression. We compared norD expression in strain MW2 grown in TSB supplemented with 400 μM dipyridyl (a ferrous iron chelator) with and without the addition of 50 μM ferrous sulfate. The presence of dipyridyl alone increased norD expression by 3.6-fold compared to that in TSB (Fig. 3A). This induction did not occur (1.2-fold) when ferrous sulfate was added together with dipyridyl. Furthermore, the addition of ferrous sulfate to TSB without dipyridyl resulted in a 2.1-fold decrease in norD expression. All of the genes in the opp1-norD operon were upregulated when dipyridyl was added to the culture medium, with increases ranging from 3.5- to 15.3-fold (Fig. 3B). These data suggest that free iron concentrations are an environmental signal for opp1-norD expression. Genes for other S. aureus efflux pumps (norA, norB, norC, and tet38) were not inducible under limiting iron conditions (data not shown).
Fig 3.
The expression of genes in the opp1-norD operon is upregulated under low free iron conditions. Dipyridyl (DIP) and FeSO4 (Fe) were added to final concentrations of 400 μM and 50 μM, respectively, when the OD600 of the S. aureus MW2 culture reached ∼0.4. Cultures were incubated at 37°C for 30 min before the total bacterial RNA was extracted. (A) Expression of norD by strain MW2 under conditions of rich and low free iron. (B) Expression of genes in opp1-norD operon in MW2 under low free iron conditions. Both the medians and the ranges of relative expression from at least two independent experiments are shown.
opp1 and norD are components of the same operon.
Because of genetic arrangement and coregulation of opp1 and norD in abscesses and in vitro under iron-limiting conditions, we determined whether the opp1ABCDF genes and norD were cotranscribed as components of an operon. Using an online tool for identifying Rho-independent terminator (http://rna.igmors.u-psud.fr/toolbox/arnold/index.php), we identified two putative Rho-independent terminators in the transcription orientation of norD; one was upstream of MW2394, and the other was downstream of MW2385 (Fig. 1), suggesting an operon extending from MW2394 to MW2385. We analyzed the transcripts of these genes in S. aureus MW2 grown in culture by synthesizing cDNA using a specific reverse primer to MW2385 and then PCR amplification using primer pairs spanning two adjacent ORFs (Fig. 1). All of the PCRs had products of the appropriate size, suggesting cotranscription of genes from MW2394 to MW2385, and so we named it the opp1-norD operon. A putative promoter element is located in the intergenic region upstream of MW2394. In addition to opp1ABCDF and norD, there are four ORFs (MW2394-MW2392 [upstream of opp1] and MW2385 [downstream of norD]), which encode hypothetical proteins of unknown function (Fig. 1). The opp1-norD operon is highly conserved in other S. aureus strains. All data presented herein indicate coregulation of these genes under different conditions, supporting their organization in a single operon.
Regulation of opp1-norD expression involves fur but not mgrA.
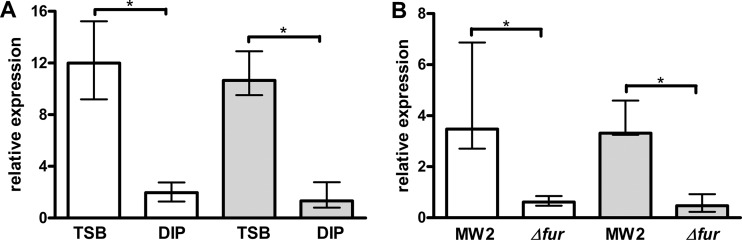
Fur is an iron-binding bacterial protein that regulates many iron-responsive genes (4, 7, 18). Because expression of S. aureus opp1-norD was responsive to environmental iron availability, Fur is a likely regulator for the operon. To test this hypothesis, we constructed a fur mutant of S. aureus MW2 and monitored the expression of opp1-norD under low free iron conditions (Fig. 4). In S. aureus MW2, dipyridyl increased expression of norD (3.5-fold) and MW2394 (3.3-fold), but induction was significantly reduced in the MW2 fur mutant, 0.6- and 0.5-fold, respectively (both P = 0.0286, Fig. 4B). Furthermore, basal levels of norD and MW2394 were 12.0-fold and 10.7-fold higher, respectively, in the MW2 fur mutant than in the parental strain under rich free iron conditions. However, when cultivated under low free iron conditions, the difference of the basal levels was significantly reduced to 2.0- and 1.3-fold (both P = 0.0286, Fig. 4A). These results indicate that Fur functions either directly or indirectly as a repressor of norD and opp1.
