Abstract
Pseudomonas aeruginosa is an opportunistic pathogen that is capable of causing both acute and chronic infections. P. aeruginosa virulence is subject to sophisticated regulatory control by two-component systems that enable it to sense and respond to environmental stimuli. We recently reported that the two-component sensor KinB regulates virulence in acute P. aeruginosa infection. Furthermore, it regulates acute-virulence-associated phenotypes such as pyocyanin production, elastase production, and motility in a manner independent of its kinase activity. Here we show that KinB regulates virulence through the global sigma factor AlgU, which plays a key role in repressing P. aeruginosa acute-virulence factors, and through its cognate response regulator AlgB. However, we show that rather than phosphorylating AlgB, KinB's primary role in the regulation of virulence is to act as a phosphatase to dephosphorylate AlgB and alleviate phosphorylated AlgB's repression of acute virulence.
INTRODUCTION
Bacteria use two-component signal transduction to sense and respond to their surrounding microenvironment. A canonical two-component system consists of a membrane-localized sensor protein that autophosphorylates itself on a conserved histidine residue in response to an environmental signal. It then transmits this phosphoryl group to a conserved aspartic acid residue on the N-terminal receiver domain of its cognate response regulator. Phosphorylation of the receiver domain of the response regulator results in a conformational change that modulates the activity of its output domain. Most response regulators are DNA binding proteins that bind to their target promoters upon phosphorylation and activate a transcriptional program. Thus, in general, the phosphorylated conformation of the response regulator has been considered the active conformation (20).
More complex variations of this basic theme exist, and two-component sensor proteins can take part in more complicated phosphorelays or interact with auxiliary regulator proteins (3). Phosphorelays are common variations of the canonical two-component transduction pathway that provide additional targets for regulation, since they involve additional phosphotransferase domains. Initiation of sporulation in Bacillus subtilis, for instance, is controlled by a multicomponent phosphorelay system (18). In addition, auxiliary regulators that modulate the activity of two-component signal transduction proteins were initially identified in B. subtilis as playing a role in sporulation and have now been found in a number of other bacterial species, including Escherichia coli, in which the protein PII inhibits the autophosphorylation activity of the sensor protein NtrB (3, 30).
It has also recently become recognized that in addition to their kinase activity, several two-component sensors are bifunctional and can both phosphorylate and dephosphorylate their cognate response regulators (20, 29). It may be beneficial for a sensor to possess both activities for a number of reasons. For example, a sensor with both phosphatase and kinase activities can more finely tune the level of phosphorylated response regulator in the cell in response to a signal(s), thus modulating the downstream output (13). Furthermore, the presence of bifunctional sensors in organisms may minimize the consumption of ATP for phosphorylating the response regulator and subsequently dephosphorylating it, in comparison to organisms that rely on separate phosphatases to exert negative feedback (43).
The E. coli two-component sensor protein, EnvZ, one of the best-studied sensors, has been shown to have both kinase and phosphatase activities (37). Phosphatase-defective envZ mutant strains show aberrant levels of porin expression, preventing them from correctly responding to changes in medium osmolarity, suggesting that EnvZ's phosphatase activity may play an important regulatory role in the cell (37). Furthermore, the phosphatase activities of several sensors have been shown to be involved in the regulation of virulence factors such as flagella, pili, and curli (22). The phosphatase activity of the Salmonella enterica serovar Typhimurium membrane-bound sensor PhoQ, which is essential for virulence in mice, is important in modulating its response to divalent cations such as Mg2+ (2). Mg2+ cations activate PhoQ's phosphatase activity, resulting in dephosphorylation of its cognate regulator, PhoP (5). Dephosphorylated PhoP represses the expression of Pho-P-activated genes that are required for survival mainly in Mg2+-limiting environments (5). The PhoQ homologue in Pseudomonas aeruginosa also modulates Mg2+ activation of PhoP and has been suggested to do so by acting as a phosphatase (27). Thus, two-component sensor phosphatase activity may be a more widespread phenomenon than previously appreciated.
One of the most complex two-component signaling networks known is found in P. aeruginosa, which encodes one of the largest sets of two-component systems in Gram-negative bacteria (35). P. aeruginosa occupies a diverse variety of environments, ranging from soil to the human lung. It is capable of causing both acute, nosocomial infections, such as in burn victims, and chronic infections in cystic fibrosis (CF) patients (39). P. aeruginosa infections are often antibiotic resistant. Its vast repertoire of two-component systems likely enables it to adapt to changing environments and finely regulate virulence and antibiotic resistance (14). We now know that P. aeruginosa two-component systems are subject to sophisticated regulatory control that can include direct protein-protein interactions in addition to the regulation of phosphorylation status (16).
We recently reported that the P. aeruginosa sensor kinase KinB is required for acute virulence in zebrafish (Danio rerio) embryos and for the regulation of acute virulence phenotypes, such as pyocyanin production and motility, and that it may play a role in mediating the switch between acute and chronic infection phenotypes (6, 7). Furthermore, it was recently reported that KinB is also required for full virulence in a murine acute pneumonia infection model (9). Interestingly, we found that KinB's regulation of acute virulence phenotypes is independent of its kinase activity, since a kinase-inactive mutant of KinB regulates acute virulence phenotypes similarly to wild-type KinB. Moreover, we found that deletion of algB, the gene encoding KinB's cognate response regulator, also resulted in acute virulence phenotypes similar to those of wild-type PA14 (7). Thus, we proposed that KinB regulates these phenotypes in a noncanonical manner.
In addition to KinB's role in inducing acute virulence phenotypes, KinB has been implicated in repressing chronic virulence phenotypes like the production of alginate (10). Deletion of kinB in PAO1 (10) and in PA14 (6) results in the overproduction of alginate. Furthermore, deletion of kinB in PAO1 results in upregulation of the global sigma factor AlgU (10). Since AlgU upregulation has been associated with defects in acute virulence phenotypes in mucoid P. aeruginosa isolated from CF patients with chronic infection (34), we hypothesized that KinB might regulate acute virulence through AlgU in order to act as a switch between acute and chronic infection phenotypes.
Here we show that KinB is required for acute virulence, acting through the global sigma factor AlgU. We also show that AlgB is required for the regulation of virulence by KinB, but not through direct phosphorylation of AlgB by KinB. Rather, we found that KinB possesses phosphatase activity and its induction of acute virulence phenotypes is dependent on its phosphatase activity. Phosphorylated AlgB acts as a repressor of acute virulence, and KinB's primary role in the induction of acute virulence appears to be to dephosphorylate AlgB.
MATERIALS AND METHODS
Strains, plasmids, and culture conditions.
The strains used in this study are listed in Table 1. All strains were struck out from frozen glycerol stocks onto LB agar plates supplemented with the appropriate antibiotic and grown overnight at 37°C. P. aeruginosa strains were grown at 37°C in LB broth or on LB agar, supplemented with an antibiotic where appropriate. Antibiotics used were as follows: Irgasan, 15 μg ml−1; kanamycin, 15 μg ml−1; and carbenicillin, 150 μg ml−1. E. coli strains were grown in LB supplemented with 100 μg ml−1 ampicillin. Oligonucleotides used are listed in Table 2.
Table 1.
Strains and plasmids used
| Strain or plasmid | Characteristic(s) or sequence | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | fhuA2 Δ(argF-lacZ)U169 phoA glnV44 ϕ80 Δ(lacZ)M15 gyrA96 recA1 relA1 endA1 thi-1 hsdR17 | Lab strain |
| SM10 | thi thr leu tonA lacY supE recA::RP4-2-Tc::Mu Km λpir | Lab strain |
| P. aeruginosa strains | ||
| PA14 | Virulent burn wound isolate of P. aeruginosa | 33 |
| PA14ΔkinB | PA14 ΔkinB (1.7-kb in-frame deletion of kinB) | 7 |
| PA14ΔalgB | ΔalgB PA14 (1.3-kb in-frame deletion of algB) | 7 |
| PA14ΔalgBΔkinB | PA14 ΔalgB ΔkinB (3.1 kb in-frame deletion of algB and kinB) | This study |
| PA14ΔalgU | PA14 ΔalgU (567 bp in-frame deletion of algU) | This study |
| PA14ΔalgUΔkinB | PA14 ΔalgU ΔkinB (1.7-kb in-frame deletion of kinB and 567-bp in-frame deletion of algU) | This study |
| PA14ΔalgR | PA14 ΔalgR (746-bp in-frame deletion of algR) | This study |
| PA14ΔalgRΔkinB | PA14 ΔalgR ΔkinB (1.7-kb in-frame deletion of kinB and a 746-bp in-frame deletion of algR) | This study |
| PA14ΔlasR | PA14 ΔlasR | 7 |
| PA14ΔkinB(+pKinB) | PA14ΔkinB with plasmid pKinB | 7 |
| PA14ΔkinB(+pKinBH385A) | PA14ΔkinB with plasmid pKinBH385A | 7 |
| PA14ΔkinB(+pKinBP390S) | PA14ΔkinB with plasmid pKinBP390S | This study |
| PA14ΔkinB(+KinBint) | PA14 ΔkinB with Plac-kinB integrated at the attB site | This study |
| PA14ΔkinB(+KinBP390Sint) | PA14ΔkinB with Plac-kinBP390S integrated at the attB site | This study |
| PA14ΔalgBΔkinB(+pAlgB) | PA14ΔalgBΔkinB with plasmid pAlgB | This study |
| PA14ΔalgBΔkinB(+pAlgBD59N) | PA14ΔalgBΔkinB with plasmid pAlgBD59N | This study |
| PA14ΔalgUΔkinB(+pAlgU) | PA14ΔalgUΔkinB with plasmid pAlgU | This study |
| Plasmids | ||
| pEXG2-attP | pEXG2-M13 with attP cassette | 7 |
| pNSC5 | pEXG2-attP with 5′ algB flanks and 3′ kinB flanks from PA14 cloned in. Used to generate ΔalgB ΔkinB PA14 | This study |
| pNSC6 | pEXG2-attP with algU flanks from PA14 cloned in. Used to generate ΔalgU PA14 and ΔalgU ΔkinB PA14 | This study |
| pNSC7 | pEXG2-attP with algR flanks from PA14 cloned in; used to generate ΔalgR ΔkinB PA14 | This study |
| pHERD20T | pUCP20T Plac replaced by a 1.3-kb AflII-EcoRI fragment of the araC-PBAD cassette (5,087bp) | 7 |
| pKinB | pHERD20T with the kinB gene from PA14 at the KpnI/HindIII site | 7 |
| pAlgB | pHERD20T with the algB gene from PA14 at the EcoRI/HindIII site; the algB gene was amplified using primers NSC14D and NSC15D | This study |
| pAlgU | pHERD20T with the algU gene from PA14 at the EcoRI/HindIII site; the algU gene was amplified using primers NSC12 and NSC13 | This study |
| pKinBH385A | pKinB with H385A | 7 |
| pKinBP390S | pKinB with P390S | This study |
| pAlgBD59N | pAlgB with D59N | This study |
| pHisAlgBD59N | pHERD20T with His-tagged algBD59N cloned at the XbaI/HindIII sites; algBD59N was amplified from pAlgBD59N with AC672 and NSC107, allowing insertion of an N-terminal His tag and XbaI/HindIII sites | This study |
| pSM95 | Plasmid that expresses a His-tagged carboxyl-terminal fragment of KinB (HC-KinB) | 26 (Ohman lab) |
| pSM95P390S | Plasmid that expresses a mutated His-tagged carboxyl terminal fragment of KinB (HC-KinB); proline 390 is mutated to serine | This study |
| pDJW403 | Plasmid that expresses a His-tagged AlgB | 25 (Wozniak lab) |
| pQF50 | pQF50 plasmid | 12 (Rahme lab) |
| pQF50algU | Plasmid containing PalgU-lacZ fusion | This study |
| pUCP18 | E. coli-P. aeruginosa shuttle vector | 38 (Rahme lab) |
| pUCP18-KinB | pUCP18 with the kinB gene from PA14 at the XbaI/HindIII site | This study |
| Mini-CTX1 | Integration-proficient plasmid for P. aeruginosa | 17 (lab stock) |
| Mini-CTX1-KinB | Mini-CTX1 with Plac-kinB from pUCP18-KinB at the BamHI/HindIII site | This study |
| Mini-CTX1-KinBP390S | Mini-CTX1-KinB with P390S | This study |
Table 2.
