Abstract
Zinc is a bivalent cation essential for bacterial growth and metabolism. The human pathogen Neisseria meningitidis expresses a homologue of the Zinc uptake regulator Zur, which has been postulated to repress the putative zinc uptake protein ZnuD. In this study, we elucidated the transcriptome of meningococci in response to zinc by microarrays and quantitative real-time PCR (qRT-PCR). We identified 15 genes that were repressed and two genes that were activated upon zinc addition. All transcription units (genes and operons) harbored a putative Zur binding motif in their promoter regions. A meningococcal Zur binding consensus motif (Zur box) was deduced in silico, which harbors a conserved central palindrome consisting of hexameric inverted repeats separated by three nucleotides (TGTTATDNHATAACA). In vitro binding of recombinant meningococcal Zur to this Zur box was shown for the first time using electrophoretic mobility shift assays. Zur binding to DNA depended specifically on the presence of zinc and was sensitive to mutations in the palindromic sequence. The Zur regulon among genes of unknown function comprised genes involved in zinc uptake, tRNA modification, and ribosomal assembly. In summary, this is the first study of the transcriptional response to zinc in meningococci.
INTRODUCTION
Trace metals, such as zinc, are essential cofactors of many enzymes and DNA-binding proteins (40). On the one hand, bacteria have to cope with decreased zinc availability during infection in the host (25), while on the other, at high concentrations, zinc can lead to the production of toxic reactive oxygen species (49). Hence, bacteria tightly control metal homeostasis by metalloregulatory proteins, which are specialized metal-sensing transcriptional regulators that change their conformation upon binding of the metal ions (5). Zinc uptake regulators (Zur) belong to the Fur protein family of transcriptional regulators that also includes Fur, Mur, and Nur, which are sensors of iron, manganese, and nickel, respectively (28). Zinc uptake systems and their regulation by Zur have been characterized for several bacteria such as Escherichia coli (45), Bacillus subtilis (13), Listeria monocytogenes (7), and Staphylococcus aureus (31). Furthermore, Zur regulons have been described for B. subtilis (14), Mycobacterium tuberculosis (32), Yersinia pestis (29), and Corynebacterium glutamicum (54). Panina et al. identified the promoter binding motifs for Zur for Gammaproteobacteria, the Agrobacteria group, the Rhodococcus group, and Gram-positive bacteria as well as for the streptococcal zinc repressor AdcR using comparative computational analysis (41).
In this study, gene regulation by zinc exposure was analyzed in the commensal Gram-negative human pathogen Neisseria meningitidis, causing septicemia and meningitis (50). Several genomes of N. meningitidis have been sequenced (4, 43, 60), which allowed for the bioinformatic prediction of a Zur binding motif for Betaproteobacteria in the RegPrecise database (39). Until now, there is experimental evidence for regulation by meningococcal Zur only for the TonB-dependent outer membrane receptor, ZnuD, which is involved in zinc acquisition upon zinc limitation (59). Stork et al. showed that expression of znuC and znuD is enhanced in a zur knockout mutant and predicted Zur binding motifs upstream of both genes based on the E. coli Zur binding sequence (59). Kumar et al. subsequently demonstrated that N. meningitidis znuD is also regulated by Fur and allows for heme capture on the cell surface when expressed in E. coli (26). Yet, even for znuD, binding of Zur to the promoter of Zur-regulated genes has not been demonstrated by in vitro experiments.
Here, we characterized the Zur regulon of N. meningitidis. The transcriptomes of strain MC58 grown under high- and low-zinc conditions were compared. A total of 15 Zur-repressed and two Zur-activated genes were found. We established a meningococcal Zur binding motif (Zur box) based on promoter sequences of these genes. The direct binding of Zur to proposed Zur boxes was verified by electrophoretic mobility shift assays (EMSAs) and shown to be zinc dependent. Our results provide the basis for further studies characterizing the molecular mechanisms of zinc adaptation in meningococci.
MATERIALS AND METHODS
Strains and mutants.
Serogroup B N. meningitidis strain MC58 (36) was kindly provided by Richard Moxon (Oxford). The zur gene (nmb1266) of MC58 was replaced with a kanamycin resistance cassette as follows: a 470-bp DNA fragment upstream of zur was amplified by PCR from strain MC58 with primers MP16 and MP17, introducing restriction sites of XbaI and EcoRI, respectively. Likewise, primers MP18 and MP19, introducing restriction sites of EcoRI and XhoI, respectively, were used to amplify 475 bp downstream of zur. Primer sequences are provided in Table 1. The upstream and downstream fragments were digested with XbaI/EcoRI and EcoRI/XhoI, respectively. A kanamycin resistance cassette was obtained from vector pUC4K (GE Healthcare) by EcoRI digestion. The flanking upstream and downstream fragments and the kanamycin cassette were cloned into the XbaI/XhoI-digested expression vector pBluescript II SK (Stratagene). The resulting plasmid pMP5 was transformed into MC58. Kanamycin-resistant clones were screened by PCR for replacement of zur by the kanamycin cassette, and zur deletion was confirmed by Southern blotting and sequencing. The resulting zur knockout strain was designated WUE4812.
Table 1.
Oligonucleotides used in this study for PCR, qRT-PCR, and EMSAa
| Assay and target gene | Forward | Sequence (5′–3′) | Reverse | Sequence (5′–3′) | Amplicon length (bp) |
|---|---|---|---|---|---|
| PCR | |||||
| zur up (nmb1267) | MP16 | GCGCGCTCTAGAGATGGCGGAATACATTTTG | MP17 | GCGCGCGAATTCTTCTCTGTTTATGCCGTCTG | 470 |
| zur down (nmb1264-1265) | MP18 | GCGCGCGAATTCATAAGCCTTTCGAAAGGAGC | MP19 | GCGCGCCTCGAGCGTTTTCACCGATAAGGAAC | 475 |
| zur(nmb1266) | MP183 | GCGCGCTCATGAAAACAAATTTCAAACAGAAAATTA | MP184 | GCGCGCAGATCTCTGCTGACATTTTTTACAAATC | 469 |
| qRT-PCR | |||||
| nmb0317 | MP141 | GCAAATCCCTGAAACTCTACCTCTT | MP142 | TGATGTTGACGCAGTCTTCATG | 72 |
| nmb0525 | MP143 | CGGCGTTCCGAATACCTTT | MP144 | GCGTAAATCGCGGCATAGAG | 66 |
| nmb0546 | MP165 | CTGCCGCCGGAATCG | MP166 | GACCAAAGAGCCGACCACTTC | 75 |
| nmb0577 | MP179 | CGGGCGCGGATAACG | MP180 | AGGCAGACGCTCCGCATA | 61 |
| nmb0586 | MP145 | GCAACTACCAAATGCAGCTCAA | MP146 | CAGCAGGGACGGCATTAAAT | 69 |
| nmb0587 | MP147 | GGCTTTGGCACGTCCTCTT | MP148 | GTGTGCCGAGGGCTTGAA | 72 |
| nmb0588 | MP149 | TCGCCCGCGAAAAAATT | MP150 | CTTGGGCGAGGTAGGGTTCT | 66 |
| nmb0817 | MP151 | AGGATGTCCTGAATGTACTCGAAAT | MP152 | CTGTCTCTGTGTCCTCCAACCTAA | 80 |
| nmb0819 | MP153 | CCCTCCTCATCCTCGACACA | MP154 | CAGCACCTGCGGCAGTAA | 69 |
| nmb0820 | MP155 | CCGCCAAAGCCCTAAACA | MP156 | GGTTGTCGGGATTTGAACGT | 72 |
| nmb0942 | MP157 | CCGCTGTTTTCGCTGGATA | MP158 | GTGTTGACGTTGCGCTGTTT | 71 |
| nmb0964 | MP117 | CCGTTCCCCGGTTTTGA | MP118 | CTGCATCGCCTGCTTTTTC | 80 |
| nmb0990 | MP171 | TGCCATTATCGCGCTTGTC | MP172 | GCCTGCTGCTTCGCAAAT | 81 |
| nmb1475 | MP159 | TCGGACAAAACTTGGAAATCG | MP160 | TTTGAAGCGTTCGCCTACGT | 68 |
| nmb1497 | MP161 | TGCCCAACATCCAAGAAATGT | MP162 | CTGGTTTTAAGGCGGTGTGAA | 68 |
| nmb2142 | MP163 | ACGACGGCGGTCATCTTTAC | MP164 | CGCCGTATGATGCACCATT | 68 |
| EMSA | |||||
| nmb0546 | MP191 | CTTTCCAAGATGTTATAATATAACATATAATCTAT | MP192 | AAATATAGATTATATGTTATATTATAACATCTTGG | 35 |
| nmb0964 | |||||
| Unaltered | MP185 | TAAAAAATGTAATGTTATATAATAACAAACTTTT | MP186 | TTTCAAAAGTTTGTTATTATATAACATTACATTT | 34 |
| TA/CA | MP193 | TAAAAAATGTAATGTTATATAACGATGAACTTTT | MP194 | TTTCAAAAGTTCATCGTTATATAACATTACATTT | 34 |
| CA | MP195 | TAAAAAATGTAATGTTATATAATAATGAACTTTT | MP196 | TTTCAAAAGTTCATTATTATATAACATTACATTT | 34 |
| TA | MP197 | TAAAAAATGTAATGTTATATAACGACAAACTTTT | MP198 | TTTCAAAAGTTTGTCGTTATATAACATTACATTT | 34 |
| C | MP199 | TAAAAAATGTAATGTTATATAATAATAAACTTTT | MP200 | TTTCAAAAGTTTATTATTATATAACATTACATTT | 34 |
| nmb1475 | |||||
| Short | MP187 | CGATACCTATTTTGTTATAACATAACAAAATCTT | MP188 | GTTAAAGATTTTGTTATGTTATAACAAAATAGGT | 34 |
| Long | MP189 | TCTTCACACGATACCTATTTTGTTATAACATAACAAAATCTTTAACCCACA | MP190 | CTCGTGTGGGTTAAAGATTTTGTTATGTTATAACAAAATAGGTATCGTGTG | 51 |
Underlined bases indicate restriction sites.
