Abstract
Based on fluorescence microscopy, the actin homolog MreB has been thought to form extended helices surrounding the cytoplasm of rod-shaped bacterial cells. The presence of these and other putative helices has come to dominate models of bacterial cell shape regulation, chromosome segregation, polarity, and motility. Here we use electron cryotomography to show that MreB does in fact form extended helices and filaments in Escherichia coli when yellow fluorescent protein (YFP) is fused to its N terminus but native (untagged) MreB expressed to the same levels does not. In contrast, mCherry fused to an internal loop (MreB-RFPSW) does not induce helices. The helices are therefore an artifact of the placement of the fluorescent protein tag. YFP-MreB helices were also clearly distinguishable from the punctate, “patchy” localization patterns of MreB-RFPSW, even by standard light microscopy. The many interpretations in the literature of such punctate patterns as helices should therefore be reconsidered.
INTRODUCTION
Just over 10 years ago, Jones and colleagues showed that the bacterial actin homologs MreB and Mbl are involved in the maintenance of rod shape in Bacillus subtilis. Based on fluorescence images, they concluded that both proteins polymerize into long helical filaments that encircle the cytoplasm (16). Using similar techniques, Shih et al. then reported that MreB (and three other proteins) formed extended helices in Escherichia coli as well (27). Numerous publications have since claimed that MreB and various other proteins polymerize into extended helices that encircle diverse rod-shaped bacteria (5, 8, 15, 22, 30, 31, 36). Data supporting these claims include fluorescence microscopy of both cells harboring fluorescent protein fusions and wild-type cells probed with fluorescently labeled antibodies. As a result, the idea of a helical cytoskeleton within rod-shaped bacteria has come to dominate the field of bacterial cell biology, playing a central role in models of not only cell shape determination but also chromosome segregation (11), cell polarity (12, 26), motility (21), and growth (32, 35). Growing confidence in the extent and ubiquity of the putative MreB helices has even led to the suggestions that every cell depends on a complete and continuous “figure 8” MreB structure that must be severed and segregated during cell division (33) and that can be used as a track to shuttle proteins from pole to pole and back (21).
During this same period, we began imaging various rod-shaped bacteria in an intact, near-native (frozen-hydrated) state to higher resolution by electron cryotomography (ECT) (9). Puzzlingly, while numerous and diverse cytoskeletal filaments were seen in the various species we (2, 7, 14, 20, 23) and others imaged, no helical filaments matching the descriptions for MreB were ever detected in either mutant or wild-type cells despite very careful inspection and extensive computational searches (reported in reference 29). Three very recent papers based on total internal reflection fluorescence (TIRF) and confocal images have also now argued that MreB does not form extended helices but rather resides in small patches or spots that move rapidly around the circumference of rod-shaped cells (6, 10, 32). It remains unclear which procedural and/or instrumental factors led to these different interpretations, but new papers continue to appear reporting MreB helices (4, 13, 35).
In an attempt to resolve these contradictions, we decided to record cryotomograms of the strain that had produced what were, in our judgment, the most compellingly helical fluorescence images: E. coli cells overexpressing an N-terminal yellow fluorescent protein (YFP)-MreB fusion (strain MC1000/pLE7) (first described in reference 27). Here we show that this strain does indeed harbor long YFP-MreB filaments; however, they are artifacts of the YFP tag, since they no longer form when the tag is removed. In contrast, cells expressing a “sandwich” MreB-mCherry fusion protein (MreB-RFPSW) (1) appeared wild type, in that they were devoid of extended helices. Light microscopy of the two tagged strains showed that, when extended helices are present, they can be clearly distinguished from punctate, nonhelical patterns, even in simple focal stacks. The many papers claiming that various proteins form helices in bacteria based on punctate fluorescence images should therefore be reconsidered.
MATERIALS AND METHODS
Cell strains and growth conditions.