Oligonucleotides used
| Primer | Sequence |
|---|---|
| PA14_72380_5pleft2 | TACAAAAAAGCAGGCTCGAGAGATTCATCACCCAGTC |
| PA14_72380_5pright3 | ACCGGTTAATTAAGCATCGTTGCTTTTTATCCTC |
| PA14_72390_3pleft3 | CGATGCTTAATTAACCGGTGTGACCGGGGCCGCT |
| PA14_72390_3pright2 | TACAAGAAAGCTGGGTATCATGCGTTCGTTCGTTCTAC |
| PA14_54430_5pleft | TACAAAAAAGCAGGCTAGTTGGGAAGGATCGAACTTG |
| PA14_54430_5pright | TCATGCTTAATTAACGAGAAGCCTGACACAGCGG |
| PA14_54430_3pleft | TCTCGTTAATTAAGCATGAAAGCTCCTCTTCGAA |
| PA14_54430_3pright | TACAAGAAAGCTGGGTTTTCCCGATTCAGCACGTAG |
| PA14_69470_5pleft | TACAAAAAAGCAGGCTCCGTTGAGTCGCTTGTTCAG |
| PA14_69470_5pright | CGCGACTTAATTAATGACGGCGGTCGGCGGTTCG |
| PA14_69470_3pleft | CGTCATTAATTAAGTCGCGCCAGAGGTTCGTCAT |
| PA14_69470_3pright | TACAAGAAAGCTGGGTAGTGGATCGTACTGCTCTCGG |
| attB1 | GGGGACAAGTTTGTACAAAAAAGCAGGCT |
| attB2 | GGGGACCACTTTGTACAAGAAAGCTGGGT |
| NSC12 | TACGAATTCGATGCTAACCCAGGAACAGGA |
| NSC13 | AACAAGCTTTCAGGCTTCTCGCAACAAAGGCTGCA |
| NSC14D | CCAGAAGCCGAATGGCAGTG |
| NSC15D | TGTCAAGCTTCGAGCTGCATCACGCTGAAC |
| NSC78 | GAACTGCGCACGTCGGTGACCGGCA |
| NSC79 | TGCCGGTCACCGACGTGCGCAGTTC |
| pAlgU1 | GGTTGTCGACTAAGTCGAGCCCTGCGACAG |
| pAlgU2 | AACAGGATCCTGAAAGCTCCTCTTCGAACCTG |
| NSC82 | ATTATCTAGAATGAGCATGCCGCTGCCGATGAAG |
| NSC83 | AATAAAAGCTTTCACACCGGCAGCAGCATGTAGAA |
| NSC94 | TTATGGATCCAGGCACCCCAGGCTTTACACT |
| NSC95 | TGCCAAGCTTTCACACCGGC |
| CTX1 | CCTCGTTCCCAGTTTGTTCC |
| attB | GTCGCCGCCGGCGATGC |
| AC672 | ATTATCTAGAATGCATCACCATCACCATCACGAAACCACTTCCGAAAAACAGG |
| NSC107 | TGTCAAGCTTCGAGCTGCATCACGC |
Mutant-strain construction.
We utilized the previously described Gateway-compatible vector pEXG2-attP to construct in-frame deletions (7). Upstream and downstream sequences (400 to 1,100 bp) flanking the target gene(s) were PCR amplified using the following sets of primers: PA14_72380_5pleft2, PA14_72380_5pright3, PA14_72390_3pleft3, and PA14_72390_3pright2 (for deletion of algB kinB); PA14_54430_5pleft, PA14_54430_5pright, PA14_54430_3pleft, and PA14_54430_3pright (for deletion of algU); and PA14_69470_5pleft, PA14_69470_5pright, PA14_69470_3pleft, and PA14_69470_3pright (for deletion of algR). The flanks were fused using crossover PCR with the primers attB1 and attB2. Primers were designed such that a PacI restriction site would be introduced at the site of the gene being deleted. The PCR products with the in-frame deletion of the target gene were cloned into our modified vector using Gateway BP Clonase (Invitrogen, Carlsbad, CA). The constructs were used to make in-frame deletions by allelic exchange. The constructs were mated with PA14 wild-type (WT) to generate PA14ΔalgBΔkinB, PA14ΔalgU, and PA14ΔalgR. The constructs were mated with PA14ΔkinB to generate PA14ΔalgUΔkinB and PA14ΔalgRΔkinB. Gentamicin and Irgasan plates were used to select the single-crossover merodiploid exconjugants. Double-crossover recombinants were selected on LB agar (lacking sodium chloride) with 10% sucrose, after overnight incubation of the merodiploids in LB broth at 37°C, to force the removal of vector DNA containing the sacB gene. The in-frame deletion of the target gene was confirmed by PCR amplification of the flanking region of the target gene and amplicon sequencing.
The algU promoter was amplified from wild-type PA14 using primers pAlgU1 and pAlgU2. The promoter region was cloned into the plasmid pQF50 (12) to generate pQF50algU. The algU gene was amplified from wild-type PA14 using primers NSC12 and NSC13, while the algB gene was amplified from wild-type PA14 using primers NSC14D and NSC15D. All primers were designed using MacVector (Apple, Cupertino, CA) from the published PA14 genome sequence (24). The gene was cloned into the shuttle vector pHERD20T (41) for complementation. The sequences of primers used are listed in Table 2. All constructs were sequence verified.
Plasmid pAlgB was custom mutagenized to generate pAlgBD59N by Genewiz Inc. (South Plainfield, NJ). Plasmid pKinB was mutagenized using primers NSC78 and NSC79 to generate pKinBP390S with a Stratagene QuikChange Lightning site-directed mutagenesis kit (Agilent, Santa Clara, CA) as per the manufacturer's protocol. All constructs were sequence verified.
The kinB gene was amplified from PA14 using primers NSC82 and NSC83. The gene was cloned into pUCP18 (38) to generate pUCP18-KinB. Plac-kinB was amplified from this plasmid using primers NSC94 and NSC95. The PCR amplicon was cloned into mini-CTX1 (17) to generate mini-CTX1-KinB. This plasmid was mutagenized using primers NSC78 and NSC79 to generate mini-CTX1-KinB-P390S with a Stratagene QuikChange Lightning site-directed mutagenesis kit (Agilent, Santa Clara, CA) as per the manufacturer's protocol. All constructs were sequence verified. Plasmid mini-CTX1-KinB and mini-CTX1-KinB-P390S were individually conjugated into PA14ΔkinB and integrated into the chromosomal attB site. Integration of the plasmids at the attB site was confirmed using primers CTX1 and attB, as previously described (42).
Miller assay.
PA14, PA14ΔkinB, and PA14ΔalgBΔkinB were transformed with plasmid pQF50algU, containing a PalgU--lacZ promoter fusion. Strains expressing the plasmid were grown to mid-log phase (OD 0.6 to 0.8) in LB supplemented with carbenicillin, and the β-galactosidase activity was determined as previously described, using chloroform and SDS to permeabilize cells and ONPG (o-nitrophenyl-β-d-galactopyranoside) as a substrate (28).
Pyocyanin quantitation assay.
Pyocyanin present in the supernatants of PA14 strains was quantified as previously described (7). All strains were grown in LB medium supplemented with 0.025% arabinose. The medium was supplemented with carbenicillin for strains carrying the pHERD20T plasmid or its derivatives.
Elastase quantitation assay.
Elastase activity was quantitated as previously described (32), with minor modifications. Briefly, strains were grown in 5 ml LB medium supplemented with 0.025% arabinose, except where otherwise indicated. The medium was supplemented with carbenicillin for strains carrying the pHERD20T plasmid or its derivatives. Strains were grown for 21 h from a starting OD600 of 0.05. A 100-μl portion of supernatant was incubated with 1 ml of buffer A (0.1 M Tris [pH 7.2], 1 mM CaCl2) and 20 mg of elastin-Congo red (ECR) (Sigma-Aldrich, St. Louis, MO) for 18 h at 37°C at 250 rpm. Insoluble ECR was removed by centrifugation of the slurry, and the A495 of the soluble Congo red in the supernatants was measured.
Motility assay.
The swimming motility assay was conducted as previously described, with minor modifications (7). Photographs of plates were taken and swim zone diameter was measured after 17 to 20 h of incubation at 30°C on 0.35% agar plates containing 0.025% arabinose and 150 μg ml−1 carbenicillin. The swim zone diameter of each strain was compared individually to that of PA14(+pHERD20T) on the same plate to enable comparison on plates containing carbenicillin. The standard error of the mean was calculated from three replicates.
Mucoidy assay.
PA14 strains were struck out on LB agar plates and allowed to grow overnight. Bacterial strains were then struck out on Pseudomonas isolation agar (PIA) plates supplemented with 150 μg ml−1 carbenicillin and 0.025% arabinose. Photographs were taken after incubation at 37°C for 24 h.
Protein purification.
