Abstract
Transcription factors regulate eukaryotic RNA polymerase II (Pol II) activity by assembling and remodeling complexes at multiple steps in the transcription cycle. In HIV, we previously proposed a two-step model where the viral Tat protein first preassembles at the promoter with an inactive P-TEFb:7SK snRNP complex and later transfers P-TEFb to TAR on the nascent transcript, displacing the inhibitory snRNP and resulting in Pol II phosphorylation and stimulation of elongation. It is unknown how the Tat:P-TEFb complex transitions to TAR to activate the P-TEFb kinase. Here, we show that P-TEFb artificially recruited to the nascent transcript is not competent for transcription but rather remains inactive due to its assembly with the 7SK snRNP. Tat supplied in trans is able to displace the kinase inhibitor Hexim1 from the snRNP and activate P-TEFb, thereby uncoupling Tat requirements for kinase activation and TAR binding. By combining comprehensive mutagenesis of Tat with multiple cell-based reporter assays that probe the activity of Tat in different arrangements, we genetically defined a transition step in which preassembled Tat:P-TEFb complexes switch to TAR. We propose that a conserved network of residues in Tat has evolved to control this transition and thereby switch the host elongation machinery to viral transcription.
INTRODUCTION
The assembly of RNA polymerase II (Pol II) transcription complexes is a dynamic process, which is controlled by transcriptional activators at multiple steps of the transcription cycle (7, 30, 98, 117). Activators that function during initiation typically possess a DNA-binding domain for promoter-specific recruitment and an activation domain (AD) that mediates interactions with the basal transcription machinery, coactivators/corepressors, or chromatin-remodeling factors (33, 63, 71, 98). Some activators function during elongation and may assemble into basal transcription complexes to generate processive complexes that elongate without premature pausing, such as Sp1 and CTF (7, 8); assemble at paused transcription complexes to stimulate subsequent elongation, such as bacteriophage λ Q protein and eukaryotic factor SII (87, 88, 115); or assemble on newly initiated transcripts to read through subsequent pause sites, such as bacteriophage λ N protein, c-Myc, and HIV Tat (14, 16, 60, 84, 86).
Studies of HIV Tat, which regulates elongation of the viral promoter (14, 29, 34, 79), have provided key insights into the host elongation machinery, largely through the discovery of one of its cofactors, positive transcription elongation factor (P-TEFb). P-TEFb, composed of a cyclin subunit (CycT1, -T2a, or -T2b) and a kinase (Cdk9) (64, 111, 121), is recognized as a global regulator that overcomes Pol II pausing at promoter-proximal regions (13, 26, 41, 72, 79, 81, 82, 94). Unlike DNA-binding transcription factors, Tat utilizes an RNA-binding domain (RBD) (residues 49 to 57) to contact the TAR stem-loop located at the 5′ end of nascent viral pre-mRNAs and uses its AD (residues 1 to 48) to recruit P-TEFb (CycT1:Cdk9) to TAR, where Cdk9 phosphorylates the Pol II carboxy-terminal domain (CTD) and elongation factors, such as negative elongation factor (NELF) and the 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB) sensitivity-inducing factor (DSIF), to stimulate processivity (79, 81, 110, 113). The timing appears to be governed by the interaction of the Tat:P-TEFb complex with RNA, similar to how pre-mRNA-processing factors are timed to function at specific RNA sites during the coupling of transcription elongation and RNA processing (21, 32, 109).
In addition to interacting with Tat and TAR, P-TEFb exists in an inactive form bound to the 7SK snRNP, which is composed of Hexim1, Larp7, and Mepce proteins and the noncoding 7SK snRNA (48, 53, 58, 66, 74, 116, 120). Tat competes with the kinase inhibitor Hexim1 to release the active form of P-TEFb (2, 66, 96) and recently has been found associated with the 7SK snRNP complex (21, 70, 101). Interestingly, these Tat:P-TEFb:7SK snRNP complexes were found assembled at the HIV promoter, with the inhibitory snRNP ejected in a Tat-TAR-dependent manner (1, 21). This led to a model in which P-TEFb is held inactive by the 7SK snRNP in paused Pol II complexes until Tat mediates the transfer of P-TEFb to TAR and subsequent kinase activation. Although the molecular details are not yet clear, there is evidence that Tat may utilize two aspects of molecular mimicry during the activation process: first, the Tat AD appears to compete with Hexim1 for a shared binding surface on CycT1, and second, the Tat RBD appears to recognize a TAR-like motif (GA-UC) within the 5′ stem-loop of 7SK snRNA, as well as an additional feature in the 3′-end stem-loop (2, 25, 53, 54, 70, 95). Thus, while Tat alone is able to displace Hexim1 and extract P-TEFb from the 7SK snRNP complex in vitro and in vivo (2, 21, 54), the Tat-TAR interaction likely also plays a significant role in the context of actively transcribing complexes in vivo. Indeed, we speculated that an intermediate step exists in which Tat does not activate the P-TEFb kinase until the Tat:P-TEFb complex is transferred to TAR, a step that has not been recapitulated in vitro (21, 54).
To uncover intermediate steps during Tat activation, we utilized a set of cell-based reporter assays designed to probe various arrangements of HIV transcription complexes, in part based on the principles of artificial recruitment (50, 83), together with systematic mutagenesis of the Tat AD to identify genetically separable activities. This approach identified residues in Tat required for the TAR-dependent and -independent steps of the viral transcription cycle, which can be interpreted in the context of the recent Tat:P-TEFb crystal structure (107). The results suggest that adaptive conformational changes in Tat:P-TEFb are used to transition HIV transcription complexes bound at the promoter into the active nascent RNA-bound form. In particular, it appears that the N-terminal region of Tat, in close proximity to the Cdk9 T loop, may stabilize the P-TEFb kinase upon CycT1 interaction, while a zinc finger motif (ZnF2) containing the evolutionarily conserved Tyr26 and acetylatable Lys28 residues (20, 22) is needed to assemble the Tat:P-TEFb complex on TAR RNA, but not at the promoter. These results are consistent with a model in which HIV transcription complexes are remodeled into an elongation-competent form via the Tat-TAR interaction and concomitant release of the 7SK snRNP.
MATERIALS AND METHODS
Cell culture, plasmids, and transcription reporter assays.
HeLa, 293, and 293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS) at 37°C with 5% CO2. Site-directed Tat mutants were made using 100 ng of the relevant plasmid, 5′-phosphorylated oligonucleotides, and High Fidelity Turbo Pfu (Stratagene). Primer sequences used in the mutagenesis experiments will be provided upon request. For the reporter assays, HeLa or 293T cells were transfected using Polyfect (Qiagen) with 25 ng of a firefly luciferase (FFL) reporter plasmid and 1 ng of a cytomegalovirus (CMV)-Renilla (RL) luciferase plasmid in 48-well plates. Activator-expressing plasmids were transfected in varying amounts, as indicated in the figures. All point mutants were tested at five plasmid concentrations to determine the linear range of the assay, and only values within the linear range were used. In all cases, reporter activities are presented as fold activation relative to reporter alone and normalized to RL and are averages of three experiments. Luciferase levels were measured 40 to 44 h posttransfection using the Dual-Luciferase Reporter Assay (Promega).
RNA interference (RNAi) assays.
HeLa HIV RRE:FFL reporter cells (23) were plated to a density of 1.5 × 105 cells per well in 12-well plates and transfected with Larp7 small interfering RNA (siRNA) (target sequence, 5′-ACAAGCGAGUAAACAUAUA-3′; Dharmacon) at a final concentration of 100 μM, or with pBS vector, using Lipofectamine 2000 (Invitrogen) for 42 h. The cells were next transfected with a Strep-tagged Tat plasmid (or empty vector) using Polyjet (SignaGen) for 42 h. Cell lysates were analyzed in luciferase reporter assays for CycT1N-Rev activation and by Western blotting for Larp7 knockdown using P-TEFb/7SK snRNP subunits and β-actin antibodies (21).
Reverse transcription (RT)-PCR.
Total RNA from transfected HeLa cells was isolated using TRIzol (Invitrogen). First-strand cDNAs were synthesized using Superscript III (Invitrogen), and PCR was performed with a KOD kit (Novagen). We modified a previously described strategy to monitor transcription initiation and elongation using primers that bind to the HIV:RRE luciferase reporter plasmid (55). To monitor initiation, we used primers P1 (5′-TCTCTCTGGTTAGACCAGTG-3′) and P4 (5′-AACGCACACCGGCCTTATTCC-3′), which amplified a promoter-proximal 122-bp fragment. To monitor elongation, we used primers P2 (5′-AGTGGGCGCAGCGTCAATGAC-3′) and P4 (5′-AACGCACACCGGCCTTATTCC-3′), which amplified a 229-bp fragment. 18S rRNA was amplified using gene-specific primers and an 18S quantum mRNA standard for internal controls (Ambion).
Affinity purifications, coimmunoprecipitation assays, and Western blots.
We used 293 cells for protein-protein interaction experiments because protein expression levels were higher than in HeLa cells. 293 cells (3.5 × 106 to 4.0 × 106) were plated into 10-cm dishes ≥24 h before transfecting 4 to 8 μg DNA using Polyjet (SignaGen). The cells were washed with 1× PBS 38 to 40 h posttransfection, suspended in 1× hypotonic lysis buffer (10 mM HEPES-KOH, pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol [DTT], cOmplete Mini EDTA-free protease inhibitor cocktail [Roche] supplemented with 0.1% Igepal CA-630), and immediately centrifuged (13,500 × g for 5 min at 4°C) to separate nuclear and cytoplasmic fractions. The nuclear pellet was extracted in 20 mM HEPES-KOH, pH 7.9, 1.5 mM MgCl2, 0.34 M NaCl, 0.2 mM EDTA, 25% (vol/vol) glycerol, 0.5 mM DTT, and cOmplete mini EDTA-free protease inhibitor cocktail (Roche) with shaking for 60 min. This crude nuclear extract was further diluted with ∼1.3 volumes of 20 mM HEPES-KOH, pH 7.9, 1.5 mM MgCl2, 0.2 mM EDTA, 0.18% NP-40, 0.5 mM DTT, and protease inhibitor cocktail; mixed for 30 min at 4°C; and centrifuged at 13,500 × g for 10 min to remove insoluble material. For the coimmunoprecipitations (IPs), nuclear extracts containing Flag-tagged or Strep-tagged proteins were incubated with 10 μl of EZview Red anti-flag M2 (Sigma) beads or Strep-Tactin Superflow beads (IBA), respectively, at 4°C for 2 h with rotation and washed 3 times with 0.5 ml wash buffer (20 mM HEPES-KOH, pH 7.9, 1.5 mM MgCl2, 0.15 M NaCl, 0.2 mM EDTA, 0.1% NP-40). Elutions were performed in 50 μl wash buffer containing 0.2 mg/ml 3× flag peptide (Sigma) or desthiobiotin (IBA), respectively, with shaking for 30 min at 4°C.
