Abstract
Cold shock proteins (CSPs) are nucleic acid binding chaperones, first described as being induced to solve the problem of mRNA stabilization after temperature downshift. Caulobacter crescentus has four CSPs: CspA and CspB, which are cold induced, and CspC and CspD, which are induced only in stationary phase. In this work we have determined that the synthesis of both CspA and CspB reaches the maximum levels early in the acclimation phase. The deletion of cspA causes a decrease in growth at low temperature, whereas the strain with a deletion of cspB has a very subtle and transient cold-related growth phenotype. The cspA cspB double mutant has a slightly more severe phenotype than that of the cspA mutant, suggesting that although CspA may be more important to cold adaptation than CspB, both proteins have a role in this process. Gene expression analyses were carried out using cspA and cspB regulatory fusions to the lacZ reporter gene and showed that both genes are regulated at the transcriptional and posttranscriptional levels. Deletion mapping of the long 5′-untranslated region (5′-UTR) of each gene identified a common region important for cold induction, probably via translation enhancement. In contrast to what was reported for other bacteria, these cold shock genes have no regulatory regions downstream from ATG that are important for cold induction. This work shows that the importance of CspA and CspB to C. crescentus cold adaptation, mechanisms of regulation, and pattern of expression during the acclimation phase apparently differs in many aspects from what has been described so far for other bacteria.
INTRODUCTION
Free-living organisms must be able to cope with a multitude of environmental changes, which include nutrient starvation, osmolality imbalance, and temperature shifts, and responses to these stresses require a fine-tuned expression of specific genes (30). Temperature downshift causes growth and cell proliferation arrest, also altering the pattern of protein expression (29). Low temperature interferes with cell physiology by affecting several molecules in their biochemical and physical properties, resulting in modifications in general metabolism. There are two classes of responses involved in cold adaptation, named low-temperature response (LTR) and cold shock response (CSR) (43). LTR depends on the absolute temperature and persists as long as the stress exists, and the proteins induced are usually involved in cellular metabolism and membrane functionality (43). CSR, on the other hand, is a quick response to temperature downshift, which is not dependent on the absolute temperature but instead is relative to the amplitude of temperature variance before and after the shift. This response is turned off by a negative-feedback control that occurs when cells become adapted to the new situation. Some of the CSR proteins are nucleic acids chaperones, helicases, and nucleoid-associated proteins (38, 39, 46, 48).
According to Horn and colleagues (26), CSR is divided into four stages: optimal growth, cold shock, acclimation phase, and recovery phase. The moment of temperature downshift is called cold shock, after which begins the growth arrest acclimation phase. Proteins involved in rapid cold acclimation, including CSPs, are highly and transiently induced during this phase, while the expression of the other cellular proteins remains unaffected or becomes repressed (29, 38, 39). At the recovery phase, the levels of proteins induced at the acclimation phase (other than those involved in LTR) decline, returning to those before cold shock or to intermediate levels (26, 46).
One of the biggest challenges imposed by low temperature is the increase of the stability of secondary structures in RNA that may prevent mRNA turnover and cause decreased translation efficiency. The CSPs then provide the necessary activity of nucleic acid chaperones and transcription antitermination (3, 28, 39, 40). This family of proteins is widespread throughout bacteria, presenting a highly divergent number of paralogues in each organism. They respond not exclusively to temperature downshift but also to osmotic and oxidative stresses, stationary phase, carbon starvation, and antibiotic resistance, and some may even be constitutively expressed (6, 9, 10, 39, 42, 54). Escherichia coli has nine csp paralogues, of which five are exclusively cold induced, one is constitutively expressed, and one is a replication inhibitor induced at stationary phase and under carbon starvation (26, 54). In other organisms such as Bacillus subtilis and Listeria monocytogenes, which have three copies of those genes, there are paralogues responding to more than one condition (22, 42).
The induction of E. coli major cold shock protein CspA is not a result of an enhancement in transcription but is due to an increased mRNA stability during low-temperature incubation (16). The stabilization of CSP mRNAs is in part due to their 5′ untranslated region (5′-UTR), which is highly structured and protects the mRNA from RNase action, increasing its half-life at low temperature (27). In addition to stabilization, translational enhancement mediated by regions of interaction of mRNA with 16S rRNA called upstream box (UB) and downstream box (DB) was also proposed as being critical for cold shock induction of the csp genes in E. coli (13, 14, 55).
Caulobacter crescentus is a free-living and oligotrophic alphaproteobacterium widespread in aquatic environments and able to grow at temperatures as low as 4°C, and it shows an outstanding resistance to freezing (33). The C. crescentus genome contains four paralogues of csp genes, two of them (cspA and cspB) being cold induced, encoding proteins containing a single cold shock domain (CSD), and the other two (cspC and cspD) being stationary phase induced, encoding proteins with two CSDs (4, 31). Recently, we demonstrated that stationary-phase induction of C. crescentus cspD is regulated by the levels of the second messenger ppGpp and by the SpdR/SpdS two-component system (7). In this work, we have characterized the regulatory mechanisms underlying cspA and cspB cold induction, finding differences in the roles of these genes in growth and cold adaptation and identifying sites in their regulatory regions important for cold induction.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains and plasmids are described in Table 1. C. crescentus strains were grown in rich peptone-yeast extract (PYE) or minimal (M2) medium (11) at 30°C or 10°C with shaking. When necessary, antibiotics were added at the following concentrations: kanamycin (5 μg/ml), tetracycline (1 μg/ml), nalidixic acid (20 μg/ml), or rifampin (20 μg/ml). E. coli strains were grown at 37°C in Luria-Bertani medium. When necessary, antibiotics were added at the following concentrations: kanamycin (50 μg/ml) or tetracycline (12.5 μg/ml). Plasmids were introduced into C. crescentus by conjugation with E. coli strain S17-1 or by direct transformation.
Table 1.
Bacterial strains and plasmids
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| S17-1λpir | E. coli strain for plasmid mobilization | 44 |
| DH5α | E. coli strain for cloning purposes | 25 |
| C. crescentus | ||
| NA1000 | Synchronizable derivative of wild-type CB15 | 15 |
| MM60 | NA1000 strain with pRM6 translational fusion integrated in the genome | This study |
| MM61 | NA1000 strain with pRM7 translational fusion integrated in the genome | This study |
| MM63 | NA1000 strain with pRM8 translational fusion integrated in the genome | This study |
| MM62 | NA1000 strain with pRM9 translational fusion integrated in the genome | This study |
| MM64 | NA1000 strain with pRM10 translational fusion integrated in the genome | This study |
| MM8 | NA1000 ΔcspA | 31 |
| MM30 | NA1000 ΔcspB | This study |
| MM26 | NA1000 ΔcspC | 4 |
| MM9 | NA1000 ΔcspD | 31 |
| MM28 | NA1000 ΔcspA ΔcspB | This study |
| Plasmids | ||
| pGEM-T Easy | Cloning vector; Ampr | Promega |
| pNPTS138 | Suicide vector containing oriT and sacB; Kanr | D. Alley |
| pRKlacZ290 | Vector for translational fusion with lacZ gene; Tetr | 21 |
| pMR20 | Broad-host-range, low-copy-number vector; Tetr | 41 |
| pJBZ281 | pRK2-derived vector with promoterless lacZ gene; Kanr | M. R. K. Alley |
| pEL4 | Fragment containing cspD promoter cloned into pRKlacZ290 | 31 |
| pEL6 | Fragment containing cspC promoter cloned into pRKlacZ290 | 31 |
| pRM1 | Fragment from −380 to +162 of cspA amplified using CSPA-B/CSPA-R4 transcriptionally fused to lacZ into pRKlacZ290 | This study |
| pRM2 | Fragment from −196 to +162 of cspA amplified using CSPA-R5/CSPA-R4 transcriptionally fused to lacZ into pRKlacZ290 | This study |
| pRM3 | Fragment from −380 to −133 upstream from cspA amplified using CSPA-B/CSPA-R8 transcriptionally fused to lacZ into pRKlacZ290 | This study |
| pRM4 | Fragment from −734 to +152 of cspB amplified using CSPB-F/CSPB-R4 transcriptionally fused to lacZ into pRKlacZ290 | This study |
| pRM5 | Fragment from −187 to +27 of cspB amplified using CSPB-R1/CSPB-C transcriptionally fused to lacZ into pRKlacZ290 | This study |
| pRM6 | Fragment from −380 to +162 of cspA amplified using CSPA-B/CSPA-R4 translationally fused in frame to lacZ into pJBZ281 | This study |
| pRM7 | Fragment from −380 to +3 of cspA amplified using CSPA-B/CSPA-R2 translationally fused in frame to lacZ into pJBZ281 | This study |
| pRM8 | Fragment from −734 to +152 of cspB amplified using CSPB-F/CSPB-R4 translationally fused in frame to lacZ into pJBZ281 | This study |
| pRM9 | Fragment from −734 to −93 fused to the fragment −17 to +3 of cspB amplified using CSPB-F/CSPB-R3 translationally fused in frame to lacZ into pJBZ281 | This study |
| pRM10 | Fragment from −356 to +3 of cspB amplified using CSPB-A/CSPB-R2 translationally fused in frame to lacZ into pJBZ281 | This study |
| pRM11 | Fragment amplified with CSPA-A/CSPA-B containing cspA gene cloned into pMR20 | This study |
| pRM12 | Fragment amplified with CSPB-A/CSPB-B containing cspB gene cloned into pRM11 | This study |
| pRM13 | Fragment from −380 to −90 fused to the fragment −15 to +3 of cspA amplified using CSPA-B/CSPA-R3 translationally fused in frame to lacZ into pJBZ281 | This study |
Fragment positions are relative to the ATG.