A previously described plasmid, pSM95, that overexpressed a His6-tagged carboxyl-terminal fragment (Gly-198 to Val-595) of KinB (HC-KinB) was obtained from Dennis Ohman (Virginia Commonwealth University) (26). This plasmid was mutagenized using primers NSC78 and NSC79 to generate pSM95KinBP390S with a Stratagene QuikChange Lightning site-directed mutagenesis kit (Agilent, Santa Clara, CA) as per the manufacturer's protocol. Plasmid pSM95KinBP390S thus overexpressed a His6-tagged carboxyl-terminal fragment of KinB containing a proline-390-to-serine mutation. Incorporation of the mutation was confirmed by sequencing. E. coli BL21(DE3) (New England BioLabs, Ipswich, MA) cells harboring pSM95 and pSM95KinBP390S were grown overnight at 37°C in 5 ml LB supplemented with kanamycin. The cells were diluted 1:1,000 into 500 ml LB, supplemented with kanamycin, and allowed to grow to an approximate OD600 of 0.85 at 37°C. At an OD600 of 0.85, 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) was added to the cultures. After overnight incubation at 25°C and 250 rpm, the cells were harvested and soluble proteins were extracted with B-PER reagent (Thermo Scientific, Waltham, MA) supplemented with lysozyme (Sigma-Aldrich, St. Louis, MO) and Benzonase (Sigma-Aldrich, St. Louis, MO), and complete mini-EDTA-free protease inhibitor tablets (Roche, Indianapolis, IN) for 20 min at room temperature. Lysates were sonicated, and proteins were purified from the lysates using nickel-nitrilotriacetic acid (Ni-NTA) beads (Qiagen, Chatsworth, CA). Proteins were eluted with 250 mM imidazole (Sigma-Aldrich, St. Louis, MO). Eluates were further purified using ion-exchange chromatography (1-ml Mono-Q column; GE Healthcare, Piscataway, NJ). Fractions were analyzed by SDS-PAGE. As estimated by SDS-PAGE and Coomassie blue staining, HC-KinB and HC-KinBP390S were 85 to 90% pure. A plasmid overexpressing His6-tagged AlgB, pDJW403, was obtained from Daniel Wozniak (Ohio State University) (25). E. coli BL21(DE3) (New England BioLabs, Ipswich, MA) cells harboring pDJW403 were grown overnight at 37°C in 5 ml LB supplemented with ampicillin. The cells were diluted 1:350 into 500 ml LB, supplemented with ampicillin, and allowed to grow to an approximate OD of 0.6 at 37°C. At an OD600 of 0.6, 1 mM IPTG was added and the cells were allowed to grow for another 4 h at 37°C. The cell pellet was frozen at −80°C and lysed the next morning as noted above. His-AlgB was purified as described above and found to be >90% pure by SDS-PAGE and Coomassie blue staining.
Autophosphorylation of KinB.
His6-tagged carboxyl-terminal fragments of KinB and KinBP390S (2.5 μM) were incubated in 80 μl kinase buffer containing 50 mM KCl, 5 mM MgCl2, 50 mM Tris (pH 8.0), 55 μM ATP and 4 μCi [γ-32P]-ATP from a stock at ∼10 Ci/mmol (Perkin Elmer). After incubation for various periods at room temperature, 10 μl of the reaction mixture was removed and added to 3 μl of a 4× SDS sample buffer, and the aliquot was kept on ice. Reactions were analyzed by 10% SDS-PAGE. The gels were dried and imaged using a phosphorimager.
In vitro phosphotransfer assays.
For phosphotransfer analysis, wild-type HC-KinB and HC-KinBP390S were autophosphorylated as described above for 2 h at room temperature in 110 μl kinase buffer. Phosphotransfer was analyzed by incubating autophosphorylated kinase with His6-tagged AlgB (H-AlgB), each at a final concentration of 2.35 μM, at room temperature for various periods. Aliquots (10 μl) of the reaction mixtures were removed as noted above and analyzed by 10% SDS-PAGE and phosphorimaging. For the experiments depicted in Fig. 6C, HC-KinBP390S was autophosphorylated as above for 2 h at room temperature. Phosphotransfer was initiated by incubating autophosphorylated HC-KinBP390S with H-AlgB, each at a final concentration of 2.35 μM, at room temperature for various time periods in 80 μl kinase buffer. One hour after the addition of H-AlgB, 4 μg of HC-KinB was added to a 20-μl aliquot of the above-described reaction mixture, and 4 μg of HC-KinBP390S was added to a second 20-μl aliquot of the mixture. A 3-μl portion of 4× SDS sample buffer was added to half of each reaction mixture at various time points, and the mixture was placed on ice. Reaction products were analyzed by 10% SDS-PAGE; gels were dried and imaged using a phosphorimager.
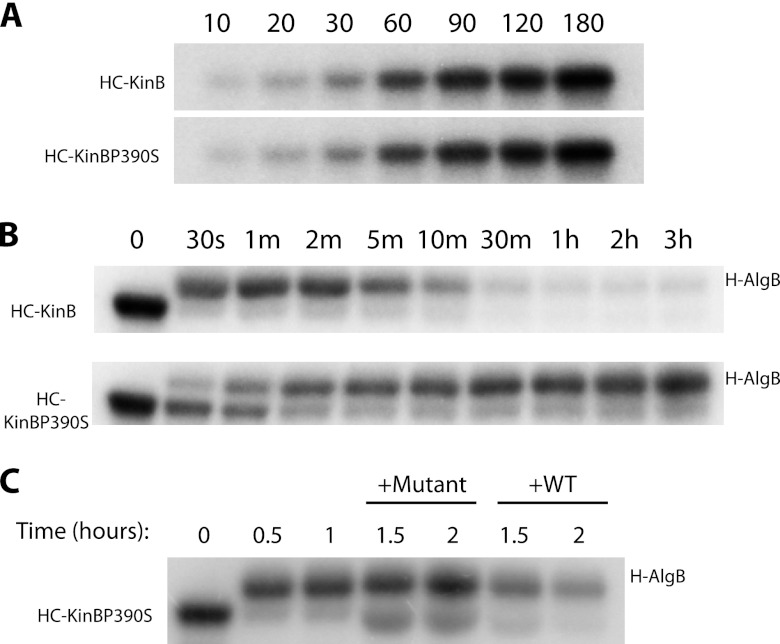
Fig 6.
KinB acts as a phosphatase to regulate AlgB activity. Autophosphorylation and phosphotransfer assays were performed in vitro with radiolabeled ATP and labeled proteins were separate by SDS-PAGE. (A) Autophosphorylation of HC-KinB and HC-KinBP390S. (B) Kinetics of phosphotransfer of the radiolabel from HC-KinB to H-AlgB (top) and from HC-KinBP390S to H-AlgB (bottom). (C) Autophosphorylated HC-KinBP390S (lane 1) was incubated with H-AlgB, resulting in phosphotransfer within 30 min to 1 h (lanes 2 and 3). Subsequent addition of unlabeled WT (HC-KinB; lanes 6 and 7) but not unlabeled mutant (HC-KinBP390S; lanes 4 and 5) KinB protein to phosphotransfer reactions resulted in loss of the radiolabel from H-AlgB. All results are representative of three replicates.
Zebrafish embryo infections.
Zebrafish embryos (AB line) were inoculated with either a P. aeruginosa mutant or the wild-type PA14 strain in the yolk circulation valley 50 h postfertilization (hpf) as previously described (8).
Statistical methods.
Prism 5 (GraphPad Inc.) was used for all statistical analyses. The data in Fig. 1, 2, 3, 4, and 5 were compared using one-way analysis of variance (ANOVA) followed by Bonferroni's multiple comparison test (set at 0.05). Kaplan-Meier survival curves were plotted for the data depicted in Fig. 7, and significance was calculated using the log-rank test.
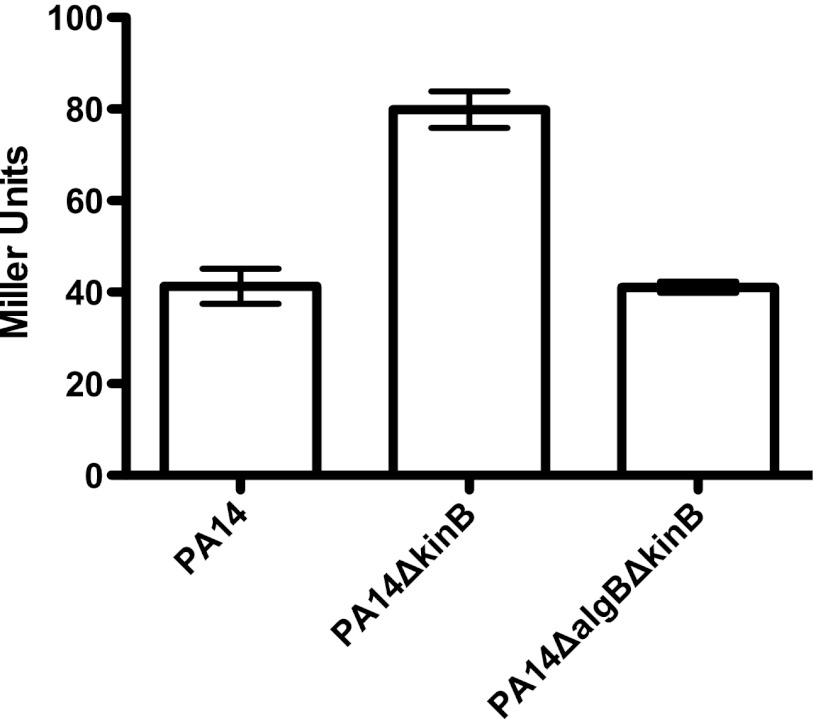
Fig 1.
AlgU expression is upregulated in PA14ΔkinB. Miller activity from PalgU-lacZ reporters expressed episomally in the indicated strains. Error bars indicate standard errors of the mean computed from three technical replicates. The data are representative of two biological replicates.