MC1000, MC1000/pLE6, and MC1000/pLE7 were grown in LB at 37°C with 50 μg/ml ampicillin when appropriate. Strain MC1000 (Δlac Δara) was obtained from the ATCC (no. 39531) and MC1000/pLE6 (Plac-mreB) and MC1000/pLE7 (Plac-yfp::mreB) were gifts from Larry Rothfield. Similar levels of overexpression of native MreB (MC1000/pLE6) and YFP-MreB (MC1000/pLE7) were obtained by growing cells to an optical density at 600 nm (OD600) of 0.2 and then adding 1 mM and 20 μM IPTG (isopropyl-β-d-thiogalactopyranoside), respectively, for 2 h (OD600 of 0.8). In order to hyperoverexpress YFP-MreB, the same growth conditions were used, but 1 mM IPTG was used to induce expression in MC1000/pLE7 cells. Strain FB76 (mreB'-mCherry-‘mreB yhdE<>cat) was a gift from Piet de Boer and was grown at 30°C with 10 μg/ml chloramphenicol to an OD600 of 0.6. Cells were then either plunge frozen for cryotomography or applied to a 1% agar pad containing LB for live fluorescence imaging.
Electron cryotomography.
Cells were applied to freshly glow-discharged Quantifoil grids (R2/2) and plunge frozen using a Vitrobot (FEI). Samples were maintained at a temperature colder than −150°C during transfer into an F30 Polara (FEI) using a field emission gun and a Gatan lens-coupled Ultracam (4,000 by 4,000 pixels) operating at 300 keV. Tilt series were collected from −64 to 64 degrees in one-degree increments using UCSF Tomo (UCSF). The total electron dose was ∼200 e−/Å2. Tomograms were reconstructed and modeled using the IMOD software package (bio3d.colorado.edu/imod/).
Fluorescence microscopy.
Epifluorescence images of cells expressing YFP-MreB (MC1000/pLE7) and MreB-RFPSW (FB76) were collected on a Nikon Eclipse 90i microscope using a 100× oil objective (numerical aperture [NA], 1.4) and a Photometrics CoolSnap HQ2 charge-coupled device (CCD). Due to very fast photobleaching, focal stacks of FB76 were collected on a Nikon Eclipse Ti using a 100× oil objective (NA, 1.49) and a Photometrics QuantEM 512sc electron-multiplying CCD (EMCCD) equipped with a photomultiplier. With both microscopes, focal stacks were collected in 120-nm increments.
Western blots.
To compare expression levels of native MreB and YFP-MreB, the amount of sample for loading onto SDS-polyacrylamide gels was normalized based on the OD600. Protein was transferred to a nitrocellulose membrane and blocked overnight at 4°C in blocking buffer (phosphate-buffered saline [PBS] and 5% dry milk powder). Membranes were incubated with anti-MreB antibodies (a gift from Larry Rothfield) for 1 h at room temperature in blocking buffer, washed with PBS, and incubated again for 1 h with goat anti-rabbit secondary antibodies conjugated to horseradish peroxidase (HRP) (AbCam). A chemiluminescent signal was then generated using a SuperSignal West Pico chemiluminescent substrate (Thermo Scientific), exposed to film, and developed.
RESULTS
Four different strains of E. coli were cultured in suspension, spread into a thin layer across carbon-coated electron microscopy grids, plunge frozen, and imaged in three dimensions (3D) by ECT (Fig. 1). Whereas no helical filaments were seen in wild-type (MC1000) cells (0 out of 10) (Fig. 1A), 90% of MC1000/pLE7 cells (40 out of 44) exhibited clear filaments next to the membrane (Fig. 1B). While segments of individual filaments could be visualized in 2D tomographic slices, the membrane and filaments of some cells were modeled to provide 3D views of all the filaments at once (Fig. 2). Many of the filaments were oriented along a helical path, but the angles varied anywhere between parallel and perpendicular to the long axis of the cell. Even though the well-understood missing wedge in cryotomographic reconstructions obscured the paths of many individual filaments over the top and bottom of the cells, large bundles could often be followed as they ran helically through these regions.
Fig 1.
