Abstract
In herpes simplex virus 1 (HSV-1), binding clusters enriched in CTCF during latency have been previously identified. We hypothesized that CTCF binding to CTCF clusters in HSV-1 would be disrupted in a reactivation event. To investigate, CTCF occupation of three CTCF binding clusters in HSV-1 was analyzed following sodium butyrate (NaB)- and explant-induced reactivation in the mouse. Our data show that the CTCF domains positioned within the HSV-1 genome, specifically around the latency-associated transcript (LAT) and ICP0 and ICP4 regions of the genome, lose CTCF occupancy following the application of reactivation stimuli in wild-type virus. We also found that CTCF binding clusters upstream of the ICP0 and ICP4 promoters both function as classical insulators capable of acting as enhancer blockers of the LAT enhancer. Finally, our results suggest that CTCF occupation of domains in HSV-1 may be differentially regulated both during latency and at early times following reactivation by the presence of lytic transcripts and further implicate epigenetic regulation of HSV-1 as a critical component of the latency-reactivation transition.
INTRODUCTION
The human alphaherpesvirus herpes simplex virus 1 (HSV-1), infects sensory neurons, where it establishes a lifelong latent infection as a circular episome associated with histones (6, 16, 38, 40, 44). During HSV-1 latency, the latency-associated transcript (LAT) is abundantly transcribed, and lytic regions are, in essence, silent (6, 38, 40). Recent studies have shown that there are epigenetic components involved in HSV-1 latency and in reactivation (1, 13, 21, 31, 36, 39). For example, during latency, the LAT regions of the LAT reactivation-critical promoter and LAT 5′ exon (containing the LAT enhancer) (8) are enriched in euchromatic histone markers compared to the promoters of the immediate-early (IE) ICP0, ICP4, and ICP27 genes (1, 13, 21, 24, 31). Further, these permissive marks established on the LAT are independent of LAT transcription (24). Additional reports indicated that the LAT region of HSV-1 also contains facultative heterochromatin (12, 25), a form of repressed chromatin that can convert to euchromatin via posttranslational histone tail modifications. Consequently, the nearby IE regions are also enriched with repressive histone marks that have been characterized predominantly as facultative heterochromatin (12, 25, 26, 45). These key findings provide evidence that HSV-1 latency is established and maintained, at least in part, by complex epigenetic mechanisms that further poise the virus for reactivation. To support this, recent data show that the enrichments of euchromatic histone marks on the HSV-1 LAT and IE promoters change in response to reactivation stimuli in both rabbit and mouse models latently infected with wild-type HSV-1 (1, 13, 26, 31, 36). Specifically, histone marks on ICP0 and ICP4 rapidly and transiently become more euchromatic in nature, while the LAT region loses enrichment of euchromatic histone marks as LAT is degraded at early times in reactivation in the two different in vivo models (1, 13, 31). Considering the reversibility of latency reactivation in vivo, it is logical that epigenetic mechanisms play a critical role in this transition in the neuron at very early times following a reactivation stressor. Therefore, thorough and systematic evaluation of functional components that might contribute to this epigenetic regulation during latency and reactivation in vivo will likely provide novel insight into future therapeutic prevention of recurrent episodes of HSV-1.
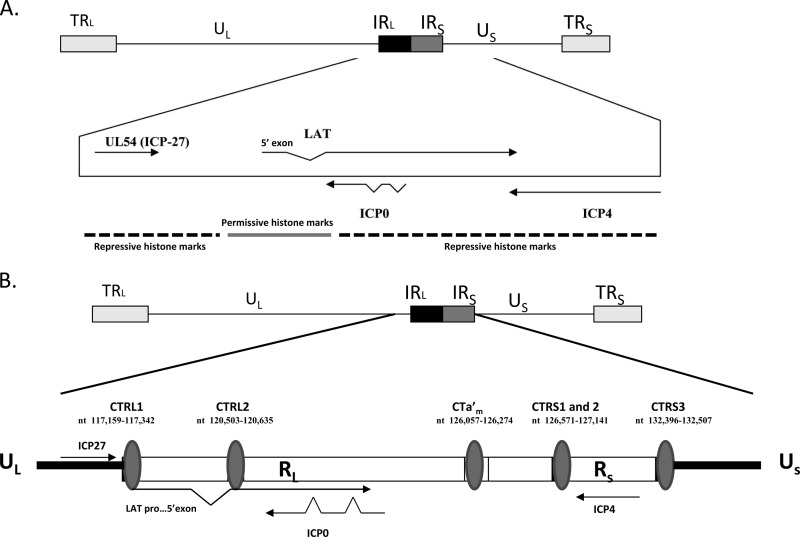
Previous studies have shown that distinct heterochromatic and euchromatic chromatin domains are likely established within the latent genome (Fig. 1A) (2). This configuration of transcriptionally permissive and transcriptionally repressed chromatin during latency implies the presence of functional barriers within the HSV-1 genome that help to maintain the integrity of a given domain (2). One logical element to consider is a chromatin insulator element, which would have the capability of preventing inappropriate gene activation and/or silencing by nearby chromatin domains during latency in HSV-1 (2, 7). Chromatin insulators are long-range cis-acting DNA sequence elements that can protect a given transcriptional domain from inappropriate signals from other domains (46). To date, insulators have predominantly been characterized in eukaryotic cells, where they have the capability of acting as classical enhancer blockers, which block an enhancer element from activating genes distal to the insulator (47), or as barrier elements, which prevent the extension of heterochromatin to areas of euchromatin, likely by recruiting histone-modifying enzymes or corepressive silencer proteins to coordinate function (19). In eukaryotes, all functional insulators have the cellular insulator protein CTCF bound to the DNA sequence elements that facilitate appropriate insulator activity (34). CTCF is an 11-zinc-finger phosphoprotein that binds to pentanucleotide motifs, such as CCCTC and CTCCC, and has been implicated in the recruitment of other coregulatory proteins to form complexes that can facilitate either gene activation or repression (34, 35). CTCF insulators likely maintain the transcriptional status of a chromatin domain through the recruitment of histone-modifying enzymes or through protein-protein interactions that span a domain (9, 28). However, more recent findings show that CTCF insulators can also be involved in the organization and regulation of higher-order chromatin structures that form loop or chromatin domains and are involved in gene regulation and expression via long-range DNA interactions (35).