Fig 4.
Relative expression of norD (white bars) and MW2394 (gray bars) in S. aureus MW2 and an MW2 fur mutant under low free iron conditions. (A) Expression of norD and MW2394 in the MW2 fur mutant relative to that in MW2. (B) Expression of norD and MW2394 in MW2 and in the MW2 fur mutant under low free iron conditions relative to that under rich free iron conditions. TSB culture was supplemented with 400 μM dipyridyl (DIP) when the culture OD600 reached about 0.4 and incubated for 30 min. Median values that were significantly different are indicated by a bar and asterisk. Both the medians and the ranges of relative expression from four independent experiments are shown.
MgrA is a global transcriptional regulator that negatively modulates the expression of norB, norC, and tet38 (33, 35). Thus, we determined whether mgrA also affects the expression of norD. The levels of expression of norD did not differ, however, between S. aureus MW2 mgrA mutant and MW2. The expression profiles of the genes in opp1 and norD mutants (see below) also revealed no autofeedback regulation of the opp1-norD operon (MW2392 [1.8-fold] and norD [1.9-fold] in MW2 opp1 mutant and opp1F [1.1-fold] in MW2 norD mutant relative to that in MW2, respectively).
norD mutants, but not opp1 mutants, exhibit a fitness defect in subcutaneous abscesses.
Selective increases in expression of transporters norD and opp1 in subcutaneous abscesses suggested that their functions may be important for bacterial fitness in the abscess milieu. To address this possibility, we constructed individual in-frame deletion mutants of norD and opp1 in S. aureus MW2 and evaluated their fitness relative to that of MW2 using competition assays in the subcutaneous abscess infection model. In order to score the survival of mutant versus parental strains, we used as a comparator a streptomycin-resistant variant of MW2 (MW2SM), which demonstrated no fitness defect in competition assays with MW2 when the two strains were injected in roughly equal numbers in mouse flanks and harvested at 48 h (CI = 0.83) or grown in vitro up to 24 h (CI = 1.52) (Fig. 5A). Thus, MW2SM is comparable to MW2 in its growth in vivo and in vitro.
In direct competition experiments with S. aureus MW2SM and MW2 norD, the norD mutant exhibited a substantial fitness defect with a median CI of 0.065 (P < 0.0001) at 48 h (Fig. 5A). Moreover, in 5 out of 14 abscesses, MW2 norD colonies were not detected among 96 colonies patched on LB plates containing 100 μg/ml streptomycin. In contrast, the MW2 norD mutant exhibited a small growth advantage over MW2SM in vitro (CI = 5.3). We also performed a set of single-strain assays to investigate the fitness difference between MW2 and MW2 norD, in which each of two strains with similar inocula was singly injected on opposite flanks of mice. At 48 h after injection, the median numbers of viable bacteria recovered from each abscess were 7.3 × 107 CFU for MW2 and 0.3 × 107 CFU for the MW2 norD mutant, a 24-fold difference (P < 0.0001). In contrast, the numbers of CFU from in vitro cultures (24 h) were similar for the two strains (1.5 × 1010 CFU ml−1 for MW2 and 1.8 × 1010 CFU ml−1 for the MW2 norD mutant). The growth curves of these strains in vitro were similar (Fig. 5B), indicating that the fitness defect of the norD mutant is specific for the abscess environment. Thus, the norD mutant exhibits a selective in vivo growth defect in both competition and single-strain assays.