Mapping of regulatory regions important for cspA and cspB expression.
Transcriptional fusions with the lacZ reporter gene were constructed in plasmid pRKplacZ290, and translational fusions were made using plasmid pJBZ281 (Table 1). The constructions obtained are illustrated below (see Fig. 5 and 6). pRKlacZ290 and pJBZ281 were introduced into C. crescentus by conjugation and electroporation, respectively. Gene expression was measured by determining β-galactosidase activity as described by Miller (34). β-Galactosidase activity assays were performed with mid-log-phase cultures from C. crescentus NA1000 and ΔcspA, ΔcspB, ΔcspC, and ΔcspD strains at 30°C and at different time points during incubation at 10°C (1 h, 2 h, and 5 h). Expression of lacZ from constructions described above was assessed either in PYE or in M2 medium.
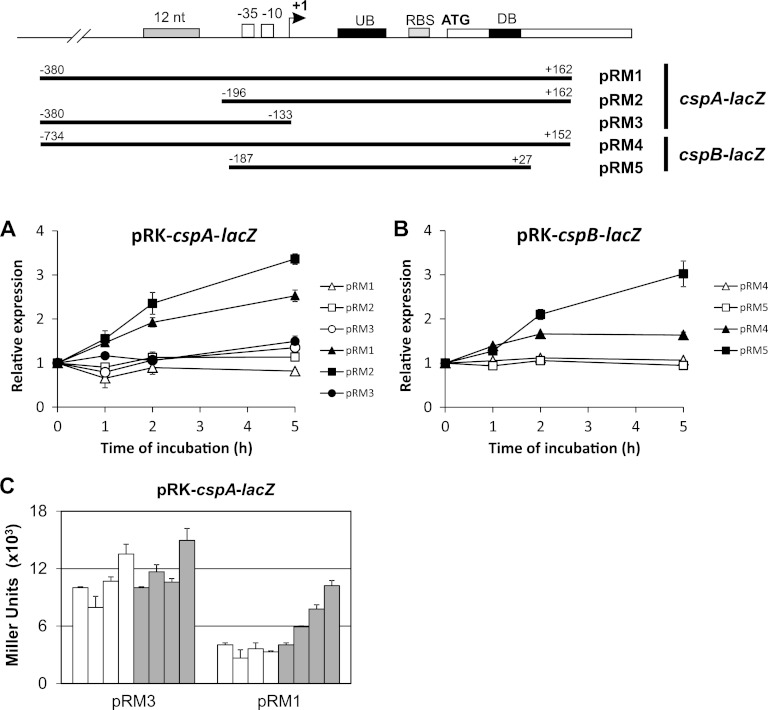
Fig 5.
Role of promoter and 5′-UTR on cold induction of cspA and cspB. Transcriptional fusions of each regulatory region with the lacZ reporter gene were constructed as indicated in the top panel. (A and B) β-Galactosidase assays were performed at 30°C (open symbols) and after 0 h, 1 h, 2 h, and 5 h at 10°C (filled symbols). (A) Relative expressions obtained by lacZ fusions were as follows: cspA promoter including its 5′-UTR region and part of the open reading frame (ORF) (pRM1), same as pRM1 without the 184 nt upstream from the promoter (pRM2); cspA promoter without the 5′-UTR region (pRM3). (B) Relative expression driven by the cspB promoter and the 5′-UTR, containing (pRM4) or not (pRM5) the region upstream of the promoter. The results shown are averages of at least three experiments. Error bars indicate standard deviations. (C) Absolute expression of transcriptional lacZ fusions of the cspA promoter with (pRM1) and without (pRM3) the 5′-UTR. β-Galactosidase assays were performed at 30°C (open bars) and after 0 h, 1 h, 2 h, and 5 h at 10°C (gray bars).
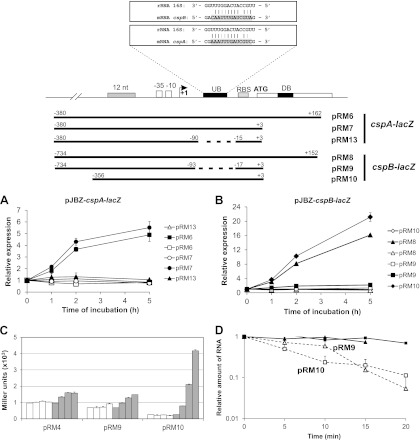
Fig 6.
Role of the upstream box on cold induction of cspA and cspB and mRNA stabilization. Translational fusions of each regulatory region with the lacZ reporter gene in pJBZ281 were constructed as indicated at the top of the figure. (A through C) β-Galactosidase assays were performed at 30°C (open symbols and bars) and prior to (0 h) and 1 h, 2 h, and 5 h after temperature downshift to 10°C (filled symbols and bars). (A) Relative expression driven by the regulatory regions of cspA containing the promoter and the 5′-UTR, with (pRM6) or without (pRM7 and pRM13) the initial portion of the coding region. In construct pRM13, the region corresponding to the UB was deleted. (B) Relative expression driven by the regulatory regions of cspB containing the promoter and the 5′-UTR with (pRM8) or without (pRM9 and pRM10) the initial portion of the coding region. In construct pRM9, the region corresponding to the UB was deleted. The results shown are the averages of at least three experiments. Error bars indicate standard deviations. (C) Absolute expression of a transcriptional lacZ fusion of cspB promoter (pRM4) and translational fusions of cspB (pRM9 and pRM10). (D) mRNA stability of lacZ carried by fusions pRM9 (triangles) and pRM10 (squares) was determined by RT-PCR from total RNAs obtained from cultures incubated at 30°C (open symbols) and after 2 h at 10°C (filled symbols). Aliquots were taken for RNA extraction every 5 min after rifampin addition. The results shown are the averages of four experiments. Lines indicate trends of decay rates for each construct.
Growth tests.
Growth of NA1000 and csp mutant strains was carried out in 10 ml of PYE medium at 30°C or 10°C with agitation. All experiments were initiated with cultures at an optical density at 600 nm (OD600) of 0.2, arising from culture dilution to OD of 0.05. Growth was monitored by measuring OD600 and by determining CFU counts. Relative CFU was obtained by dividing the CFU at each time point after incubation at 10°C by the CFU of the same culture immediately prior to transfer to low temperature.
Construction of csp mutant and complemented strains.
The cspA, cspC, and cspD mutant strains were constructed previously (4, 31). cspB mutant and cspA cspB double mutant strains were obtained by allelic replacement using the NA1000 and cspA mutant strains, respectively, as background. All plasmids used in cloning are listed in Table 1 and primers in Table 2. A 760-bp fragment upstream and an 805-bp fragment downstream from CCNA_00701 (cspB) were amplified by PCR using genomic DNA from NA1000 as the template with primers CSPB-F/CSP1-C and CSP1-D/CSP1-E. Both PCR products were cloned in tandem into the pNPTS138 suicide vector, and the construction was used for deletion of cspB, generating strains MM30 (ΔcspB) and MM28 (ΔcspA ΔcspB). Gene deletions were confirmed by PCR (not shown).
Table 2.