Fig 1.
The latent HSV-1 genome. (A) Regions of repressed (heterochromatin) and permissive (euchromatin) chromatin juxtaposed within the HSV-1 genome. TRL, terminal repeat long; UL, unique long region; US, unique short region; IRL, inverted long repeat; IRS, inverted short repeat; TRS, terminal repeat short. (B) Previously identified regions of the HSV-1 genome containing CCCTC and CTCCC repeats for CTCF binding flanking the LAT reactivation-critical region (CTRL1 and CTRL2), ICP0 (CTa′m), and ICP4 (CTRS1/2 and CTRS3) (2). Note that CTRS1/2 is actually 2 CTCF binding clusters separated by less than 100 nucleotides. An additional CTCF motif (CTUS1) was identified by Amelio et al. (2) (not shown).
In addition to their characterization in eukaryotes, functional CTCF insulators have recently been identified in a number of gammaherpesviruses, including Epstein-Barr virus (EBV) and Kaposi's sarcoma-associated herpesvirus (KSHV) (27, 42). For example, during EBV latency, CTCF insulators block the epigenetic silencing of an essential promoter, Qp, while preventing transcription of genes surrounding the Qp promoter (43). More recent studies have shown that CTCF insulators play an integral role in chromatin loop formation in latent EBV (41). Similarly, in KSHV, CTCF cohesion complexes mediate chromatin loops that regulate lytic and latent gene expression (23). Further, it has been shown that in the alphaherpesvirus family, reiterated sequences of the CTCF binding motifs are conserved and are arranged in clusters that partition the IE genes into separate domains (2). Further genome-wide analyses of the HSV-1 genome identified seven clusters of CTCF binding motifs (2). Significantly, these seven CTCF binding clusters are not only organized around each of the IE genes, they also flank the reactivation-critical region of LAT (Fig. 1B). During latency, each of the seven CTCF binding clusters of HSV-1 is occupied by the protein CTCF in the dorsal root ganglia (DRG) (2, 11). Finally, the HSV-1 CTCF binding motif identified as CTRL2 by Amelio et al. has been characterized as an insulator element with both enhancer-blocking and -silencing capabilities (2).
We and others have previously reported that transcriptionally permissive histone marks, specifically acetyl H3 K9, K14, and dimethyl H3 K4, accumulate on the repressed IE regions of HSV-1 by 2 to 4 h following the application of reactivation stimuli (1, 13, 31). This provides evidence that the distinct chromatin domains observed during latency are no longer intact at early times following reactivation stressors. Considering these findings, as well as the role that CTCF plays in gene regulation of eukaryotes and the multifaceted role that CTCF insulators play in gammaherpesvirus gene regulation, we hypothesized that a critical component of HSV-1 reactivation would be a loss of CTCF binding to the genome. The loss of CTCF binding could result in scenarios such as activation of the IE regions by the LAT enhancer of HSV-1 or the spread of heterochromatin associated with the IE regions to the reactivation-critical regions of the LAT. On the other hand, the HSV-1 CTCF domains could act in a much more complex and multifaceted role during latency and reactivation by displaying both enhancer-blocking and barrier functions. In this study, we explored aspects of the first possibility. We sought to determine whether CTCF binding to HSV-1 is disrupted following application of reactivation stimuli at early times and whether additional CTCF binding domains in HSV-1 have the capability of acting as functional enhancer blocker insulator elements. We focused on three of the seven previously identified CTCF binding domains of HSV-1, including the CTRL2 region downstream of the LAT enhancer, the CTa′m region upstream of the ICP0 promoter, and the CTRS3 region upstream of ICP4 promoter. These domains were of interest primarily because the LAT, ICP0, and ICP4 regions have been shown to undergo significant epigenetic changes in response to reactivation stimuli (1, 13, 26, 31). Our studies showed that CTCF occupation of each of these three CTCF binding motifs in HSV-1 decreased by at least 3-fold following both sodium butyrate (NaB)- and explant-induced reactivation in mice latently infected with the wild-type HSV-1 strain 17Syn+. Further, CTCF binding was essentially abolished at the CTCF binding domains upstream of the ICP0 and ICP4 promoters by 6 h post-NaB reactivation. Consequently, the time frame for loss of CTCF binding at these sites in HSV-1 is consistent with previously reported changes in the histone marks on the IE regions following reactivation in mouse and rabbit models (1, 31). Additional analyses of the CTa′m and CTRS3 sites showed both sites were functional enhancer blockers of the LAT enhancer, with enhancer-blocking activity similar to that of the CTRL2 domain (2). Finally, to begin to tease out the mechanisms responsible for changes in CTCF occupation of HSV-1 at early times in reactivation, we analyzed two recombinant viruses, both lacking IE transcription during latency, to establish the role of IE gene transcription in CTCF binding during HSV-1 latency and reactivation. Our analyses showed that CTCF occupation of the HSV-1 genome is attenuated in the absence of IE transcription in a site-specific manner. Our data also confirm that in the absence of IE transcription, CTCF binding is not lost following the application of reactivation stimuli. Taken together, these data indicate that CTCF binding to the HSV-1 genome is involved in the maintenance of latency and that a loss of CTCF binding, possibly facilitated by IE transcription, is fundamental to HSV-1 reactivation in vivo.