To complement the fitness defect of S. aureus MW2 norD mutant in vivo, we constructed pNorD carrying the entire norD ORF from strain MW2 cloned into the NdeI and BamHI sites of pOS1-Plgt (27, 37). norD was strongly expressed from pNorD as determined by qRT-PCR, and pNorD was stable in MW2 and its derivatives when grown in TSB for up to 72 h in the absence of chloramphenicol and during infection for up to 48 h (data not shown). The in vitro growth rate of MW2(pNorD) was similar to that of the wild-type strain for up to 24 h (Fig. 5C). In competition assays (Fig. 5D), MW2(pNorD) showed only a modest decrease in fitness compared to MW2 (CI of 0.73; P = 0.02). As noted above, the CI of the MW2 norD mutant was 0.065, and thus, the in vivo fitness defect of the norD mutant was substantially complemented by pNorD. As noted above, there is no autofeedback regulation related to norD as suggested by the comparable transcript level of opp1F (1.1-fold) in MW2(pNorD) relative to that in MW2.
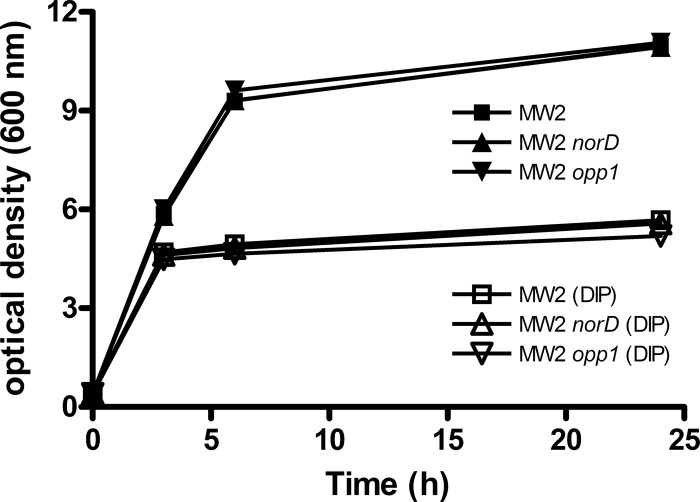
In contrast to the striking fitness defect of the norD mutant in vivo (CI of 0.065), an in-frame opp1 deletion mutant exhibited only a slight defect (CI of 0.53; P = 0.0002) (Fig. 5A). Compared to strain MW2, neither the opp1 nor the norD mutant showed any difference in their growth in vitro in rich or low free iron medium (Fig. 6).
Fig 6.
Growth of S. aureus MW2 and opp1 and norD mutants in TSB broth at 37°C after the addition of 0.4 mM DIP when the OD600 reached 0.4. Representative results of repeated experiments are shown.
DISCUSSION
In this study, we have identified and characterized the opp1-norD operon of S. aureus based on its selective overexpression in a murine subcutaneous abscess model. The operon spans from MW2394 to MW2385, and it includes an opp1 locus and norD, a newly recognized gene encoding an efflux pump. Opp1 is a member of the ABC transporter family that is energized by ATP. NorD is a member of the MFS transporter family that is energized by hydrogen and other ion gradients across the bacterial cytoplasmic membrane. In strain RN6390, the transcription of these genes was confirmed to be linked, extending from upstream SAOUHSC_02770 to SAOUHSC_02762 and corresponding to MW2394 to norD in strain MW2 (15). We have further characterized the arrangement of these two transporters and determined their relative contributions to fitness in a mouse infection model.
NorD is related to other MFS efflux transporters, such as NorA, NorB, NorC, and Tet38. Like norD, norB and tet38 are also selectively upregulated in the abscess environment, and norB and tet38 mutants showed selective fitness defects in murine abscesses (12). Overexpression of norB confers increased resistance to quinolones, such as ciprofloxacin and moxifloxacin, and overexpression of tet38 confers increased resistance to tetracycline (33). In contrast, overexpression of norD in S. aureus MW2 produced no detectable change in susceptibility to quinolones (ciprofloxacin, norfloxacin, moxifloxacin, delafloxacin, and levofloxacin), tetracyclines, polymyxin B, daptomycin, nalidixic acid, trimethoprim, ethidium bromide, or triclosan (data not shown). Thus, the substrates of NorD are not known, and it is likely that the natural substrates of NorB and Tet38 that contribute to their ability to confer fitness in an abscess environment are also not directly related to their antibiotic resistance properties but reflect instead the broad substrate profiles of MFS transporters. MFS and other efflux transporters may function to remove toxic metabolites from the cytoplasm or environmental toxins from the bacterial cytoplasm or cell membrane; for example, AcrAB-TolC of E. coli is induced by and provides resistance to bile salts, likely contributing to the ability of E. coli to colonize the intestinal tract (29). It is, thus, plausible that NorD removes an as yet unidentified toxin present in abscesses, thereby enhancing S. aureus survival in the neutrophil-rich environment of the abscess.