Bacterial growth and RNA isolation.
Strains MC58 and MC58 Δzur were grown overnight at 37°C and 5% CO2 on GC agar plates (BD). Colonies of each overnight culture were resuspended in 20 ml of RPMI medium (Invitrogen) to an optical density at 600 nm (OD600) of 0.2 (2 × 108 CFU/ml). The medium was supplemented with 100 μM FeCl3 to improve bacterial growth. The cultures were incubated at 37°C with shaking at 200 rpm and grown to mid-log phase (OD600, 1.0). Both liquid cultures were then diluted to an OD600 of 0.2 with RPMI medium plus 100 μM FeCl3. The MC58 culture was consecutively divided into two parts of 20 ml each. To one culture vial, 0.5 μM ZnSO4 was added to establish a high-zinc condition according to Stork et al. (59). The other culture and the MC58 Δzur culture were further grown without zinc supplementation. All three 20-ml cultures were grown at 37°C and 200 rpm for 2 h, resulting in an OD600 of about 1.0. Bacterial cultures were then mixed with twice the volume of RNAprotect Bacteria Reagent (Qiagen) to minimize RNA degradation. Total RNA was isolated using the RNeasy Mini Kit (Qiagen). Remaining DNA was digested with RNase-free recombinant DNase I (Invitrogen), and RNA samples were stored at −80°C. RNA integrity was verified with the Agilent 2100 Bioanalyzer (Agilent Technologies).
According to Invitrogen's data sheet, the RPMI medium used here does not contain any source of zinc. However, we assumed that this RPMI contains a minimal concentration of zinc because RPMI distributed by Sigma was shown to comprise at least ∼1.69 μM zinc (59). We therefore refer to “low” (RPMI) and “high” (RPMI+ZnSO4) zinc conditions in this study.
Furthermore, we decided against depletion of zinc, e.g., by an ion chelator such as N,N,N′,N′-tetrakis-(2-pyridylmethyl)-ethylenediamine (TPEN), prior to zinc supplementation as conducted in studies of iron response (18, 56) because the probable chelation of ions other than zinc could have an influence on the general transcriptional response.
cDNA microarray analysis.
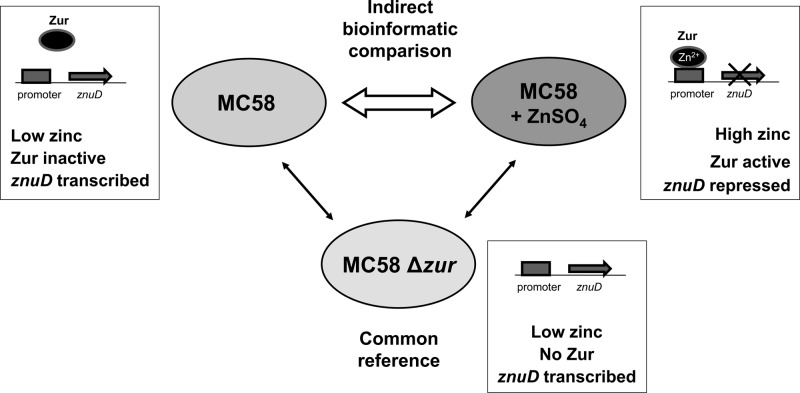
Microarray analysis was performed using whole-genome DNA microarrays based on 70-mer oligonucleotides covering four meningococcal genomes (MC58, Z2491, FAM18, and α14) as described previously (55). In order to characterize the Zur regulon, we used a common reference design that allows for future extension of the data set (65). As outlined in Fig. 1, cDNA obtained from strain MC58 grown with or without zinc supplementation (low- and high-zinc conditions) was hybridized separately along with the common reference, i.e., cDNA obtained from strain MC58 Δzur. MC58 Δzur was used as the common reference under the assumption that Zur mostly represses gene expression. In the zur knockout strain, mRNA of most of the Zur-regulated genes was therefore expected to be present, which is a prerequisite of valid comparisons. The transcriptome of MC58 grown without zinc supplementation was consecutively indirectly compared to that of MC58 grown in the presence of high zinc.
Fig 1.
Common reference design of microarray comparison used in this study. cDNAs obtained from MC58 grown without addition of zinc (low zinc) and from MC58 grown with addition of zinc for 2 h (high zinc) were hybridized along with cDNA of the MC58 zur knockout strain grown under low-zinc conditions as a common reference. MC58 Δzur was used as a common reference under the assumption that total mRNA of this mutant will contain mRNA of Zur-repressed genes. The zinc-dependent promoter repression by Zur is visualized by cartoons using the example of znuD.
Ten micrograms of RNA obtained from each culture was transcribed into cDNA and labeled with either Cy3-dCTP or Cy5-dCTP (GE Healthcare) using SuperScript II (Invitrogen) and random nonamer primer (Sigma-Aldrich). Remaining RNA was digested with DNase-free RNase (Roche). The labeled cDNA was purified using illustra AutoSeq G50 columns (GE Healthcare). For each slide, two differentially labeled cDNA probes were combined, and probes were then hybridized to a prehybridized microarray slide. Hybridization was carried out in a Tecan HS4800TM Pro hybridization station. Three biological replicates (independent bacterial cultures and RNA isolations) with at least two technical replicates (independent slides including dye swap) were used for microarray hybridization. The hybridized slides were scanned with the Genepix 4200 scanner, and the raw data were acquired using Genepix Pro 4.0. The raw gpr files from the microarray slides were processed with the Limma package (58) implemented in R (47) to identify significantly differentially regulated genes. Only genes with a false-discovery rate (FDR) of <0.01 and a B statistic value of >0 were considered for further analyses.
qRT-PCR.
Quantitative real-time PCR (qRT-PCR) analysis of the RNA samples described above was performed as described previously using the StepOnePlus system and the Power SYBR green Master Mix (53). nmb1567, encoding a putative membrane-associated peptidyl-prolyl isomerase, which was not differentially regulated in our microarray, was applied as endogenous control for data normalization. The oligonucleotides used are given in Table 1. The threshold cycle (ΔΔCT) and relative quantity (RQ) values of qRT-PCR were obtained using the StepOne software v2.0. RQ values represent the fold change of expression of the investigated gene in MC58 grown under low zinc compared to that in MC58 grown under high-zinc conditions. Microarray results were accepted as differentially regulated only if comparison of the same RNA by qRT-PCR analysis yielded RQ values of at least 1.5.
Computational analysis of Zur binding sites.