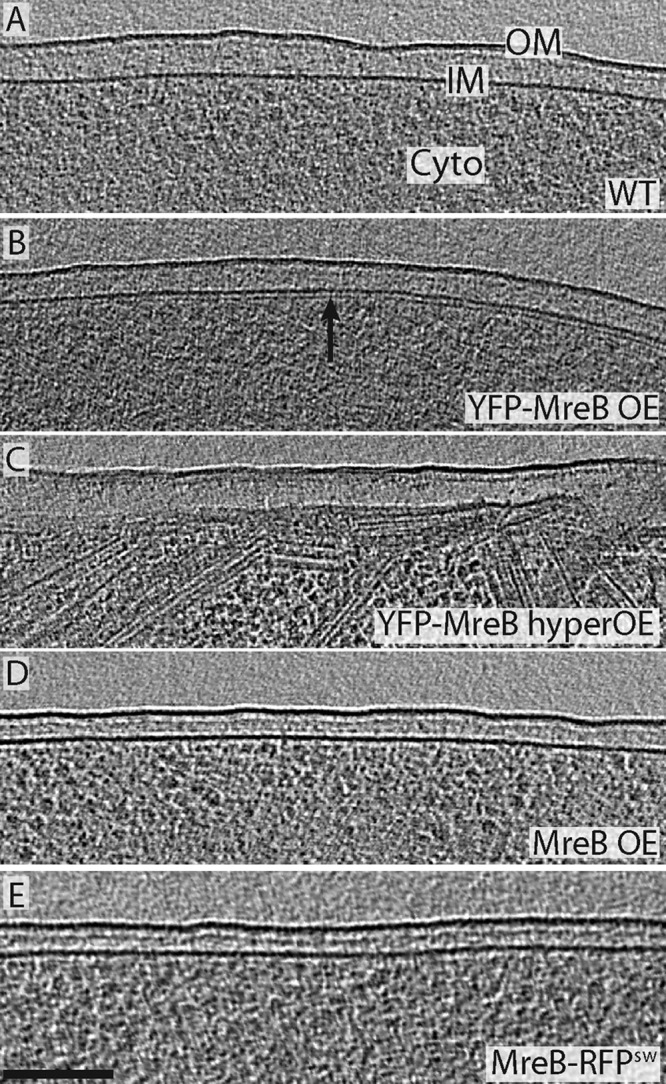
MreB helices are an artifact of the YFP tag. (A to E) Tomographic slices through the edge of five representative E. coli cells. No filaments were seen near the membranes of wild-type cells (A), cells overexpressing native MreB (D), or cells harboring MreB-RFPSW (E), but many helical and other filaments were found just inside the membranes of cells overexpressing YFP-MreB (strain pLE7) (B). We confirmed that these filaments are YFP-MreB by their dramatically increased abundance in cells overexpressing YFP-MreB at high levels (C). The outer membrane (OM), inner membrane (IM), and cytoplasm (Cyto) are labeled in panel A. Bar, 100 nm.
Fig 2.

YFP-MreB filaments exhibit a range of lengths and helical pitches. (Top) 3D model of a pLE7 cell's inner membrane (blue) and YFP-MreB filaments (white) (1 to 3). Tomographic slices through the cell modeled in the top panel. The slice numbers correspond to the filaments of the same number in the top panel. Insets depict the angle of the slice through the cell and arrowheads point to the visible ends of the filament. The outer membrane (OM), inner membrane (IM), and cytoplasm (Cyto) are labeled in slice 1. The scale bar represents 100 nm.
To confirm that these filaments were in fact YFP-MreB, we dramatically increased expression levels and again recorded cryotomograms. In this case, 10 out of the 13 cells imaged were now filled with hundreds of filaments of the same diameter (∼5 nm) and same distance from the membrane (∼7 nm center to center), wrapping helically around the inside of the cell membrane (Fig. 1C; see also Movie S1 in the supplemental material).
To test whether the helical filaments were a result of either overexpression of MreB from a plasmid or an artifact of the YFP tag, we imaged MC1000/pLE6 cells overexpressing wild-type (untagged) MreB to similar levels (see Fig. S1 in the supplemental material). Not a single filament was found in any of the 15 cells imaged (Fig. 1D), so we conclude that the helical filaments are artifacts of the tag.