Four polycistronic opp operons (including opp1) are present in the S. aureus genome, along with one monocistronic opp5A gene. Although these operons are annotated as oligopeptide permeases, only opp3 has been shown to have a role in the uptake of oligopeptides. Furthermore, opp3 alone appears to be sufficient for this function (15). opp2 and opp5A function in nickel transport and are attenuated in a staphylococcal urinary tract infection model (16). None of the opp operons other than opp1 has a downstream coregulated transporter gene like norD. Thus, the function of opp1 remains unknown. opp1, although coregulated with norD, appears not to be a major contributor to fitness in the abscess model, unlike norD. A previous study demonstrated that opp1 transposon mutants were attenuated in abscess, wound, and systemic infection models (9). Given our findings that an opp1 in-frame deletion mutant exhibited little attenuation in an abscess infection model, it is likely that opp1 attenuation seen in the prior study reflected the polar effects of the opp1 transposon insertion on norD expression. The lack of a fitness defect in the opp1 mutant could reflect redundancy of function, consistent with multiple opp operons present in the staphylococcal genome.
A condition known to be present in the abscess environment that triggered the increased expression of opp1 and norD in vitro was low free iron. Other conditions found in abscesses, such as oxidative stress and low pH, did not affect in vitro expression of opp1 or norD, but low pH has been shown to increase the expression of norB (31). Because of their upregulation in response to iron, opp1 and norD could be directly involved in iron acquisition or metabolism. However, since neither mutant exhibited a growth defect in vitro under iron-limiting conditions, it is more likely that low iron may be an in vivo signal for S. aureus, allowing increased gene expression.
Bacteria sense iron-restricted environments in part through the Fur protein (4, 7, 14, 18), which binds ferrous ions, dimerizes, and binds to Fur box DNA sequences (18, 25, 30, 38) upstream of iron-regulated genes. In S. aureus, it is estimated that the Fur regulon comprises ∼20 operons involving ∼50 genes. A recent study revealed that the S. aureus efflux pump NorA is a member of the Fur regulon and may function to export a siderophore for acquisition of environmental ferric ions (10). Fur positively modulates norA expression by interacting directly with a Fur box in the norA promoter region. By regulating norA expression, Fur thus contributes to quinolone resistance in S. aureus. In the MW2 fur mutant, opp1-norD gene upregulation diminished under iron-restricted conditions, suggesting that Fur regulates expression of the opp1-norD genes. Our data and a previous study suggest that the promoter of the opp1-norD operon is located in the intergenic region upstream of MW2395 (15). However, we could not identify the Fur binding site in the region by gel shift assays with purified Fur protein (data not shown), suggesting the possibility of indirect regulation of opp1-norD by a separate Fur-regulated gene product.
MgrA regulates expression of genes encoding other Nor efflux pumps and virulence factors (8, 20, 22–24, 32–34), but it does not affect regulation of the opp1-norD operon. Our data show that opp1-norD expression was comparable in S. aureus MW2 and MW2 mgrA mutant in both rich medium and under low free iron conditions (data not shown), suggesting that regulation of opp1-norD is independent of MgrA. In preliminary studies, an mgrA mutant showed only a 2-fold defect in fitness in vivo, confirming that it does not regulate opp1-norD in vivo.
In summary, we have identified in S. aureus MW2 a putative efflux pump designated NorD. Its expression is polycistronic with upstream opp1ABCDF, thus forming the opp1-norD operon. The operon is ubiquitous among staphylococci, and the genes in the operon are coregulated. opp1-norD expression is selectively increased within subcutaneous abscesses relative to growth in vitro. fur-dependent upregulation of opp1 and norD was observed in S. aureus MW2 grown in a low-iron environment in vitro, a condition that is characteristic of infection sites within mammalian hosts. The efflux pump NorD, but not Opp1, significantly contributed to staphylococcal fitness in subcutaneous abscesses, a common form of S. aureus infection.