Sequences of genes and upstream regions of the identified candidate genes of strain MC58 were retrieved from the NeMeSys database (51). Similarity searches were performed using the BLAST program of the National Center for Biotechnology Information (NCBI) (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Operons were predicted using RNAseq data (B. Joseph and C. Schoen, unpublished data) and DOOR database (35). Annotation data were retrieved from the NeMeSys (51) and Uniprot (62) databases. Upstream sequences were screened for the presence of a Zur box by alignments using ClustalW in BioEdit 7.0.9 (20) based on the in silico predicted motif for Betaproteobacteria as implemented in the RegPrecise database (39) and the hypothetical motif for znuD (nmb0964), which was recently proposed (59). Furthermore, upstream regions were searched for conserved motifs with the GIBBS Motif Sampler (61). The meningococcal Zur binding motif was visualized using WebLogo 3.2 (6).
Expression and purification of recombinant Zur.
The Zur protein was recombinantly expressed for electrophoretic mobility shift assays. For gene cloning and protein expression, the QIAexpressionist kit (Qiagen) was used according to the manufacturer's instructions. Briefly, the entire coding region of the zur gene without the start and stop codons (469 bp) was amplified from MC58 using primers MP183 and MP184 harboring a BspHI and a BglII restriction site, respectively (Table 1). The PCR product was cloned between the NcoI and BglII sites of pQE-60 (Qiagen) containing a C-terminal His tag. E. coli M15(pREP4) was transformed with the recombinant plasmid, and clones were verified by DNA sequencing. Expression and purification of Zur were performed under native conditions. Protein expression was induced by 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 4.5 h at 37°C, and cells were harvested, lysed, and broken by ultrasonic treatment.
For purification, cell extracts were loaded on a HIS GraviTrap nickel Sepharose column (GE Healthcare) equilibrated with lysis buffer (50 mM NaH2PO4, 0.3 M NaCl, 10 mM imidazole [pH 8.0]). The column was washed twice with wash buffer (50 mM NaH2PO4, 0.3 M NaCl, 20 mM imidazole [pH 8.0]) before elution of the His-tagged Zur protein with elution buffer (50 mM NaH2PO4, 0.3 M NaCl, 250 mM imidazole [pH 8.0]). The Zur eluate from the nickel Sepharose column was dialyzed against Tris-borate (TB) buffer (44.5 mM Tris, 44.5 mM boric acid [pH 8.0]) to remove imidazole and nickel. The purity of Zur was verified by SDS-PAGE with Coomassie blue staining, and its concentration was determined using the Pierce BCA protein assay kit (ThermoScientific). Protein sample aliquots were stored at −20°C.
Electrophoretic mobility shift assays to visualize DNA-Zur binding.
EMSAs were performed using the digoxigenin (DIG) Gel Shift kit (Roche) to determine the ability of Zur to interact with short double-stranded DNA (dsDNA) fragments containing the Zur box. Oligonucleotides were purchased from Sigma-Aldrich (Table 1) and resuspended in TEN buffer (10 mM Tris, 1 mM EDTA, 0.1 M NaCl [pH 8.0]). Complementary oligonucleotides in equal concentrations were annealed to yield dsDNA fragments by heating the oligonucleotide mixture for 10 min at 95°C and subsequently slowly cooling it down to room temperature. dsDNA fragments were labeled with digoxigenin (DIG Gel Shift kit; Roche). Binding reactions were performed in a final volume of 10 μl with 0.4 ng (15.5 fmol) of the labeled dsDNA fragment and 15 pmol (250 ng) of purified Zur protein (if not stated differently) in the binding buffer [20 mM Tris-HCl (pH 8.0), 50 mM KCl, 1 mM dithiothreitol (DTT), 5% glycerol, 0.1 μg/μl poly(dI-dC), 0.01 μg/μl poly-l-lysine] modified after the method of Shin et al. (57). If not stated otherwise, the binding buffer was supplemented with 100 μM ZnSO4 according to the concentration used by Gaballa and Helmann for B. subtilis (13). We anticipated that zinc is required for Zur-DNA interaction as already shown for other bacteria (14, 29, 32, 54, 57) and therefore omitted EDTA from the binding buffer as well as from the running buffer to avoid chelation of zinc. Reaction mixtures were incubated at room temperature for 20 min. A native 8% polyacrylamide gel was prerun at 120 V for 30 min, and samples were then applied to the gel and separated at 120 V for 110 min at 4°C. After electroblotting onto a nylon membrane in TB buffer, chemiluminescent detection of the bound dsDNA fragments was carried out.
CaCl2, CoCl2, CuSO4, FeSO4, MgCl2, MnSO4, and NiSO4 were purchased from Merck or Sigma-Aldrich and added to the binding buffer at a final concentration of 100 μM. The ion chelators EDTA and TPEN were used at a final concentration of 312.5 μM. For binding competition experiments, 125- to 1,000-fold excess of the unlabeled dsDNA probe was added to the reactions.
Statistics.
The Wilcoxon rank sum test with continuity correction was applied to analyze the impact of mismatches in the Zur box palindromic sequence on gene expression.
Microarray data accession number.
The microarray data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus database (12) under accession no. GSE38033.
RESULTS
The aim of this study was to provide a data set of genes regulated by exposure to zinc. Microarray comparison between MC58 grown with and without zinc supplementation (high- and low-zinc condition, respectively) was employed using a common reference design (65) that is schematically represented in Fig. 1.
Growth conditions.
Growth of MC58 Δzur in the chemically defined medium RPMI supplemented with FeCl3 was indistinguishable from that of the parental wild-type strain (data not shown). This indicates that deactivation of Zur does not alter growth in RPMI. Growth conditions for microarray analysis were validated by qRT-PCR of znuD expression, which previously was shown to be increased in a zur knockout mutant (59). Strains were first grown in RPMI medium under low-zinc conditions, followed by addition of 0.5 μM ZnSO4 to the medium for 1, 2, or 3 h.
After 1 h of zinc addition, MC58 showed 64.8-fold-reduced expression levels of znuD by qRT-PCR compared to MC58 Δzur. However, znuD expression in MC58 grown under low-zinc conditions still was 27.3-fold reduced compared to MC58 Δzur, probably due to intracellular zinc remains of the overnight culture on GC agar. After 2 h of zinc addition, compared to the zur knockout, expression of znuD in MC58 was 48.8-fold reduced, whereas MC58 grown under low-zinc conditions showed similar expression. After 3 h, znuD expression of MC58 grown at high zinc was 42.5-fold reduced compared to the zur knockout. Based on these qRT-PCR results, exposure to zinc for 2 h was selected for microarray analysis.
Transcriptome analysis of zinc-dependent genes.
Gene expression profiles of strain MC58 grown under low-zinc and high-zinc conditions were compared by a common reference design using the MC58 zur knockout strain as the common reference (Fig. 1). Sixteen genes were upregulated, whereas three genes were downregulated, in MC58 grown at low zinc compared to high zinc. The genes of this data set were analyzed for operon structures. To confirm microarray analysis, we directly compared expression differences between MC58 (low zinc) and MC58 (high zinc) by qRT-PCR for the first gene of predicted operon structures and all single genes. All genes except two were confirmed to be differentially expressed. Therefore, the regulon verified in this study comprised 15 genes upregulated and only 2 downregulated in MC58 grown at low zinc compared to high zinc (Table 2). Nine genes were organized in four transcriptional units: nmb0317 and nmb0316 encode a 7-cyano-7-deazaguanine reductase and an integral membrane protein, respectively; nmb0817 and nmb0818 code for hypothetical proteins that belong to the DUF723 family and may have a role in DNA-binding (62); nmb0942 and nmb0941 encode paralogues of 50S ribosomal proteins; and nmb0588 and nmb0587, together with one single gene, nmb0586, encode the components of the putative ABC transporter for zinc, ZnuCBA (59). The single genes nmb0819 and nmb0820 both encode proteins that contain putative DNA-binding helix-turn-helix motifs. Highly differential expression of the TonB-dependent outer membrane receptor znuD (nmb0964), involved in zinc uptake (59), represents a positive control for our analysis, as znuD was used for optimization of our microarray conditions as indicated above. Expression of another putative TonB-dependent receptor of yet-unknown function, nmb1497, was also increased at low zinc. Strongly increased at low zinc were expression of nmb0525 (queC), encoding a zinc-binding 7-cyano-7-deazaguanine synthase, and expression of nmb1475, coding for a conserved hypothetical periplasmic protein with similarities to the acetate kinase AckA of Bacillus spp. Only two genes were repressed at low zinc: nmb0546 (adhP), which codes for a zinc-containing alcohol dehydrogenase, and nmb0577, which shows similarities to the Haemophilus influenzae pfkA, encoding a 6-phosphofructokinase.