RNA was extracted from the immunoprecipitated material by proteinase K (Roche) treatment at 55°C for 2 h, phenol-chloroform extraction, and ethanol precipitation. The RNA was analyzed by SDS-PAGE and ethidium bromide staining. CycT1 immunoprecipitations showed one detectable RNase-sensitive species of about 330 nucleotides (nt) (7SK snRNA) as previously described (116). The identity of the immunoprecipitated RNA was verified by transferring the ethidium bromide-stained gel to zeta probe membranes (Bio-Rad) for 2 h at 500 mA and hybridizing to a 7SK probe corresponding to the first 100 nt of stem-loop I.
For Western blots, immunoprecipitated samples were electrophoresed on 4 to 15% gradient TGX gels (Bio-Rad) and transferred to polyvinylidene difluoride (PVDF) (Bio-Rad) or 0.45 μM nitrocellulose (Bio-Rad) membranes, blocked in PBS containing 0.2% Tween 20 and 5% nonfat milk for 1 h, and incubated with primary antibodies from 1 h to overnight at 4°C. The monoclonal anti-Strep antibody was horseradish peroxidase (HRP) conjugated (Millipore). Primary antibodies were reported previously (21), except Larp7 (AV40847; Sigma). Secondary antibodies coupled to HRP, donkey anti-rabbit IgG–HRP (sc-2313), goat anti-mouse IgG–HRP (sc-2005), and donkey anti-goat IgG–HRP (sc-2020) (Santa Cruz Biotechnologies) were incubated at 1:10,000 dilutions for 1 h; the blots were developed using Supersignal West Pico or West Femto chemiluminescent substrates (ThermoFisher).
ChIP assays.
Chromatin immunoprecipitation (ChIP) assays were performed as previously described (21) using a HeLa cell line containing the HIV:RRE promoter driving FFL expression transfected with CycT1N-Rev in the absence or presence of Tat AD using calcium phosphate. The cells were incubated for 36 h and washed in PBS before cross-linking and lysis (23).
Clustering analysis.
Heat maps representing the activities of Tat mutants in each transcription reporter assay were generated using MATLAB version 7.4.0 (MathWorks, Natick, MA). Data were clustered using a Euclidean-distance metric, and centroid linkage was performed with the program Cluster 3.0 (18). Clusters were formed among nodes with correlation thresholds of 0.90 or greater. Residue identity was calculated using an in-house Python script based on the multiple-sequence alignment of Tat proteins from 1,496 HIV/SIVcpz isolates (http://www.hiv.lanl.gov; 2009). Pearson correlation coefficients (r) between all pairwise assays were calculated using Excel software (Microsoft).
RESULTS
Reporter assays to detect intermediates during Tat activation.
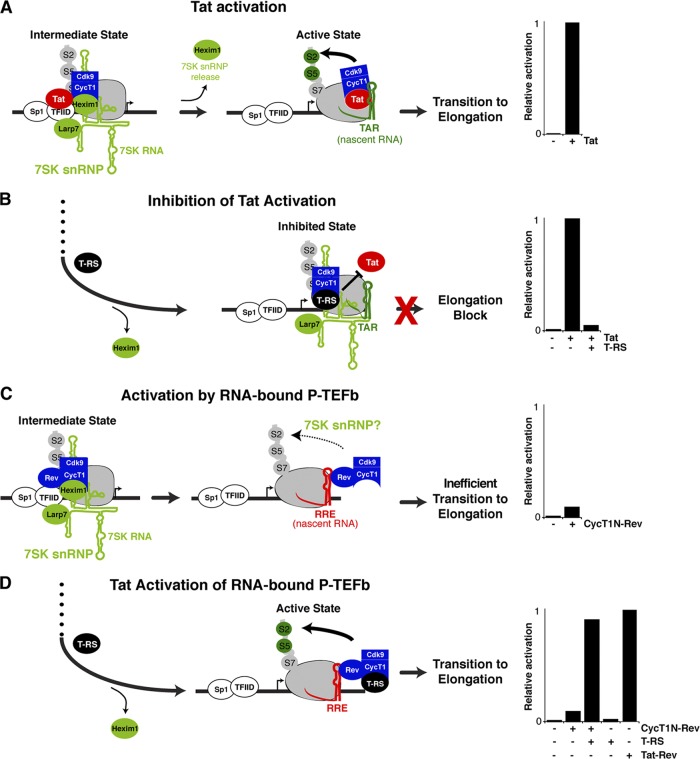
Our current model of Tat activation proposes that P-TEFb is initially recruited to the HIV promoter in a kinase-inactive state due to its association with the 7SK snRNP. Following Tat:P-TEFb preassembly at the promoter, the kinase is subsequently activated upon P-TEFb transfer to TAR on the nascent RNA. During this transition, release of inhibitory factors, Hexim1 and 7SK snRNA, triggers Cdk9 phosphorylation of the Pol II CTD and elongation factors, facilitating escape into productive transcription elongation (Fig. 1A) (21, 35, 94). Transcription is an inherently dynamic process requiring the coupling of various steps; therefore, it is difficult to define intermediate states that may arise as Tat:P-TEFb complexes assembled at the promoter transfer to TAR to form elongation-competent complexes. To address this problem, we have devised additional cell-based reporter assays that complement the original Tat activation assay (Fig. 1A). These alternate assays examine Tat:P-TEFb complex assembly at the promoter and at the nascent RNA in TAR-independent contexts, thereby decoupling the Tat requirements for P-TEFb kinase activation from TAR association during transcriptional activation (Fig. 1B to D). Although the Tat activation assay constitutes all the steps required for transcription activation, we propose that differences in activities between the assays may help delineate discrete steps.
Fig 1.
Reporter assays and proposed models for Tat activation in several tethered arrangements. (A) Tat activation refers to the activity of Tat in its wild-type arrangement using an HIV long terminal repeat (LTR) FFL reporter. The model shown is based on a recently proposed mechanism in which Tat activates the Cdk9 kinase by transferring P-TEFb from an inactive promoter-bound state, together with Sp1 and TFIID, to the active TAR-bound state in which Cdk9 phosphorylates the CTD of Pol II at serines 5 and 2 (S5 and S2), as well as other elongation factors (not shown for simplicity) (21). The bar graphs (A to D, right) show activation levels normalized to wild-type Tat activity (see Materials and Methods). (B) Inhibition of Tat activation refers to assays in which the Tat AD, alone or as part of the T-RS fusion, functions as a dominant-negative inhibitor that prevents P-TEFb transfer to the nascent RNA and consequently blocks elongation (23). (C) Activation by RNA-bound P-TEFb refers to assays in which P-TEFb is directly tethered to the nascent RNA by a heterologous protein-RNA (Rev-RRE) interaction (57), in this case, the N-terminal domain of CycT1 fused to Rev (CycT1N-Rev) on an HIV:RRE reporter (Table 1). The activity in this case is substantially lower than expected, possibly reflecting association with the 7SK snRNP complex (see the text). (D) Tat activation of RNA-bound P-TEFb refers to assays in which Tat AD or T-RS in trans is able to stimulate the activity of the CycT1N-Rev fusion used in panel C to wild-type Tat levels. The same reporter can be activated by Tat-Rev (a fusion of the Tat AD to the Rev protein) artificially tethered to the nascent RNA through the Rev:RRE protein-RNA interaction (23, 97, 102).
In the first assay (inhibition of Tat activation) (Fig. 1B), an RBD-deficient Tat protein (T-RS) excludes wild-type Tat from the promoter by preferentially assembling with P-TEFb through the Tat AD but cannot facilitate transfer of P-TEFb to TAR, thus blocking transition to elongation (23). Therefore, this reporter assay monitors Tat:P-TEFb preassembly at the promoter irrespective of TAR and Tat RNA-binding activity. Comparison between inhibition of Tat activation and the standard Tat activation assay (Fig. 1A) allows the uncoupling of Tat:P-TEFb requirements for promoter assembly versus transition to the nascent RNA and thus may define a step prior to TAR binding (21, 23, 43, 68).
A second assay (activation by RNA-bound P-TEFb) (Fig. 1C) was designed to test if the elongation block imposed by loss of the Tat RBD can be overcome by artificially tethering P-TEFb to the nascent RNA (Tat bypass), thereby completely eliminating the need for Tat, as well as TAR. For this assay, the N-terminal cyclin box of CycT1 (the minimal domain required for Tat activity [40]) was fused to HIV Rev (CycT1N-Rev) and used to activate an HIV promoter derivative (HIV:RRE) in which TAR was replaced by RRE (Table 1). Relatively weak activity (≤5-fold activation) (Fig. 1C) suggested that Tat is indispensable for activation, although P-TEFb is bound to the nascent RNA.
Table 1.
Transcription reporter and activator combinations used in all cell-based assays
| Promoter reporter | DNA or RNA element | TAR-dependent Tat:P-TEFb activation | Activator | Cell-based assay |
|---|---|---|---|---|
| HIV | HIV TAR | Yes | Tat | Tat activation |
| HIV | HIV TAR | Yes | Tat + T-RS, Tat AD | Inhibition of Tat activation |
| HIV:RRE | HIV RRE | No | Tat-Rev | TAR-independent Tat activation |
| HIV:RRE | HIV RRE | No | P-TEFb–Rev | Activation by RNA-bound P-TEFb |
| HIV:RRE | HIV RRE | No | P-TEFb–Rev + Tat, Tat AD or T-RS | Tat activation of RNA-bound P-TEFb |
| HIV:MS2 | MS2 phage | No | P-TEFb–MS2cp | Activation by RNA-bound P-TEFb |
| Gal4:HIV:RRE | Yeast Gal4 and HIV RRE | No | Gal4-Tat, Gal4–P-TEFb, Tat-Rev | |
| HIV | HIV TAR | Yes | HJ Tat | P-TEFb-dependent TAR-binding Tat activation |
| HIV:BIV | BIV TAR | No | HJ Tat | P-TEFb-dependent TAR-binding Tat activation |
Since RNA-bound P-TEFb alone only modestly activated transcription (Fig. 1C), in a third assay (Tat activation of RNA-bound P-TEFb) (Fig. 1D), we coexpressed T-RS with CycT1N-Rev or Cdk9-Rev and found that T-RS no longer functioned as an inhibitor, but rather, synergistically stimulated transcription when P-TEFb was tethered to RNA (Fig. 1D and data not shown). Tat is known to activate transcription when tethered to the nascent transcript through another protein (Rev), using a heterologous protein-RNA interaction (Tat-Rev–RRE) (23, 102). In this context, the Tat-Rev fusion activates transcription to a level comparable to CycT1N-Rev and T-RS, thus highlighting the importance of the Tat AD, but not the RBD, during Tat activation of RNA-bound P-TEFb (Fig. 1D). Since full-length Tat or the Tat AD also stimulated transcription of RNA-bound P-TEFb to the same level as T-RS (data not shown), we conclude that the RS moiety does not alter Tat AD function and stimulation is not a consequence of the artificial T-RS fusion (23); therefore, we use fused and unfused Tat interchangeably throughout the paper.