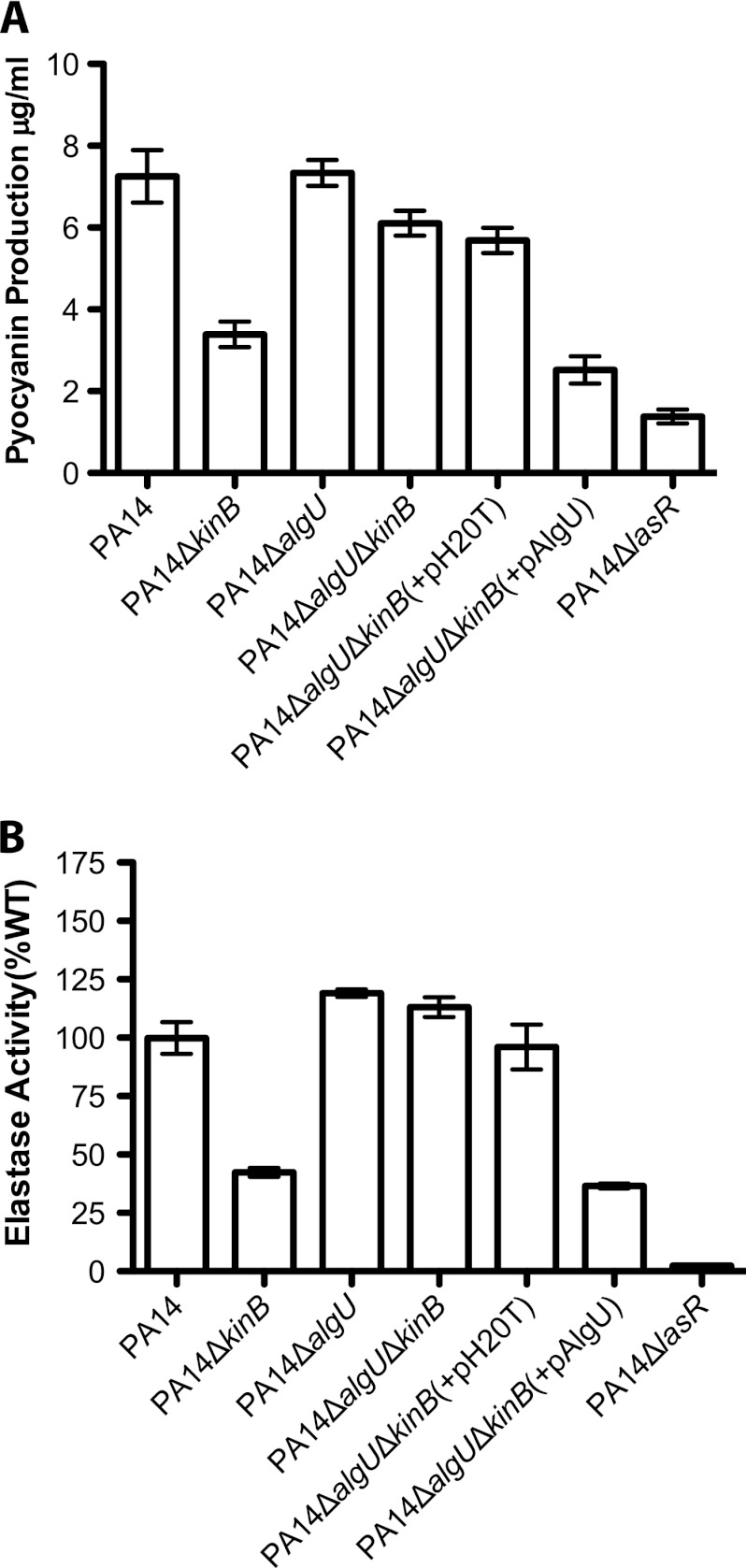
Fig 2.
AlgU is required for the repression of acute virulence phenotypes in PA14ΔkinB. (A) Pyocyanin produced by different P. aeruginosa strains after 22 h of growth at 37°C and 250 rpm. Error bars indicate standard errors of the means computed from three technical replicates. The data are representative of two biological replicates. (B) Elastase activities from culture fluids from 21-h cultures in LB medium measured in elastin-Congo red assays. The A495 for each strain is shown as a percentage of the mean wild-type absorbance. Error bars indicate standard errors of the means computed from three technical replicates. The data are representative of two biological replicates.
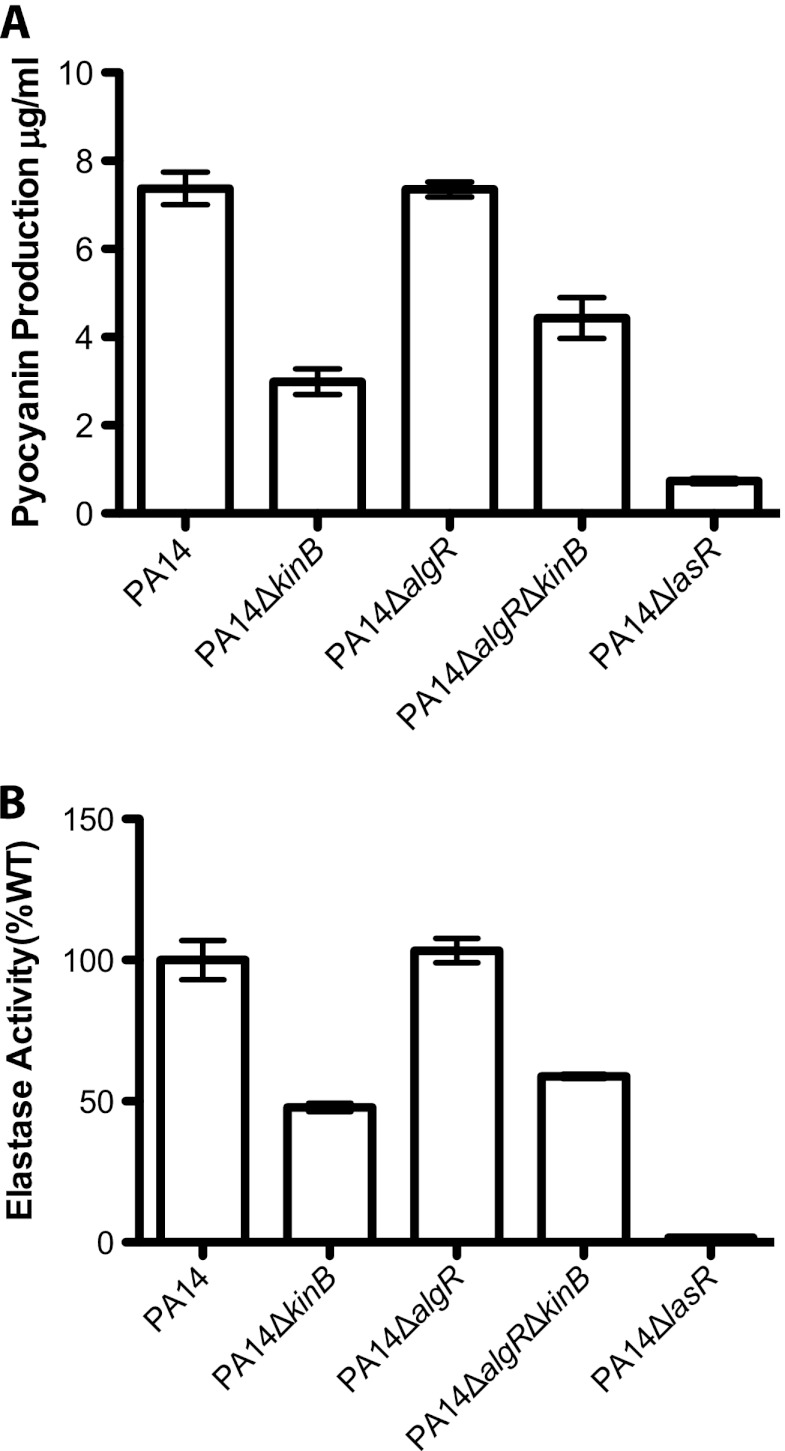
Fig 3.
AlgR does not play a key role in the repression of acute virulence phenotypes in PA14ΔkinB. (A) Pyocyanin produced by different P. aeruginosa strains after 22 h of growth at 37°C and 250 rpm. Error bars indicate standard errors of the means computed from three technical replicates. The data are representative of two biological replicates. (B) Elastase activities from culture fluids from 21 h cultures in LB medium measured in elastin-Congo red assays. The A495 for each strain is shown as a percentage of the mean wild-type absorbance. Error bars indicate standard errors of the means computed from three technical replicates. The data are representative of two biological replicates.
Fig 4.
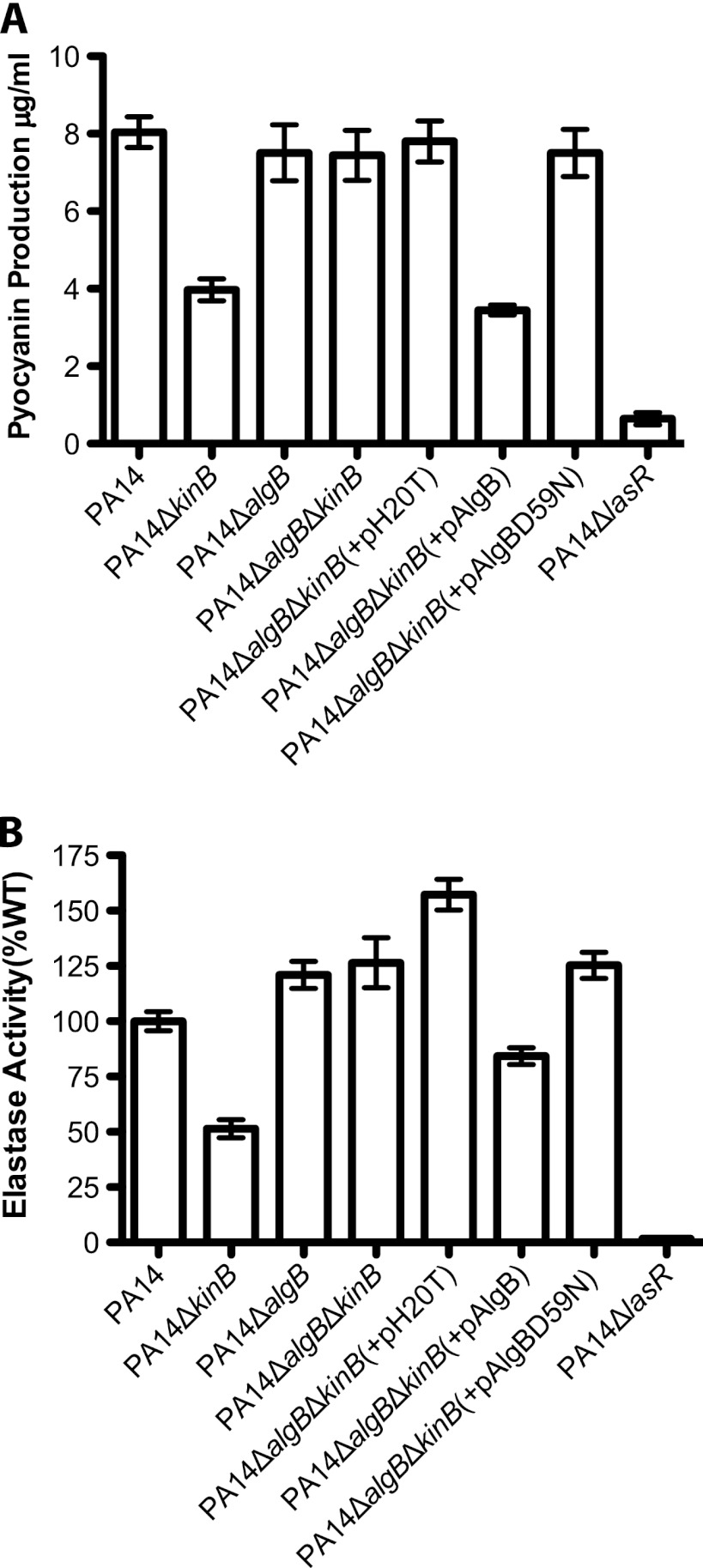
Phosphorylatable AlgB is required for the repression of acute virulence phenotypes in PA14ΔkinB. (A) Pyocyanin produced by different P. aeruginosa strains after 22 h of growth at 37°C and 250 rpm. Error bars indicate standard errors of the means computed from three technical replicates. The data are representative of two biological replicates. (B) Elastase activities from culture fluids from 21 h cultures in LB medium measured in elastin-Congo red assays. The A495 for each strain is shown as a percentage of the mean wild-type absorbance. Error bars indicate standard errors of the means computed from three technical replicates. The data are representative of two biological replicates.