MATERIALS AND METHODS
Viruses and cells.
All ChIP experiments were performed using ganglia from mice infected with the HSV-1 strains 17Syn+, 17Δ348 (a LAT enhancer mutant with a 348-bp deletion of nucleotides [nt] 119007 to 119355 [NC_001806]), and KD6 (an ICP4-null recombinant virus obtained from David C. Bloom, University of Florida). The KD6 recombinant used was generated from the parent KOS(M), with a 3.1-kb deletion within the coding region of ICP4 generated by digesting the plasmid pATD51 with SphI, which liberated a 3.8-kb fragment (lacking the HincII region of ICP4). This fragment was electroeluted and cotransfected with KOS(M) DNA into E5 cells. The virus was plaque purified and screened for the capacity to hybridize to the HSV-1 HindIII A fragment, but not the HincII fragment. The isolates were then tested for the ability to replicate on E5 cells and not rabbit skin cells (18, 37). All other viruses were amplified and titrated on rabbit skin cells using Eagle's minimal essential medium (Life Technologies) supplemented with 5% calf serum, 250 U of penicillin/ml, 250 μg of streptomycin/ml, and 292 μg of l-glutamine/ml (Life Technologies). Neither recombinant virus used contains deletions of nucleotides within 1.0 kb of the CTRL2, CTa′m, or CTRS3 core CTCF binding motifs (Fig. 1B).
Mouse ocular infections.
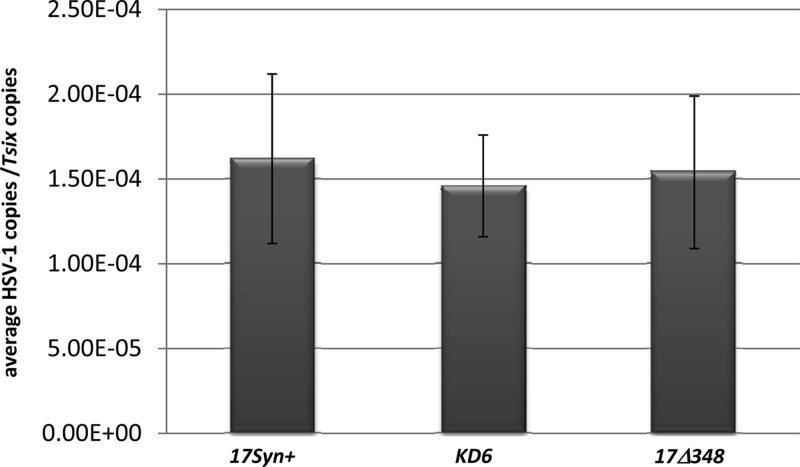
Female BALB/c mice 4 to 6 weeks of age (obtained from the National Cancer Institute [NCI]) were anesthetized using intramuscular injections of ketamine and xylazine (at a dose of 200 mg/kg ketamine:10 mg/kg xylazine). Using a 27-gauge needle, a light 2-by-2 crosshatch pattern was made on the corneal epithelium. Viral suspensions of 150,000 PFU/eye in a 5-μl volume were added to each eye for 17Syn+ and 17Δ348. In the case of KD6, a viral suspension of 1 × 106 PFU/eye was used to ensure comparable amounts of viral DNA were present in the trigeminal ganglia (TG) of the latent mice. Slit lamp examination (SLE) was performed 3 days postinfection to confirm infections. Animals were considered latent at >28 days postinfection, at which time no lesions could be observed following SLE (31). To verify that equivalent infections were established following infection with each virus, genome copies per ganglion were measured by quantitative real-time PCR (qRT-PCR) using primers and a probe specific for HSV-1 DNA polymerase (Fig. 2).
Fig 2.
HSV-1 genome copies per TG latent mouse. The number of HSV-1 genomes per sample was determined to show equivalent infections among the viruses. The relative numbers of copies of HSV-1 DNA polymerase and mouse Tsix were determined by PCR using 5 mice (10 TG) that were latently infected with 17Syn+, 17Δ348, or KD6. TG were harvested at 28 days postinfection, and DNA was extracted. The data are presented as a ratio of HSV-1 DNA pol (polymerase) to Tsix. One-way ANOVA of the individual ratios of DNA pol to Tsix showed no significant difference in HSV-1 copies between the viruses. The error bars indicate standard deviations from the mean.