Our results indicate that P-TEFb recruitment to the nascent transcript is insufficient to recapitulate Tat activation (5, 36, 42), as the Tat AD is required to fully activate the P-TEFb kinase. These results support a model in which inactive P-TEFb complexes are loaded at the HIV promoter and inhibitory factors are released upon Tat-TAR binding, upon delivery of the Tat AD to the nascent RNA through a heterologous protein-RNA interaction, or by the Tat AD in trans through protein-protein interactions with P-TEFb bound to the nascent RNA (Fig. 1D) (21). We present additional experimental evidence demonstrating that Tat-mediated disassembly of 7SK snRNP from P-TEFb underlies activation by RNA-bound P-TEFb (see below).
Tat activates transcription elongation in trans through P-TEFb bound to the nascent RNA.
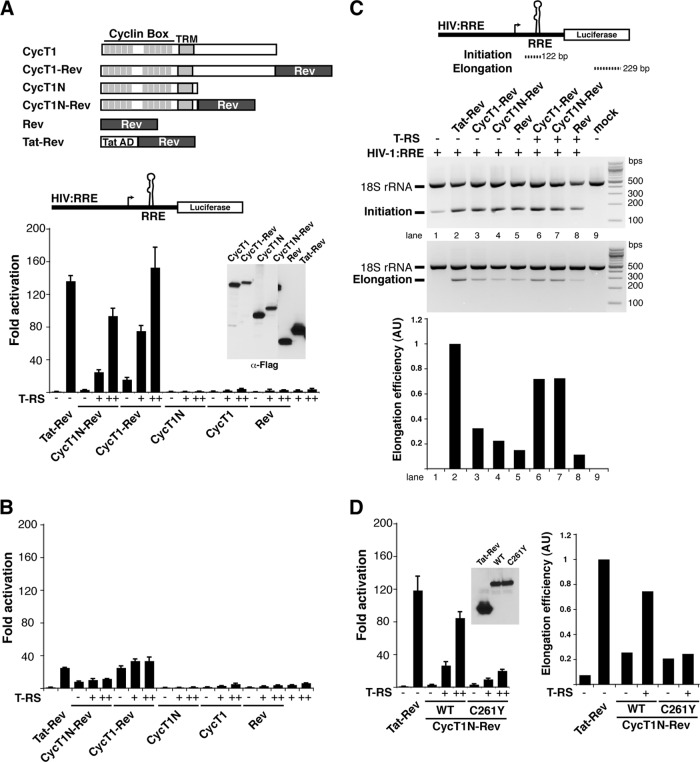
Two previous reports suggested that the sole function of Tat is to recruit P-TEFb to the nascent RNA (5, 85); however, we observed only weak activity with P-TEFb artificially recruited to the nascent RNA alone (Fig. 1C) and, surprisingly, substantial stimulation with the addition of Tat AD (Fig. 1D). Since our experiments were performed in HeLa cells using the N-terminal domain of CycT1 fused to Rev (CycT1N-Rev) whereas both previous studies measured activities in 293T cells using full-length CycT1-Rev fusions, we compared CycT1N-Rev and CycT1-Rev activation in HeLa cells and observed only slightly higher activity with full-length CycT1 alone (∼5-fold) and, as with the N-terminal fragment, substantial stimulation with Tat AD (Fig. 2A). Conversely, Tat and P-TEFb artificially recruited to the nascent RNA through the Rev-RRE interaction demonstrated much lower activation in 293T cells than in HeLa cells (Fig. 2B), likely reflecting activation from the more tightly repressed basal state in HeLa cells (∼35-fold less luciferase for the reporter alone), which have low NF-κB levels (4, 114). Since P-TEFb artificially tethered to the transcript still requires Tat for efficient activation, we explored the mechanism in greater detail to uncover TAR-dependent and -independent steps utilized by the Tat:P-TEFb complex.
Fig 2.
Tat activates RNA-bound P-TEFb at the transcription elongation step. (A) Schematic of the CycT1 and Rev fusions and their activities on an HIV:RRE luciferase reporter. N in the CycT1 nomenclature refers to the fully active N-terminal cyclin box (40). Also shown is a Western blot probed against the C-terminal Flag tag of each protein to show relative steady-state expression levels (α-Flag). HeLa cells were transfected with 25 ng of reporter plasmid, 1 ng of the CycT1 or Rev plasmid, and 0.1 ng (+) or 1 ng (++) of T-RS. The error bars indicate standard deviations. (B) The same experiment as in panel A, conducted in 293T cells. Activation in the 293T cells is substantially lower than in HeLa cells and reflects the higher basal activity of the promoter (41,000 ± 6,700 versus 1,200 ± 21 relative luciferase units, which represents ∼25-fold activation versus ∼135-fold). (C) Schematic of RT-PCR products used to assess initiation and elongation. The gel shows the products resulting from transfection of HeLa cells with the indicated constructs, and the graph plots the calculated elongation efficiencies expressed in arbitrary units (AU) (22). An 18S rRNA fragment was coamplified as an internal control. (D) Activity of a CycT1 mutant (C261Y) that weakens the interaction with Tat (11, 17, 39) in HeLa cells, using the same activation and RT-PCR assays as for panels A and C.
To test whether Tat activation of RNA-bound P-TEFb recapitulates the known function of Tat in elongation, we measured initiating and elongating transcripts and found that both CycT1N-Rev and CycT1-Rev alone only slightly stimulated elongation (Fig. 2C). Only simultaneous expression of Tat AD and CycT1N-Rev or CycT1-Rev stimulated elongation to levels comparable to those of the Tat-Rev control on the same HIV:RRE derivative promoter (Fig. 2C). Mutation of the Tat-TAR recognition motif (TRM) of CycT1 at Cys261 (17, 22, 40, 107), a known region of Tat AD interaction that affects direct protein-protein interactions (11, 17, 22, 40), decreased transcriptional stimulation by the Tat AD ∼4-fold and transcription elongation rates (Fig. 2D), corresponding to inefficient displacement of Hexim1, the P-TEFb kinase inhibitor, from 7SK snRNP complexes (see below). Thus, although bypassing the Tat-TAR interaction (22), Tat activation of RNA-bound P-TEFb mirrors native transcription elongation by Tat (29) and thus constitutes a unique system to uncover Tat:P-TEFb requirements during complex preassembly and transfer to TAR RNA.
RNA and promoter requirements for Tat activation of RNA-bound P-TEFb are identical to those for native Tat activation.
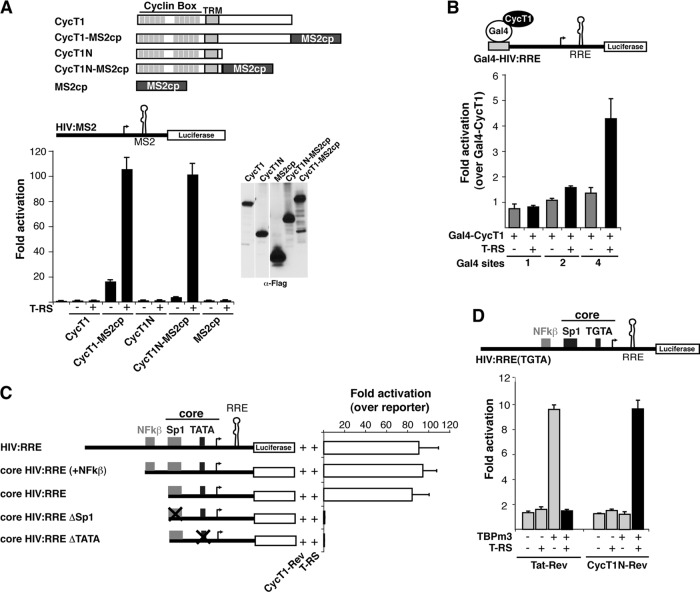
Since Tat activation of RNA-bound P-TEFb significantly reconfigures the mechanism of activation, we investigated whether the same requirements for activation still applied. First, to examine the RNA requirements for Tat activation of P-TEFb when bound to the nascent RNA, we replaced the Rev-RRE interaction used to tether P-TEFb to the RNA in the previous reporters with the MS2cp-MS2 (MS2 coat protein-MS2 RNA) interaction and observed modest (3- to 10-fold) activation by CycT1-MS2cp chimeras without Tat, none by the unfused constructs, and substantial stimulation by the Tat AD (Fig. 3A). These results are identical to those observed using Rev-RRE tethering (Fig. 1 and 2), suggesting that the nature of the protein-RNA interaction used to recruit P-TEFb to the nascent RNA is irrelevant for activation by the Tat AD in trans.
Fig 3.
Tat activates P-TEFb bound to RNA but not DNA and uses basal transcription elements. (A) Activation of RNA-bound P-TEFb through the MS2 protein-RNA interaction by T-RS in HeLa cells. The cells were transfected with 25 ng of an HIV:MS2 reporter plasmid, 1 ng of the CycT1 or MS2cp plasmid, and 1 ng of T-RS. The Western blot shows steady-state expression levels of C-terminally Flag-tagged proteins. The error bars indicate standard deviations. (B) Activation of DNA-bound P-TEFb, using Gal4:HIV:RRE reporters containing 1, 2, or 4 Gal4-binding sites and Gal4-CycT1 and T-RS under the same conditions as for panel A. Tat-Rev activates this reporter through the RRE by 85-fold ± 4.7-fold (data not shown). (C) Sp1 and TATA elements, but not NF-κB sites, are required for Tat activation of RNA-bound P-TEFb. The HIV:RRE reporters shown were cotransfected with 1 ng of CycT1N-Rev and 1 ng of T-RS, and activation levels were normalized to a cotransfected CMV-RL plasmid. (D) Requirement for TBP using an altered-specificity mutant. Activation assays were performed as for panel C using an HIV:RRE reporter containing a mutant TATA box (TGTA) complemented with a TBP mutant (TBPm3) that has enhanced binding specificity for the mutated TATA box (105).
While tethering P-TEFb to the promoter appears to be an important component of Tat activation of RNA-bound P-TEFb, we asked whether the specific nature of P-TEFb recruitment was crucial for Tat activation. When CycT1 was directly bound to the DNA template via a Gal4 DNA-binding domain fusion (85, 103), we observed only ∼3.5-fold stimulation by the Tat AD even when four Gal4 sites were present (Fig. 3B), whereas tethering Tat to the nascent transcript via the Rev-RRE interaction on the same reporter resulted in strong activation (∼80-fold) (data not shown). Thus, Tat shows strong activation only when P-TEFb is recruited to the nascent RNA but not to DNA, consistent with the hypothesis that Tat activates the P-TEFb kinase when transferred to nascent RNA, but not when it is still bound to the initiating and/or preelongation Pol II complex (21). Similarly, Tat delivered as Gal4-Tat to the viral promoter requires multiple Gal4-binding sites to achieve levels of activation above basal activity, albeit much less than native Tat activation (reference 103 and data not shown), and thus apparently does not activate the P-TEFb kinase well in this artificial context (see Discussion).