As another example of a cell harboring a tagged MreB, E. coli strain FB76 was also imaged. In this cell, the chromosomal MreB gene was replaced with MreB-RFPSW, where the fluorescent protein is inserted into an internal loop rather than the N terminus, and expressed from the wild-type promoter (1). No filaments were seen along the membrane in 13 cells (Fig. 1E).
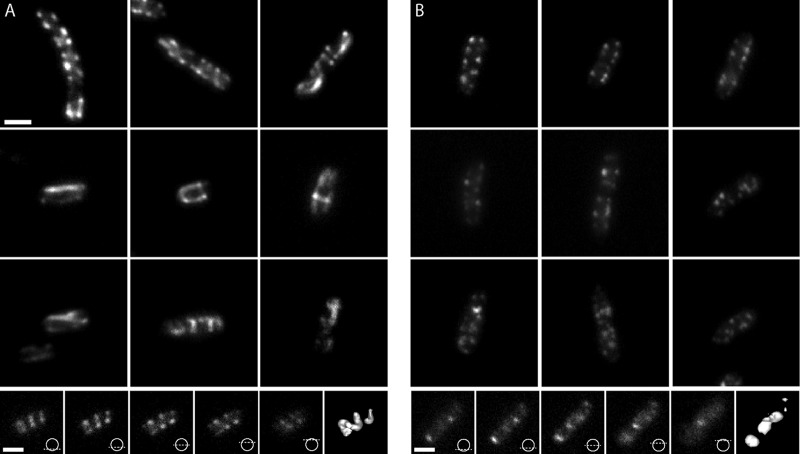
The possession of one tagged strain that produced extended filaments and a second tagged strain that did not allowed us to explore how well these patterns could be distinguished by standard fluorescence microscopy (Fig. 3). MC1000/pLE7 showed a variety of fluorescence patterns, including straight and curved streaks in a wide variety of orientations as well as puncta and diffuse patches. Perhaps most importantly, focal stacks revealed unambiguous helical patterns along the membranes of many cells. While the majority was only partial helices (helical arcs not completely surrounding the cell), others completed one or more full rotations around the cell, and a small number extended the full length of the cell (Fig. 3A, bottom row; see also Movie S2 in the supplemental material). In such cases, the vertical portions of the helix appeared as puncta along the membrane, but they were clearly connected by cross-streaks at the top and bottom of the focal stack, oriented in different directions. Without any deconvolution or image processing, a simple “isosurface” rendering of the fluorescent signal in a focal stack revealed the helix in 3D (Fig. 3A, last panel). While these results largely agree with previous reports (27, 33), unbroken helical structures extending the length of the cell were rare, and in more than one hundred cells, no continuous loops (described previously as “figure 8” structures) were observed. The pitch of the helices and filaments, with respect to the long axis of the cell, was also inconsistent. The cell in the focal stack of Fig. 3A depicts relatively tight helical turns, for instance, but there were filaments with different pitches as well as orientations either parallel or perpendicular to the long axis of the cell (see Movie S2). In contrast, live-cell fluorescence microscopy of strain FB76 showed only disconnected patches of fluorescence distributed mostly around the periphery of cells, with no clear helices (Fig. 3B; see also Movie S2).
Fig 3.
Comparison of fluorescence patterns in strains MC1000/pLE7 and FB76. (A) Epifluorescence images of various MC1000/pLE7 cells overexpressing YFP-MreB. The top row shows the familiar punctate and helical streaks commonly shown in the literature. The two middle rows show a variety of other patterns seen. The bottom row shows the individual images of a focal stack through a cell with an extended helical fluorescent pattern. The diagrams illustrate where the focal plane was in the cell for each image. The rightmost panel is a 3D isosurface of the fluorescent signal in the focal stack, which is clearly helical. (B) Epifluorescence images of E. coli strain FB76. Individual cells in the top rows illustrate the puncta along the membrane that often look similar to cells in the top row of panel A. The middle two rows show a variety of patchy distributions seen in epifluorescence images of FB76. The patchy signal is seen consistently, and no long filamentous structures are visible as seen in MC1000/pLE7. For further comparison, the bottom row shows a focal stack through one cell and its corresponding isosurface, which is obviously not helical. Scale bars represent 1 μm.