Primers used in this study
| Name | Sequence (5′→ 3′)a | Annealing position |
|---|---|---|
| CSPB-A | ATAGGATCCGATCGCTCACCGCGAGAC | Forward primer, anneals 356 bp upstream from cspB translation start site |
| CSPB-B | GAAAAGCTTCAAGGCCTCACGCTGCGC | Reverse primer, anneals 487 bp downstream from cspB translation start site |
| CSPB-C | AAAAAGCTTGAACCACTTTACGGTGCCG | Reverse primer, anneals 26 bp downstream from cspB translation start site |
| CSPB-D | AAAAAGCTTCCGGGCAGTTGACCGCAGCG | Forward primer, anneals 187 bp downstream from cspB translation start site |
| CSPB-E | AAAGGTACCGAATCTGGCAGGTGTGGCC | Reverse primer, anneals 992 bp downstream from cspB translation start site |
| CSPB-F | GGATCCGCTCATTGCCGATGCTGG | Forward primer, anneals 734 bp upstream from cspB translation start site |
| CSPB-R1 | AGGATCCCCTGAAAATAAAACAATTGTGG | Forward primer, anneals 187 bp upstream from cspB translation start site |
| CSPB-R2 | AAAGCTTCATGTTTGTATCTTTCAGATGTG | Reverse primer, anneals 3 bp downstream from cspB translation start site |
| CSPB-R3 | AAAGCTTCATGTTTGTATCTTTCAGATAAGAGCAAGCGTGGAGAGCGAC | Reverse primer, anneals 93 bp upstream from cspB translation start site containing the region −17 to +3 |
| CSPB-R4 | ATAAAGCTTCTCGTAGTTGAGCTTCTGACC | Reverse primer, anneals 152 bp downstream from cspB translation start site |
| CSPA-A | CGAAACGGCTCGAGCGATG | Reverse primer, anneals 330 bp downstream from cspA translation start site |
| CSPA-B | ATAGGATCCGTCGTTCTCAGAACCATC | Forward primer, anneals 380 bp upstream from cspA translation start site |
| CSPA-R2 | AAAGCTTCATGGGGATGTTCCTTGTAGG | Reverse primer, anneals 3 bp downstream from cspA translation start site |
| CSPA-R3 | AAAGCTTCATGGGGATGTTCCTTGTAGCTCCTGGTCCAGGAGCAAGG | Reverse primer, anneals 90 bp upstream from cspA translation start site containing the region −15 to +3 |
| CSPA-R4 | TTTAAGCTTGACTTCGGGCTCGTACGAGATC | Reverse primer, anneals 162 bp downstream from cspA translation start site |
| CSPA-R5 | AGGATCCACGGTATTTCTCTTGATATCCCC | Forward primer, anneals 196 bp upstream from cspA translation start site |
| CSPA-R8 | TTTAAGCTTATTCGATACAACCATAAACAGG | Reverse primer, anneals 133 bp upstream from cspA translation start site |
| RicLacFow | GTTTTACAACGTCGTGACTGG | Forward primer, anneals 31 bp downstream from lacZ translation start site |
| RicLacRev | GATGGGCGCATCGTAACC | Reverse primer, anneals 300 bp downstream from lacZ translation start site |
Boldface letters indicate restriction enzyme recognition sites, used for cloning purposes.
For the complementation of the mutations, a 720-bp DNA fragment containing the cspA gene was amplified by PCR with CSPA-A/CSPA-B primers and cloned into pMR20 plasmid, generating pRM11. An 842-bp DNA fragment containing the cspB gene was amplified with CSP1-A/CSP1-B and cloned into pRM11 plasmid for complementation of the double mutant, generating pRM12.
Protein pulse-labeling and 2-D electrophoresis.
NA1000 and ΔcspA and ΔcspB strain cultures were grown at 30°C in M2 medium to mid-log phase. Proteins were pulse-labeled for 10 min at 30°C with 20 μCi l-[35S]methionine followed by chasing with 10 mM nonradioactive l-methionine. The remaining culture was shifted to 10°C, and 2-ml aliquots were taken after 5, 20, 50, and 110 min for pulse-labeling for 10 min, as described above. Proteins were extracted by trichloroacetic acid (TCA)-acetone precipitation and solubilized in rehydration buffer (8 M urea, 2% Triton X-100, 0.002% bromophenol blue, 0.28% dithiothreitol [DTT], 2% IPG 3-10 NL buffer [GE Healthcare]), and an equal amount of labeled protein (2 × 106 cpm) was separated by two-dimensional (2-D) gel electrophoresis. In the first dimension, 18-cm gels with a nonlinear pH gradient ranging from pH 3 to 10 (Immobiline DryStrip pH 3-10 NL, 18 cm; GE Healthcare) were rehydrated overnight with each protein sample, and the isoelectric focusing was carried out for 28,000 V · h at a maximum of 3,500 V in the Multiphor II system (GE Healthcare). After the isoelectric focusing, the strips were subjected to 2 steps of incubation, for 15 min each, in SDS equilibration buffer (6 M urea, 75 mM Tris-HCl [pH 8.8], 29.3% glycerol, 2% SDS, 0.002% bromophenol blue) supplemented with 1% DTT in the first incubation and 2.5% iodoacetamide in the following one. In the second dimension, SDS-PAGE was carried out in continuous 13.5% polyacrylamide gels. Gels were incubated for 30 min with Amplify Fluorographic Reagent (GE Healthcare) before drying. The radiolabeled proteins were detected by exposing the gels to Hyperfilm (GE Healthcare) for 72 h at −80°C.
mRNA decay analysis by quantitative reverse transcriptase PCR (qRT-PCR).
In order to determine mRNA levels, 10-ml cultures were harvested from mid-log cultures of C. crescentus NA1000 growing in PYE medium at 30°C and after 2 h at 10°C. RNA was isolated by TRIzol extraction (Invitrogen) according to the manufacturer's instructions and treated with DNase I (Fermentas) to remove contaminating DNA. As a negative control for genomic DNA contamination, PCR was performed in the absence of reverse transcriptase, under the following conditions: 5 min at 95°C, 30 cycles of 5 min at 95°C, 30 s at 55°C, and 30 s at 72°C, followed by an additional incubation of 7 min at 72°C. An aliquot of 5 μg of RNA was used as a template for total cDNA synthesis in 20-μl reaction mixtures with the Superscript First-Strand synthesis system for RT-PCR (Invitrogen). cDNA synthesis conditions were as follows: 10 min at 25°C, 50 min at 50°C for reverse transcription, 5 min at 85°C, followed by 20 min at 37°C for RNase H treatment. PCR was performed using the following primers for cspB and cspA: qRT-CSPBf/qRT-CSPBr and qRT-CSPAf/qRT-CSPAr, respectively (Table 2), at a concentration of 100 nM. For quantitative PCR, an amount of cDNA corresponding to 50 ng of input RNA was used for each reaction. Reactions were performed with the Platinum SYBR green qPCR SuperMix-UDG (Invitrogen) and analyzed in the ABI 7500 real-time PCR system. The measured threshold cycle (CT) for each sample was determined using second-derivative analysis carried out by the instrument software. The measured cspA and cspB transcript CT values were normalized by subtracting the CT for the rho transcript, which was also measured for each sample, as a reference. To determine the mRNA degradation rates under each condition, the zero time point ΔCT was used as a reference to calculate the relative mRNA levels in samples removed at time points after addition of 20 μg/ml rifampin.
In order to determine mRNA levels of lacZ carried by pRM10 and pRM9, RNA was extracted and treated as described above from C. crescentus NA1000 carrying either one or the other construction integrated into its chromosome. PCR was performed with 1 μg of cDNA using primers RicLacFow/RicLacRev at a concentration of 1 μM for amplification of the lacZ coding region. PCRs were carried out as follows: 5 min at 95°C, 15 cycles of 5 min at 95°C, 30 s at 55°C, and 30 s at 72°C, followed by an additional incubation of 7 min at 72°C for templates obtained from cultures incubated at 10°C or by 22 amplification cycles for templates obtained from cultures incubated at 30°C. The PCR products were separated in a 2% agarose Tris-borate-EDTA (TBE) gel, and after ethidium bromide staining the intensity of bands visualized under UV were calculated using ImageJ software (ImageJ 1.44p; Wayne Rasband, National Institutes of Health, Bethesda, MD). To determine the mRNA degradation rates under each condition, the zero time point band density was used as a reference to calculate the relative mRNA levels in samples removed at time points after addition of 20 μg/ml rifampin.
RESULTS
CspA and CspB expression during C. crescentus cold shock.