Fig 5.
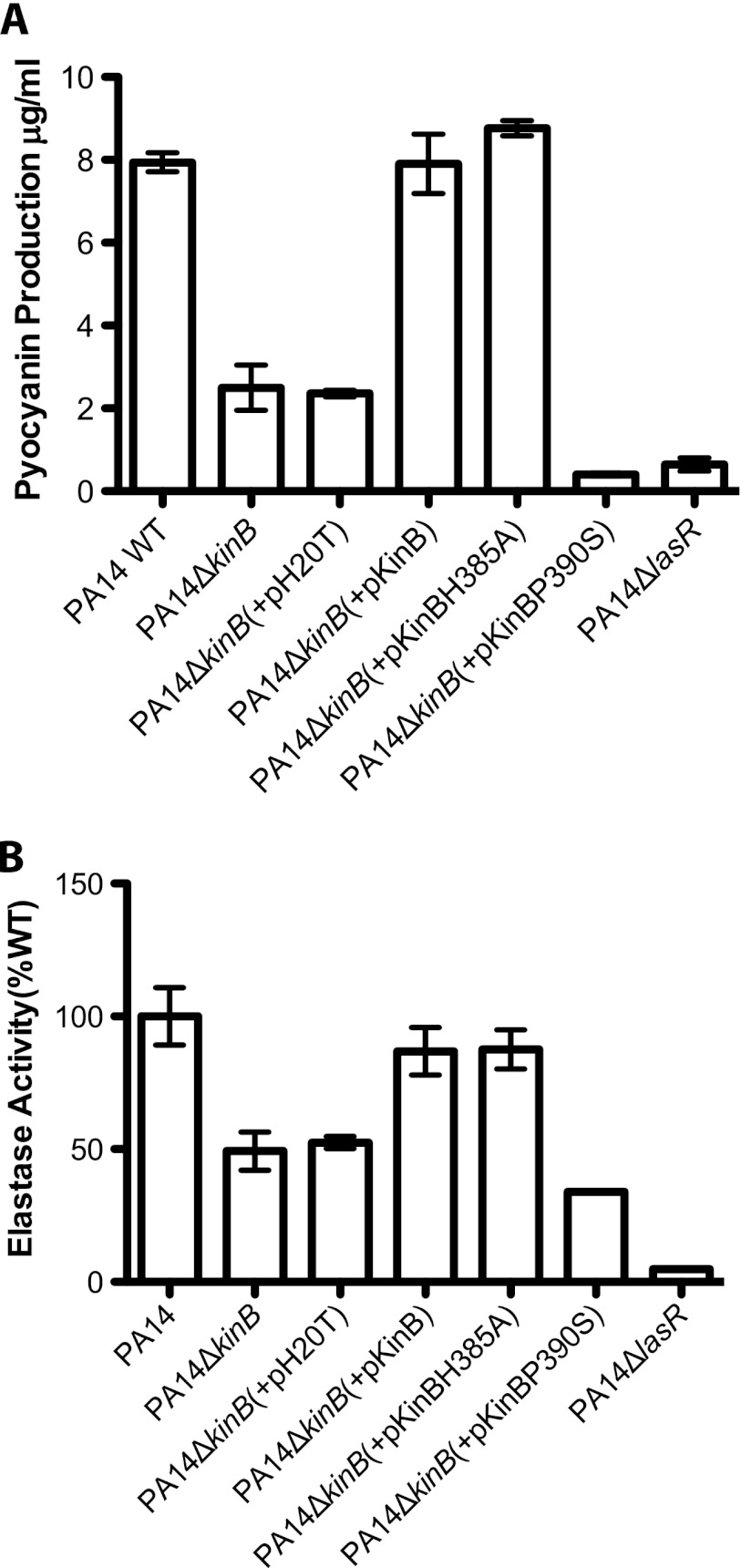
KinB acts as a phosphatase to regulate P. aeruginosa acute virulence. (A) Pyocyanin produced by different P. aeruginosa strains after 22 h of growth at 37°C and 250 rpm. Error bars indicate standard errors of the means computed from three technical replicates. The data are representative of two biological replicates. (B) Elastase activities from culture fluids from 21-h cultures in LB medium measured in elastin-Congo red assays. The A495 for each strain is shown as a percentage of the mean wild-type absorbance. Error bars indicate standard errors of the means computed from three technical replicates. The data are representative of two biological replicates.
Fig 7.
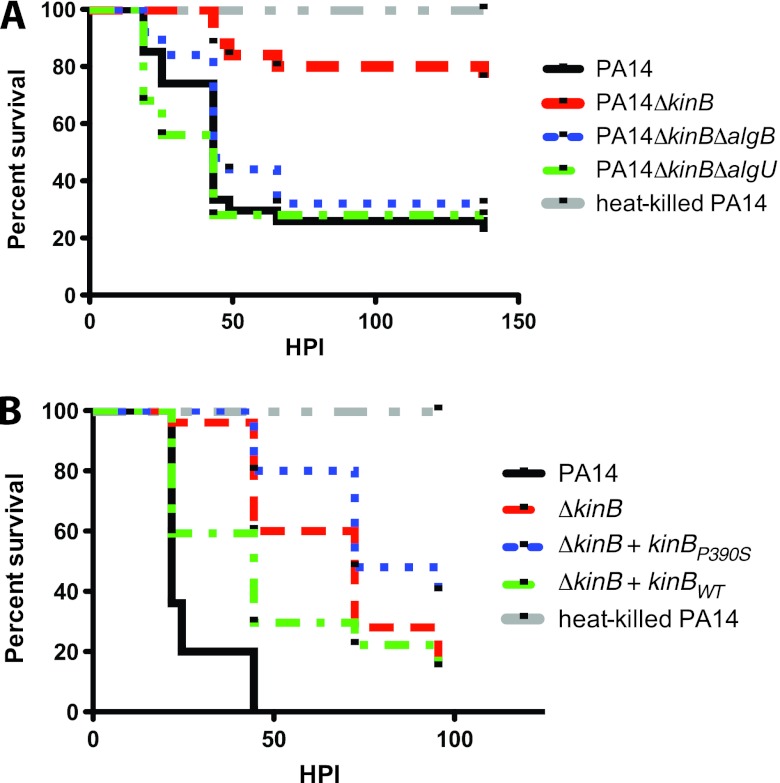
KinB acts as a phosphatase to regulate AlgB activity in vivo. Kaplan-Meier embryo survival curves following infection of 50-hpf embryos with PA14 (8,540 ± 556 CFU), PA14ΔkinB (10,930 ± 415 CFU), PA14ΔkinBΔalgB (10,047 ± 473 CFU), PA14ΔkinBΔalgU (12,445 ± 289 CFU), or heat-killed PA14 (A) or with PA14 (11,205 ± 2,272 CFU), PA14ΔkinB (8,450 ± 901 CFU), the phosphatase-inactive KinB mutant, PA14ΔkinB(+kinBP390S) (9,400 ± 472 CFU), PA14ΔkinB(+kinB) (11,730 ± 1,149 CFU), or heat-killed PA14 (B). The data are representative of two replicates with 20 to 30 embryos per condition per replicate.
RESULTS
AlgU expression is upregulated in PA14ΔkinB.
It was recently reported that algU promoter activity is upregulated in PAO1ΔkinB (10). We found that algU expression is also induced in strain PA14 when kinB is deleted by comparing activity of a transcriptional fusion of the algU promoter to lacZ in wild-type PA14 and a PA14ΔkinB strain. algU promoter activity is upregulated in PA14ΔkinB (∼2-fold) compared to that in the wild-type strain (Fig. 1; P < 0.05; P = 0.0002 for the overall data set).
AlgU plays a role in the repression of virulence phenotypes in PA14ΔkinB.
Given that acute virulence phenotypes such as pyocyanin, elastase production, and motility are repressed in PA14ΔkinB (7), that algU expression is induced in PA14ΔkinB, and that increased AlgU expression is associated with defects in acute virulence phenotypes, such as motility (34), we wondered whether the repression of acute virulence factors in PA14ΔkinB is mediated by AlgU. To test this hypothesis, we constructed a PA14ΔalgUΔkinB strain. Deletion of algU in a ΔkinB background restores wild-type levels of pyocyanin and elastase production in this strain (Fig. 2A, P > 0.05 and P < 0.0001 for the overall data set; Fig. 2B, P > 0.05 and P < 0.0001 for the overall data set; P values are reported for PA14 in comparison to PA14ΔalgUΔkinB). Additionally, this double deletion strain is as motile as wild-type PA14 (Table 3). Thus, wild-type production of pyocyanin and elastase in PA14ΔalgUΔkinB is similar to that in PA14ΔalgU but contrasts with that in PA14ΔkinB (and PA14ΔlasR [7], a quorum-sensing mutant known to be defective in pyocyanin and elastase production included throughout as a negative control). Complementation of algU in PA14ΔalgUΔkinB [PA14ΔalgUΔkinB(+pAlgU)] restored the reduced pyocyanin, elastase, and motility phenotypes observed in PA14ΔkinB, unlike PA14ΔalgUΔkinB(+pH20T), where PA14ΔalgUΔkinB is complemented with the empty vector [Fig. 2A, P < 0.05; Fig. 2B, P < 0.05 and Table 3; P values are reported for PA14ΔalgUΔkinB in comparison to PA14ΔalgUΔkinB(+pAlgU)]. We also found that deletion of algU in PA14ΔkinB results in a nonmucoid strain and that episomal complementation of algU in PA14ΔalgUΔkinB restores mucoidy (Table 4). These results suggest that the repression of acute virulence phenotypes in PA14ΔkinB is mediated by AlgU.
Table 3.