Table 2.
Differentially expressed genes in N. meninigitidis MC58 observed by comparison of low- to high-zinc conditionsa
| Locus | Gene | Predicted function | Predicted localization | Size (bp) | Differential gene expression |
|
|---|---|---|---|---|---|---|
| Fold change by cDNA microarray hybridization | RQ by qRT-PCR | |||||
| nmb0546 | adhP | Alcohol dehydrogenase, propanol preferring | Cytoplasmic | 1,047 | −4.1 | −21.7 |
| nmb0577 | NosR-related protein | Unknown | 351 | −1.8 | −1.8 | |
| nmb0588 | znuC | ABC transporter, ATP-binding protein | Cytoplasmic | 756 | 1.4 | 4.4 |
| nmb0586 | znuA | Putative ABC transporter substrate-binding protein | Cytoplasmic membrane | 915 | 1.5 | 6.3 |
| nmb0820 | Hypothetical protein | Unknown | 198 | 1.6 | 1.9 | |
| nmb1497 | Putative TonB-dependent receptor | Outer membrane | 2,766 | 1.6 | 1.8 | |
| nmb0818 | Hypothetical protein | Unknown | 411 | 1.9 | ||
| nmb0817 | Hypothetical protein | Unknown | 384 | 2.0 | 3.1 | |
| nmb0587 | znuB | Putative ABC transporter permease protein | Cytoplasmic membrane | 876 | 2.1 | 3.3 |
| nmb0942 | rpmE2 | 50S ribosomal protein L31 type B | Cytoplasmic | 276 | 2.2 | 41.6 |
| nmb0819 | Hypothetical protein | Unknown | 393 | 2.3 | 1.8 | |
| nmb0525 | queC | 7-Cyano-7-deazaguanine synthase | Cytoplasmic | 660 | 2.5 | 3.9 |
| nmb0941 | rpmJ | 50S ribosomal protein L36 | Cytoplasmic | 126 | 2.7 | |
| nmb0317 | queF | NADPH-dependent 7-cyano-7-deazaguanine reductase | Cytoplasmic | 474 | 3.4 | 5.8 |
| nmb1475 | Conserved hypothetical periplasmic protein | Periplasmic | 807 | 3.6 | 96.4 | |
| nmb0964 | znuD | TonB-dependent receptor | Outer membrane | 2,277 | 4.4 | 51.1 |
| nmb0316 | Conserved hypothetical integral membrane protein | Cytoplasmic membrane | 687 | 4.6 | ||
In silico prediction of promoter organization and the Zur box.
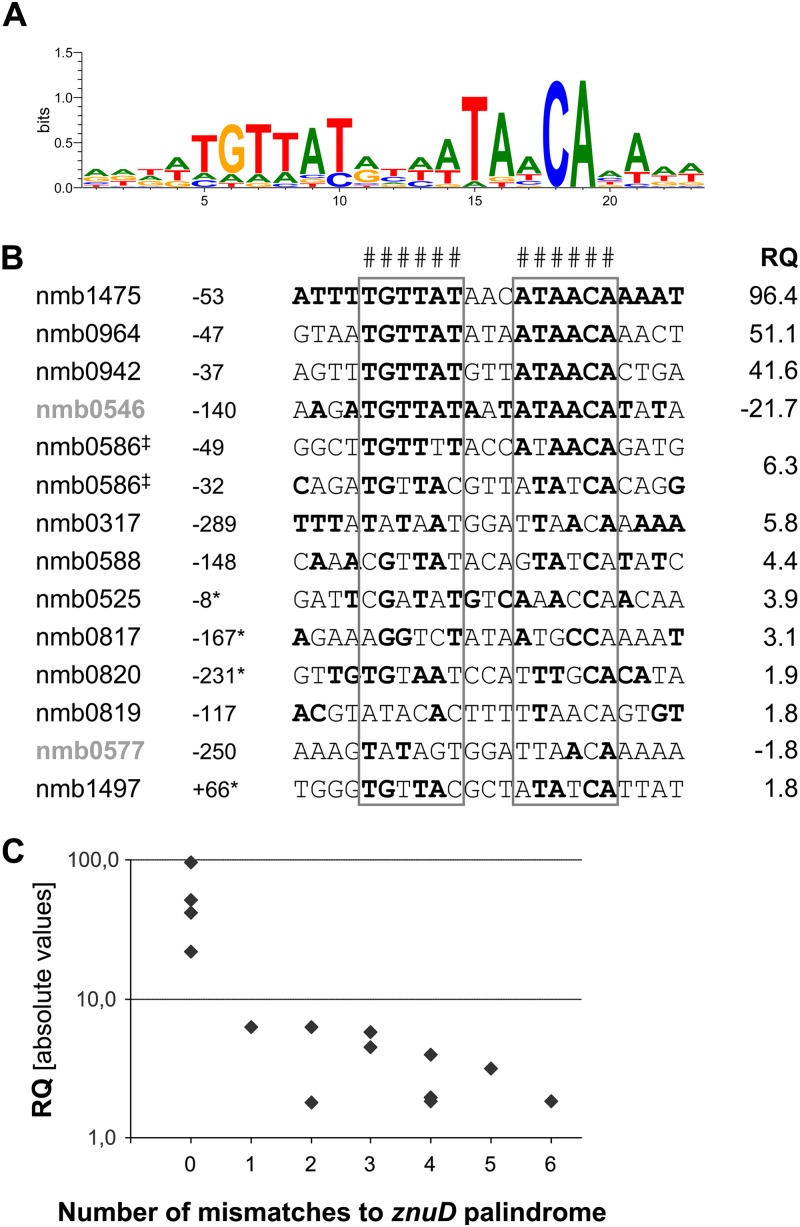
Zur binding to a palindromic sequence upstream of znuD has been previously postulated (59). The published sequence resembles the in silico-predicted Zur binding motif for Betaproteobacteria in the RegPrecise database (39). We analyzed the promoter regions of the confirmed zinc-responsive genes for a conserved Zur box. Homologies to the postulated Zur binding motif were found in the upstream regions of all regulated transcription units (genes or operons). This finding suggests that the approach employed here at least partially elucidated the Zur regulon and that zinc treatment of the bacteria did not deregulate genes other than Zur-controlled ones. The deduced meningococcal Zur binding motif was visualized as the consensus motif and graphically displayed by WebLogo 3.2 (Fig. 2A). It is a 23-bp motif with a central palindromic part comprising hexameric inverse repeats separated from each other by three nucleotides, which is also found in the predicted motif for znuD published by Stork et al. (59). The alignment of all putative Zur motifs indicated that the strength of gene regulation by Zur was dependent on the precision and length of the palindrome (Fig. 2B). The extent of zinc-dependent gene regulation (as deduced from RQ values) was reduced if at least one mismatch occurred in the palindrome of the putative Zur box (Fig. 2C). Gene expression changes as calculated based on the absolute ΔΔCT values obtained by qRT-PCR were significantly higher in genes with a perfect Zur box than in genes with imperfect Zur boxes having at least one mismatch (Wilcoxon rank sum test with continuity correction, P < 0.01). Extension of the length of the palindromic sequence may compensate for mismatches in the central palindrome, e.g., in the case of nmb0317 or nmb0588. The longest palindrome was found for nmb1475 with a decameric inverse repeat that probably represents the perfect Zur box, because this gene showed the highest repression of gene expression upon zinc exposure (RQ = 96.4). We detected two putative Zur boxes upstream of nmb0586. However, this duplication did not affect the strength of gene expression.
Fig 2.