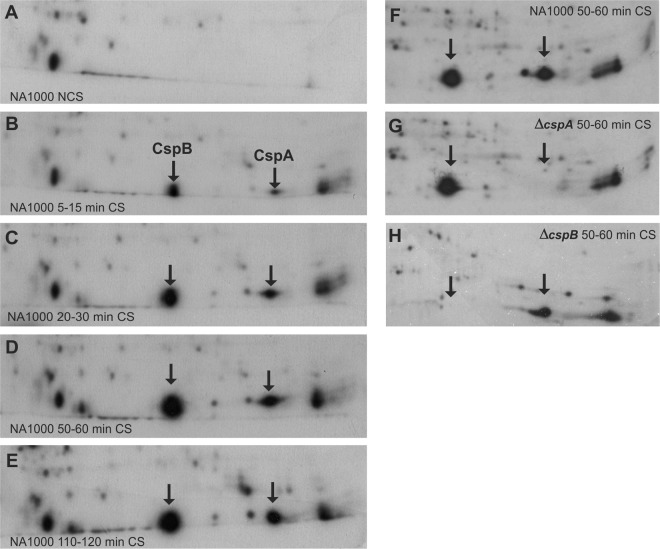
We have previously shown that cspA and cspB expression is cold induced in C. crescentus (31). To characterize the protein expression profile at the acclimation phase, we analyzed CspA and CspB protein synthesis kinetics after cold shock (Fig. 1A to E). In general, overall protein synthesis was repressed, while some proteins had their synthesis increased after cold shock. Among these, two proteins were particularly highly induced under this condition, corresponding to CspA, with predicted mass of 7.3 kDa and pI 6.54, and CspB, with predicted mass of 7.3 kDa and pI 5.64. The identification of the spots corresponding to each protein was possible by comparison to extracts from the null mutant strains (Fig. 1F through H). The induction of these proteins begins early, within the first 15 min of temperature downshift (Fig. 1B), and reaches the highest levels of synthesis around 1 h at 10°C (Fig. 1D). After 2 h at 10°C, the majority of proteins induced during acclimation phase had returned to the initial levels of synthesis, but a small set of proteins, including CspA and CspB, still showed a more intense synthesis than at 30°C (Fig. 1E). We have previously determined that the growth lag period following cold shock is about 4 h (33). These results suggest that the maximum expression of CSPs occurs during the early acclimation phase.
Fig 1.
CspA and CspB cold-induced synthesis. The autoradiograms show the regions of CspA and CspB in 2-D gels containing l-[35S]methionine-labeled proteins from C. crescentus NA1000 cultures incubated at 30°C (A) or at different time points at 10°C (B through E). Proteins were pulse-labeled for 10 min, from 5 to 15 min (B), 20 to 30 min (C), 50 to 60 min (D, F), and 110 to 120 min (E) at 10°C. Protein spots corresponding to CspA and CspB are indicated by arrows and are not present in C. crescentus ΔcspA (G) and ΔcspB (H) null mutant backgrounds, respectively.
CspA and CspB are required for appropriate cold adaptation of C. crescentus.
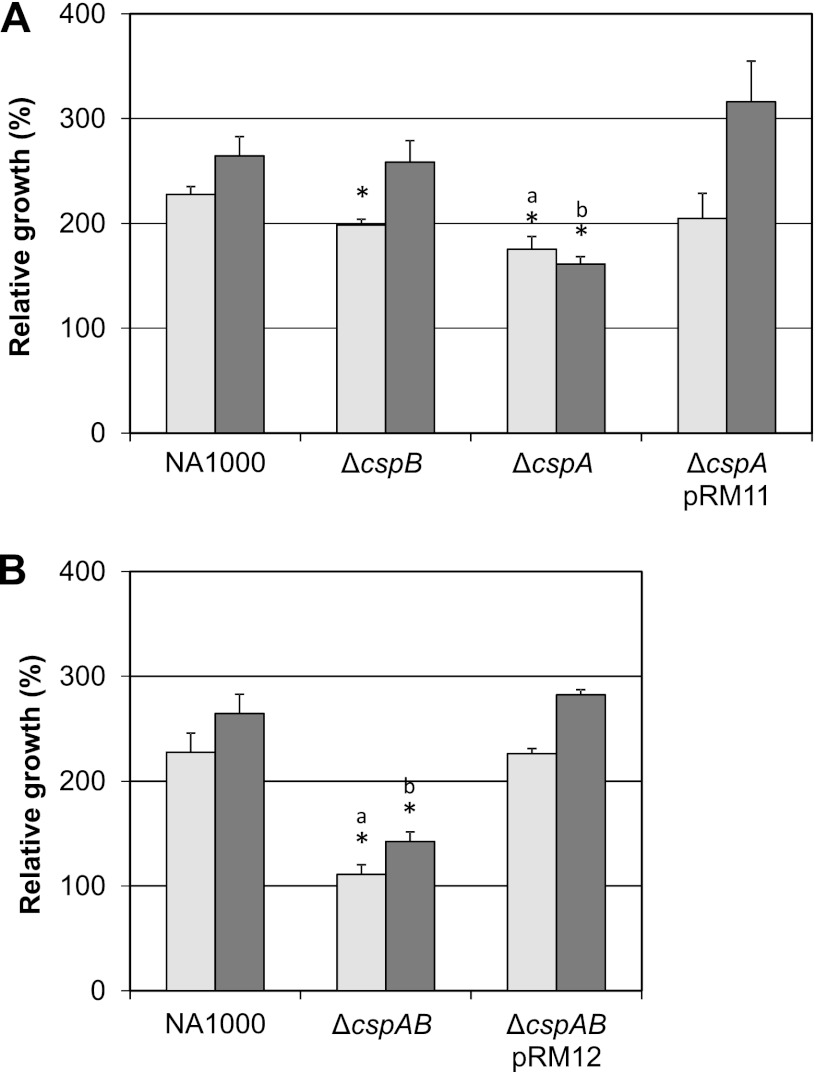
In order to investigate the contribution of each cold-induced C. crescentus csp paralogue for cold adaptation, mutant strains for cspA or cspB and a ΔcspA ΔcspB double mutant strain were constructed, as well as complemented strains. All strains were incubated at 10°C for 48 h, and viability was determined by comparing CFU counts before and after low-temperature incubation.
The ΔcspA strain presented a more pronounced defect in cold adaptation, while the cspB strain showed a slight defect in growth only at the 24-h time point. Although this difference was statistically significant, this phenotype was no longer observed at the 48-h time point, suggesting that the cspB contribution is minor. The cold-related phenotype of the ΔcspA strain was completely reverted when a copy of cspA was inserted in trans (Fig. 2A). The double mutant strain presented a more severe cold-related phenotype, showing no growth after 24 h (Fig. 2B) and very subtle growth after 48 h at 10°C. Despite the fact that the absence of cspB did not cause a persistent cold-related growth phenotype, its absence in the ΔcspA ΔcspB strain showed some additive effect on growth phenotype compared to the ΔcspA strain.
Fig 2.
Relative growth of csp mutant strains at low temperature. (A) Cultures of C. crescentus strains were incubated in PYE medium at 30°C under agitation. When cultures reached mid-log phase (OD, 0.2), samples were taken to determine CFU counts and cultures were transferred to 10°C. After 24 h (light gray bars) and 48 h (dark gray bars) of incubation at 10°C, aliquots of each culture were taken for CFU determination. Relative growth percentage was obtained by dividing the CFU from each incubation time by the respective CFU at 30°C and multiplying by 100. The results shown are the averages of at least three experiments. Error bars indicate standard deviations. (A) Single mutant strains (as indicated) carrying either the empty vector pMR20 or pRM11 (in the case of the ΔcspA strain). (B) Double mutant ΔcspA ΔcspB strain carrying either the empty vector pMR20 or pRM12. Asterisks indicate statistical differences relative to the wild-type strain according to parametric unpaired t test (P < 0.05). Letters a and b indicate statistical comparisons of the respective means of ΔcspA and ΔcspA ΔcspB mutant strains by parametric unpaired t test (P < 0.05).