Swimming motilities on 0.35% LB agar plates
| Strain | Swim diam (% of WT; mean ± SEM)a |
|---|---|
| PA14ΔkinB (+pHERD20T) | 38.0 ± 3.6 |
| PA14ΔalgU(+pHERD20T) | 101.4 ± 4.1 |
| PA14ΔalgUΔkinB (+pHERD20T) | 94.0 ± 3.4 |
| PA14ΔalgUΔkinB (+pAlgU) | 28.5 ± 4.9 |
| PA14ΔalgB(+pHERD20T) | 102.5 ± 3.3 |
| PA14ΔalgBΔkinB(+pHERD20T) | 107.4 ± 1.6 |
| PA14ΔalgBΔkinB(+pAlgB) | 46.9 ± 2.8 |
| PA14ΔalgBΔkinB(+pAlgBD59N) | 104.7 ± 2.6 |
| PA14ΔkinB (+pKinB) | 110.1 ± 4.9 |
| PA14ΔkinB (+pKinBH385A) | 89.4 ± 3.1 |
| PA14ΔkinB (+pKinBP390S) | 53.4 ± 6.9 |
| PA14ΔalgR(+pHERD20T) | 87.5 ± 3.3 |
| PA14ΔalgRΔkinB(+pHERD20T) | 56.3 ± 2.4 |
| PA14ΔflgK(transposon mutant) | NDb |
The swim zone diameter of each strain was compared individually to that of PA14(+pHERD20T) to enable comparison on plates containing carbenicillin. The standard error of the mean was calculated from three replicates.
ND, not determined [PA14ΔflgK(transposon mutant) was not motile].
Table 4.
Mucoidy phenotype of different strains on PIA plates
| Strain | Phenotypea |
|---|---|
| PA14 | NM |
| PA14ΔkinB (+pHERD20T) | M |
| PA14ΔalgU(+pHERD20T) | NM |
| PA14ΔalgUΔkinB(+pHERD20T) | NM |
| PA14ΔalgUΔkinB(+pAlgU) | M |
| PA14ΔalgB(+pHERD20T) | NM |
| PA14ΔalgBΔkinB(+pHERD20T) | NM |
| PA14ΔalgBΔkinB(+pAlgB) | M |
| PA14ΔalgBΔkinB(+pAlgBD59N) | NM |
| PA14ΔkinB (+pKinB) | NM |
| PA14ΔkinB (+pKinBH385A) | NM |
| PA14ΔkinB (+pKinBP390S) | M |
| PA14ΔalgR(+pHERD20T) | NM |
| PA14ΔalgRΔkinB(+pHERD20T) | NM |
M, mucoid; NM, nonmucoid.
AlgR does not play a key role in the repression of virulence phenotypes in PA14ΔkinB.
Jones et al. recently reported that loss-of-function mutations in the anti-sigma factor gene mucA result in activation of MucA's corresponding sigma factor, AlgU, indirectly resulting in the downregulation of acute virulence factors, such as type III secretion toxins and exotoxin A production, in a manner dependent on the transcription factor AlgR, which is thought to act downstream of AlgU in this pathway (21). Since we observed that the repression of acute virulence factors such as pyocyanin and elastase is mediated by AlgU, we wondered whether AlgR also acts downstream of AlgU in PA14ΔkinB. To test this hypothesis, we constructed a PA14ΔalgRΔkinB strain. Deletion of algR in PA14ΔkinB does not restore wild-type levels of pyocyanin and elastase production or wild-type motility (Fig. 3A, P < 0.05; Fig. 3B, P < 0.05; Table 3; P values are reported for PA14 in comparison to PA14ΔalgRΔkinB). This suggests that in PA14, AlgU acts through an alternative pathway to repress the production of pyocyanin and elastase. However, PA14ΔalgRΔkinB is nonmucoid (Table 4), suggesting that AlgR is required for the enhanced production of alginate in this strain.
AlgB plays a role in the repression of virulence phenotypes in PA14ΔkinB.
We had previously observed that PA14ΔalgB produced wild-type levels of acute virulence factors, suggesting that AlgB might not play a role in virulence regulation (7). However, because Damron et al. (10) had observed in their study of alginate biosynthesis that an increase in algU expression in PAO1ΔkinB requires AlgB, we constructed the ΔalgB ΔkinB double deletion in PA14 and tested for expression of algU. Deletion of algB in the ΔkinB background restores wild-type levels of algU expression, suggesting that AlgB is required for KinB regulation of algU (Fig. 1). Similarly, algB deletion in PA14ΔkinB also restores wild-type levels of pyocyanin and elastase (Fig. 4A, P > 0.05 and P < 0.0001 for the overall data set; Fig. 4B, P > 0.05 and P < 0.0001 for the overall data set; P values are reported for PA14 in comparison to PA14ΔalgBΔkinB) and motility (Table 3). Episomal complementation of the algB deletion in the PA14ΔalgBΔkinB strain restores the reduced pyocyanin, elastase, and motility phenotypes of the PA14ΔkinB strain [Fig. 4A, P < 0.05; Fig. 4B, P < 0.05; Table 3; P values are reported for PA14ΔalgBΔkinB in comparison to PA14ΔalgBΔkinB(+pAlgB)]. Thus, PA14ΔalgBΔkinB phenocopies PA14ΔalgB and wild-type PA14 with respect to these acute virulence phenotypes, in contrast to PA14ΔkinB (Fig. 4A and B and Table 3). We also found that PA14ΔalgBΔkinB is nonmucoid, while episomal expression of AlgB in this strain restores mucoidy (Table 4). These data suggest that AlgB is, in fact, required in the absence of KinB for the phenotypes we observed, and thus, KinB signals through AlgB, albeit in a noncanonical manner, since deletion of algB alone has no phenotype.
Phosphorylatable AlgB is required for repression of virulence in PA14ΔkinB.
To determine whether the phosphorylation of AlgB is required for the repression of acute virulence phenotypes in the absence of KinB, we constructed a plasmid expressing a mutated AlgBD59N protein. The aspartate at position 59 is the critical residue that is typically phosphorylated by a histidine sensor kinase, while an asparagine mutant cannot be phosphorylated (25). Episomal expression of AlgBD59N in PA14ΔalgBΔkinB fails to restore the PA14ΔkinB phenotype of repressed acute virulence and increased mucoidy (Fig. 4A and B and Tables 3 and 4), unlike wild-type AlgB, which phenocopies the repressed pyocyanin and elastase production and increased mucoidy observed in PA14ΔkinB. Since PA14ΔalgBΔkinB(+pAlgBD59N) phenocopies PA14ΔalgBΔkinB, it remained possible that AlgBD59N was not expressed. We eliminated this possibility by confirming that an N-terminally His-tagged AlgBD59N is expressed inPA14ΔalgBΔkinB(+pHisAlgBD59N) (see Fig. S1 in the supplemental material) and by confirming that this strain phenocopies its untagged counterpart, PA14ΔalgBΔkinB(+pAlgBD59N), in acute virulence factor production (data not shown). These data suggest that in the absence of KinB, AlgB must be phosphorylated in order to repress virulence. Further, they suggest that in PA14ΔkinB, AlgB can be phosphorylated in a KinB-independent manner, perhaps by alternative sensor kinases.
KinB acts as a phosphatase to regulate AlgB activity.
Previously, a bioinformatics study had predicted that KinB might act as a bifunctional sensor with possible phosphatase activity, based on structural features determined by homology modeling techniques (1). Given our findings that KinB regulates acute virulence independent of its kinase activity (7) but dependent on the phosphorylation state of its cognate response regulator, AlgB, we hypothesized that KinB might act as a phosphatase to dephosphorylate and inactivate AlgB. To test this hypothesis, we engineered an allele of KinB containing a point mutation at a key residue predicted to play a role in the phosphatase activity of two-component sensors. We aligned the sequence of the KinB H box with the sequence of the H box from the prototypical sensor kinase/phosphatase EnvZ and found that KinB contains a conserved proline (P390) residue that has been shown to be required for EnvZ's phosphatase activity (4, 19). We thus constructed the mutant allele kinB(P390S), in which the corresponding proline (P390) is mutated to a serine. Interestingly, we found that episomal introduction of the proline mutant allele (pKinBP390S) into PA14ΔkinB is unable to complement the pyocyanin and elastase defects of the PA14ΔkinB strain, unlike the wild-type allele (pKinB) and the catalytically inactive kinase allele (pKinBH385A) [Fig. 5A, P < 0.05 and P < 0.0001, for the overall data set; Fig. 5B, P < 0.05 and P < 0.0001 for the overall data set; P values are reported for PA14 in comparison to PA14ΔkinB(+pKinBP390S)]. Furthermore, the proline mutant allele is unable to complement the motility defect of the PA14ΔkinB strain (Table 3). PA14ΔkinB(+pKinBP390S) is as mucoid as PA14ΔkinB (Table 4). Further, PA14ΔkinB(+pKinBP390S) has an even more severe defect in pyocyanin production than PA14ΔkinB (Fig. 5A, P < 0.05 and P < 0.0001 for the overall data set) but while PA14ΔkinB(+pKinBP390S) also displays a reduction in elastase activity compared to PA14ΔkinB, this difference is not statistically significant. These findings suggest that KinB induces acute virulence by dephosphorylating AlgB and that in the absence of KinB's phosphatase activity, phosphorylated AlgB acts as a repressor of acute virulence and inducer of mucoidy.
To verify that mutating KinB's proline residue did not result in a misfolded protein, we purified the His-tagged, carboxyl-terminal domain of KinB (HC-KinB) (26) and mutant KinB (HC-KinBP390S). It has been shown that this domain of KinB retains autophosphorylation activity (26). We found that mutating the proline (P390) residue to serine has no significant effect on KinB's autophosphorylation activity and both proteins are autophosphorylated at comparable rates (Fig. 6A).
To directly demonstrate that KinB dephosphorylates AlgB, we performed biochemical studies with purified HC-KinB and AlgB. We purified recombinant HC-KinB and AlgB from E. coli and found that incubation of autophosphorylated HC-KinB with purified His-tagged AlgB (H-AlgB) results in transfer of the radiolabeled phosphate to AlgB in 30 s (Fig. 6B). However, 30 min after incubation with HC-KinB, there is a clear loss of labeled phosphate from H-AlgB (Fig. 6B). In contrast, when AlgB was incubated with the mutant HC-KinBP390S, which is predicted to be phosphatase deficient, AlgB retained the labeled phosphate even after 3 h (Fig. 6B). Furthermore, following incubation of H-AlgB for 1 h with the mutant HC-KinBP390S, we then added back HC-KinB and found that there was clear loss of labeled phosphate from H-AlgB 30 min after incubation, unlike addition of HC-KinBP390S, which resulted in H-AlgB's retention of the radiolabel (Fig. 6C). Together, these data argue that KinB exhibits phosphatase activity and can directly dephosphorylate AlgB.
KinB acts as a phosphatase to regulate AlgB activity in vivo.