HSV-1 reactivation.
Establishment of a latent viral infection was confirmed at 28 days postinfection through the absence of corneal lesions upon SLE. In order to stimulate viral reactivation, either chemically induced or explant-induced viral-reactivation strategies were utilized. For chemically induced viral reactivation, mice were injected intraperitoneally with a single 1,200-mg/kg dose of NaB, as previously described (32). Following NaB treatment, the mice were euthanized at 1 h, 2 h, 3 h, 4 h, or 6 h post-NaB treatment, and the TG were rapidly removed and homogenized in cold phosphate-buffered saline (PBS) with protease inhibitors for chromatin immunoprecipitation (ChIP) procedures (for the method, see below). Latent TG samples were not treated with NaB prior to homogenization for ChIP processing. In explant-induced-reactivation experiments, the TG were harvested from latently infected mice; placed in Dulbecco's modified Eagle's medium (DMEM) supplemented with 1% fetal bovine serum, 2% penicillin-streptomycin; and placed in an incubator at 37°C with 5% CO2 for 1 h, 3 h, or 6 h. Following incubation, the TG were removed from DMEM, placed in cold PBS with protease inhibitors, and processed for ChIP assays.
ChIP assays.
Chromatin immunoprecipitation assays were performed as previously described for mice using the specific antibody anti-CTCF (Millipore) (31). Each ChIP assay mixture contained 3 mice pooled (6 TG). The TG were rapidly removed following euthanasia, the ganglia were homogenized, and the chromatin was cross-linked in 1% formaldehyde (Sigma-Aldrich). The cross-linked cell lysates were sonicated to shear the chromatin into fragments of between 300 and 800 bp. The fragment size following sonication was confirmed by agarose gel electrophoresis using a 1.5% gel. The sheared chromatin was precleared with salmon sperm DNA-protein A-agarose beads (Millipore) prior to antibody incubation. An aliquot representing 1/5 of the total sample volume was removed as a sample input (I). The remaining sample was incubated in a cold room overnight with shaking with 2 μg of CTCF antibody per 1 ml of sample. The chromatin-antibody complexes were collected with salmon sperm DNA-protein A-agarose beads, and the complexes were eluted from the beads, representing the bound (B) fraction. Both the bound and input fractions were treated with 5 M NaCl, RNase A, and proteinase K, and the DNA was finally purified using a Qiaquick PCR purification kit (Qiagen).
Real-Time PCR analysis of ChIP experiments.
All real-time PCR experiments were performed with TaqMan universal PCR master mix and No AmpErase uracil N-glycolase on a single-color MyiQ real-time PCR Detection System Version 2 (Bio-Rad) using custom-designed primer-probe mixtures (Applied Biosystems) with a general protocol as follows: 95°C for 10 min (1 time) and then 95°C for 15 s followed by 60°C for 1 min (45 times). Threshold values used for PCR analyses were set within the linear range of PCR target amplification based on a standard curve generated for each plate. The primer-probe sequences for host controls were as follows: Tsix imprinting/choice center CTCF site A (AJ421479) forward (CGCAGGGCAGCCAGAA) and reverse (TCTGGTGTTATCCCTTCCTGTCTT) and probe (CAGCCATTCACAATCC); mouse MT498 (NT_039554) forward (TACAAGATGCAAGTCTAGATATTTTAAGTCTATGTAT) and reverse (ACACACACACACACACACACA) and probe (CACACACACACAAACAC); and mouse adenine phosphoribosyltransferase (APRT) forward (CTCAAGAAATCTAACCCCTGACTCA) and reverse (GCGGGACAGGCTGAGA) and probe (CCCCACACACACCTC). The primer and probe sequences used for each viral target are listed in Table 1.
Table 1.
Real-time PCR primers and probe sequences for HSV-1 targets
| Target | Sequence (5′–3′) | Accession no. (nucleotide no.) |
|---|---|---|
| HSV-1 CTRL2 | NC_001806 (120455–120643) | |
| Forward | CCG CGG CTC TGT GGT TA | |
| Reverse | GGA TGC GTG GGA GTG GG | |
| Probe | ACA CCA GAG CCT GCC CAA CAT GGC A | |
| HSV-1 CTa'm | NC_001806 (126368–126560) | |
| Forward | GCT GCC ACA GGT GAA ACC | |
| Reverse | TGT AGC AGG AGC GGT GTG | |
| Probe | ACC TGC CCA ACA ACA CAA CT | |
| HSV-1 CTRS3 | NC_001806 (132247–132385) | |
| Forward | ATC GCA TCG GAA AGG GAC ACG | |
| Reverse | CCA AGG TGC TTA CCC GTG CAA A | |
| Probe | ACA GAA ACC CAC CGG TCC GCC TTT | |
| HSV-1 gC | NC_001806 (96331–96643) | |
| Forward | CCT TGC CGT GGT CCT GTG GA | |
| Reverse | GGT GGT GTT GTT CTT GGG TTT G | |
| Probe | CCC CAC GTC CAC CCC CGA CC | |
| HSV-1 DNA polymerase | NC_001806 (65880–65953) | |
| Forward | AGA GGG ACA TCC AGG ACT TTG T | |
| Reverse | CAG GCG CTT GTT GGT GTA C | |
| Probe | ACC GCC GAA CTG AGC A |
Determination of bound/input ratios from ChIP.