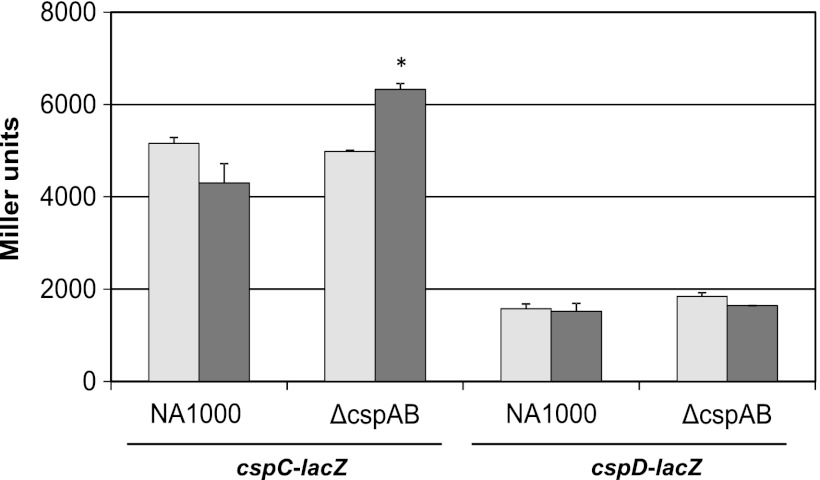
In order to determine if cspC and cspD would be differentially expressed in the ΔcspA ΔcspB strain, as an attempt to compensate for the absence of cspA and cspB during cold shock response, β-galactosidase assays using cspC and cspD transcriptional fusions were performed. There was no alteration on cspD expression in NA1000 or in the ΔcspA ΔcspB strain after 3 h at 10°C in these assays. On the other hand, cspC was partially repressed in NA1000 cultures after temperature downshift, but its transcription was higher in ΔcspA ΔcspB under the same conditions (Fig. 3).
Fig 3.
Expression of cspC and cspD in the cspAB mutant strain at low temperature. Promoters of cspC and cspD cloned into pRKlacZ290 (pEL6 and pEL4, respectively) were introduced into C. crescentus NA1000 and ΔcspA ΔcspB. Gene expression was determined by measuring β-galactosidase activity at 30°C (light gray bars) and after 3 h at 10°C (dark gray bars). The results shown are the averages of at least three experiments. Error bars indicate standard deviations. Asterisks indicate statistical differences relative to the wild-type strain according to parametric unpaired t test (P < 0.05).
Effects of each csp paralogue on cspA and cspB expression.
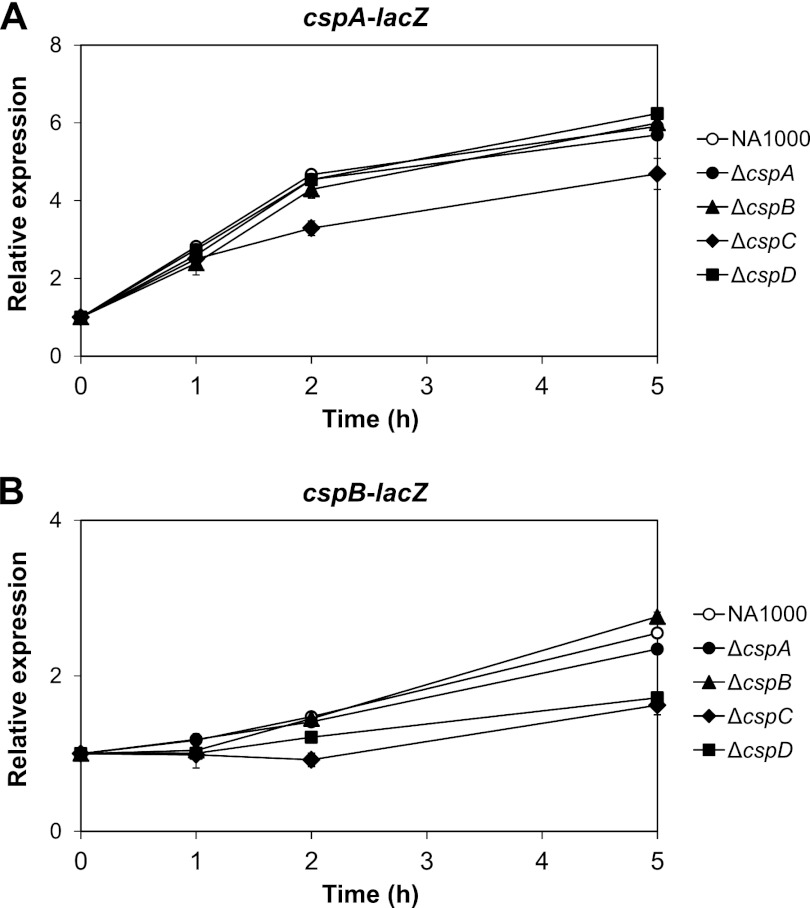
It has been reported that some CSPs may affect their own expression and that of other csp genes (1, 2, 12, 17, 55); therefore, it would be interesting to evaluate the importance of each csp paralogue on the expression of cspA and cspB during cold acclimation. Transcriptional fusions of the promoters and 5′-UTR regions from both cold shock genes to the lacZ reporter gene were introduced into the ΔcspA, ΔcspB, ΔcspC, and ΔcspD mutant strains and assayed prior to and after cold shock (Fig. 4), showing that neither cspA nor cspB is self-regulated and their absence had no effect on each other's expression at least during 5 h of incubation at 10°C. The absence of cspC resulted in a consistent decrease in the expression of cspA and cspB from the late stage of the acclimation phase, reducing expression levels of cspA (by about 15%) and of cspB (by about 40%) after 5 h at 10°C. Interestingly, the absence of cspD compromised only the expression of cspB to levels similar to those generated in the absence of cspC during cold acclimation. These results indicate that CspC and CspD have an important role in cspB cold induction whereas cspA expression is affected only in the absence of CspC.
Fig 4.
Analysis of the contribution of each CSP to cspA and cspB cold induction. The promoter regions of cspA (pRM1) (A) and cspB (pRM4) (B) were cloned into pRKlacZ290 in front of a lacZ reporter gene, and the constructs were introduced into C. crescentus NA1000, ΔcspA, ΔcspB, ΔcspC, and ΔcspD strains. Expression was determined by measuring β-galactosidase activity at 30°C and 1 h, 2 h, and 5 h at 10°C. Relative expression was obtained by dividing Miller units from each time point at 10°C by the units at 30°C. The results shown are the averages of at least three experiments. Error bars indicate standard deviations. Expressions of cspA in the ΔcspC mutant and of cspB in the ΔcspC and ΔcspD mutants after 2 h were statistically different relative to the wild-type strain according to parametric unpaired t test (P < 0.05).
Characterization of cspA and cspB regulatory regions and their role in expression at optimal temperature and after cold shock.
Initially, in order to map the regulatory regions important for cspA and cspB expression, the 250-nucleotide (nt) regions upstream from the start codon of each gene were aligned in an attempt to identify conserved regions in the two sequences that could indicate a common regulatory mechanism. Two regions with noticeable conservation were found: the first is 12 nt long and is located 35 nt upstream of the predicted −35 region of both promoters, and the other is 13 nt long and is located on the 5′-UTR on both mRNAs, approximately 30 nt upstream of the respective ribosome binding site (RBS) (Fig. 5).
The relevance of the 12-nt-long region for the expression of each gene was evaluated at 30°C and 10°C (Fig. 5A and B). At optimal temperature, no changes in expression profile were observed for either gene, but after temperature downshift, expression of cspA (pRM1 in Fig. 5A) increased gradually up to 5 h at 10°C, while cspB expression (pRM4 in Fig. 5B) reached a maximum level 2 h after cold shock. The absence of the putative 12-nt regulatory region upstream of both promoters had no effect on expression at optimal growth temperature but generated higher levels of expression at longer times at low temperature than fusions containing this putative regulatory region (compare pRM1 with pRM2 in Fig. 5A and pRM4 with pRM5 in Fig. 5B). These results indicate that this 12-nt region has a negative role in transcription, and considering its location one may suggest that it affects transcription initiation.
Another transcriptional fusion (pRM3) was constructed to evaluate if the induction seen after temperature downshift is due to transcriptional enhancement or is a result of posttranscriptional mechanisms. This fusion of the cspA promoter differs from the others by the absence of the 5′-UTR, and the results showed that the induction of expression at 10°C is lost (Fig. 5A). However, the absolute expression values in Miller units of this shorter fusion are 5 times higher than those of pRM1 at 30°C, with values similar to pRM1 expression after 5 h following temperature downshift (Fig. 5C). Taken together, these results indicate that cold induction is due to posttranscriptional mechanisms and that cspA expression at 30°C is under 5′-UTR-dependent negative regulation.
The main mechanism described for cold shock induction of genes in E. coli and B. subtilis is the decrease of their mRNA turnover rate (12, 16, 26, 27). Thus, we investigated if C. crescentus cspA and cspB cold induction could be an effect of mRNA stabilization during cold acclimation. We have determined cspA and cspB mRNA half-lives both at 30°C and after temperature downshift. The stability of both mRNAs increased 8-fold at 10°C relative to 30°C (data not shown). We have also determined the half-lives at 30°C and 10°C for five genes not cold induced (phoH, ftsZ, cspD, aceB, and sodA), and the average increase in stability for these genes was 3-fold. These results suggest that the cspA and cspB mRNAs undergo preferential stabilization compared to the other mRNAs in the cell. However, given the narrow spectrum of genes evaluated, it is not possible to confirm whether mRNA stabilization is an attribute specific to csp cold induction or if this is a more widespread effect.