To verify that AlgB and AlgU are required for the repression of acute virulence in PA14ΔkinB in vivo, we infected zebrafish embryos with PA14ΔalgUΔkinB and PA14ΔalgBΔkinB. In contrast to PA14ΔkinB, both PA14ΔalgUΔkinB and PA14ΔalgBΔkinB were as virulent as wild-type PA14 (Fig. 7A). Thus, KinB regulates P. aeruginosa virulence through AlgU and AlgB in vivo. Furthermore, when zebrafish embryos were infected with PA14ΔkinB(+KinBP390Sint), which expresses the phosphatase-inactive form of KinB, a significantly greater fraction of embryos survived than when infection was with PA14 (P < 0.0001), PA14ΔkinB(+KinBint) (P = 0.0016), and even PA14ΔkinB (P = 0.0417) (Fig. 7B). Thus, the proline mutant allele of kinB is unable to complement the virulence defect of PA14ΔkinB, unlike wild-type kinB (Fig. 7B) or the kinase-inactive form of kinB (7), and is even less virulent than PA14ΔkinB itself. Thus, KinB's key role in the regulation of acute virulence is likely to dephosphorylate AlgB, which represses AlgU expression and thus results in the activation of virulence. In the absence of KinB, phosphorylated AlgB induces AlgU expression to repress acute virulence.
DISCUSSION
P. aeruginosa is an extremely flexible pathogen that can survive in a variety of niches both inside and outside the human host. During human infection, P. aeruginosa isolates from chronically and acutely infected patients are often phenotypically different. Isolates from chronically infected CF patients commonly overproduce alginate (and thus are mucoid) and are nonmotile and/or resistant to antibiotics but do not express type III secretory proteins. In contrast, strains isolated from acute infections generally express type III secretory proteins, are motile, and do not overproduce alginate (are nonmucoid) (31, 36). Clearly, these opposing phenotypes are required for optimal survival of the organism in acute versus chronic infection settings, and P. aeruginosa has evolved tight regulatory mechanisms to control the switch between these two lifestyles.
It is clear that TCSs help to regulate this switch between acute and chronic infection lifestyles, and this is perhaps best exemplified by the TCS sensors GacS, RetS, and LadS. The sensors GacS and RetS are thought to physically interact in the membrane (16) and, in this way, negatively regulate the transcription of the small RNAs rsmZ and rsmY, which ultimately promotes the expression of virulence genes required for acute infection (e.g., T3SS genes) (15, 16, 40). During chronic infection, on the other hand, GacS and LadS positively regulate the expression of rsmZ/Y and thus promote the expression of the pel genes important for biofilm formation and repress genes important during acute infection (T3SS genes) (14). As demonstrated by GacS, RetS, and LadS, the regulation of the transition between acute and chronic infection lifestyles is complex, requires more than just one TCS sensor, and the TCS signaling involved in this transition can be described as noncanonical insofar as a physical interaction between two sensors (RetS and GacS) is important for signal transduction to occur rather than simple phosphorylation of a response regulator by its cognate sensor.
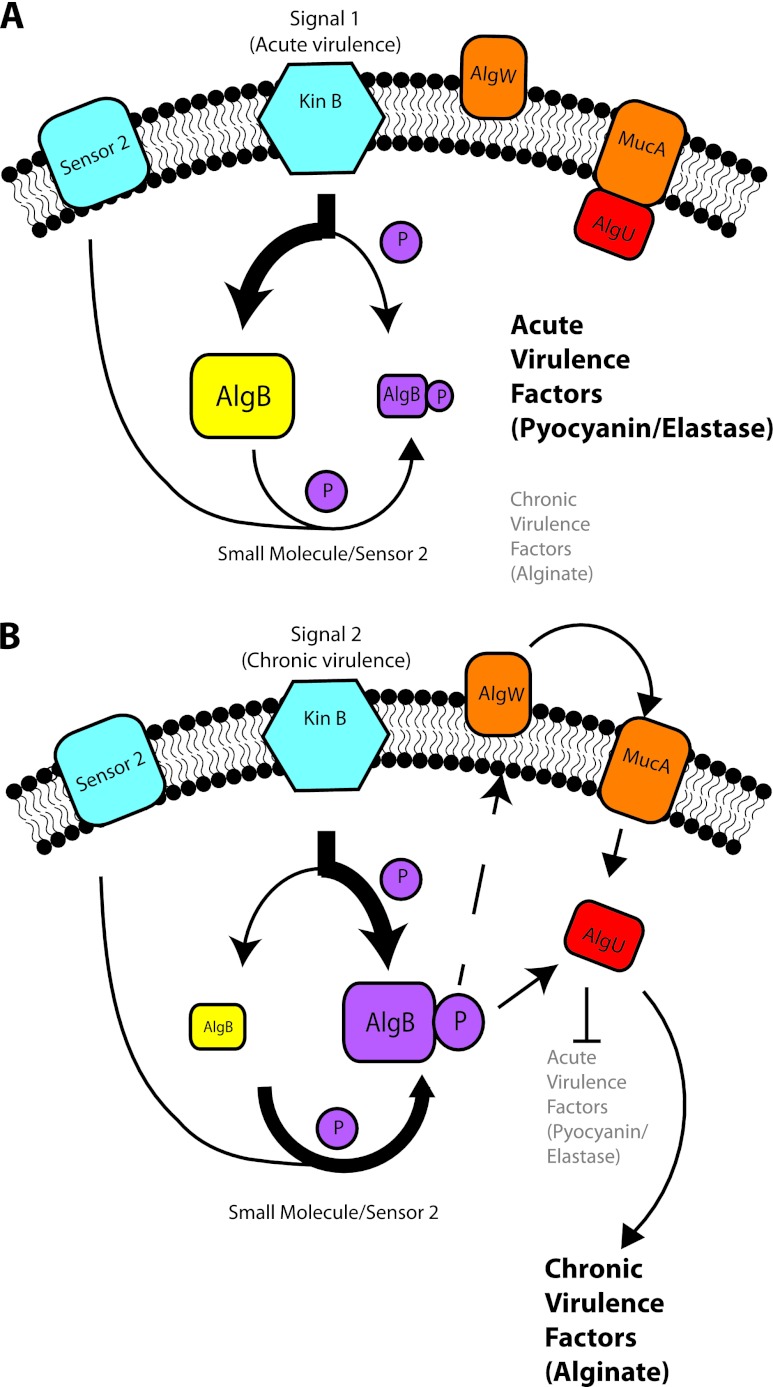
Here we add further complexity to the regulation of the transition between acute and chronic infection lifestyles by TCSs in P. aeruginosa. We find that the TCS sensor KinB also plays a role in the switch between acute and chronic infection phenotypes and signals through its cognate response regulator AlgB and the sigma factor AlgU. Interestingly however, KinB's phosphatase activity rather than its kinase activity on AlgB is important for its regulation of acute virulence phenotypes since a KinB kinase-inactive mutant is still functional (7) during acute infection while a KinB phosphatase-inactive mutant is attenuated. Further, an AlgB mutant that cannot be phosphorylated cannot repress acute virulence phenotypes in a ΔkinB background unlike wild-type AlgB, demonstrating that the presence of phosphorylated AlgB is important for the repression of acute virulence. These findings suggest a model where phosphorylated AlgB acts to upregulate AlgU activity, which results in the repression of acute virulence and promotes chronic phenotypes such as alginate production (Fig. 8). In acute infection, in contrast, KinB acts as a phosphatase, dephosphorylating AlgB and enabling the expression of pyocyanin, elastase, and motility genes that are important during acute infection.
Fig 8.
Model of KinB's role in the regulation of P. aeruginosa virulence. (A) Acute virulence. Acute virulence factors are expressed while chronic virulence factors are repressed. In this state, AlgU is sequestered by MucA, and AlgB exists predominantly in the dephosphorylated state. On detecting appropriate signals, KinB acts predominantly as a phosphatase to maintain AlgB in the dephosphorylated state. (B) Chronic virulence. Chronic virulence factors are expressed while acute virulence factors are repressed. AlgB exists predominantly in the phosphorylated state. AlgB may be phosphorylated by KinB itself (in WT cells), or AlgB may be phosphorylated by another sensor (sensor 2) or small molecules such as acetyl phosphate. AlgB may activate MucA-degrading proteases such as AlgW. AlgW activation could result in the degradation of MucA and the release of AlgU. Alternatively, phosphorylated AlgB may directly upregulate AlgU expression.
An example of a sensor acting as a phosphatase to regulate virulence phenotypes has been reported. QseC/QseB's regulation of type 1 pili, curli, and flagella in uropathogenic E. coli mirrors KinB/AlgB's regulation of virulence through dephosphorylation (22). Similar to ΔkinB strains, null mutation of the sensor QseC results in its cognate response regulator, QseB, being locked in an active (phosphorylated) form, which negatively regulates transcription of pili, curli, and flagella. Our work suggests that this mode of regulation of virulence by two-component sensors may be more widespread.
Our work suggests that in the absence of KinB, AlgB must be phosphorylated; however, the question of how it is phosphorylated remains. Promiscuous phosphorylation of response regulators such as AlgB can occur by sensor kinases other than their respective cognate sensor or by small molecules such as acetyl phosphate (23). A given sensor's phosphatase activity may be important to help prevent cross talk and ensure fidelity of signaling (23). The background activation of a response regulator that occurs through the promiscuous kinase activity of a noncognate sensor or through small molecules like acetyl phosphate can be minimized by its cognate sensor's phosphatase activity. In rapidly changing environments, more rapid switches from one phenotype to another may be desired than can occur through spontaneous phosphate hydrolysis. Thus, coupling a sensor's kinase activity with phosphatase activity allows tight control of the phosphorylation state of the response regulator by allowing rapid reversal of overactivated or inappropriately activated response regulator (23).
In this study, we show that signaling through KinB/AlgB is ultimately mediated through the sigma factor AlgU. The repression of acute virulence phenotypes in PA14ΔkinB requires AlgU, since wild-type virulence is restored in PA14ΔalgUΔkinB (Fig. 2A and B and Table 3) and AlgU expression is upregulated in PA14ΔkinB (Fig. 1). Previous work demonstrated that AlgU represses some acute virulence phenotypes (type III secretion and exotoxin A production in strain PAO1) through the response regulator AlgR (21). Here, we found that the KinB-mediated regulation of different acute virulence phenotypes (motility and pyocyanin and elastase production) is not mediated through AlgR, since algR deletion in PA14ΔkinB did not restore wild-type production of pyocyanin and elastase or wild-type motility (Fig. 3A and B and Table 3). This suggests that AlgU may act through an alternative AlgB-mediated pathway to repress the production of pyocyanin and elastase compared to type III secretion and exotoxin A production or that strain PA14 and PAO1 may simply differ in their regulation.