Given the importance of the Sp1 and TATA elements within the HIV promoter for Tat activation (4, 21, 103), we individually mutated these elements and lost activation by CycT1-Rev and Tat AD, as well as Tat-Rev (Fig. 3C and data not shown); in contrast, deletion of the NF-κB sites had no effect. As loss of Sp1 and TATA elements also affected basal transcription levels, this potentially links the basal machinery to Tat activity during elongation (4). The requirement for TBP/TFIID at the TATA box was further confirmed using a mutated TATA box (TGTA) and a compensatory TBP mutant (TBPm3) with altered specificity (Fig. 3D) (105, 112). Thus, Tat activation of RNA-bound P-TEFb recapitulates the known HIV promoter requirements for Tat activation and indicates the importance of assembling the correct basal transcription machinery.
P-TEFb chimeras assemble into 7SK snRNP complexes, and Tat activation correlates with Hexim1 displacement.
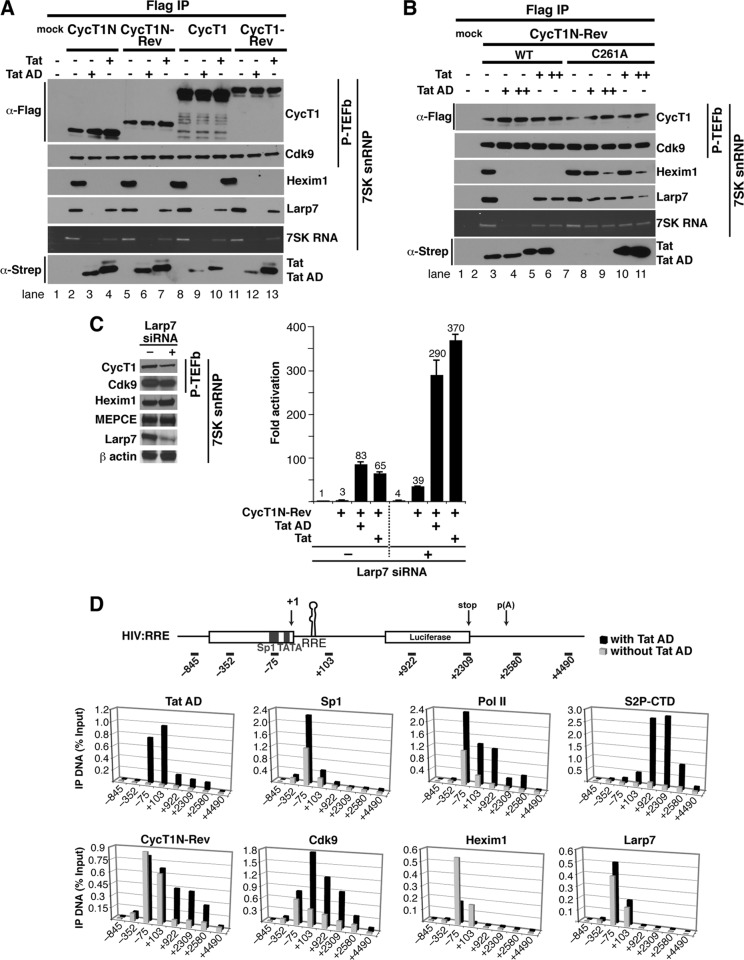
Based on our model that inactive P-TEFb complexes initially assemble at the HIV promoter and that Tat remodels the complex by ejecting Hexim1 and completely displacing 7SK snRNP when Tat:P-TEFb transfers to TAR, we hypothesized that RNA-bound P-TEFb may similarly be held in an inactive state by 7SK snRNP until Tat displaces the kinase inhibitor or induces conformational changes that eject 7SK snRNP. In support of this hypothesis, we found that both full-length and N-terminal CycT1 and CycT1-Rev chimeras assemble with Cdk9, as well as components of the 7SK snRNP complex (Hexim1, Larp7, and the 7SK RNA). Hexim1 and Larp7 are displaced by wild-type Tat and the RNA-binding-deficient Tat mutants (Tat AD and T-RS), albeit at different levels, during Tat:P-TEFb complex formation (Fig. 4A and data not shown). Mutation of the Tat interaction surface of CycT1 (TRM; C261A) (17, 22, 40) leads to less efficient Tat or Tat AD displacement of Hexim1 (Fig. 4B), while Tat mutants that cannot interact with CycT1 do not eject Hexim1 (data not shown). Notably, the degree of Hexim1 displacement (Fig. 4B) correlates well with activation of the P-TEFb kinase and the transition to elongation (Fig. 2D).
Fig 4.
RNA-bound P-TEFb assembles with 7SK snRNP complexes, and Tat displaces Hexim1 at the viral promoter. (A) Flag-tagged CycT1N or full-length CycT1, unfused or fused to Rev, was expressed in 293 cells with or without Strep-tagged Tat or Tat AD and immunoprecipitated using Flag beads. Mock refers to a Flag immunoprecipitation of cells transfected with an empty vector. The composition of P-TEFb/7SK snRNP components in the immunoprecipitation was analyzed by Western blotting using Cdk9, Hexim1, and Larp7 antibodies, while 7SK snRNA was visualized directly as a 330-nt RNA species whose identity was confirmed by Northern blotting. (B) Flag-tagged CycT1N-Rev (the wild type and a C261A mutant) was transfected into 293 cells with or without Strep-tagged Tat or Tat AD, and the P-TEFb/7SK snRNP composition was evaluated as for panel A. (C) Larp7 depletion by RNAi. Western blots show that Larp7 is reduced by ∼70% but that the other 7SK snRNP subunits or β-actin used as a control are unaffected. The plot shows reporter assays in HeLa cells transfected with an HIV:RRE reporter (with [+] or without [−] Larp7 siRNA) in the presence of CycT1N-Rev (with or without Tat and Tat AD). Fold activation represents values normalized to the activity of the reporter alone without siRNA. The error bars indicate standard deviations. (D) ChIP assays were performed with chromatin extracts prepared from the HIV:RRE reporter cell line 48 h after a Flag-tagged CycT1N-Rev transfection alone (gray bars) or cotransfected with Strep-tagged Tat AD (black bars) using the indicated antibodies. The values represent the percentages of input DNA immunoprecipitated and are the averages of two independent PCR assays from two separate experiments.
Since our model proposes that Tat activation of RNA-bound P-TEFb relies on 7SK snRNP displacement (Fig. 1C and D), we asked whether depleting endogenous Larp7 could mechanistically replace Tat, rendering activation of the HIV:RRE reporter Tat independent. Indeed, 7SK snRNP depletion (∼70% efficiency) increases both basal levels (∼4-fold) and activation by RNA-bound P-TEFb (∼3-fold) in the absence of Tat (Fig. 4C). Addition of Tat further activates transcription, either with or without Larp7 depletion, suggesting that incomplete Larp7 knockdown may leave residual 7SK-bound complexes for Tat disassembly or that 7SK snRNP removal may not phenocopy all aspects of Tat function. Further investigation is needed to more precisely define the effects of 7SK snRNP depletion.
To examine the dynamics of complex assembly between Tat and P-TEFb bound to the nascent RNA at the HIV promoter and transcribed regions, we monitored occupancy of CycT1N-Rev, components of the 7SK snRNP complex, as well as Sp1 and Pol II in ChIP experiments using a HeLa cell line with an integrated HIV:RRE reporter in the absence or presence of Tat AD (Fig. 4D). As previously seen with Tat and an HIV reporter, Tat activation of RNA-bound P-TEFb is accompanied by increased Sp1 and Pol II occupancy at the promoter and Pol II CTD serine 2 phosphorylation (S2P-CTD) throughout the gene body (21, 85), consistent with enhancing the transition into elongation. In agreement, the P-TEFb chimera (CycT1N-Rev and Cdk9) is found upstream of the transcription start site (TSS) (+1) with increased occupancy within the gene only in the presence of Tat AD, consistent with P-TEFb traveling with elongating complexes (51, 80). Importantly, Hexim1 occupancy is observed at the promoter (−75) and in the region of the nascent RNA (+103) without Tat AD but is significantly decreased in both regions upon Tat AD expression. Near-complete loss of Hexim1 at the nascent RNA (+103) is consistent with increased Hexim1 displacement upon Tat-TAR binding (21). However, in the context of the integrated reporter, we observed that Larp7 occupancy did not change in the presence of Tat AD, a result that partially conflicts with the immunoprecipitation experiments in which Tat AD, but not Tat, completely ejects Hexim1 from P-TEFb (Fig. 4B). Thus, in this chimeric context, it appears that Hexim1 displacement, not Larp7/7SK RNA, is required for kinase activation and transition to elongation (Fig. 2 and 4D). In agreement with this, it was recently shown that Tat stably associates with the 7SK snRNP complex by competitively displacing Hexim1 and, unexpectedly, that the Tat:7SK snRNP complex displays lower CTD kinase activity than P-TEFb assembled on the super elongation complex (21, 45, 101). These results indicate that Tat activates transcription through the RNA-bound P-TEFb complex without completely dismantling the Tat-7SK RNA interaction. It remains to be determined if these results reflect sequential displacement of the Hexim1 and Larp7/7SK snRNA inhibitory components.
Genetic dissection of Tat activation steps and Tat:P-TEFb complex assembly.