ChIP data are presented as a ratio of the relative numbers of copies of the PCR target in two fractions (bound and input), where the bound fractions are representative of aliquots incubated overnight with the antibody, anti-CTCF, and the input fractions are precleared chromatin only (no specific antibody incubation). Standard curves were generated for each PCR run for each gene region analyzed using serial dilutions of purified HSV-1 DNA or purified mouse DNA with primers/probes specific for that region. Each ChIP assay was analyzed as follows. First, duplicate PCRs were performed using DNA purified from either the bound or input fraction as the target. The average cycle threshold (Ct) for the bound fraction and the average Ct for the input fraction were then used to determine the relative quantity of target DNA in each fraction by using the equation for the standard curve specific to the primer and probe set used. The quantity was expressed as a ratio of the relative bound quantity to the relative input quantity (B/I ratio). All relative B/I ratios from each gene region were normalized to the B/I ratio for the cellular-control Tsix site A B/I for that ChIP assay.
ChIP validation.
Prior to real-time PCR analyses of viral targets, all ChIP assays were validated by determining the B/I ratios of the cellular controls Tsix imprinting/choice center CTCF site A (positive control) and MT498 (negative control) (10). Only assays having a >2-fold abundance of CTCF bound to Tsix imprinting/choice center CTCF site A relative to MT498 were used in further analyses. The normalized B/I ratios for the viral gC gene was used as a negative viral control due to the fact that it lies ∼15 kb away from the nearest CTCF binding domain in HSV-1 and therefore should not be associated with CTCF occupation in ChIP analyses.
Determination of relative amounts of viral DNA in latently infected mice and mice subjected to reactivation.
Real-time primer-probe sets specific to HSV-1 DNA polymerase were used on all input fractions from ChIP samples to determine equivalent infections during latency and following HSV-1 reactivation stimuli through 6 h. Real-time PCR was performed in duplicate, as described above. The relative number of HSV-1 copies for each sample was further normalized to the relative number of copies of the host control APRT or Tsix imprinting/choice center CTCF site A.
Statistical analysis.
One-way analysis of variance (ANOVA) was performed using StatPlus Professional version 2009 for Windows XP (AnalystSoft) by analyzing the individual normalized B/I ratio for a specific gene region during latency compared to the normalized B/I ratio for the same gene region following NaB treatment (or explant-induced reactivation) at each individual time point analyzed by ChIP. The P values are indicated on the graphs presented in the figures above the time point at which a significant change was determined by one-way ANOVA.
Plasmid constructs and enhancer-blocking assays.
The HSV-1 LAT enhancer (nt 118888 to 119477) was directionally cloned into the 5′ KpnI and 3′ MluI polylinker sites of the pGL3-simian virus 40 (SV40) promoter control vector (Promega) using PCR-generated KpnI and MluI linkers to generate the pGL3-control/LTE enhancer plasmid, as previously described (2). An additional enhancer-blocking test construct was generated containing the 2.1-kb fragment (nt 118888 to 120942) with the LAT enhancer and CTRL2 insulator in their native HSV-1 configurations. This fragment was directionally cloned into the 5′ KpnI and 3′ XhoI polylinker sites of the pGL3-SV40 promoter using PCR-generated KpnI and XhoI linkers to create pGL3-control/LTE+CTRL2 (2). This construct was then used as a positive control in all transient-transfection assays. Additional test constructs were generated to test the enhancer-blocking abilities of the CTa′m and CTRS3 regions of HSV-1. First, a 1.3-kb fragment (nt 125013 to 126375) containing sequences flanking the 217-bp core CTCF binding cluster of CTa′m (Fig. 1B) was generated by PCR with NheI and XhoI linkers. The pGL3-control/CTa′m and pGL3-control/LTE+CTa′m plasmids were generated by directionally cloning the CTa′m fragment into the 5′ NheI and 3′ XhoI sites of the pGL3-SV40 promoter control vector or the pGL3-control/LTE enhancer, respectively, using PCR-generated NheI and XhoI linkers. Next, a 1.6-kb fragment (nt 131000 to 132600) containing sequences flanking the core 111-bp cluster of CTCF, the binding motif CTRS3 (Fig. 1B), was generated by PCR with NheI and XhoI linkers. The pGL3-control/CTRS3 and pGL3-control/LTE+CTRS3 plasmids were generated by directionally cloning the CTRS3 into the 5′ NheI and 3′ XhoI sites using PCR-generated NheI and XhoI linkers to create the indicated plasmids. Transient transfections were set up with 1 μg of each luciferase test construct described above in 24-well dishes seeded 24 h earlier with rabbit skin cells at 2 × 105 cells/ml (CCL-68; ATCC). Five hundred nanograms of phRL-TK Renilla construct was cotransfected as a control for transfection efficiency. Each luciferase construct was tested in triplicate and repeated 5 times. Transfections were carried out using 5 μl of SuperFect Transfection Reagent (Qiagen) per reaction well following the manufacturer's protocols. Following a 24-h incubation, the cells were lysed, and luciferase levels were tested using a Dual Luciferase Reporter Assay kit (Promega) following the manufacturer's protocols, and the levels were measured using a BioTek Synergy 2 Luminometer with the Gen5 software package.