Prediction of the putative meningococcal Zur box. (A) Graphical display of the Zur box for meningococci based on the consensus sequence of predicted Zur binding sites for all zinc-responsive genes (generated with WebLogo 3.2). (B) Nucleotide sequence alignment of putative Zur boxes upstream of zinc-regulated genes. Distances of the Zur motif to the start codon of translation and expression differences determined by qRT-PCR are given for each gene (RQ; low-zinc to high-zinc conditions). The order of motifs is based on the RQ values. The two genes upregulated by Zur are in bold gray letters. The strongly conserved palindrome composed of two hexameric inverted repeats separated by three nucleotides is indicated (#) and boxed for all motifs. Bold nucleotides signify complementary nucleotide pairs between the two sides of the motifs. Symbols: *, an alternative start for the gene is possible; ‡, two different motifs were found in the same promoter region. (C) Comparison of the absolute RQ values of gene expression with the number of mismatches in the palindrome of each Zur box compared to the znuD (nmb0964) palindrome sequence (TGTTATDNHATAACA). Gene expression changes were significantly higher in genes with a perfect Zur box than in genes with at least one mismatch in the Zur box (Wilcoxon rank sum test with continuity correction, P < 0.01).
Verification of Zur binding to predicted DNA motifs by electrophoretic mobility shift assay.
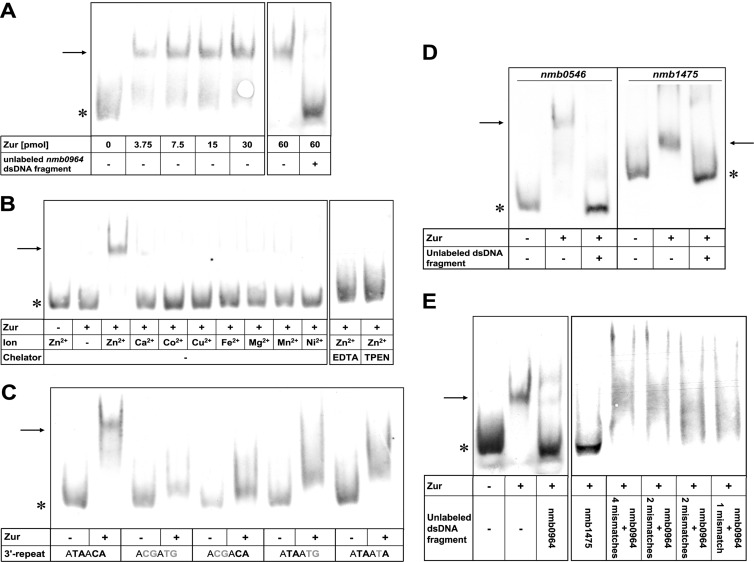
To demonstrate that Zur binds to in silico-predicted DNA motifs, we performed EMSAs with a His-tagged recombinant Zur protein and synthetic dsDNA fragments. A mobility shift of a 34-bp dsDNA fragment comprising the nmb0964 Zur box was observed upon incubation with the recombinant protein (Fig. 3A). The shift of the nmb0964 dsDNA fragment was competed by 125-fold excess of the unlabeled nmb0964 dsDNA fragment, which confirms binding specificity (Fig. 3A). A band shift was observed only in the presence of 100 μM ZnSO4, whereas CaCl2, CoCl2, CuSO4, FeSO4, MgCl2, MnSO4, and NiSO4 did not support binding of Zur to the dsDNA fragment, indicating that zinc is needed specifically to mediate Zur-DNA interaction (Fig. 3B). Addition of EDTA and TPEN, separately, blocked the zinc effect by chelation, underlining the importance of zinc (Fig. 3B).
Fig 3.
Zur binding to dsDNA fragments in EMSA. Band shifts are indicated with an arrow, unbound dsDNA fragments with an asterisk. (A) Binding of Zur to the dsDNA fragment comprising the nmb0964 Zur box. Addition of different amounts of Zur from 3.75 pmol to 60 pmol led to a band shift of the 34-mer dsDNA fragment. The unlabeled nmb0964 dsDNA fragment in 125-fold excess acted as a specific competitor that abrogated the band shift. (B) Effect of divalent ions and chelators on Zur binding to the nmb0964 dsDNA fragment. Different divalent ions (100 μM) were added to the reactions. Only Zn2+ mediated Zur binding to the nmb0964 dsDNA fragment. Addition of the chelator (312.5 μM) EDTA or TPEN reverted the band shift. (C) Mutational analysis of the Zur box. Mutation of conserved nucleotides in the 3′-inverse repeat of the palindrome of the nmb0964 Zur box inhibited the band shift. The four highly conserved nucleotides are displayed in bold. Mutated nucleotides are marked in gray. (D) Binding of Zur to dsDNA fragments comprising the Zur boxes of nmb0546 and nmb1475. Zur led to a band shift of the 34-mer nmb0546 dsDNA fragment and the 51-mer nmb1475 dsDNA fragment covering the respective Zur boxes. Addition of 1,000-fold excess of the respective unlabeled dsDNA fragment led to competition of the shift. (E) Competition studies with the nmb0964 dsDNA fragment. Incubation with a 125-fold excess of the unlabeled nmb0964 and nmb1475 dsDNA fragments abrogated Zur binding; 125-fold excess of unlabeled mutated nmb0964 dsDNA fragments comprising one to four mismatches within the 3′-inverse repeat of the palindrome (panel C) was unable to fully compete the shift.
Mutations of the four most conserved nucleotides in the 3′-inverse repeat of the palindrome (ATAACA) (where the conserved nucleotides are in boldface) of the nmb0964 dsDNA fragment abrogated or reduced Zur binding (Fig. 3C). However, interference with shifts was less clear upon mutation of marginal nucleotides or in the case of a single nucleotide mutation. This indicates that (i) all four conserved bases are important for Zur binding but the inner bases (TA) of the palindrome are more essential than the marginal ones (CA) and (ii) only one mutated nucleotide is not sufficient to completely abrogate the shift.
We further investigated the binding of Zur to the in silico-predicted Zur motifs of nmb0546, one of the two genes activated in response to zinc. A clear band shift that could be inhibited by the unlabeled nmb0546 dsDNA fragment was seen. For reasons unknown, competition of the nmb0546 shift required higher concentrations of the competing DNA than nmb0964. For nmb1475, the gene with an extended perfect palindrome in the Zur box, no shift was observed with a 34-mer dsDNA fragment (data not shown). Upon extending the surrounding region by 8 and 9 bp on each side of the extended perfect palindrome, a clear shift became also visible (Fig. 3D). We can only speculate that Zur binding to the perfect oversized palindrome may be possible only if the DNA structure is stabilized by extended flanking DNA sequences.
To furthermore affirm binding specificity, we performed EMSA with the nmb0964 dsDNA fragment and attempted to compete the shift with unlabeled nmb0964 or nmb1475 dsDNA fragments or mutated versions of the nmb0964 sequence. Indeed, the unlabeled nmb0964 dsDNA fragment itself and the unlabeled nmb1475 dsDNA fragment comprising the same palindrome abrogated Zur binding, whereas the nmb0964 dsDNA fragment comprising mismatches within the 3′-inverse repeat palindrome (ATAACA) was not able to completely compete the shift (Fig. 3E).
In summary, we detected Zur boxes upstream of all genes that were verified by qRT-PCR to be differentially expressed in response to zinc. Our EMSA results prove that Zur binds to the predicted binding motifs of three selected genes regulated in response to zinc. Therefore, we infer that all transcriptional units with Zur boxes identified here were indeed regulated by Zur. Zinc was indispensable for interaction of Zur with the DNA. The palindromic part of the motif was essential for the binding as demonstrated by mutational analysis. In vitro binding of dsDNA fragments harboring a Zur box was observed for genes irrespective of their activation or repression by zinc.
DISCUSSION
In the present study, we analyzed the role of the meningococcal zinc uptake regulator Zur in transcriptional regulation using a comparative microarray approach. We defined the Zur box for meningococci and demonstrated in vitro interaction of recombinantly expressed Zur protein with the predicted Zur boxes upstream of three selected target genes.