DISCUSSION
While the first fluorescence images of MreB interpreted as helices were of Bacillus subtilis, the images of E. coli strain MC1000/pLE7 that soon followed were so compellingly helical that they came to be regarded by many as conclusive proof. Later images of MreB, for instance, in Caulobacter crescentus and Vibrio cholerae, have been less decisive (8, 28), but were nevertheless interpreted as helical, probably in part because of the strength of the E. coli data. More recent papers that claim MreB forms a helical cytoskeleton (including the striking helices seen by the superresolution imaging method RESOLFT [13, 35]) have continued to use strain MC1000/pLE7. Here we have shown that, at least in this case, MreB does in fact form extended helices, but they are artifacts of YFP fusion.
The helices were not simply made detectable to ECT by the additional mass of the YFP, since filaments were not seen in the MreB-RFPSW fusion (where not just many copies but every copy of MreB was fused to a fluorescent protein), and we and others have previously resolved other actin homologs and thin cytoskeletal filaments within intact bacteria by ECT (17, 18, 20, 23, 25, 29). While it is true that if MreB filaments were embedded within the membrane they would be masked to ECT, MreB is known to bind directly to the membrane and be clearly visible by ECT, at least in vitro (24).
In a very recent review by Chastanet and Carballido-Lopéz (3), the authors hypothesize that the reason MreB was first reported to form helices but then later reported to form moving patches was that MreB only formed long filaments when overexpressed or in late stages of cell growth. It was also suggested that image deconvolution may have made disconnected puncta or patches appear connected. Our findings suggest instead that the key is whether and where MreB is tagged. Deconvolution and the fact that punctate patterns can resemble cross sections through 3D helices has also probably added significantly to the confusion, however, especially in the interpretation of the immunofluorescence images that should have revealed the artifact (26, 33). The conclusions of many papers need to be revisited in light of our findings that punctate patterns are not reliable evidence of helices.
The N-terminal YFP tag clearly disrupts function, since the YFP-MreB fusion only partially recovers cell shape in MreB mutant cells (27), but it remains unclear why it causes MreB to form long helical filaments. Based on the recent finding that fluorescent proteins can themselves multimerize, and thereby artifactually localize proteins into foci (19), one possibility is that YFP itself promotes polymerization through self-association. If so, the effect is not YFP specific, however, because when the YFP tag was replaced by a reversibly photoswitchable enhanced GFP (rsEGFP) in the RESOLFT study (13), extended helices still formed. Instead, the YFP tag likely disrupts MreB's natural interactions with the membrane and this somehow promotes polymerization. In MC1000/pLE7, YFP is attached to the N terminus, and recent work has established that E. coli MreB interacts directly with the membrane via an N-terminal amphipathic helix (24). When the helix was truncated, MreB again formed long filaments, but they were straight and unassociated with the membrane. With the N-terminal tag, the filaments in this study were found ∼7 nm away from the membrane (center to center). Assuming MreB and the membrane were in direct contact as modeled by Salje et al. (24), one would expect a center-to-center distance of ∼5 nm. It is therefore reasonable to imagine that a single YFP molecule (diameter, 2.5 nm) could have further displaced the filament, but in a way that does not completely disassociate it from the membrane. In this context, it is interesting to note that in the early study claiming MreB formed extended helices in B. subtilis, “the clearest and most reproducible” helical patterns were of N-terminally tagged (c-myc) MreB (16).
The structure and function of native MreB polymers inside cells unfortunately remain unclear. While we previously ruled out the possibility of MreB forming extended helices (29), the electron and light microscopy data presented here of E. coli strain FB76 are compatible with the idea that native MreB exists in small patches or short filaments that move around the cell circumference as part of the cell wall synthetic machinery (6, 10, 32). MreB may link other proteins in the cell wall synthesis complex together, perhaps creating bridges between nascent glycan strands that add rigidity to the cell (34).