Analysis of mouse TG for LAT, ICP0, and ICP4 RNA by qRT-PCR.
Latent mouse TG or mouse TG subjected to explant-induced reactivation was used in all RNA experiments. The TG were isolated immediately following euthanasia, and RNA was extracted by adding TRIzol reagent (Sigma-Aldrich) to each sample. Briefly, each TG was homogenized in 1.0 ml TRIzol, and following the addition of 0.2 volume of chloroform, samples were centrifuged for phase separation. RNA was precipitated from the aqueous phase using 0.7 volume of isopropanol, followed by DNase treatment using DNA-free (Ambion), according to the manufacturer's directions. Reverse transcription using random primers was performed with a High Capacity cDNA Reverse Transcription Kit (ABI), according to the manufacturer's instructions. Briefly, 20-μl reaction mixtures contained DNase-treated RNA, manufacturer-supplied buffer, deoxynucleoside triphosphate mixture, 1 μM random hexamer primer, 1 U RNase inhibitor (Ambion), and Multiscribe reverse transcriptase (Applied Biosystems; ABI). In the case of ICP0, a 10-μl aliquot of purified RNA was used with the strand-specific primer for the ICP0 transcript (LAT I-1, GACACGGATTGGCTGGTGTAGTGGG; nucleotides 120797 to 120820) (1). All reaction conditions followed the manufacturer's instructions. Real-time PCRs were performed on cDNA according to the above-described procedures and protocols.
RESULTS
CTCF motifs found in HSV-1 are occupied by CTCF during latency in the mouse TG following ocular inoculation.
Using dorsal root ganglia from mice infected with HSV-1 strain 17Syn+ via footpad inoculation, Amelio et al. identified 7 clusters of CTCF binding motifs, all occupied by the cellular insulating protein CTCF during latency (2). To establish that CTCF occupies the CTCF binding motifs of HSV-1 in mouse TG following the establishment of latency via an ocular route of infection, female BALB/c mice were ocularly infected with HSV-1 17Syn+ and allowed to establish a latent infection, as previously reported (>28 days postinfection [p.i.]) (31). We performed ChIP assays (described in Materials and Methods) using anti-CTCF (Millipore) and combined them with real-time PCR to determine the relative enrichments of CTCF bound to three CTCF binding motifs in HSV-1, namely, the CTRL2 region downstream of the LAT enhancer, the CTa′m region upstream of the ICP0 promoter, and the CTRS3 region upstream of the ICP4 promoter in latent TG (2, 31). We chose to focus on these loci first, due to the fact that we and others have shown significant epigenetic changes to the LAT, ICP0, and ICP4 regions of HSV-1 following explant-induced and NaB-stimulated reactivation in mouse models (1, 31).
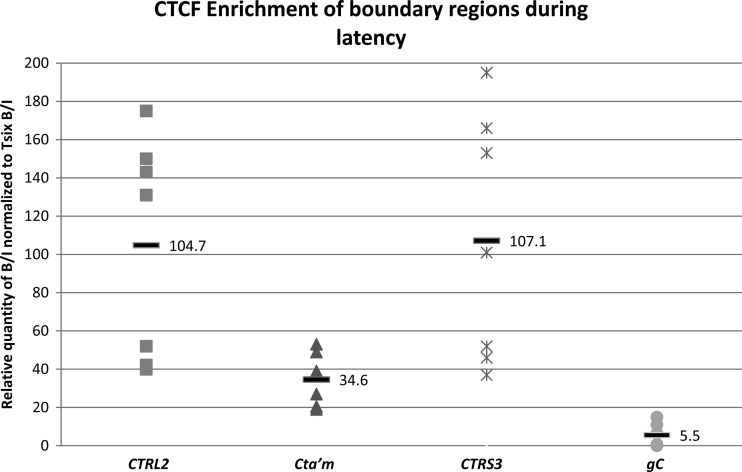
ChIP data are presented as a ratio of the relative number of copies of the PCR target in bound and input fractions (B/I), where the bound fractions are representative of aliquots incubated overnight with the anti-CTCF antibody and the input fractions are representative of samples removed prior to antibody incubation. Prior to real-time PCR analyses of viral targets, all ChIP assays were validated by determining the B/I ratios of the cellular controls Tsix imprinting/choice center CTCF site A (positive control) and MT498 (negative control), and only assays with a 2-fold or greater abundance of CTCF bound to Tsix imprinting/choice center CTCF site A relative to MT498 were used for PCR of viral targets (10). All B/I ratios determined from PCR of viral targets were normalized to the B/I ratio of Tsix imprinting/choice center CTCF site A for that assay [(B/I) for viral target/(B/I) for Tsix]. Finally, all normalized B/I ratios determined for CTRL2, CTa′m, and CTRS3 were compared to normalized HSV-1 gC B/I as a negative viral control due to the fact that it lies ∼15 kb away from the nearest CTCF binding domain in HSV-1 and therefore should not be associated with CTCF occupation in ChIP analyses. ChIP and real-time PCR of the CTRL2, CTa′m, and CTRS3 binding domains in HSV-1 showed that each of these three sites is significantly enriched in CTCF during latency in the TG relative to gC (Fig. 3). Specifically, the CTRL2 motif is ∼15-fold more enriched in CTCF during latency (P < 0.001), the CTa′m motif is ∼6-fold more enriched in CTCF during latency (P < 0.001), and the CTRS3 motif is ∼16-fold more enriched in CTCF during latency (P < 0.005) relative to the negative viral control, gC. Our findings are consistent with previously reported results showing that the HSV-1 genome is occupied by CTCF during latency in sensory neurons (2).