Previous studies of B. subtilis, Y. pestis, M. tuberculosis, and C. glutamicum applied direct microarray comparisons of a wild-type strain with a zur knockout mutant to elucidate the Zur regulon (14, 29, 32, 54). The advantage of our approach of analyzing the response to zinc was that pleiotropic effects of a constitutive zur knockout might be avoided. Indeed, the meningococcal Zur regulon deduced from transcriptome data and consecutive Zur box analysis is remarkably small. We identified 15 genes downregulated and two genes upregulated at high zinc using microarray analysis and qRT-PCR. All transcriptional units harbored a Zur box. The entity of 17 regulated genes is comparable to the number found in microarray analyses of Zur regulons of other bacteria, i.e., 18 genes and 32 genes upregulated in a zur deletion mutant of C. glutamicum (54) and M. tuberculosis (32), respectively. Only in a comparative microarray analysis of a Y. pestis wild-type strain and its zur knockout mutant, both grown under zinc-rich conditions, were a much higher number of Zur-regulated genes found (154 genes). However, as only four genes had a Zur box, regulation of most genes presumably has not been a direct result of Zur binding to promoters but reflects a general alteration of gene regulation (29). Following several studies of the iron response (18, 56), we could have applied zinc depletion, achieved by addition of an ion chelator such as TPEN, to compare with a zinc repletion condition. However, it cannot be excluded that TPEN chelates ions other than zinc and interferes with the membrane integrity.
Among the 17 genes regulated by zinc exposure, we detected 2 that are upregulated, i.e., nmb0546 and nmb0577. Gene activation by Zur has been reported in only two other studies. In a microarray analysis of the B. subtilis Zur regulon, two genes were upregulated in the wild type compared to a zur deletion mutant. However, Zur motifs were not detected in the genes' regulatory regions and it was assumed that their upregulation is probably due to indirect effects of the altered zinc homeostasis (14). This can be excluded in this study, as we identified Zur boxes in the upstream regions of both Zur-activated genes. Only in one other study, of the phytopathogen Xanthomonas campestris, did Zur act as a direct activator of one gene (22).
Based on the prediction of the Zur binding motif for znuD by Stork et al. (59), we analyzed the promoter regions of all genes found to be zinc regulated in our study. We established a binding motif for meningococcal Zur with the ideal palindromic sequence TGTTATDNHATAACA, which is identical to the Zur binding motif proposed for znuD (59) and consistent with the palindrome in the previously bioinformatically predicted motif for the related Gammaproteobacteria.
We found an ideal palindrome only within the promoters of the genes nmb0546, nmb0942, znuD, and nmb1475. These genes showed the highest differences in expression when low- and high-zinc conditions were compared. Mismatches in the palindrome reduced gene expression alteration. This finding was confirmed by EMSA, also in line with previous investigations of the Zur box of C. glutamicum (54).
The zinc-activated gene nmb0546 was previously shown to be Hfq repressed in N. meningitidis (37, 42). Hfq is an RNA chaperone that stabilizes small regulatory RNAs (sRNAs) and mediates their binding to their mRNA targets, which leads to subsequent repression of mRNA translation (37). If nmb0546 and nmb0577 were targets of a yet-unknown sRNA transcribed at zinc depletion and downregulated by Zur upon zinc repletion, activation of the genes would occur indirectly due to Zur-mediated downregulation of the putative sRNA, which otherwise leads to degradation of both mRNAs. However, RNA sequencing needs to be applied to analyze the regulation of sRNAs by zinc.
Functions of regulated genes.
The previously identified meningococcal genes carrying homologues of the E. coli znuCBA operon encoding an ABC transporter for high-affinity zinc uptake, i.e., nmb0588-nmb0587-nmb0586, and nmb0964 (znuD), coding for a TonB-dependent receptor mediating zinc uptake at low zinc concentration (59), were shown to be repressed at high zinc in this study. This finding is reminiscent of E. coli (44), M. tuberculosis (32), B. subtilis (14), C. glutamicum (54), Y. pestis (29), and Streptomyces coelicolor (57). We demonstrated by EMSA that meningococcal Zur binds to the in silico predicted Zur box first proposed by Stork et al. for znuD (59).
The most strongly regulated gene in our study was nmb1475. It codes for a conserved hypothetical periplasmic protein that shows 34% similarity to the acetate kinase AckA of Bacillus spp. AckA is an enzyme involved in the conversion of acetate to acetyl coenzyme A (acetyl-CoA) (62), and its E. coli homologue was shown to bind zinc (24). The protein also harbors a conserved domain that is similar to the CbiK (COG5266) domain of the periplasmic component of an ABC-type Co2+ transport system as identified by NCBI Blast. Thus, NMB1475 may be involved in the uptake of zinc and/or other transition metals.
It has been suggested that zinc uptake systems are important for bacterial survival and virulence upon infection, as the access to zinc is limited within the human host (21, 59). Of note, expression of NMB0586 (designated MntC by van Alen et al. [63] or ZnuA by Stork et al. [59]) and NMB1475 was upregulated in biofilms, and deletion of nmb0586 reduced biofilm formation (63). Extenuated biofilm formation has previously also been seen upon deletion of znuA in gonococci (30) and nmb0587 (znuB) in E. coli (19). A gonococcal znuA (mntC) mutant in vitro also was more sensitive to hydrogen peroxide than the wild-type strain and showed reduced invasion to primary human cervical epithelial cells (64). Furthermore, znuA contributes to the in vivo pathogenicity of Salmonella enterica (3). Moreover, ZnuABC and ZnuD were lately shown to contribute to bacterial adhesion to epithelial cells in E. coli (15) and N. meningitidis (26), respectively.
Two meningococcal genes, nmb0317 and nmb0525, that were repressed at high zinc and harbor an upstream Zur box are possibly involved in queuosine biosynthesis as deduced from protein similarity analysis. nmb0525 encodes the zinc-binding 7-cyano-7-deazaguanine synthase QueC, and nmb0317 codes for the 7-cyano-7-deazaguanine reductase QueF (62). To date, queuosine biosynthesis in bacteria is still not fully understood, but the pathways have been studied in E. coli and B. subtilis (16, 27). Queuosine is one of the most complex modified nucleosides that are incorporated at the wobble position of a subset of tRNAs (10). Such modification of tRNAs was shown to improve the efficiency and correctness of translation (9). Dineshkumar et al. demonstrated a natural defect in queuosine biosynthesis of E. coli and noticed reduced fitness under nutrient limitation (8). Lack of queuosine also attenuated Shigella flexneri virulence (9, 10). In meningococci, queuosine biosynthesis seems to be strongly Zur regulated, since two enzymes of the biosynthesis are upregulated under zinc-limiting conditions as they occur in the host (25). It will be of interest for future studies to address the question of whether enhanced queuosine modification of tRNAs might be the result of zinc depletion and contribute to increased expression of virulence factors and thus support fitness of N. meningitidis upon infection.
Concordant with what has been observed in E. coli (17, 41), Y. pestis (29), M. tuberculosis (32), S. coelicolor (57), and B. subtilis (2), the expression of several genes encoding ribosomal proteins (r-proteins) was also Zur repressed in N. meningitidis. Several bacterial genomes code for two paralogous forms of r-proteins (34). The first form (C+) contains a metal-binding Zn2+ ribbon usually consisting of four conserved cysteines, whereas in the second form (C−) this zinc ribbon is degenerated (52). For the paralogous pair of the L31 r-protein in B. subtilis, RpmE (C+) and YtiA (C−), it was shown that YtiA (C−) expression is repressed by Zur and YtiA liberates RpmE (C+) from the ribosome under zinc-deficient conditions (2, 29). Panina et al. identified candidate binding sites for different zinc repressors upstream of genes coding for C− paralogues of the r-proteins L31, L33, L36, and S14 of a range of different bacterial species and confirmed that upon existence of a C+ and a C− copy of the r-protein the gene encoding the C− copy is regulated by a zinc-dependent repressor (41). The replacement of zinc-containing r-proteins by non-zinc-containing paralogues upon zinc depletion might liberate zinc for maintaining zinc homeostasis and may enhance bacterial survival when facing zinc-restrictive conditions in vivo (25, 41). A potential evolutionary explanation for the duplication of r-proteins with different zinc contents might be that ribosomal assembly in any case needs to be maintained under zinc-restrictive conditions. In the meningococcal genome, the genes for the 50S ribosomal proteins L31 and L36 are duplicated. Based on sequence similarity to B. subtilis RpmE (C+) and YtiA (C−), the paralogous pairs of N. meningitidis r-proteins L31 and L36 are NMB1956 (C+)/NMB0942 (C−) and NMB0164 (C+)/NMB0941 (C−), respectively (34, 51). The nmb0942-nmb0941 (rpmEJ) operon was shown to be repressed at high zinc in this study, and we identified a Zur box upstream of nmb0942. Therefore, we assume that also in meningococci in the absence of zinc the enzyme lacking the zinc ribbon takes over the function of the C+ protein.