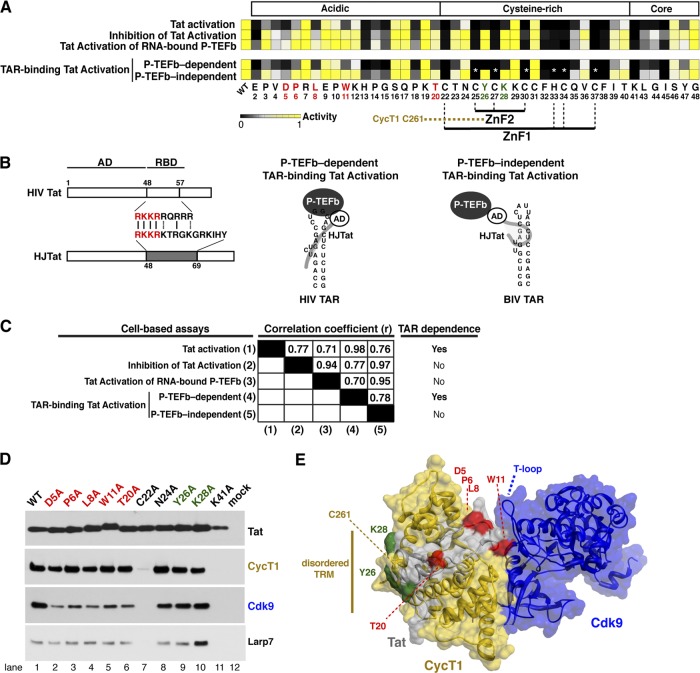
Having established assays reflecting different aspects of Tat activation, we proceeded to define the unique molecular surfaces of the Tat AD governing Tat:P-TEFb preassembly and transfer to TAR. For this, we introduced Ala substitutions at 45 positions within the Tat AD and assessed their activities in the three previously described assays: (i) standard Tat activation, (ii) inhibition of Tat activation, and (iii) Tat activation of RNA-bound P-TEFb (Fig. 1 and 5A). For Tat activation, which monitors the entire activation process on an HIV luciferase reporter, 30 full-length Tat mutants decreased activity by at least 4-fold (Fig. 5A), including at least 15 that are zinc-coordinating residues or otherwise contribute to the structural stability of Tat within the P-TEFb complex (20, 91, 92, 107). In inhibition of Tat activation, which monitors Tat:P-TEFb preassembly at the promoter and precedes Tat-TAR interaction (23), 21 mutations engineered into the TAR-binding-deficient Tat mutant decreased inhibition ≥4-fold (Fig. 5A). The mutations resulting in the most striking differences in activity between these two assays reside in the N-terminal acidic and cysteine-rich regions. A nearly identical activity pattern was observed during Tat activation of RNA-bound P-TEFb by these same TAR-binding-deficient Tat mutants. However, this assay monitors P-TEFb kinase activation on the nascent RNA independent of TAR (Fig. 1D and 5A). The reduced requirement for Tat AD residues in the last two TAR-independent assays most likely reflects the bypassed requirement for RNA binding during Tat:P-TEFb assembly and transcription activation (Fig. 1). For example, Lys28 acetylation by p300/CBP-associated factor (PCAF) was previously shown to enhance the affinity of Tat:TAR:P-TEFb complexes by affecting Tat interaction with CycT1 TRM (21); however, since mutation of Lys28 does not affect inhibition of Tat activation, acetylation probably does not play a role in the early stages of Tat:P-TEFb preassembly but may be important for ejection of the 7SK snRNP complex or transfer of P-TEFb to the nascent RNA. An adjacent residue, Tyr26, shows an identical phenotype and may be functionally coupled to Lys28.
Fig 5.
Activities of single-site Tat Ala mutants in reporter and coprecipitation assays. (A) Heat map of Tat activities (wild type or single-site Ala mutants) across the entire AD (residues 1 to 48), except the initiating Met1 and preexisting Ala residues 21 and 42 in the HXB2 reference sequence. The basis for each of the five assays—Tat activation, inhibition of Tat activation, Tat activation of RNA-bound P-TEFb, P-TEFb-dependent TAR-binding Tat activation, and P-TEFb-independent TAR-binding Tat activation—is described in Fig. 1B. Quantitative raw activity data for all assays (data not shown) were used to derive the relative activity value for each mutant (0 to 1) represented in the heat map (black to yellow). In the HJTat context, Cys25, Cys27, Cys30, His33, Cys34, and Cys37 were not mutated, since they completely disrupted protein folding in the other three assays. The black boxes with white asterisks represent the expected phenotype and not actual data. P-TEFb-dependent TAR-binding Tat activation and P-TEFb-independent TAR-binding Tat activation are assays in which HJTat activates an HIV reporter or an engineered HIV promoter with HIV TAR replaced by BIV, respectively (Table 1). (B) Schematic of HIV Tat and the HJTat chimera composed of the HIV Tat AD and the JDV RBD, which is able to bind to both HIV and BIV TARs in P-TEFb-dependent and P-TEFb-independent modes, respectively (22, 99). Conserved residues between the HIV and JDV Tat RBDs used for HIV TAR binding (P-TEFb-dependent TAR-binding Tat activation) are highlighted in red. Schematic of ternary complexes showing that HJTat binding to HIV TAR is dependent on interactions between Tat and CycT1/P-TEFb while binding to BIV TAR is P-TEFb independent. (C) Pearson correlation coefficients (r) for pairwise comparisons of all assays. (D) Affinity-purified Strep-tagged HJTat (wild-type and select Tat AD Ala mutants) was analyzed by Western blotting for interactions with endogenous CycT1, Cdk9, and Larp7. Mock refers to an empty-vector-transfected control. (E) Surface representation of Tat (gray), CycT1 (gold), and Cdk9 (blue) using the coordinates of the Tat:P-TEFb structure (107). Mutated Tat residues examined in panel D map to the CycT1 TRM (green) or are positioned to interact with Cdk9 (red). The Cdk9 T loop essential for kinase activity is indicated.
To confirm the differential requirements of Tat AD residues during the transition of preassembled Tat:P-TEFb complexes to TAR RNA, we constructed a parallel set of mutations in the context of a chimeric Tat protein referred to as HJTat. This Tat variant contains the RBD of Jembrana disease virus (JDV) Tat instead of the native HIV RBD (22, 99) and can therefore bind two orthologous TAR elements (HIV and bovine immunodeficiency virus [BIV]) (Fig. 5B and Table 1). While the P-TEFb kinase is needed to activate both reporters, P-TEFb is needed only for Tat:P-TEFb complex assembly on HIV TAR (referred to as P-TEFb-dependent TAR-binding Tat activation). Hence, this comparison identifies Tat AD residues that are specifically required for P-TEFb-dependent and -independent RNA-binding mechanisms and can further refine models in which preassembled Tat:P-TEFb complexes are remodeled as they transition from the inactive promoter-bound state to the active TAR-bound state (Fig. 1A) (21). Indeed, residues indispensable for Tat activation through BIV TAR (P-TEFb-independent TAR-binding Tat activation) are virtually identical to those for the TAR-independent assays (Fig. 5A). Conversely, the indispensability of N-terminal residues, as well as Tyr26 and Lys28, for activation through HIV TAR indicated that these residues are needed for interactions in the context of the Tat:TAR:P-TEFb ternary complex, but not Tat:P-TEFb. A high Pearson correlation coefficient (r) distinctly clusters the two TAR-dependent assays (Tat activation and P-TEFb-dependent TAR-binding Tat activation) and the three TAR-independent assays (inhibition of Tat activation, Tat activation of RNA-bound P-TEFb, and P-TEFb-independent TAR-binding Tat activation), whereas the values calculated between dissimilar assays display much lower coefficients (Fig. 5C). Strong correlation between the two TAR-dependent assays reinforces a model in which specific residues and Tat acetylation are needed for the transfer of preassembled Tat:P-TEFb complexes from the promoter to TAR.
To better define the roles of N-terminal Tat residues, as well as Tyr26 and Lys28, we affinity purified Strep-tagged HJTat mutants representative of the residue classifications defined below and examined their association with endogenous P-TEFb and 7SK snRNP subunits (Fig. 5D). All of the N-terminal mutants tested showed reduced interaction with Cdk9, but not CycT1 (Fig. 5D), consistent with the Tat:P-TEFb structure, in which residues Leu8 through Trp11 form a 310 helix positioned near the T loop of Cdk9 (Fig. 5E) that may stabilize the complex (107). CycT1 and Cdk9 interactions were abolished by mutation of known structural residues, Cys22 and Lys41, but were unaffected by mutation of Asn24, Tyr26, and Lys28. As expected, Hexim1 did not copurify with wild-type or mutant Tat proteins (2, 21), while Larp7 (7SK snRNP) copurified to similar levels, except with the Lys28 mutant, which, interestingly, showed increased Larp7 association. In light of our previous studies showing that Lys28 is needed to assemble high-affinity Tat:P-TEFb complexes on TAR (22), this observation suggests potentially coupled roles for Lys28 in both Tat-7SK snRNP disassembly and Tat:P-TEFb:TAR complex formation.
An evolutionarily conserved network of residues in the Tat:P-TEFb complex.
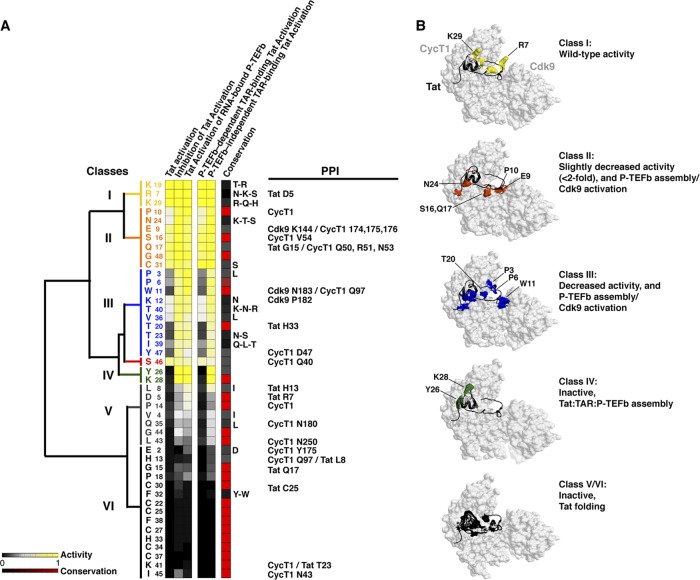
With the large functional data sets generated for all Tat AD mutants using multiple cell-based reporter assays (Fig. 5A), it becomes possible to cluster residues based on their activities and to map them onto the recent Tat:P-TEFb structure (107). Moreover, this comprehensive analysis permits us to define functional interfaces in the context of the two-step model of activation, where Tat first preassembles with inactive P-TEFb/7SK complexes at the promoter and is then transferred to TAR (21). By hierarchical clustering, we categorized six classes of residues showing strong functional correlations, high evolutionary conservation in most cases (Fig. 6A), and physical clustering within the structure (Fig. 6B). Class I residues (yellow) do not disrupt Tat activity in any assay when mutated and are not evolutionarily conserved; conversely, at the other extreme, class VI residues (black) show severe defects in all assays when mutated and are strictly evolutionarily conserved. This is likely the consequence of disrupting Tat folding and is exemplified by the cysteine-rich domain, which folds into a compact structure composed of two α-helices mediating the coordination of two Zn ions (20, 107). Class V residues (gray) show moderate defects when mutated but also are likely to affect folding or perhaps interactions with other transcription factors. Mutation of class II residues (orange) only slightly (<2-fold) decreases activity in the TAR-dependent Tat activation assays and does not affect activation in assays not operating through TAR (inhibition of Tat activation and Tat activation of RNA-bound P-TEFb). These residues are positioned in the structure to perturb P-TEFb assembly or Cdk9 activation. Similarly, class III residues (blue) show pronounced decreases in Tat activation when mutated, with 3 residues (Pro3, Pro6, and Trp11) possibly involved in Cdk9 interactions (107). Single-site Ala mutagenesis has been generally useful in defining protein-protein interactions, protein-folding residues, and energetic parameters (15, 67, 89, 108); however, in cases where the substitutions are nonconservative, such as Pro3, Pro6, and Trp11, changes in structure or other long-range effects may also underlie the functional disruptions observed.