Fig 3.
CTCF occupation of three CTCF binding motifs in HSV-1 during latency in mouse TG. ChIP data are presented as a ratio of the relative numbers of copies of the PCR target in two fractions (B/I), where the bound fractions are representative of aliquots incubated overnight with the antibody anti-CTCF and the input fractions are representative of the total DNA present. All PCR was performed on a single-color MyiQ real-time PCR Detection System Version 2 (Bio-Rad). Standard curves were generated for each gene region analyzed using purified HSV-1 DNA or purified mouse DNA. Relative copy numbers in the B or I fractions of ChIP samples were determined from the equation for the standard curve specific to the primer-probe set used. Prior to real-time PCR analyses of viral targets, all ChIP assays were validated by determining the B/I ratios of the cellular controls Tsix imprinting/choice center CTCF site A (positive control) and MT498 (negative control). Each data point represents a normalized B/I ratio for 3 mice (6 TG pooled). Each time point contains a minimum of 6 data points (n = 6). All relative B/I ratios from each gene region were normalized to the B/I ratio for the cellular control Tsix site A B/I for that ChIP assay. The horizontal bars represent the average normalized B/I ratios for a given gene region from all ChIP assays at that time point. Statistical data were determined by one-way analysis of variance by comparing the normalized B/I ratios of each gene region to the normalized B/I ratio for the negative control, gC. All three sites analyzed were significantly enriched in CTCF relative to gC.
CTCF binding to CTRL2, CTa′m, and CTRS3 is disrupted at early times following NaB as a reactivation stimulus.
Previous studies have shown that the HSV-1 genome is likely separated into and maintained as euchromatic and heterochromatic domains by CTCF insulator elements during latency (2). It has also been shown that at early times following the application of a reactivation stimulus, these distinct domains are no longer intact, as chromatin associated with LAT becomes transiently less euchromatic and chromatin associated with ICP0 and ICP4 becomes transiently (more) euchromatic (1, 31). Considering that in eukaryotes, CTCF insulators can function to prevent the extension of heterochromatin to areas of euchromatin (19), presumably through the recruitment of histone-modifying enzymes, we hypothesized that during a reactivation event, CTCF binding may be disrupted at these CTCF binding motifs. This could, in turn, allow the transcriptional activation of lytic genes by allowing a change in the chromatin environment associated with IE genes during latency. To begin testing this hypothesis, we first needed to determine if CTCF binding was diminished or disrupted following a reactivation stressor. We subjected mice latently infected with 17Syn+ to i.p. treatment with NaB as an HSV-1 reactivation method. We chose NaB as our method of reactivation for several reasons. First, NaB has been effectively used as a herpesvirus reactivator in EBV and KSHV cell culture models (14, 23), as well as in HSV-1 quiescence models (15). Second, NaB is a highly efficient method of in vivo reactivation in the mouse, yielding >75% reactivation frequency in mice latently infected with 17Syn+ (32), making the method ideal in combination with the less efficient ChIP process. Even though NaB is known to have pleiotropic effects in vivo, including histone deacetylase inhibition, we have successfully used this method of reactivation to show that IE promoter regions of HSV-1 become transiently enriched in the permissive histone mark acetyl H3 K9, K14, while the LAT region undergoes deacetylation simultaneously (1, 31, 32). These epigenetic data were later confirmed in an alternative in vivo model of HSV-1 reactivation using transcorneal iontophoresis of epinephrine as the reactivation stimulus, providing evidence that the epigenetic changes observed at early times during a reactivation episode are not dependent on the method of reactivation (13).