The importance of the initial translated regions for the expression of each gene was evaluated using translational fusions in β-galactosidase activity assays (Fig. 6A and B). Absence of these regions caused no changes in the pattern of expression at 30°C or at 10°C (Fig. 6A and B), indicating that the translated region has no regulatory role, in contrast to the downstream box described for E. coli csp genes (13, 14). The 13-nt region on both 5′-UTRs shows partial complementarity to a portion of 16S mRNA from C. crescentus (from positions 1059 to 1083) and will be referred to here as upstream box (UB) in analogy to the UB described for E. coli (55). When the putative UB was deleted in translational fusions of cspA and cspB (constructs pRM13 and pRM9, respectively, in Fig. 6), the cold induction of both genes was completely lost (Fig. 6A and B). Moreover, the absolute levels in Miller units generated by pRM9 at 30°C were about 3-fold higher than those generated by pRM10 under the same conditions (Fig. 6C), but this was not observed when we compared pRM13 and pRM7 (data not shown). These results suggest that UB is necessary for the induction of cspA and cspB expression at 10°C and that it also interferes negatively with cspB expression at 30°C.
Interestingly, the relative expression of the translational fusion pRM9 shows a pattern similar to that of the transcriptional fusion pRM4 (Fig. 6C), both at 30°C and at 10°C, suggesting that deletion of UB eliminates a translational component of cspB regulation. In order to ascertain that the reduced cold induction of pRM9 was not a result of poor cold-dependent mRNA stabilization, the mRNA decay rates for lacZ from pRM10 and pRM9 were evaluated (Fig. 6D). The absence of a UB region on cspB mRNA slightly increases its stability at initial times after transcription arrest at 30°C but has no effect on cold-dependent stabilization. Therefore, the reduction in the cold induction is due to a translational effect, and the increase in the absolute levels of expression at 30°C is probably a result of a small increase in mRNA stability.
DISCUSSION
Cells at different stages of cold shock response are at different physiological states. Thus, temporal definition of acclimation and recovery/adaptive phases after cold shock is necessary to understand the regulatory mechanisms and importance of CSPs for cold response. The most relevant set of proteins transiently induced immediately after cold shock, during the acclimation phase of CSR in E. coli, can be loosely categorized in two groups: (i) CSPs, RNA helicases, ribonucleases, and ribosomal factors that are expressed in low levels at optimal growth temperature and dramatically induced after temperature downshift; and (ii) RecA, GyrA, IF-2, and H-NS, which are abundant at optimal temperature and show a small induction after cold shock (29, 38, 39, 46, 48, 49).
The characterization of the CSR phases in C. crescentus was achieved by two-dimensional electrophoresis of NA1000 proteins at different time points after shifting cultures from 30°C to 10°C, using the cold-induced CspA and CspB proteins as indicators to define the kinetic pattern of CSR protein expression. The peak of CspA/CspB expression is reached about 1 h after cold shock (this is also true for many other proteins [data not shown]) (Fig. 1). This differs slightly from what is observed in E. coli, where CSPs reach the maximum expression about 2 to 3 h after cold shock, during the late 4-h-long acclimation phase (29). Up- or downregulation of individual proteins in a time-dependent manner may reflect strictly controlled regulatory networks with precisely defined roles, which may suggest that the CSRs from either bacterium may be different, the expression of CSPs being required earlier for Caulobacter. Attesting to the diversity of CSRs, Yersinia enterocolitica restarts growth immediately following the downregulation after cold induction of the major cold shock protein during acclimation phase (36). Caulobacter, on the other hand, remains in lag phase even after the decrease of CSP expression during acclimation phase.
The contribution of CSPs in maintaining cell viability and allowing growth at low temperature was previously analyzed in several bacteria belonging to the Firmicutes (e.g., L. monocytogenes, Lactobacillus plantarum, Staphylococcus aureus, Lactococcus lactis, and B. subtilis) and Gammaproteobacteria (e.g., E. coli) divisions (9, 10, 22, 42, 48, 50, 52). On the other hand, little is known about the importance and regulation of CSPs among Alphaproteobacteria, although many genera in this group may be exposed to temperature extremes due to their free-living lifestyle. The absence of cspA in C. crescentus produces a cold-related phenotype; however, deletion of cspB causes only a subtle and transient decrease in growth after 24 h at 10°C, indicating that CspA is more important for cold adaptation than CspB. However, when both cold-induced CSPs are lacking, there is a manifestation of a more severe phenotype than that observed for the ΔcspA strain, indicating that CspB also plays a role in cold adaptation. Furthermore, the absence of cold-induced CSPs does not lead to a decrease in cell viability but instead causes a period of growth arrest through the initial hours at low temperature, after which growth resumes at a lower rate than for the wild type. In contrast, in E. coli a more severe cold-related phenotype is manifested when four of five cold-induced CSPs are absent, with cultures being unable to form colonies at low temperature (53). In L. monocytogenes and B. subtilis, a growth deficiency at low temperature was observed when a specific cold-induced CSP was absent, but in this case no additive effect on phenotype was seen when more than one cold-induced CSPs were deleted (42, 48, 53).
It was reported for E. coli and B. subtilis that single or combined deletions of cold shock genes cause changes in the expression of remaining CSPs, which usually become more expressed (48, 53). Thus, it may be possible that higher expression of the remaining CSPs in the cspA and cspB mutants could lead to an attenuation of the cold-related phenotype by a compensatory effect. The results showed that cspD is equally expressed in mutant and wild-type strains irrespective of temperature (Fig. 3). cspC expression, on the other hand, was diminished in the wild type during cold acclimation but increased in the ΔcspA ΔcspB mutant, reaching about 40% higher levels than those of the wild-type strain. However, despite being induced, CspC was not sufficient to prevent the cold-related growth phenotype, suggesting that either it does not perform all CspA and CspB functions or the protein concentration was not sufficient to compensate for deprivation of CspA and CspB. Caulobacter CspA and CspB have only one CSD and share 78% identity, while CspC has two CSDs, with the first and second domains sharing 73% and 77% of identity, respectively, with CspA. Despite such high similarity, CspA, CspB, and CspC apparently do not have equivalent roles in the cell.
The fact that only cspC but not cspD was induced in the absence of the other two paralogues suggests that CSPs can affect the expression of the other csp genes in a specific way. We showed that CspC affects positively the expression of both cspA and cspB, but in distinct situations. CspC affects cspA expression mainly at late acclimation phase (after 2 h hours of temperature downshift) and during adaptation phase (after 4 h of temperature downshift). The effect of CspC on cspB expression is apparently more prominent than on cspA, since the β-galactosidase units generated by the cspB-lacZ transcriptional fusion in the ΔcspC mutant are one-half of those shown in the wild-type strain even at 30°C (data not shown). Moreover, cspB cold induction is consistently diminished in the ΔcspC background. CspD, in turn, affects positively solely the expression of cspB to an extent similar to that of CspC but is restricted to low-temperature induction. CspC and/or CspD could be acting by decreasing transcription antitermination or RNA polymerase pausing during cspA and cspB expression. In E. coli, cspA expression is transcriptionally regulated by the RNA polymerase pausing time, which is affected directly by CspE (at 37°C) and CspA (during late acclimation phase) through interactions with a region downstream from the cold box at the initial part of the 5′-UTR (2). Mutational studies of cspA 5′-UTR in E. coli suggested that this region is not important for mRNA stability at 37°C but instead is involved in premature transcription termination (3, 17). The transcriptional fusion of the C. crescentus cspA promoter alone, without the 5′-UTR (pRM3), drives a constitutively high expression even at optimal temperature (Fig. 5C). This result suggests that for C. crescentus cspA the 5′-UTR plays an important role on transcription attenuation at optimal temperature. However, in contrast to what happens in E. coli (1, 5), cspA and cspB are not negatively regulated by their own products, nor are they regulated by each other (Fig. 4). This also confirms that the biochemical roles of each CSP in cell physiology are quite different.
cspA and cspB share a 12-nt sequence region located 30 nt upstream of the −35 element of the previously identified promoters (31). While this region is not necessary for the cold induction of either gene (Fig. 5A), it appears to be important for negative regulation of both genes at the recovery phase, possibly acting as an important element for turning off the cold shock response. This would be different from what has been reported for other bacteria, where this is done by increase of RNA polymerase pausing or mRNA destabilization. However, due to its location and distance to the promoter elements, this sequence probably does not constitute an operator site. Fis and H-NS regulate cspA of E. coli in antagonistic ways, being important only at 37°C but not after cold shock (1, 5). H-NS from E. coli binds to AT-rich sequences, including the cspA promoter region and consequently inhibiting cspA transcription, thus acting as a repressor. Although no orthologue of H-NS was found on C. crescentus CB15 or NA1000 genomes (32, 37), it is possible that another nucleoid-associated protein (e.g., HU) could be responsible for this repression.