ACKNOWLEDGMENTS
We are grateful to Taeok Bae and Olaf Schneewind for kindly providing plasmid pKOR1 and technical advice and to Dominique Missiakas, Chuan He, and Xin Deng for kindly providing strain Newman fur::tet and phage 85.
This project was supported in part by Public Health Service grant R37-AI023988 from the National Institutes of Health (to D.C.H.).
Footnotes
Published ahead of print 5 October 2012
REFERENCES
- 1. Anonymous 1999. Four pediatric deaths from community-acquired methicillin-resistant Staphylococcus aureus–Minnesota and North Dakota, 1997–1999. JAMA 282:1123–1125 [PubMed] [Google Scholar]
- 2. Baba T, et al. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819–1827 [DOI] [PubMed] [Google Scholar]
- 3. Bae T, Schneewind O. 2006. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55:58–63 [DOI] [PubMed] [Google Scholar]
- 4. Baichoo N, Helmann JD. 2002. Recognition of DNA by Fur: a reinterpretation of the Fur box consensus sequence. J. Bacteriol. 184:5826–5832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Borezee-Durant E, Hiron A, Piard JC, Juillard V. 2009. Dual role of the oligopeptide permease Opp3 during growth of Staphylococcus aureus in milk. Appl. Environ. Microbiol. 75:3355–3357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bunce C, Wheeler L, Reed G, Musser J, Barg N. 1992. Murine model of cutaneous infection with Gram-positive cocci. Infect. Immun. 60:2636–2640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Carpenter BM, Whitmire JM, Merrell DS. 2009. This is not your mother's repressor: the complex role of Fur in pathogenesis. Infect. Immun. 77:2590–2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen PR, et al. 2006. An oxidation-sensing mechanism is used by the global regulator MgrA in Staphylococcus aureus. Nat. Chem. Biol. 2:591–595 [DOI] [PubMed] [Google Scholar]
- 9. Coulter SN, et al. 1998. Staphylococcus aureus genetic loci impacting growth and survival in multiple infection environments. Mol. Microbiol. 30:393–404 [DOI] [PubMed] [Google Scholar]
- 10. Deng X, et al. 2012. Expression of multidrug resistance efflux pump gene norA is iron responsive in Staphylococcus aureus. J. Bacteriol. 194:1753–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ding Y, Davis BM, Waldor MK. 2004. Hfq is essential for Vibrio cholerae virulence and downregulates sigma expression. Mol. Microbiol. 53:345–354 [DOI] [PubMed] [Google Scholar]
- 12. Ding Y, Onodera Y, Lee JC, Hooper DC. 2008. NorB, an efflux pump in Staphylococcus aureus MW2, contributes to bacterial fitness in abscesses. J. Bacteriol. 190:7123–7129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ford CW, Hamel JC, Stapert D, Yancey RJ. 1989. Establishment of an experimental model of a Staphylococcus aureus abscess in mice by use of dextran and gelatin microcarriers. J. Med. Microbiol. 28:259–266 [DOI] [PubMed] [Google Scholar]
- 14. Hantke K. 1981. Regulation of ferric iron transport in Escherichia coli K12: isolation of a constitutive mutant. Mol. Gen. Genet. 182:288–292 [DOI] [PubMed] [Google Scholar]
- 15. Hiron A, Borezee-Durant E, Piard JC, Juillard V. 2007. Only one of four oligopeptide transport systems mediates nitrogen nutrition in Staphylococcus aureus. J. Bacteriol. 189:5119–5129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hiron A, et al. 2010. A nickel ABC-transporter of Staphylococcus aureus is involved in urinary tract infection. Mol. Microbiol. 77:1246–1260 [DOI] [PubMed] [Google Scholar]
- 17. Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51–59 [DOI] [PubMed] [Google Scholar]
- 18. Horsburgh MJ, Ingham E, Foster SJ. 2001. In Staphylococcus aureus, Fur is an interactive regulator with PerR, contributes to virulence, and is necessary for oxidative stress resistance through positive regulation of catalase and iron homeostasis. J. Bacteriol. 183:468–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61–68 [DOI] [PubMed] [Google Scholar]