Fig 6.
Conserved networks of functional residues in the Tat:P-TEFb complex. (A) Activity data for all Tat AD mutants in all cell-based assays were clustered as described in Materials and Methods and reordered in the heat map according to their values relative to wild-type Tat activation (0 to 1; black to yellow). Six functional classes were identified (I to VI; color coded), with the bars representing calculated relationships between the classes. The conservation of each residue, based on an alignment of 1,496 sequences, is plotted for comparison (low to high on a 0 to 1 scale; black to red). PPI lists the observed protein-protein interactions involving Tat intramolecular hydrogen bonds or intermolecular interactions with CycT1 or Cdk9 (107). (B) Surface representation of the Tat:P-TEFb complex, displayed using UCSF Chimera (65), with the six classes of residues color coded as in panel A, and their associated phenotypes for reporter activity and interaction profiles.
Class IV residues Tyr26 and Lys28 (green) show the most severe defects in the TAR-dependent assays yet are not apparently associated with structural defects, as Tat:P-TEFb complex formation is not abolished (Fig. 5D). Indeed, unlike the class V and VI folding residues, these mutants show no defect in activation assays operating in a TAR-independent manner and thus may not be required for the early stage of Tat:P-TEFb preassembly at the promoter. Tyr26 and Lys28 are located within the ZnF2 motif, near the CycT1 TRM, which is disordered in the Tat:P-TEFb structure due to the absence of TAR and acetylated Lys28 (Fig. 5 and 6) (20, 39, 107). Therefore, it will be especially interesting to reevaluate these mutational data once a cocrystal structure with TAR becomes available. TAR may well be important for interacting with the two residues, either directly or through conformational changes in P-TEFb or Tat upon RNA binding (22). Alternatively, because they cluster in the middle of the CycT1 TRM interface, they may be energetically important in the cooperative assembly of Tat:P-TEFb on the RNA, perhaps coordinating communication between protein and RNA conformations to increase affinity (9, 46).
DISCUSSION
Since the seminal discovery that HIV Tat enhances transcription elongation (29, 49, 56, 93), many contributing host factors have been identified, including P-TEFb (64, 79, 80, 121), 7SK snRNA (74, 116), Hexim1 (58, 66, 96), 7SK snRNP (21, 70, 101), components of the super elongation complex (44, 45, 100, 101), and the basal transcription machinery (85, 98). However, the mechanism and timing of assembly of these factors at the viral promoter during active elongation and their requirement for TAR RNA are poorly understood. Our recent results point to an activation model having at least two discrete steps, where Tat and P-TEFb are initially assembled with the inhibitory 7SK snRNP into transcription complexes at the promoter and later transferred to TAR on the nascent transcript, triggering release of the snRNP and activation of Cdk9 (21) (Fig. 1A). The crystal structure of the Tat AD bound to P-TEFb clearly shows how CycT1 and Cdk9 act as a template to fold Tat, possibly explaining the need to preassemble the complex before TAR binding (20, 107). P-TEFb enhances the affinity and specificity of Tat for TAR, while Cdk9 autophosphorylation apparently renders a P-TEFb conformation more favorable for TAR recognition (31, 38, 39, 118), thus providing further rationale for complex preassembly at the promoter and Tat:P-TEFb cooperativity for TAR.
By devising a set of cell-based reporter assays with different permutations of Tat and P-TEFb, we have been able to probe the two steps of the model in a functional context: Tat:P-TEFb preassembly at the promoter and subsequent recruitment of the complex to TAR on the nascent RNA. The reporter assays (Fig. 1) rely in part on tethering (or artificial recruitment), an instrumental strategy in defining discrete molecular events during transcription, splicing, RNA processing, and translation (10, 12, 19, 27, 50, 52, 57, 59, 62, 73, 83, 104). While these systems are somewhat artificial, the results can be validated by recapitulating phenotypes in the wild-type setting or, in our case, activity measurements using an extensive set of Tat mutants in each context and coimmunoprecipitation assays. The most surprising result is that Tat, without the need for Tat RBD or TAR binding (23), activates P-TEFb tethered directly to the nascent RNA (Fig. 1D). For the first time, we are able to uncouple the requirements for Tat:P-TEFb preassembly from Tat:TAR:P-TEFb complex formation in a comprehensive manner.
Although the role of molecular mimicry between TAR and 7SK RNA during activation remains unclear, one can envisage that Larp7 and 7SK RNA are completely ejected from the promoter immediately before the Tat-TAR interaction (21). ChIP experiments showing the presence of 7SK snRNP at the promoter regardless of TAR argue against Hexim1 recruitment to the viral promoter through TAR binding (75, 96) and indicate that the kinase inhibitor is released from 7SK snRNP by Tat–P-TEFb interactions on the nascent RNA (Fig. 4D), as predicted by our model (21). In this two-step model of Tat activation, interaction between the Tat AD and the TRM of CycT1 might initially eject Hexim1 from preassembled 7SK snRNP complexes. Upon Tat-mediated transfer of P-TEFb to the nascent RNA, 7SK snRNP disassembles and the P-TEFb kinase is activated. Supporting this model, Larp7 depletion, which destabilizes 7SK snRNA and consequently disassembles Hexim1 from P-TEFb (53), increases the activity of P-TEFb bound to the nascent RNA (Fig. 4C). In vitro systems will be required to more precisely define the kinetics of assembly and displacement of these factors and the activity of the P-TEFb kinase alone, bound to Tat, and in the context of the Tat:7SK snRNP complex (38, 45, 79, 101, 110, 119). In apparent disagreement with our model, a previous observation that P-TEFb is easily extracted from chromatin with high-salt treatment (6) suggested that 7SK snRNP complexes do not stably associate with chromatin (76, 78); however, recent genome-wide experiments examining the assembly of noncoding RNAs with chromatin demonstrated that 7SK snRNA is enriched several thousandfold in the chromatin fraction (69), raising the possibility that 7SK complexes selectively assemble at specific promoters (28).
It is particularly instructive to compare the activities of Tat and P-TEFb recruited to the HIV promoter by other means. For example, recruiting Tat through a heterologous DNA-protein interaction (Gal4-Tat) requires multiple Gal4-binding sites and still results in weaker activation than TAR-mediated recruitment (reference 103 and data not shown); this is also true for P-TEFb recruitment through DNA (Fig. 3B). Similarly, recruiting Gal4–P-TEFb to the hsp70 promoter in Drosophila activates transcription in the absence of heat shock to a much lower level than by heat shock factor 1 (61), suggesting that these artificial recruitment mechanisms only partially recapitulate function. Even Tat activation in the context of heterologous protein-RNA interactions (Fig. 1D) (5, 97, 102) does not fully recapitulate activity. For example, acetylation of Tat Lys28 is dispensable in heterologous systems but is important in the native HIV Tat-TAR context to modulate the affinity for TAR and fine tune escape into productive elongation (22). Interestingly, we also find that mutation of Lys28 reduces 7SK snRNP dismantling in the context of Tat activation by RNA-bound P-TEFb (Fig. 5D). Thus, these heterologous systems can be especially informative compared with the wild-type mechanism of Tat activation. Our results provide strong evidence that Tat does not bind TAR directly at the promoter and that a preassembly step is required. The generation of an in vitro system that recapitulates the results observed in vivo will be needed to further define the role of Tat residues in the two-step model.
Comprehensive Tat mutagenesis coupled with analyses using multiple cell-based reporter assays defined networks of functional residues whose evolutionary constraints are not readily explained by the Tat:P-TEFb structure (107). Evolutionarily conserved residues constitute molecular surfaces for essential interactions, allosteric communication, or transmission of conformational changes into functional behavior (3, 90, 106). Many proteins undergo conformational changes upon transient interactions, often involving disorder-to-order transitions (47, 77). The intrinsic disorder and structural flexibility of Tat may be relevant for interactions with more than one binding partner and may allow competing interactions to occur in a sequential manner (24, 37), such as switching from unbound to RNA-bound states during activation. While further investigation is needed to uncover the molecular details, the observed functional and structural clustering of residues suggests that Tat:P-TEFb complexes have evolved networks of residues to orchestrate the switch from transcription initiation to elongation, RNA binding, and the assembly/disassembly of 7SK snRNP complexes.
ACKNOWLEDGMENTS
We thank Robert Nakamura and members of the Frankel laboratory for critical reading of the manuscript and James Fraser for helpful discussions. We thank K.-T. Jeang for the TBPm3 construct.
This work was supported by NIH grants AI29135 and P50 GM082250 (HARC Center) and funding from the California Center for Antiviral Drug Discovery to A.D.F., NIH grants K99A112185 and R00AI083087 and Welch Foundation grant I-1782 to I.D., UC GREAT fellowship 2004-21 to A.W.P., NIH postdoctoral fellowship GM077868 to G.M.J., NIH Research supplement to promote diversity in Health Related Research to E.Q., and NIH fellowships R25GM56847 and F31GM09535 to D.S.B.