Comparison to other regulons.
In a study by Wu et al., the manganese-responsive PerR regulon has been described (64). Gonococcal PerR is a member of the Fur family and shows 96% protein identity to meningococcal Zur. Wu et al. conducted a microarray comparison of a Neisseria gonorrhoeae wild-type strain versus its perR knockout mutant. They found 11 genes upregulated and 1 gene downregulated in the perR mutant. Except for three genes, all genes do have meningococcal homologues with 93% to 98% protein identity. All of those homologues also have been identified in our microarray analysis of the zinc-responsive regulon of Neisseria meningitidis: nmb0586-nmb0588 (ng0170-ng0168), nmb0941-nmb0942 (ng0931-ng0930), nmb0964 (ng1205), nmb1475 (ng1049), nmb1497 (ng0952), and nmb0546 (ng1442) (51, 64). This suggests that the meningococcal zinc-dependent Zur regulation is related to the gonococcal manganese-dependent PerR regulation.
Several of the Zur-regulated proteins were also regulated differentially in a microarray study that compared transcriptional profiles of N. meningitidis grown with different host iron binding proteins (i.e., hemoglobin, transferrin, and lactoferrin) as the sole iron source (23). The genes nmb0941, nmb0942, nmb1475, and nmb1497 were downregulated, and nmb0546 was upregulated upon exposure to lactoferrin (compared to hemoglobin or transferrin) (23). Lactoferrin, which is present in secretions and on mucosal surfaces of the human host, can be used by meningococci employing a lactoferrin receptor to catch the iron from lactoferrin (23, 38). Therefore, regulation of these genes upon lactoferrin exposure may additionally be accomplished by a second regulator that senses iron. This was shown recently for znuD, whose expression also was iron induced (26). Besides its upstream Zur binding site, znuD also harbors a Fur binding site where Fur in vitro binds to, independently of Zur (26). However, we did not detect any Fur box upstream of nmb0942-nmb0941, nmb1475, nmb1497, and nmb0546, nor has znuD been regulated upon lactoferrin exposure in the study by Jordan and Saunders (23). Hence, we favor a second hypothesis to explain the overlap of the zinc and lactoferrin regulons. As lactoferrin has been reported to also loosely bind zinc (1), meningococci might use it as an additional source of zinc. Zinc could then act as a cofactor for Zur, leading to Zur-mediated regulation of the genes mentioned above as also seen in this study.
The transcriptomic response of N. meningitidis to whole-blood exposure was recently recorded using microarrays (11). Interestingly, several of the genes deregulated by this approach were also found to be changed in their expression upon zinc exposure. This finding suggests that zinc depletion upon transfer of bacteria from liquid culture to whole blood is responsible for a part of the transcriptional changes observed. Furthermore, this finding also reinforces that changes of zinc concentration mimic an important environmental signal encountered by bacteria during pathogen-host interaction.
In summary, we elucidated the transcriptional adaptation of N. meningitidis to zinc using strain MC58. The regulon is assumed to cover at least a significant portion of the Zur regulon, as all transcriptional units (genes/operons) were preceded by promoters harboring a Zur box. The functionality of representative motifs was confirmed by EMSA. It will be of interest to investigate the concerted action of genes derepressed at low-zinc conditions in vivo, as low zinc is encountered by the bacteria upon infection (25).
ACKNOWLEDGMENTS
The study was funded by a grant to U.V. provided by the German Federal Ministry of Education and Research (reference number 0315434) via the ERA-NET PathoGenoMics program (2nd call).
Footnotes
Published ahead of print 5 October 2012
REFERENCES
- 1. Ainscough EW, Brodie AM, Plowman JE. 1980. Zinc transport by lactoferrin in human milk. Am. J. Clin. Nutr. 33:1314–1315 [DOI] [PubMed] [Google Scholar]
- 2. Akanuma G, Nanamiya H, Natori Y, Nomura N, Kawamura F. 2006. Liberation of zinc-containing L31 (RpmE) from ribosomes by its paralogous gene product, YtiA, in Bacillus subtilis. J. Bacteriol. 188:2715–2720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ammendola S, et al. 2007. High-affinity Zn2+ uptake system ZnuABC is required for bacterial zinc homeostasis in intracellular environments and contributes to the virulence of Salmonella enterica. Infect. Immun. 75:5867–5876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bentley SD, et al. 2007. Meningococcal genetic variation mechanisms viewed through comparative analysis of serogroup C strain FAM18. PLoS Genet. 3:e23 doi:10.1371/journal.pgen.0030023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen PR, He C. 2008. Selective recognition of metal ions by metalloregulatory proteins. Curr. Opin. Chem. Biol. 12:214–221 [DOI] [PubMed] [Google Scholar]
- 6. Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dalet K, Gouin E, Cenatiempo Y, Cossart P, Hechard Y. 1999. Characterisation of a new operon encoding a Zur-like protein and an associated ABC zinc permease in Listeria monocytogenes. FEMS Microbiol. Lett. 174:111–116 [DOI] [PubMed] [Google Scholar]
- 8. Dineshkumar TK, Thanedar S, Subbulakshmi C, Varshney U. 2002. An unexpected absence of queuosine modification in the tRNAs of an Escherichia coli B strain. Microbiology 148:3779–3787 [DOI] [PubMed] [Google Scholar]
- 9. Durand JM, Bjork GR. 2003. Putrescine or a combination of methionine and arginine restores virulence gene expression in a tRNA modification-deficient mutant of Shigella flexneri: a possible role in adaptation of virulence. Mol. Microbiol. 47:519–527 [DOI] [PubMed] [Google Scholar]
- 10. Durand JM, Dagberg B, Uhlin BE, Bjork GR. 2000. Transfer RNA modification, temperature and DNA superhelicity have a common target in the regulatory network of the virulence of Shigella flexneri: the expression of the virF gene. Mol. Microbiol. 35:924–935 [DOI] [PubMed] [Google Scholar]
- 11. Echenique-Rivera H, et al. 2011. Transcriptome analysis of Neisseria meningitidis in human whole blood and mutagenesis studies identify virulence factors involved in blood survival. PLoS Pathog. 7:e1002027 doi:10.1371/journal.ppat.1002027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Edgar R, Domrachev M, Lash AE. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30:207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gaballa A, Helmann JD. 1998. Identification of a zinc-specific metalloregulatory protein, Zur, controlling zinc transport operons in Bacillus subtilis. J. Bacteriol. 180:5815–5821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gaballa A, Wang T, Ye RW, Helmann JD. 2002. Functional analysis of the Bacillus subtilis Zur regulon. J. Bacteriol. 184:6508–6514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gabbianelli R, et al. 2011. Role of ZnuABC and ZinT in Escherichia coli O157:H7 zinc acquisition and interaction with epithelial cells. BMC Microbiol. 11:36 doi:10.1186/1471-2180-11-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gaur R, Varshney U. 2005. Genetic analysis identifies a function for the queC (ybaX) gene product at an initial step in the queuosine biosynthetic pathway in Escherichia coli. J. Bacteriol. 187:6893–6901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Graham AI, et al. 2009. Severe zinc depletion of Escherichia coli: roles for high affinity zinc binding by ZinT, zinc transport and zinc-independent proteins. J. Biol. Chem. 284:18377–18389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Grifantini R, et al. 2003. Identification of iron-activated and -repressed Fur-dependent genes by transcriptome analysis of Neisseria meningitidis group B. Proc. Natl. Acad. Sci. U. S. A. 100:9542–9547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gunasekera TS, Herre AH, Crowder MW. 2009. Absence of ZnuABC-mediated zinc uptake affects virulence-associated phenotypes of uropathogenic Escherichia coli CFT073 under Zn(II)-depleted conditions. FEMS Microbiol. Lett. 300:36–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. 41:95–98 [Google Scholar]
- 21. Hantke K. 2001. Bacterial zinc transporters and regulators. Biometals 14:239–249 [DOI] [PubMed] [Google Scholar]
- 22. Huang DL, et al. 2008. The Zur of Xanthomonas campestris functions as a repressor and an activator of putative zinc homeostasis genes via recognizing two distinct sequences within its target promoters. Nucleic Acids Res. 36:4295–4309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jordan PW, Saunders NJ. 2009. Host iron binding proteins acting as niche indicators for Neisseria meningitidis. PLoS One 4:e5198 doi:10.1371/journal.pone.0005198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Katayama A, Tsujii A, Wada A, Nishino T, Ishihama A. 2002. Systematic search for zinc-binding proteins in Escherichia coli. Eur. J. Biochem. 269:2403–2413 [DOI] [PubMed] [Google Scholar]
- 25. Kehl-Fie TE, Skaar EP. 2010. Nutritional immunity beyond iron: a role for manganese and zinc. Curr. Opin. Chem. Biol. 14:218–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kumar P, Sannigrahi S, Tzeng YL. 2012. The Neisseria meningitidis ZnuD zinc receptor contributes to interactions with epithelial cells and supports heme utilization when expressed in Escherichia coli. Infect. Immun. 80:657–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee BW, Van Lanen SG, Iwata-Reuyl D. 2007. Mechanistic studies of Bacillus subtilis QueF, the nitrile oxidoreductase involved in queuosine biosynthesis. Biochemistry 46:12844–12854 [DOI] [PubMed] [Google Scholar]
- 28. Lee JW, Helmann JD. 2007. Functional specialization within the Fur family of metalloregulators. Biometals 20:485–499 [DOI] [PubMed] [Google Scholar]
- 29. Li Y, et al. 2009. Characterization of Zur-dependent genes and direct Zur targets in Yersinia pestis. BMC Microbiol. 9:128 doi:10.1186/1471-2180-9-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lim KH, et al. 2008. Metal binding specificity of the MntABC permease of Neisseria gonorrhoeae and its influence on bacterial growth and interaction with cervical epithelial cells. Infect. Immun. 76:3569–3576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lindsay JA, Foster SJ. 2001. zur: a Zn(2+)-responsive regulatory element of Staphylococcus aureus. Microbiology 147:1259–1266 [DOI] [PubMed] [Google Scholar]
- 32. Maciag A, et al. 2007. Global analysis of the Mycobacterium tuberculosis Zur (FurB) regulon. J. Bacteriol. 189:730–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reference deleted.