Yamanaka and colleagues (55) performed a mutational analysis of the 5′-UTR of E. coli cspA mRNA and showed that specific regions affect mRNA stability, while others were responsible for posttranscriptional and potentially translational regulation. Moreover, it has been described for E. coli that preferential translation of the csp genes occurs at low temperature (18, 19, 20, 24). The 5′-UTRs of cspA and cspB mRNAs from C. crescentus share a region of sequence similarity of 13 nt (called upstream box), whose relevance for regulation was evaluated in expression assays using translational fusions. According to the results from the expression assays (compare Fig. 5 and 6), the cold induction of cspA and cspB is largely due to translation regulation, this feature being more relevant to cspB expression. The translational component of cold induction was due to the putative upstream box, as determined by translational fusions of both genes lacking that region (pRM9 and pRM13 in Fig. 6).
That region, as well as the upstream box (UB) from E. coli, which is present in mRNAs from the majority of cold-induced CSPs in this organism (CspA, CspB, CspG, and CspI) (35), shows partial complementarity to RNA from the 16S ribosomal subunit (Fig. 6), suggesting that it could play a role as a translational enhancer. In contrast to the UB of cold-induced csp genes in E. coli, which are located 4 to 11 nucleotides upstream from the RBS (27, 55), the UB of C. crescentus cspA and cspB 5′-UTRs are located 34 nt and 32 nt upstream from the RBS, respectively. According to the three-dimensional structure prediction for 16S RNA (51), a putative anti-UB sequence from C. crescentus (positions 1059 to 1083) and one from E. coli (positions 1023 to 1035) are predicted to be both localized in a similar region of the 3′ major domain (not shown). However, as the anti-UB sequence is located in a short hairpin with low base pairing, we cannot discard the possibility of the UB region being involved in the conformational changes of mRNA after cold shock without ribosome interaction. It was described for E. coli cspA that the mRNA changes its conformation after cold shock, freeing the RBS region that was unavailable for ribosome recognition before temperature downshift (20). In a similar way, the UB region in C. crescentus csp mRNAs could engage in base pairing within the 5′-UTR, leaving the RBS free for ribosome recognition only after cold shock, and thus modulate the translation of CspA and CspB. Sequences similar to this putative UB were used for an in silico screen of the C. crescentus genome using RSAT (regulatory sequence analysis tools) (47) in order to find genes that could share the same type of regulation. The results indicated that there are other genes that contain this 13-nt sequence up to 100 nt upstream of the annotated start codon, suggesting that this mechanism of regulation is not restricted to cspA and cspB (data not shown). Additionally, in E. coli a second region called downstream box was also proposed to act as a translational enhancer in the same way as UB (13, 14). However, in cspA and cspB from C. crescentus, no equivalent DB region was identified.
The removal of the UB region of cspB was evaluated as to whether it could affect mRNA decay (Fig. 6D). Constructs pRM9 and pRM10 present very similar mRNA decay rates at 10°C, indicating that the cold-dependent stabilization was not affected. Nevertheless, pRM9 was more stable at 30°C than pRM10 during the first minutes after rifampin addition, which agrees with β-galactosidase assays, in which pRM9 showed 3 times more Miller units than pRM10 at 30°C (Fig. 6C). According to mRNA secondary structure predictions (data not shown), the UB is part of a hairpin in cspB mRNA formed at 30°C, which is not predicted to exist at 10°C. This hairpin could be a target for degradation by ribonucleases that recognize double-stranded RNA (dsRNA), such as RNase III. The main endonuclease described as responsible for E. coli cspA mRNA degradation is RNase E (16), which apparently cleaves single-stranded RNA (ssRNA) more efficiently than dsRNA, but RNase III is another key endonuclease to bacterial mRNA decay (23, 45). In E. coli, the overexpression of the S1 ribosomal protein, which is believed to bind preferentially at single-stranded regions in the cspE 5′-UTR, inhibits RNase E-dependent decay of cspE mRNA (8). This decay apparently occurs by an alternative pathway, strengthening the hypothesis of the involvement of other RNases in csp mRNA turnover (8). Therefore, it is possible that RNase III would be responsible for quick mRNA decay during the first minutes after its synthesis, which would render the pRM9 mRNA more stable than pRM10 mRNA. The difference between half-lives of mRNAs from pRM9 and pRM10 fusions is more apparent at the beginning of the assay and is lost after longer times of incubation with rifampin. That could be explained by the progressive decrease of the amount of total RNA in the cell and the consequent increase in the availability of free RNAses for an effective degradation of highly structured RNAs without RNase III participation.
Surprisingly, as a psychrotolerant free-living bacterium, and hence subjected more easily to temperature changes, C. crescentus has less than one-half the number of cold-induced CSPs than E. coli, and that is enough to allow it to undergo this lifestyle. Despite the fact that regulation of CspA and CspB shares some similarities with cold induction of CSPs from E. coli, their importance to C. crescentus cold adaptation, mechanisms of regulation, and pattern of expression during the acclimation phase apparently differs in many aspects from what has been described so far for other bacteria. The biochemical and physiological roles of these proteins in Caulobacter CSR are currently being investigated.
ACKNOWLEDGMENTS
We thank Regina L. Baldini for critical reading of the manuscript and Gianlucca G. Nicastro for valuable help with 2-D assays.
This work was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). During the course of this work, R.R.M and C.A.P.T.S. were supported by fellowships from FAPESP. M.V.M. was partially supported by CNPq.