Footnotes
Published ahead of print 24 September 2012
REFERENCES
- 1.Barboric M, Lenasi T. 2010. Kick-sTARting HIV-1 transcription elongation by 7SK snRNP deporTATion. Nat. Struct. Mol. Biol. 17:928–930 [DOI] [PubMed] [Google Scholar]
- 2.Barboric M, et al. 2007. Tat competes with HEXIM1 to increase the active pool of P-TEFb for HIV-1 transcription. Nucleic Acids Res. 35:2003–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barua B, Pamula MC, Hitchcock-DeGregori SE. 2011. Evolutionarily conserved surface residues constitute actin binding sites of tropomyosin. Proc. Natl. Acad. Sci. U. S. A. 108:10150–10155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berkhout B, Gatignol A, Rabson AB, Jeang KT. 1990. TAR-independent activation of the HIV-1 LTR: evidence that tat requires specific regions of the promoter. Cell 62:757–767 [DOI] [PubMed] [Google Scholar]
- 5.Bieniasz PD, Grdina TA, Bogerd HP, Cullen BR. 1999. Recruitment of cyclin T1/P-TEFb to an HIV type 1 long terminal repeat promoter proximal RNA target is both necessary and sufficient for full activation of transcription. Proc. Natl. Acad. Sci. U. S. A. 96:7791–7796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biglione S, et al. 2007. Inhibition of HIV-1 replication by P-TEFb inhibitors DRB, seliciclib and flavopiridol correlates with release of free P-TEFb from the large, inactive form of the complex. Retrovirology 4:47–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blau J, et al. 1996. Three functional classes of transcriptional activation domain. Mol. Cell. Biol. 16:2044–2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown SA, Weirich CS, Newton EM, Kingston RE. 1998. Transcriptional activation domains stimulate initiation and elongation at different times and via different residues. EMBO J. 17:3146–3154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calabro V, Daugherty MD, Frankel AD. 2005. A single intermolecular contact mediates intramolecular stabilization of both RNA and protein. Proc. Natl. Acad. Sci. U. S. A. 102:6849–6854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carey M, Lin YS, Green MR, Ptashne M. 1990. A mechanism for synergistic activation of a mammalian gene by GAL4 derivatives. Nature 345:361–364 [DOI] [PubMed] [Google Scholar]
- 11.Chen D, Fong Y, Zhou Q. 1999. Specific interaction of Tat with the human but not rodent P-TEFb complex mediates the species-specific Tat activation of HIV-1 transcription. Proc. Natl. Acad. Sci. U. S. A. 96:2728–2733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clement SL, Lykke-Andersen J. 2008. A tethering approach to study proteins that activate mRNA turnover in human cells. Methods Mol. Biol. 419:121–133 [DOI] [PubMed] [Google Scholar]
- 13.Core LJ, Lis JT. 2008. Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science 319:1791–1792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cullen BR. 1990. The HIV-1 Tat protein: an RNA sequence-specific processivity factor? Cell 63:655–657 [DOI] [PubMed] [Google Scholar]
- 15.Cunningham BC, Wells JA. 1989. High-resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis. Science 244:1081–1085 [DOI] [PubMed] [Google Scholar]
- 16.Das A. 1992. How the phage lambda N gene product suppresses transcription termination: communication of RNA polymerase with regulatory proteins mediated by signals in nascent RNA. J. Bacteriol. 174:6711–6716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Das C, Edgcomb SP, Peteranderl R, Chen L, Frankel AD. 2004. Evidence for conformational flexibility in the Tat-TAR recognition motif of cyclin T1. Virology 318:306–317 [DOI] [PubMed] [Google Scholar]
- 18.de Hoon MJ, Imoto S, Nolan J, Miyano S. 2004. Open source clustering software. Bioinformatics 20:1453–1454 [DOI] [PubMed] [Google Scholar]
- 19.Dorris DR, Struhl K. 2000. Artificial recruitment of TFIID, but not RNA polymerase II holoenzyme, activates transcription in mammalian cells. Mol. Cell. Biol. 20:4350–4358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.D'Orso I, Frankel AD. 2010. HIV-1 Tat: its dependence on host factors is crystal clear. Viruses 2:2226–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.D'Orso I, Frankel AD. 2010. RNA-mediated displacement of an inhibitory snRNP complex activates transcription elongation. Nat. Struct. Mol. Biol. 17:815–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.D'Orso I, Frankel AD. 2009. Tat acetylation modulates assembly of a viral-host RNA-protein transcription complex. Proc. Natl. Acad. Sci. U. S. A. 106:3101–3106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.D'Orso I, Grunwell JR, Nakamura RL, Das C, Frankel AD. 2008. Targeting tat inhibitors in the assembly of human immunodeficiency virus type 1 transcription complexes. J. Virol. 82:9492–9504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dunker AK, Silman I, Uversky VN, Sussman JL. 2008. Function and structure of inherently disordered proteins. Curr. Opin. Struct. Biol. 18:756–764 [DOI] [PubMed] [Google Scholar]
- 25.Durney MA, D'Souza VM. 2010. Preformed protein-binding motifs in 7SK snRNA: structural and thermodynamic comparisons with retroviral TAR. J. Mol. Biol. 404:555–567 [DOI] [PubMed] [Google Scholar]
- 26.Espinosa JM. 2010. The meaning of pausing. Mol. Cell 40:507–508 [DOI] [PubMed] [Google Scholar]
- 27.Farrell S, Simkovich N, Wu Y, Barberis A, Ptashne M. 1996. Gene activation by recruitment of the RNA polymerase II holoenzyme. Genes Dev. 10:2359–2367 [DOI] [PubMed] [Google Scholar]
- 28.Faust T, Frankel AD, D'Orso I. 2012. Transcription control by long non-coding RNAs. Transcription 3:78–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feinberg MB, Baltimore D, Frankel AD. 1991. The role of Tat in the human immunodeficiency virus life cycle indicates a primary effect on transcriptional elongation. Proc. Natl. Acad. Sci. U. S. A. 88:4045–4049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fishburn J, Mohibullah N, Hahn S. 2005. Function of a eukaryotic transcription activator during the transcription cycle. Mol. Cell 18:369–378 [DOI] [PubMed] [Google Scholar]
- 31.Fong YW, Zhou Q. 2000. Relief of two built-in autoinhibitory mechanisms in P-TEFb is required for assembly of a multicomponent transcription elongation complex at the human immunodeficiency virus type 1 promoter. Mol. Cell. Biol. 20:5897–5907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fong YW, Zhou Q. 2001. Stimulatory effect of splicing factors on transcriptional elongation. Nature 414:929–933 [DOI] [PubMed] [Google Scholar]
- 33.Frankel AD, Kim PS. 1991. Modular structure of transcription factors: implications for gene regulation. Cell 65:717–719 [DOI] [PubMed] [Google Scholar]
- 34.Frankel AD, Young JA. 1998. HIV-1: fifteen proteins and an RNA. Annu. Rev. Biochem. 67:1–25 [DOI] [PubMed] [Google Scholar]
- 35.Fuda NJ, Ardehali MB, Lis JT. 2009. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature 461:186–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fujinaga K, et al. 1998. The ability of positive transcription elongation factor B to transactivate human immunodeficiency virus transcription depends on a functional kinase domain, cyclin T1, and Tat. J. Virol. 72:7154–7159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ganguly D, Chen J. 2009. Atomistic details of the disordered states of KID and pKID. Implications in coupled binding and folding. J. Am. Chem. Soc. 131:5214–5223 [DOI] [PubMed] [Google Scholar]
- 38.Garber ME, et al. 2000. CDK9 autophosphorylation regulates high-affinity binding of the human immunodeficiency virus type 1 tat-P-TEFb complex to TAR RNA. Mol. Cell. Biol. 20:6958–6969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garber ME, Wei P, Jones KA. 1998. HIV-1 Tat interacts with cyclin T1 to direct the P-TEFb CTD kinase complex to TAR RNA. Cold Spring Harb. Symp. Quant. Biol. 63:371–380 [DOI] [PubMed] [Google Scholar]
- 40.Garber ME, et al. 1998. The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev. 12:3512–3527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gilchrist DA, et al. 2010. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell 143:540–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gold MO, Yang X, Herrmann CH, Rice AP. 1998. PITALRE, the catalytic subunit of TAK, is required for human immunodeficiency virus Tat transactivation in vivo. J. Virol. 72:4448–4453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Green M, Ishino M, Loewenstein PM. 1989. Mutational analysis of HIV-1 Tat minimal domain peptides: identification of trans-dominant mutants that suppress HIV-LTR-driven gene expression. Cell 58:215–223 [DOI] [PubMed] [Google Scholar]
- 44.He N, et al. 2011. Human polymerase-associated factor complex (PAFc) connects the super elongation complex (SEC) to RNA polymerase II on chromatin. Proc. Natl. Acad. Sci. U. S. A. 108:E636–E645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He N, et al. 2010. HIV-1 Tat and host AFF4 recruit two transcription elongation factors into a bifunctional complex for coordinated activation of HIV-1 transcription. Mol. Cell 38:428–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hobson D, Uhlenbeck OC. 2006. Alanine scanning of MS2 coat protein reveals protein-phosphate contacts involved in thermodynamic hot spots. J. Mol. Biol. 356:613–624 [DOI] [PubMed] [Google Scholar]
- 47.Janin J, Bahadur RP, Chakrabarti P. 2008. Protein-protein interaction and quaternary structure. Q. Rev. Biophys. 41:133–180 [DOI] [PubMed] [Google Scholar]
- 48.Jeronimo C, et al. 2007. Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol. Cell 27:262–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kao SY, Calman AF, Luciw PA, Peterlin BM. 1987. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature 330:489–493 [DOI] [PubMed] [Google Scholar]
- 50.Keaveney M, Struhl K. 1998. Activator-mediated recruitment of the RNA polymerase II machinery is the predominant mechanism for transcriptional activation in yeast. Mol. Cell 1:917–924 [DOI] [PubMed] [Google Scholar]
- 51.Keen NJ, Gait MJ, Karn J. 1996. Human immunodeficiency virus type-1 Tat is an integral component of the activated transcription-elongation complex. Proc. Natl. Acad. Sci. U. S. A. 93:2505–2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koh SS, Ansari AZ, Ptashne M, Young RA. 1998. An activator target in the RNA polymerase II holoenzyme. Mol. Cell 1:895–904 [DOI] [PubMed] [Google Scholar]
- 53.Krueger BJ, et al. 2008. LARP7 is a stable component of the 7SK snRNP while P-TEFb, HEXIM1 and hnRNP A1 are reversibly associated. Nucleic Acids Res. 36:2219–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krueger BJ, Varzavand K, Cooper JJ, Price DH. 2010. The mechanism of release of P-TEFb and HEXIM1 from the 7SK snRNP by viral and cellular activators includes a conformational change in 7SK. PLoS One 5:e12335 doi:10.1371/journal.pone.0012335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kurosu T, Peterlin BM. 2004. VP16 and ubiquitin; binding of P-TEFb via its activation domain and ubiquitin facilitates elongation of transcription of target genes. Curr. Biol. 14:1112–1116 [DOI] [PubMed] [Google Scholar]
- 56.Laspia MF, Rice AP, Mathews MB. 1989. HIV-1 Tat protein increases transcriptional initiation and stabilizes elongation. Cell 59:283–292 [DOI] [PubMed] [Google Scholar]
- 57.Li Q, Peterlin BM. 2009. Genetic analysis of P-TEFb function via heterologous nucleic acid tethering systems. Methods 48:375–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li Q, et al. 2005. Analysis of the large inactive P-TEFb complex indicates that it contains one 7SK molecule, a dimer of HEXIM1 or HEXIM2, and two P-TEFb molecules containing Cdk9 phosphorylated at threonine 186. J. Biol. Chem. 280:28819–28826 [DOI] [PubMed] [Google Scholar]