Supplementary Material
ACKNOWLEDGMENTS
We thank L. Rothfield for E. coli strains MC1000/pLE7 and MC1000/pLE6. We also thank Piet De Boer for E. coli strain FB76.
This work was supported by NIH grant R01 GM094800B to G.J.J.
Footnotes
Published ahead of print 17 August 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Bendezú FO, Hale CA, Bernhardt TG, de Boer PAJ. 2009. RodZ (YfgA) is required for proper assembly of the MreB actin cytoskeleton and cell shape in E. coli. EMBO J. 28:193–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Briegel A, et al. 2006. Multiple large filament bundles observed in Caulobacter crescentus by electron cryotomography. Mol. Microbiol. 62:5–14 [DOI] [PubMed] [Google Scholar]
- 3. Chastanet A, Carballido-Lopéz R. 2012. The actin-like MreB proteins in Bacillus subtilis: a new turn. Front. Biosci. (Schol. Ed.) 4:1582–1606 [DOI] [PubMed] [Google Scholar]
- 4. Dempwolff F, Reimold C, Reth M, Graumann PL. 2011. Bacillus subtilis MreB orthologs self-organize into filamentous structures underneath the cell membrane in a heterologous cell system. PLoS One 6:e27035 doi:10.1371/journal.pone.0027035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Divakaruni AV, Baida C, White CL, Gober JW. 2007. The cell shape proteins MreB and MreC control cell morphogenesis by positioning cell wall synthetic complexes. Mol. Microbiol. 66:174–188 [DOI] [PubMed] [Google Scholar]
- 6. Domínguez-Escobar J, et al. 2011. Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science 333:225–228 [DOI] [PubMed] [Google Scholar]
- 7. Draper O, et al. 2011. MamK, a bacterial actin, forms dynamic filaments in vivo that are regulated by the acidic proteins MamJ and LimJ. Mol. Microbiol. 82:342–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Figge RM, Divakaruni AV, Gober JW. 2004. MreB, the cell shape-determining bacterial actin homologue, co-ordinates cell wall morphogenesis in Caulobacter crescentus. Mol. Microbiol. 51:1321–1332 [DOI] [PubMed] [Google Scholar]
- 9. Gan L, Jensen GJ. 2012. Electron tomography of cells. Q. Rev. Biophys. 45:27–56 [DOI] [PubMed] [Google Scholar]
- 10. Garner EC, et al. 2011. Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science 333:222–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gitai Z, Dye NA, Reisenauer A, Wachi M, Shapiro L. 2005. MreB actin-mediated segregation of a specific region of a bacterial chromosome. Cell 120:329–341 [DOI] [PubMed] [Google Scholar]
- 12. Gitai Z, Dye N, Shapiro L. 2004. An actin-like gene can determine cell polarity in bacteria. Proc. Natl. Acad. Sci. U. S. A. 101:8643–8648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Grotjohann T, et al. 2011. Diffraction-unlimited all-optical imaging and writing with a photochromic GFP. Nature 478:204–208 [DOI] [PubMed] [Google Scholar]
- 14. Ingerson-Mahar M, Briegel A, Werner JN, Jensen GJ, Gitai Z. 2010. The metabolic enzyme CTP synthase forms cytoskeletal filaments. Nat. Cell Biol. 12:739–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jennings PC, Cox GC, Monahan LG, Harry EJ. 2010. Super-resolution imaging of the bacterial cytokinetic protein FtsZ. Micron [Epub ahead of print.] doi:10.1016/j.micron.2010.09.003 [DOI] [PubMed] [Google Scholar]
- 16. Jones LJ, Carballido-López R, Errington J. 2001. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104:913–922 [DOI] [PubMed] [Google Scholar]
- 17. Komeili A, Li Z, Newman DK, Jensen GJ. 2006. Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science 311:242–245 [DOI] [PubMed] [Google Scholar]