Footnotes
Published ahead of print 21 September 2012
REFERENCES
- 1. Bae W, Jones PG, Inouye M. 1997. 1997. CspA, the major cold shock protein of Escherichia coli, negatively regulates its own gene expression. J. Bacteriol. 179:7081–7088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bae W, Phadtare S, Severinov K, Inouye M. 1999. Characterization of Escherichia coli cspE, whose product negatively regulates transcription of cspA, the gene for the major cold shock protein. Mol. Microbiol. 31:1429–1441 [DOI] [PubMed] [Google Scholar]
- 3. Bae W, Xia B, Inouye M, Severinov K. 2000. Escherichia coli CspA-family RNA chaperones are transcription antiterminators. Proc. Natl. Acad. Sci. U. S. A. 97:7784–7789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Balhesteros H, Mazzon RR, da Silva CA, Lang EA, Marques M. 2010. CspC and CspD are essential for Caulobacter crescentus stationary phase survival. Arch. Microbiol. 192:747–758 [DOI] [PubMed] [Google Scholar]
- 5. Brandi A, Spurio R, Gualerzi CO, Pon CL. 1999. Massive presence of the Escherichia coli major cold-shock protein CspA under non-stress conditions. EMBO J. 18:1653–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chanda PK, Mondal R, Sau K, Sau S. 2009. Antibiotics, arsenate and H2O2 induce the promoter of Staphylococcus aureus cspC gene more strongly than cold. J. Basic Microbiol. 49:205–211 [DOI] [PubMed] [Google Scholar]
- 7. da Silva CA, Balhesteros H, Mazzon RR, Marques M. 2010. SpdR, a response regulator required for stationary-phase induction of Caulobacter crescentus cspD. J. Bacteriol. 192:5991–6000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Delvillani F, Papiani G, Dehò G, Briani F. 2011. S1 ribosomal protein and the interplay between translation and mRNA decay. Nucleic Acids Res. 39:7702–7715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Derzelle S, Hallet B, Ferain T, Delcour J, Hols P. 2003. Improved adaptation to cold-shock, stationary-phase, and freezing stresses in Lactobacillus plantarum overproducing cold-shock proteins. Appl. Environ. Microbiol. 69:4285–4290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Duval BD, Mathew A, Satola SW, Shafer WM. 2010. Altered growth, pigmentation, and antimicrobial susceptibility properties of Staphylococcus aureus due to loss of the major cold shock gene cspB. Antimicrob. Agents Chemother. 54:2283–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ely B. 1991. Genetics of Caulobacter crescentus. Methods Enzymol. 204:372–384 [DOI] [PubMed] [Google Scholar]
- 12. Ermolenko DN, Makhatadze GI. 2002. Bacterial cold-shock proteins. Cell. Mol. Life Sci. 59:1902–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Etchegaray JP, Inouye M. 1999. A sequence downstream of the initiation codon is essential for cold shock induction of cspB of Escherichia coli. J. Bacteriol. 181:5852–5854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Etchegaray JP, Inouye M. 1999. Translational enhancement by an element downstream of the initiation codon in Escherichia coli. J. Biol. Chem. 274:10079–10085 [DOI] [PubMed] [Google Scholar]
- 15. Evinger M, Agabian N. 1977. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J. Bacteriol. 132:294–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fang L, Jiang W, Bae W, Inouye M. 1997. Promoter-independent cold-shock induction of cspA and its derepression at 37 degrees C by mRNA stabilization. Mol. Microbiol. 23:355–364 [DOI] [PubMed] [Google Scholar]
- 17. Fang L, Xia B, Inouye M. 1999. Transcription of cspA, the gene for the major cold-shock protein of Escherichia coli, is negatively regulated at 37 degrees C by the 5′-untranslated region of its mRNA. FEMS Microbiol. Lett. 176:39–43 [DOI] [PubMed] [Google Scholar]
- 18. Giuliodori AM, Brandi A, Giangrossi M, Gualerzi CO, Pon CL. 2007. Cold-stress-induced de novo expression of infC and role of IF3 in cold-shock translational bias. RNA 13:1355–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Giuliodori AM, Brandi A, Gualerzi CO, Pon CL. 2004. Preferential translation of cold-shock mRNAs during cold adaptation. RNA 10:265–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Giuliodori AM, et al. 2010. The cspA mRNA is a thermosensor that modulates translation of the cold-shock protein CspA. Mol. Cell 37:21–33 [DOI] [PubMed] [Google Scholar]
- 21. Gober JW, Shapiro L. 1992. A developmentally regulated Caulobacter flagellar promoter is activated by 3′ enhancer and IHF biding elements. Mol. Biol. Cell 3:913–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Graumann PL, Marahiel MA. 1996. Cold shock stress-induced proteins in Bacillus subtilis. J. Bacteriol. 178:4611–4619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Grunberg-Manago M. 1999. Messenger RNA stability and its role in control of gene expression in bacteria and phages. Annu. Rev. Genet. 33:193–227 [DOI] [PubMed] [Google Scholar]
- 24. Gualerzi CO, Giuliodori AM, Pon CL. 2003. Transcriptional and post-transcriptional control of cold-shock genes. J. Mol. Biol. 331:527–539 [DOI] [PubMed] [Google Scholar]
- 25. Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557–580 [DOI] [PubMed] [Google Scholar]
- 26. Horn G, Hofweber R, Kremer W, Kalbitzer HR. 2007. Structure and function of bacterial cold-shock proteins. Cell. Mol. Life Sci. 64:1457–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jiang W, Fang L, Inouye M. 1996. The role of the 5′-end untranslated region of the mRNA for CspA, the major cold-shock protein of Escherichia coli, in cold-shock adaptation. J. Bacteriol. 178:4919–4925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jiang W, Hou Y, Inouye M. 1997. CspA, the major cold-shock protein of Escherichia coli, is a RNA chaperone. J. Biol. Chem. 272:196–202 [DOI] [PubMed] [Google Scholar]
- 29. Jones PG, Vanbogelen RA, Neidhardt FC. 1987. Induction of proteins in response to low temperature in Escherichia coli. J. Bacteriol. 169:2092–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Klinkert B, Narberhaus F. 2009. Microbial thermosensors. Cell. Mol. Life Sci. 66:2661–2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lang EAS, Marques M. 2004. Identification and transcriptional control of Caulobacter crescentus genes encoding proteins containing a cold shock domain. J. Bacteriol. 186:5603–5613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Marks ME, et al. 2010. The genetic basis of laboratory adaptation in Caulobacter crescentus. J. Bacteriol. 192:3678–3688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mazzon RR, Lang EAS, Braz S, Marques M. 2008. Characterization of Caulobacter crescentus response to low temperature and identification of genes involved in freezing resistance. FEMS Microbiol. Lett. 288:178–185 [DOI] [PubMed] [Google Scholar]
- 34. Miller JH. 1972. Experiments in molecular genetics. A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 35. Mitta M, Fang L, Inouye M. 1997. Deletion analysis of cspA of Escherichia coli: requirement of the AT-rich Up element for cspA transcription and the downstream box in the coding region for its shock induction. Mol. Microbiol. 26:321–335 [DOI] [PubMed] [Google Scholar]
- 36. Neuhaus K, Rapposch S, Francis KP, Scherer S. 2000. Restart of exponential growth of cold-shocked Yersinia enterocolitica occurs after down-regulation of cspA1/A2 mRNA. J. Bacteriol. 182:3285–3288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nierman WC, et al. 2001. Complete genome sequence of Caulobacter crescentus. Proc. Natl. Acad. Sci. U. S. A. 98:4136–4141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Phadtare S. 2004. Recent developments in bacterial cold-shock response. Curr. Issues Mol. Biol. 6:125–136 [PubMed] [Google Scholar]
- 39. Phadtare S, Alsina J, Inouye M. 1999. Cold-shock response and cold-shock proteins. Curr. Opin. Microbiol. 2:175–180 [DOI] [PubMed] [Google Scholar]
- 40. Phadtare S, Severinov K. 2005. Nucleic acid melting by Escherichia coli CspE. Nucleic Acids Res. 33:5583–5590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Roberts RC, et al. 1996. Identification of a Caulobacter crescentus operon encoding hrcA, involved in negatively regulating heat-inducible transcription, and chaperone gene grpE. J. Bacteriol. 178:1829–1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schmid B, et al. 2009. Role of cold shock proteins in growth of Listeria monocytogenes under cold and osmotic stress conditions. Appl. Environ. Microbiol. 75:1621–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schumann W. 2009. Temperature sensors of eubacteria. Adv. Appl. Microbiol. 67:213–256 [DOI] [PubMed] [Google Scholar]
- 44. Simon R, Priefer U, Puhler A. 1983. A broad host range immobilization system for in vivo engineering: transposon mutagenesis in Gram negative bacteria. Biotechnology 1:784–790 [Google Scholar]
- 45. Takata R, Izuhara M, Akiyama K. 1992. Processing in the 5′ region of the pnp transcript facilitates the site-specific endonucleolytic cleavages of mRNA. Nucleic Acids Res. 20:847–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Thieringer HA, Jones PG, Inouye M. 1998. Cold shock and adaptation. Bioessays 20:49–53 [DOI] [PubMed] [Google Scholar]
- 47. van Helden J. 2003. Regulatory sequence analysis tools. Nucleic Acids Res. 31:3593–3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Weber MH, Marahiel MA. 2003. Bacterial cold shock responses. Sci. Prog. 86:9–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. White-Ziegler CA, Davis TR. 2009. Genome-wide identification of H-NS-controlled, temperature-regulated genes in Escherichia coli K-12. J. Bacteriol. 191:1106–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Willimsky G, Bang H, Fischer G, Marahiel MA. 1992. Characterization of cspB, a Bacillus subtilis inducible cold shock gene affecting cell viability at low temperatures. J. Bacteriol. 174:6326–6335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wimberly BT, et al. 2000. Structure of the 30S ribosomal subunit. Nature 407:327–339 [DOI] [PubMed] [Google Scholar]
- 52. Wouters JA, Frenkiel H, De Vos WM, Kuipers OP, Abee T. 2001. Cold shock proteins of Lactococcus lactis MG1363 are involved in cryoprotection and in the production of cold-induced proteins. Appl. Environ. Microbiol. 67:5171–5178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Xia B, Ke H, Inouye M. 2001. Acquirement of cold sensitivity by quadruple deletion of the cspA family and its suppression by PNPase S1 domain in Escherichia coli. Mol. Microbiol. 40:179–188 [DOI] [PubMed] [Google Scholar]
- 54. Yamanaka K, Inouye M. 1997. Growth-phase-dependent expression of cspD, encoding a member of the CspA family in Escherichia coli. J. Bacteriol. 179:5126–5130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yamanaka K, Mitta M, Inouye M. 1999. Mutation analysis of the 5′ untranslated region of the cold shock cspA mRNA of Escherichia coli. J. Bacteriol. 181:6284–6291 [DOI] [PMC free article] [PubMed] [Google Scholar]