- 59.Lim SR, Hertel KJ. 2004. Commitment to splice site pairing coincides with A complex formation. Mol. Cell 15:477–483 [DOI] [PubMed] [Google Scholar]
- 60.Lis J, Wu C. 1993. Protein traffic on the heat shock promoter: parking, stalling, and trucking along. Cell 74:1–4 [DOI] [PubMed] [Google Scholar]
- 61.Lis JT, Mason P, Peng J, Price DH, Werner J. 2000. P-TEFb kinase recruitment and function at heat shock loci. Genes Dev. 14:792–803 [PMC free article] [PubMed] [Google Scholar]
- 62.Majello B, Napolitano G, Lania L. 1998. Recruitment of the TATA-binding protein to the HIV-1 promoter is a limiting step for Tat transactivation. AIDS 12:1957–1964 [DOI] [PubMed] [Google Scholar]
- 63.Malik S, Roeder RG. 2010. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat. Rev. Genet. 11:761–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mancebo HS, et al. 1997. P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev. 11:2633–2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Meng EC, Pettersen EF, Couch GS, Huang CC, Ferrin TE. 2006. Tools for integrated sequence-structure analysis with UCSF Chimera. BMC Bioinformatics 7:339–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Michels AA, et al. 2004. Binding of the 7SK snRNA turns the HEXIM1 protein into a P-TEFb (CDK9/cyclin T) inhibitor. EMBO J. 23:2608–2619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Milla ME, Brown BM, Sauer RT. 1994. Protein stability effects of a complete set of alanine substitutions in Arc repressor. Nat. Struct. Biol. 1:518–523 [DOI] [PubMed] [Google Scholar]
- 68.Modesti N, Garcia J, Debouck C, Peterlin M, Gaynor R. 1991. Trans-dominant Tat mutants with alterations in the basic domain inhibit HIV-1 gene expression. New Biol. 3:759–768 [PubMed] [Google Scholar]
- 69.Mondal T, Rasmussen M, Pandey GK, Isaksson A, Kanduri C. 2010. Characterization of the RNA content of chromatin. Genome Res. 20:899–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Muniz L, Egloff S, Ughy B, Jady BE, Kiss T. 2010. Controlling cellular P-TEFb activity by the HIV-1 transcriptional transactivator Tat. PLoS Pathog. 6:e1001152 doi:10.1371/journal.ppat.1001152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Naar AM, Lemon BD, Tjian R. 2001. Transcriptional coactivator complexes. Annu. Rev. Biochem. 70:475–501 [DOI] [PubMed] [Google Scholar]
- 72.Nechaev S, Adelman K. 2011. Pol II waiting in the starting gates: regulating the transition from transcription initiation into productive elongation. Biochim. Biophys. Acta 1809:34–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nevado J, Gaudreau L, Adam M, Ptashne M. 1999. Transcriptional activation by artificial recruitment in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 96:2674–2677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nguyen VT, Kiss T, Michels AA, Bensaude O. 2001. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature 414:322–325 [DOI] [PubMed] [Google Scholar]
- 75.Nilson KA, Price DH. 2011. The role of RNA polymerase II elongation control in HIV-1 gene expression, replication, and latency. Genet. Res. Int. 2011:726901 doi:10.4061/2011/726901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ott M, Geyer M, Zhou Q. 2011. The control of HIV transcription: keeping RNA polymerase II on track. Cell Host Microbe 10:426–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Perkins JR, Diboun I, Dessailly BH, Lees JG, Orengo C. 2010. Transient protein-protein interactions: structural, functional, and network properties. Structure 18:1233–1243 [DOI] [PubMed] [Google Scholar]
- 78.Peterlin BM, Brogie JE, Price DH. 2011. 7SK snRNA: a noncoding RNA that plays a major role in regulating eukaryotic transcription. Wiley Interdiscip. Rev. RNA 3:92–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Peterlin BM, Price DH. 2006. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell 23:297–305 [DOI] [PubMed] [Google Scholar]
- 80.Ping YH, Rana TM. 1999. Tat-associated kinase (P-TEFb): a component of transcription preinitiation and elongation complexes. J. Biol. Chem. 274:7399–7404 [DOI] [PubMed] [Google Scholar]
- 81.Price DH. 2000. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell. Biol. 20:2629–2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Price DH. 2008. Poised polymerases: on your mark.get set.go! Mol. Cell 30:7–10 [DOI] [PubMed] [Google Scholar]
- 83.Ptashne M, Gann A. 1997. Transcriptional activation by recruitment. Nature 386:569–577 [DOI] [PubMed] [Google Scholar]
- 84.Purnell BA, Emanuel PA, Gilmour DS. 1994. TFIID sequence recognition of the initiator and sequences farther downstream in Drosophila class II genes. Genes Dev. 8:830–842 [DOI] [PubMed] [Google Scholar]
- 85.Raha T, Cheng SW, Green MR. 2005. HIV-1 Tat stimulates transcription complex assembly through recruitment of TBP in the absence of TAFs. PLoS Biol. 3:e44 doi:10.1371/journal.pbio.0030044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rahl PB, et al. 2010. c-Myc regulates transcriptional pause release. Cell 141:432–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rappaport J, Reinberg D, Zandomeni R, Weinmann R. 1987. Purification and functional characterization of transcription factor SII from calf thymus. Role in RNA polymerase II elongation. J. Biol. Chem. 262:5227–5232 [PubMed] [Google Scholar]
- 88.Reines D, Chamberlin MJ, Kane CM. 1989. Transcription elongation factor SII (TFIIS) enables RNA polymerase II to elongate through a block to transcription in a human gene in vitro. J. Biol. Chem. 264:10799–10809 [PubMed] [Google Scholar]
- 89.Rennell D, Bouvier SE, Hardy LW, Poteete AR. 1991. Systematic mutation of bacteriophage T4 lysozyme. J. Mol. Biol. 222:67–88 [DOI] [PubMed] [Google Scholar]
- 90.Reynolds KA, McLaughlin RN, Ranganathan R. 2011. Hot spots for allosteric regulation on protein surfaces. Cell 147:1564–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rice AP, Carlotti F. 1990. Mutational analysis of the conserved cysteine-rich region of the human immunodeficiency virus type 1 Tat protein. J. Virol. 64:1864–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rice AP, Carlotti F. 1990. Structural analysis of wild-type and mutant human immunodeficiency virus type 1 Tat proteins. J. Virol. 64:6018–6026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rice AP, Mathews MB. 1988. Transcriptional but not translational regulation of HIV-1 by the tat gene product. Nature 332:551–553 [DOI] [PubMed] [Google Scholar]
- 94.Saunders A, Core LJ, Lis JT. 2006. Breaking barriers to transcription elongation. Nat. Rev. Mol. Cell Biol. 7:557–567 [DOI] [PubMed] [Google Scholar]
- 95.Schulte A, et al. 2005. Identification of a cyclin T-binding domain in Hexim1 and biochemical analysis of its binding competition with HIV-1 Tat. J. Biol. Chem. 280:24968–24977 [DOI] [PubMed] [Google Scholar]
- 96.Sedore SC, et al. 2007. Manipulation of P-TEFb control machinery by HIV: recruitment of P-TEFb from the large form by Tat and binding of HEXIM1 to TAR. Nucleic Acids Res. 35:4347–4358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Selby MJ, Peterlin BM. 1990. Trans-activation by HIV-1 Tat via a heterologous RNA binding protein. Cell 62:769–776 [DOI] [PubMed] [Google Scholar]
- 98.Sikorski TW, Buratowski S. 2009. The basal initiation machinery: beyond the general transcription factors. Curr. Opin. Cell Biol. 21:344–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Smith CA, Calabro V, Frankel AD. 2000. An RNA-binding chameleon. Mol. Cell 6:1067–1076 [DOI] [PubMed] [Google Scholar]
- 100.Smith E, Lin C, Shilatifard A. 2011. The super elongation complex (SEC) and MLL in development and disease. Genes Dev. 25:661–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sobhian B, et al. 2010. HIV-1 Tat assembles a multifunctional transcription elongation complex and stably associates with the 7SK snRNP. Mol. Cell 38:439–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Southgate C, Zapp ML, Green MR. 1990. Activation of transcription by HIV-1 Tat protein tethered to nascent RNA through another protein. Nature 345:640–642 [DOI] [PubMed] [Google Scholar]
- 103.Southgate CD, Green MR. 1991. The HIV-1 Tat protein activates transcription from an upstream DNA-binding site: implications for Tat function. Genes Dev. 5:2496–2507 [DOI] [PubMed] [Google Scholar]
- 104.Stargell LA, Struhl K. 1996. A new class of activation-defective TATA-binding protein mutants: evidence for two steps of transcriptional activation in vivo. Mol. Cell. Biol. 16:4456–4464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Strubin M, Struhl K. 1992. Yeast and human TFIID with altered DNA-binding specificity for TATA elements. Cell 68:721–730 [DOI] [PubMed] [Google Scholar]
- 106.Suel GM, Lockless SW, Wall MA, Ranganathan R. 2003. Evolutionarily conserved networks of residues mediate allosteric communication in proteins. Nat. Struct. Biol. 10:59–69 [DOI] [PubMed] [Google Scholar]
- 107.Tahirov TH, et al. 2010. Crystal structure of HIV-1 Tat complexed with human P-TEFb. Nature 465:747–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tsiang M, et al. 1995. Functional mapping of the surface residues of human thrombin. J. Biol. Chem. 270:16854–16863 [DOI] [PubMed] [Google Scholar]
- 109.Ujvari A, Luse DS. 2006. RNA emerging from the active site of RNA polymerase II interacts with the Rpb7 subunit. Nat. Struct. Mol. Biol. 13:49–54 [DOI] [PubMed] [Google Scholar]
- 110.Wada T, Takagi T, Yamaguchi Y, Watanabe D, Handa H. 1998. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 17:7395–7403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wei P, Garber ME, Fang SM, Fischer WH, Jones KA. 1998. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92:451–462 [DOI] [PubMed] [Google Scholar]
- 112.Xiao H, Lis JT, Jeang KT. 1997. Promoter activity of Tat at steps subsequent to TATA-binding protein recruitment. Mol. Cell. Biol. 17:6898–6905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yamaguchi Y, et al. 1999. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97:41–51 [DOI] [PubMed] [Google Scholar]
- 114.Yang L, et al. 1997. Distinct transcriptional pathways of TAR-dependent and TAR-independent human immunodeficiency virus type-1 transactivation by Tat. Virology 235:48–64 [DOI] [PubMed] [Google Scholar]
- 115.Yang XJ, Roberts JW. 1989. Gene Q antiterminator proteins of Escherichia coli phages 82 and lambda suppress pausing by RNA polymerase at a rho-dependent terminator and at other sites. Proc. Natl. Acad. Sci. U. S. A. 86:5301–5305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yang Z, Zhu Q, Luo K, Zhou Q. 2001. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature 414:317–322 [DOI] [PubMed] [Google Scholar]
- 117.Yankulov K, Blau J, Purton T, Roberts S, Bentley DL. 1994. Transcriptional elongation by RNA polymerase II is stimulated by transactivators. Cell 77:749–759 [DOI] [PubMed] [Google Scholar]
- 118.Zhang J, et al. 2000. HIV-1 TAR RNA enhances the interaction between Tat and cyclin T1. J. Biol. Chem. 275:34314–34319 [DOI] [PubMed] [Google Scholar]
- 119.Zhou M, et al. 2000. Tat modifies the activity of CDK9 to phosphorylate serine 5 of the RNA polymerase II carboxyl-terminal domain during human immunodeficiency virus type 1 transcription. Mol. Cell. Biol. 20:5077–5086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhou Q, Yik JH. 2006. The Yin and yang of P-TEFb regulation: implications for human immunodeficiency virus gene expression and global control of cell growth and differentiation. Microbiol. Mol. Biol. Rev. 70:646–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhu Y, et al. 1997. Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev. 11:2622–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]