How does KinB/AlgB signal through AlgU? We find that AlgB promotes enhanced AlgU expression (Fig. 1). Thus, phosphorylated AlgB may directly upregulate the transcription of AlgU by binding at the promoter. Previous attempts to show such an interaction were unsuccessful (10), although we note that this may be because AlgB needs to be in a phosphorylated state to bind at the AlgU promoter; response regulator phosphorylation is required for DNA binding and transcriptional control (13). Furthermore, it has been demonstrated that AlgU activity is elevated in PAO1ΔkinB, and this may be due to the enhanced activity of proteases like AlgW that degrade the anti-sigma factor MucA, resulting in release and activation of AlgU (Fig. 8) (10, 11). (We did find that algW deletion in PA14ΔkinB restored wild-type levels of pyocyanin [data not shown].) AlgU then activates the production of alginate by promoting transcription of the algD gene, which encodes a GDP-mannose dehydrogenase that plays a critical role in alginate biosynthesis (41). AlgU also activates expression of the transcription factors AlgR and AlgB, which further enhances expression of AlgD.
While the mechanism by which AlgU promotes chronic infection phenotypes is clearer, since it can directly activate the production of alginate, it is not clear how it might repress acute virulence phenotypes, though there is evidence that higher AlgU activity is correlated with a reduction in acute virulence phenotypes. Mutations in AlgU's anti-sigma factor MucA have a reduction in acute-virulence-related phenotypes, such as the production of rhamnolipids, and an attenuation of virulence in the Caenorhabditis elegans infection model (34). As mentioned above, elevated AlgU activity has also been shown to reduce expression of acute virulence factors such as type III secretion and exotoxin A in strain PAO, though this is mediated by AlgR (21). The molecular basis for AlgU's inhibition of acute virulence phenotypes will no doubt form the basis of future work.
It seems likely that P. aeruginosa may use the two-component sensor KinB to switch between chronic and acute phenotypes in the appropriate microenvironment. Given the recognition that other two-component sensors such as RetS and GacS also regulate this transition (15), it will be interesting to understand how KinB interfaces with other regulators and how the complexity of such overlapping systems benefits P. aeruginosa survival in changing environments. Some of the complexity may lie in their ability to respond to different extracellular signals, though identifying these signals for KinB and other two-component sensors remains a significant challenge. Nevertheless, two-component sensors clearly play critical roles in regulating the behavior of bacterial pathogens and a more detailed understanding of their signaling networks could enable the design of new therapeutics that target these networks.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grant AI 97613-01 (D.T.H.).
We thank Jack Tsai for helpful discussions and technical assistance. We thank Laurence Rahme for providing the pQF50 and pUCP18 plasmids and Daniel J. Wozniak (Ohio State University) and Dennis J. Ohman (Virginia Commonwealth University) for providing the pDJW403 and pSM95 plasmids, respectively.
Footnotes
Published ahead of print 28 September 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Alves R, Savageau MA. 2003. Comparative analysis of prototype two-component systems with either bifunctional or monofunctional sensors: differences in molecular structure and physiological function. Mol. Microbiol. 48:25–51 [DOI] [PubMed] [Google Scholar]
- 2. Bader MW, et al. 2005. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell 122:461–472 [DOI] [PubMed] [Google Scholar]
- 3. Buelow DR, Raivio TL. 2010. Three (and more) component regulatory systems—auxiliary regulators of bacterial histidine kinases. Mol. Microbiol. 75:547–566 [DOI] [PubMed] [Google Scholar]
- 4. Capra EJ, et al. 2010. Systematic dissection and trajectory-scanning mutagenesis of the molecular interface that ensures specificity of two-component signaling pathways. PLoS Genet. 6:e1001220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Castelli ME, Garcia Vescovi E, Soncini FC. 2000. The phosphatase activity is the target for Mg2+ regulation of the sensor protein PhoQ in Salmonella. J. Biol. Chem. 275:22948–22954 [DOI] [PubMed] [Google Scholar]
- 6. Chand NS, Hung DT. 2011. The two-component sensor kinase KinB acts as a non-canonical switch between acute and chronic infection. Virulence 2:553–558 [DOI] [PubMed] [Google Scholar]
- 7. Chand NS, et al. 2011. The sensor kinase KinB regulates virulence in acute Pseudomonas aeruginosa infection. J. Bacteriol. 193:2989–2999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Clatworthy AE, et al. 2009. Pseudomonas aeruginosa infection of zebrafish involves both host and pathogen determinants. Infect. Immun. 77:1293–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Damron FH, et al. 2012. Analysis of the Pseudomonas aeruginosa regulon controlled by the sensor kinase KinB and sigma factor RpoN. J. Bacteriol. 194:1317–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Damron FH, Qiu D, Yu HD. 2009. The Pseudomonas aeruginosa sensor kinase KinB negatively controls alginate production through AlgW-dependent MucA proteolysis. J. Bacteriol. 191:2285–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Damron FH, Yu HD. 2011. Pseudomonas aeruginosa MucD regulates the alginate pathway through activation of MucA degradation via MucP proteolytic activity. J. Bacteriol. 193:286–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Farinha MA, Kropinski AM. 1990. Construction of broad-host-range plasmid vectors for easy visible selection and analysis of promoters. J. Bacteriol. 172:3496–3499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gao R, Stock AM. 2009. Biological insights from structures of two-component proteins. Annu. Rev. Microbiol. 63:133–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gooderham WJ, Hancock RE. 2009. Regulation of virulence and antibiotic resistance by two-component regulatory systems in Pseudomonas aeruginosa. FEMS Microbiol. Rev. 33:279–294 [DOI] [PubMed] [Google Scholar]
- 15. Goodman AL, et al. 2004. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev. Cell 7:745–754 [DOI] [PubMed] [Google Scholar]
- 16. Goodman AL, et al. 2009. Direct interaction between sensor kinase proteins mediates acute and chronic disease phenotypes in a bacterial pathogen. Genes Dev. 23:249–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hoang TT, Kutchma AJ, Becher A, Schweizer HP. 2000. Integration-proficient plasmids for Pseudomonas aeruginosa: site-specific integration and use for engineering of reporter and expression strains. Plasmid 43:59–72 [DOI] [PubMed] [Google Scholar]
- 18. Hoch JA. 2000. Two-component and phosphorelay signal transduction. Curr. Opin. Microbiol. 3:165–170 [DOI] [PubMed] [Google Scholar]
- 19. Hsing W, Silhavy TJ. 1997. Function of conserved histidine-243 in phosphatase activity of EnvZ, the sensor for porin osmoregulation in Escherichia coli. J. Bacteriol. 179:3729–3735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huynh TN, Stewart V. 2011. Negative control in two-component signal transduction by transmitter phosphatase activity. Mol. Microbiol. 82:275–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jones AK, et al. 2010. Activation of the Pseudomonas aeruginosa AlgU regulon through mucA mutation inhibits cyclic AMP/Vfr signaling. J. Bacteriol. 192:5709–5717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kostakioti M, Hadjifrangiskou M, Pinkner JS, Hultgren SJ. 2009. QseC-mediated dephosphorylation of QseB is required for expression of genes associated with virulence in uropathogenic Escherichia coli. Mol. Microbiol. 73:1020–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Laub MT, Goulian M. 2007. Specificity in two-component signal transduction pathways. Annu. Rev. Genet. 41:121–145 [DOI] [PubMed] [Google Scholar]
- 24. Lee DG, et al. 2006. Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial. Genome Biol. 7:R90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ma S, et al. 1998. Phosphorylation-independent activity of the response regulators AlgB and AlgR in promoting alginate biosynthesis in mucoid Pseudomonas aeruginosa. J. Bacteriol. 180:956–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ma S, Wozniak DJ, Ohman DE. 1997. Identification of the histidine protein kinase KinB in Pseudomonas aeruginosa and its phosphorylation of the alginate regulator algB. J. Biol. Chem. 272:17952–17960 [DOI] [PubMed] [Google Scholar]
- 27. Macfarlane EL, Kwasnicka A, Ochs MM, Hancock RE. 1999. PhoP-PhoQ homologues in Pseudomonas aeruginosa regulate expression of the outer-membrane protein OprH and polymyxin B resistance. Mol. Microbiol. 34:305–316 [DOI] [PubMed] [Google Scholar]
- 28. Miller JH. 1972. Beta-galactosidase assay, p 352–355 In Miller JH. (ed), Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 29. Nakano MM, Zhu Y. 2001. Involvement of ResE phosphatase activity in down-regulation of ResD-controlled genes in Bacillus subtilis during aerobic growth. J. Bacteriol. 183:1938–19344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ninfa AJ, Jiang P. 2005. PII signal transduction proteins: sensors of alpha-ketoglutarate that regulate nitrogen metabolism. Curr. Opin. Microbiol. 8:168–173 [DOI] [PubMed] [Google Scholar]
- 31. Oliver A, Canton R, Campo P, Baquero F, Blazquez J. 2000. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288:1251–1254 [DOI] [PubMed] [Google Scholar]
- 32. Pearson JP, Pesci EC, Iglewski BH. 1997. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J. Bacteriol. 179:5756–5767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rahme LG, et al. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899–1902 [DOI] [PubMed] [Google Scholar]
- 34. Rau MH, et al. 2010. Early adaptive developments of Pseudomonas aeruginosa after the transition from life in the environment to persistent colonization in the airways of human cystic fibrosis hosts. Environ. Microbiol. 12:1643–1658 [DOI] [PubMed] [Google Scholar]
- 35. Rodrigue A, Quentin Y, Lazdunski A, Mejean V, Foglino M. 2000. Two-component systems in Pseudomonas aeruginosa: why so many? Trends Microbiol. 8:498–504 [DOI] [PubMed] [Google Scholar]
- 36. Roy-Burman A, et al. 2001. Type III protein secretion is associated with death in lower respiratory and systemic Pseudomonas aeruginosa infections. J. Infect. Dis. 183:1767–1774 [DOI] [PubMed] [Google Scholar]
- 37. Russo FD, Silhavy TJ. 1993. The essential tension: opposed reactions in bacterial two-component regulatory systems. Trends Microbiol. 1:306–310 [DOI] [PubMed] [Google Scholar]
- 38. Schweizer HP. 1991. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene 97:109–121 [DOI] [PubMed] [Google Scholar]
- 39. Van Delden C, Iglewski BH. 1998. Cell-to-cell signaling and Pseudomonas aeruginosa infections. Emerg. Infect. Dis. 4:551–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ventre I, et al. 2006. Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc. Nat. Acad. Sci. U. S. A. 103:171–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wozniak DJ, Ohman DE. 1994. Transcriptional analysis of the Pseudomonas aeruginosa genes algR, algB, and algD reveals a hierarchy of alginate gene expression which is modulated by algT. J. Bacteriol. 176:6007–6014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wyckoff TJ, Wozniak DJ. 2001. Transcriptional analysis of genes involved in Pseudomonas aeruginosa biofilms. Methods Enzymol. 336:144–151 [DOI] [PubMed] [Google Scholar]
- 43. Yeo W-S, Zwir I, Huang HV, Shin D, Kato A, Groisman EA. 2012. Intrinsic negative feedback governs activation surge in two-component regulatory systems. Mol. Cell 45:409–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.