- 34. Makarova KS, Ponomarev VA, Koonin EV. 2001. Two C or not two C: recurrent disruption of Zn-ribbons, gene duplication, lineage-specific gene loss, and horizontal gene transfer in evolution of bacterial ribosomal proteins. Genome Biol. 2:research 0033–research0033.14 doi:10.1186/gb-2001-2-9-research0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mao F, Dam P, Chou J, Olman V, Xu Y. 2009. DOOR: a database for prokaryotic operons. Nucleic Acids Res. 37:D459–D463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McGuinness BT, et al. 1991. Point mutation in meningococcal porA gene associated with increased endemic disease. Lancet 337:514–517 [DOI] [PubMed] [Google Scholar]
- 37. Mellin JR, et al. 2010. Role of Hfq in iron-dependent and -independent gene regulation in Neisseria meningitidis. Microbiology 156:2316–2326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Morgenthau A, Livingstone M, Adamiak P, Schryvers AB. 2012. The role of lactoferrin binding protein B in mediating protection against human lactoferricin. Biochem. Cell Biol. 90:417–423 [DOI] [PubMed] [Google Scholar]
- 39. Novichkov PS, et al. 2010. RegPrecise: a database of curated genomic inferences of transcriptional regulatory interactions in prokaryotes. Nucleic Acids Res. 38:D111–D118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. O'Halloran TV. 1993. Transition metals in control of gene expression. Science 261:715–725 [DOI] [PubMed] [Google Scholar]
- 41. Panina EM, Mironov AA, Gelfand MS. 2003. Comparative genomics of bacterial zinc regulons: enhanced ion transport, pathogenesis, and rearrangement of ribosomal proteins. Proc. Natl. Acad. Sci. U. S. A. 100:9912–9917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pannekoek Y, et al. 2009. Molecular characterization and identification of proteins regulated by Hfq in Neisseria meningitidis. FEMS Microbiol. Lett. 294:216–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Parkhill J, et al. 2000. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature 404:502–506 [DOI] [PubMed] [Google Scholar]
- 44. Patzer SI, Hantke K. 2000. The zinc-responsive regulator Zur and its control of the znu gene cluster encoding the ZnuABC zinc uptake system in Escherichia coli. J. Biol. Chem. 275:24321–24332 [DOI] [PubMed] [Google Scholar]
- 45. Patzer SI, Hantke K. 1998. The ZnuABC high-affinity zinc uptake system and its regulator Zur in Escherichia coli. Mol. Microbiol. 28:1199–1210 [DOI] [PubMed] [Google Scholar]
- 46.Reference deleted.
- 47.R Development Core Team 2008. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 48.Reference deleted.
- 49. Reyes-Caballero H, Campanello GC, Giedroc DP. 2011. Metalloregulatory proteins: metal selectivity and allosteric switching. Biophys. Chem. 156:103–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rosenstein NE, Perkins BA, Stephens DS, Popovic T, Hughes JM. 2001. Meningococcal disease. N. Engl. J. Med. 344:1378–1388 [DOI] [PubMed] [Google Scholar]
- 51. Rusniok C, et al. 2009. NeMeSys: a biological resource for narrowing the gap between sequence and function in the human pathogen Neisseria meningitidis. Genome Biol. 10:R110 doi:10.1186/gb-2009-10-10-r110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sankaran B, et al. 2009. Zinc-independent folate biosynthesis: genetic, biochemical, and structural investigations reveal new metal dependence for GTP cyclohydrolase IB. J. Bacteriol. 191:6936–6949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schielke S, et al. 2011. Characterization of FarR as a highly specialized, growth phase-dependent transcriptional regulator in Neisseria meningitidis. Int. J. Med. Microbiol. 301:325–333 [DOI] [PubMed] [Google Scholar]
- 54. Schröder J, Jochmann N, Rodionov DA, Tauch A. 2010. The Zur regulon of Corynebacterium glutamicum ATCC 13032. BMC Genomics 11:12 doi:10.1186/1471-2164-11-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schwarz R, et al. 2010. Evaluation of one- and two-color gene expression arrays for microbial comparative genome hybridization analyses in routine applications. J. Clin. Microbiol. 48:3105–3110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shaik YB, et al. 2007. Expression of the iron-activated nspA and secY genes in Neisseria meningitidis group B by Fur-dependent and -independent mechanisms. J. Bacteriol. 189:663–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Shin JH, Oh SY, Kim SJ, Roe JH. 2007. The zinc-responsive regulator Zur controls a zinc uptake system and some ribosomal proteins in Streptomyces coelicolor A3(2). J. Bacteriol. 189:4070–4077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Smyth G. 2005. Limma: linear models for microarray data, p 397–420. In Gentleman R, Carey V, Dudoit SR, Irizarry R, Huber W. (ed), Bioinformatics and computational biology solutions using R and Bioconductor. Springer, New York, NY [Google Scholar]
- 59. Stork M, et al. 2010. An outer membrane receptor of Neisseria meningitidis involved in zinc acquisition with vaccine potential. PLoS Pathog. 6:e1000969 doi:10.1371/journal.ppat.1000969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tettelin H, et al. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287:1809–1815 [DOI] [PubMed] [Google Scholar]
- 61. Thompson WA, Newberg LA, Conlan S, McCue LA, Lawrence CE. 2007. The Gibbs centroid sampler. Nucleic Acids Res. 35:W232–W237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.UniProt Consortium 2012. Reorganizing the protein space at the Universal Protein Resource (UniProt). Nucleic Acids Res. 40:D71–D75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. van Alen T, et al. 2010. Comparative proteomic analysis of biofilm and planktonic cells of Neisseria meningitidis. Proteomics 10:4512–4521 [DOI] [PubMed] [Google Scholar]
- 64. Wu HJ, et al. 2006. PerR controls Mn-dependent resistance to oxidative stress in Neisseria gonorrhoeae. Mol. Microbiol. 60:401–416 [DOI] [PubMed] [Google Scholar]
- 65. Yang YH, Speed T. 2002. Design issues for cDNA microarray experiments. Nat. Rev. Genet. 3:579–588 [DOI] [PubMed] [Google Scholar]