- 20. Kaatz GW, Thyagarajan RV, Seo SM. 2005. Effect of promoter region mutations and mgrA overexpression on transcription of norA, which encodes a Staphylococcus aureus multidrug efflux transporter. Antimicrob. Agents Chemother. 49:161–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lowy FD. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520–532 [DOI] [PubMed] [Google Scholar]
- 22. Luong TT, Dunman PM, Murphy E, Projan SJ, Lee CY. 2006. Transcription profiling of the mgrA regulon in Staphylococcus aureus. J. Bacteriol. 188:1899–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Luong TT, Lee CY. 2006. The arl locus positively regulates Staphylococcus aureus type 5 capsule via an mgrA-dependent pathway. Microbiology 152:3123–3131 [DOI] [PubMed] [Google Scholar]
- 24. Manna AC, Cheung AL. 2006. Expression of SarX, a negative regulator of agr and exoprotein synthesis, is activated by MgrA in Staphylococcus aureus. J. Bacteriol. 188:4288–4299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mazmanian SK, Ton-That H, Su K, Schneewind O. 2002. An iron-regulated sortase anchors a class of surface protein during Staphylococcus aureus pathogenesis. Proc. Natl. Acad. Sci. U. S. A. 99:2293–2298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mei JM, Nourbakhsh F, Ford CW, Holden DW. 1997. Identification of Staphylococcus aureus virulence genes in a murine model of bacteraemia using signature-tagged mutagenesis. Mol. Microbiol. 26:399–407 [DOI] [PubMed] [Google Scholar]
- 27. Schneewind O, Mihaylova-Petkov D, Model P. 1993. Cell wall sorting signals in surface proteins of Gram-positive bacteria. EMBO J. 12:4803–4811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Speziali CD, Dale SE, Henderson JA, Vines ED, Heinrichs DE. 2006. Requirement of Staphylococcus aureus ATP-binding cassette-ATPase FhuC for iron-restricted growth and evidence that it functions with more than one iron transporter. J. Bacteriol. 188:2048–2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Thanassi DG, Cheng LW, Nikaido H. 1997. Active efflux of bile salts by Escherichia coli. J. Bacteriol. 179:2512–2518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Torres VJ, et al. 2010. Staphylococcus aureus fur regulates the expression of virulence factors that contribute to the pathogenesis of pneumonia. Infect. Immun. 78:1618–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Truong-Bolduc QC, et al. 2011. Implication of the NorB efflux pump in the adaptation of Staphylococcus aureus to growth at acid pH and in resistance to moxifloxacin. Antimicrob. Agents Chemother. 55:3214–3219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Truong-Bolduc QC, Ding Y, Hooper DC. 2008. Posttranslational modification influences the effects of MgrA on norA expression in Staphylococcus aureus. J. Bacteriol. 190:7375–7381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Truong-Bolduc QC, Dunman PM, Strahilevitz J, Projan SJ, Hooper DC. 2005. MgrA is a multiple regulator of two new efflux pumps in Staphylococcus aureus. J. Bacteriol. 187:2395–2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Truong-Bolduc QC, Hooper DC. 2007. Transcriptional regulators NorG and MgrA modulate resistance to both quinolones and β-lactams in Staphylococcus aureus. J. Bacteriol. 189:2996–3005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Truong-Bolduc QC, Strahilevitz J, Hooper DC. 2006. NorC, a new efflux pump regulated by MgrA of Staphylococcus aureus. Antimicrob. Agents Chemother. 50:1104–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vandecasteele SJ, Peetermans WE, Merckx R, Van Eldere J. 2001. Quantification of expression of Staphylococcus epidermidis housekeeping genes with Taqman quantitative PCR during in vitro growth and under different conditions. J. Bacteriol. 183:7094–7101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wardenburg JB, Williams WA, Missiakas D. 2006. Host defenses against Staphylococcus aureus infection require recognition of bacterial lipoproteins. Proc. Natl. Acad. Sci. U. S. A. 103:13831–13836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Xiong AM, Singh VK, Cabrera G, Jayaswal RK. 2000. Molecular characterization of the ferric-uptake regulator, Fur, from Staphylococcus aureus. Microbiology 146:659–668 [DOI] [PubMed] [Google Scholar]