- 18. Kühn J, et al. 2010. Bactofilins, a ubiquitous class of cytoskeletal proteins mediating polar localization of a cell wall synthase in Caulobacter crescentus. EMBO J. 29:327–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Landgraf D, Okumus B, Chien P, Baker TA, Paulsson J. 2012. Segregation of molecules at cell division reveals native protein localization. Nat. Methods 9:480–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li Z, Trimble MJ, Brun YV, Jensen GJ. 2007. The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. EMBO J. 26:4694–4708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nan B, Zusman DR. 2011. Uncovering the mystery of gliding motility in the myxobacteria. Annu. Rev. Genet. 45:21–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Peters PC, Migocki MD, Thoni C, Harry EJ. 2007. A new assembly pathway for the cytokinetic Z ring from a dynamic helical structure in vegetatively growing cells of Bacillus subtilis. Mol. Microbiol. 64:487–499 [DOI] [PubMed] [Google Scholar]
- 23. Pilhofer M, Ladinsky MS, McDowall AW, Petroni G, Jensen GJ. 2011. Microtubules in bacteria: ancient tubulins build a five-protofilament homolog of the eukaryotic cytoskeleton. PLoS Biol. 9:e1001213 doi:10.1371/journal.pbio.1001213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Salje J, van den Ent F, de Boer P, Löwe J. 2011. Direct membrane binding by bacterial actin MreB. Mol. Cell 43:478–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Scheffel A, et al. 2006. An acidic protein aligns magnetosomes along a filamentous structure in magnetotactic bacteria. Nature 440:110–114 [DOI] [PubMed] [Google Scholar]
- 26. Shih Y-L, Kawagishi I, Rothfield L. 2005. The MreB and Min cytoskeletal-like systems play independent roles in prokaryotic polar differentiation. Mol. Microbiol. 58:917–928 [DOI] [PubMed] [Google Scholar]
- 27. Shih Y-L, Le T, Rothfield L. 2003. Division site selection in Escherichia coli involves dynamic redistribution of Min proteins within coiled structures that extend between the two cell poles. Proc. Natl. Acad. Sci. U. S. A. 100:7865–7870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Srivastava P, Demarre G, Karpova TS, McNally J, Chattoraj DK. 2007. Changes in nucleoid morphology and origin localization upon inhibition or alteration of the actin homolog, MreB, of Vibrio cholerae. J. Bacteriol. 189:7450–7463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Swulius MT, et al. 2011. Long helical filaments are not seen encircling cells in electron cryotomograms of rod-shaped bacteria. Biochem. Biophys. Res. Commun. 407:650–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Taghbalout A, Rothfield L. 2008. RNaseE and RNA helicase B play central roles in the cytoskeletal organization of the RNA degradosome. J. Biol. Chem. 283:13850–13855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Taghbalout A, Rothfield L. 2007. RNaseE and the other constituents of the RNA degradosome are components of the bacterial cytoskeleton. Proc. Natl. Acad. Sci. U. S. A. 104:1667–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. van Teeffelen S, et al. 2011. The bacterial actin MreB rotates, and rotation depends on cell-wall assembly. Proc. Natl. Acad. Sci. U. S. A. 108:15822–15827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vats P, Rothfield L. 2007. Duplication and segregation of the actin (MreB) cytoskeleton during the prokaryotic cell cycle. Proc. Natl. Acad. Sci. U. S. A. 104:17795–17800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang S, Arellano-Santoyo H, Combs PA, Shaevitz JW. 2010. Actin-like cytoskeleton filaments contribute to cell mechanics in bacteria. Proc. Natl. Acad. Sci. U. S. A. 107:9182–9185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang S, Furchtgott L, Huang KC, Shaevitz JW. 2012. Helical insertion of peptidoglycan produces chiral ordering of the bacterial cell wall. Proc. Natl. Acad. Sci. U. S. A. 109:E595–E604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. White CL, Kitich A, Gober JW. 2010. Positioning cell wall synthetic complexes by the bacterial morphogenetic proteins MreB and MreD. Mol. Microbiol. 76:616–633 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.