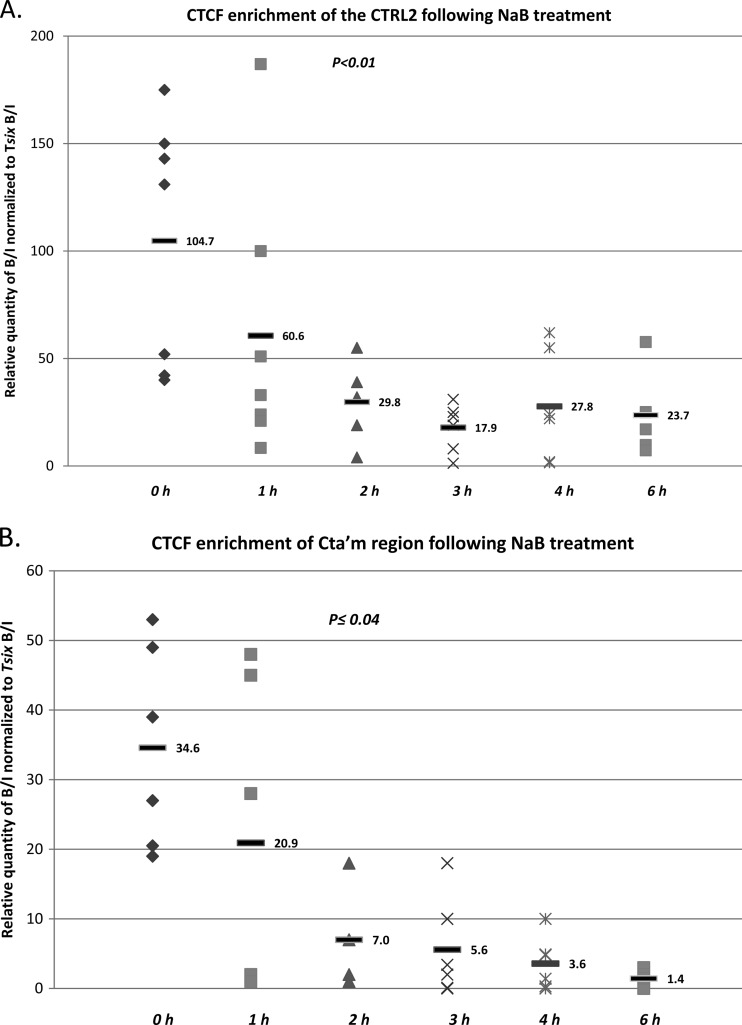
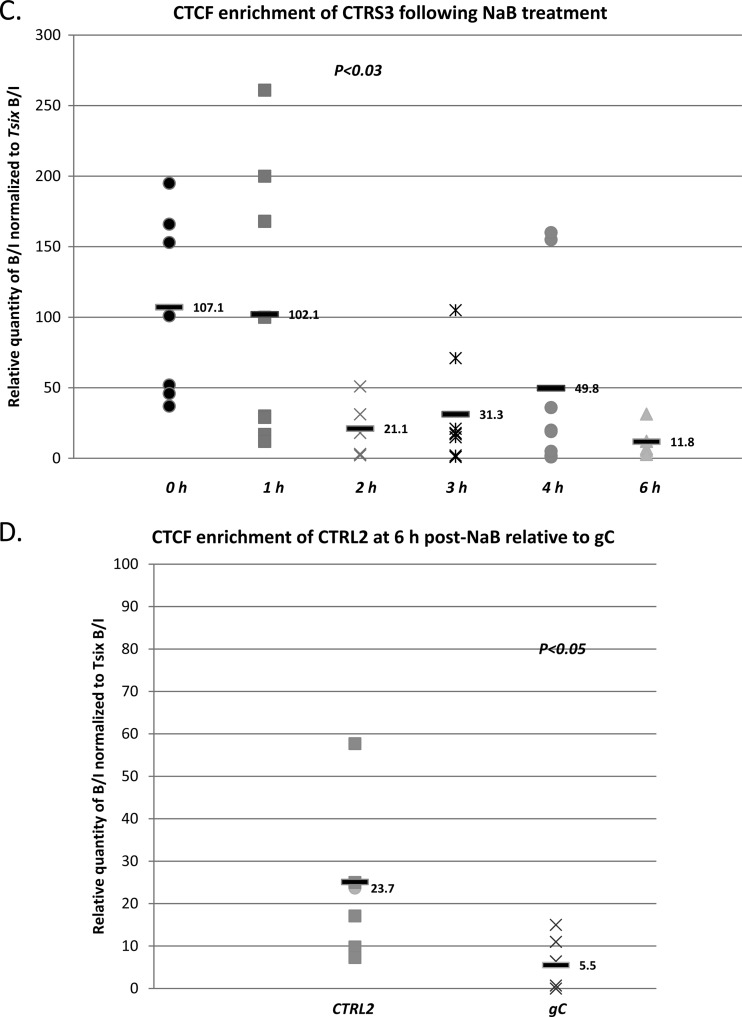
Following NaB treatment, the mouse TG were harvested at 1 h, 2 h, 3 h, 4 h, and 6 h. ChIP assays combined with real-time PCR analyses of the TG from NaB-treated mice showed that by 2 h post-NaB treatment, the CTRL2 motif downstream of the LAT enhancer, the CTa′m motif upstream of the ICP0 promoter, and the CTRS3 motif upstream of the ICP4 promoter were all >3-fold less enriched in bound CTCF than was observed in latent TG (P < 0.01, P < 0.04, and P < 0.03, respectively) (Fig. 4A, B, and C). Finally, the enrichments of CTCF at 6 h post-NaB treatment for each of the binding clusters were also determined. While each of the three CTCF binding motifs analyzed remained underenriched in bound CTCF by 6 h post-NaB treatment relative to latency, binding at the CTa′m and CTRS3, both upstream of IE promoters, was essentially abolished, as determined by one-way ANOVA comparing the normalized B/I ratios for each domain to the normalized B/I ratio for the negative control, gC (P > 0.1). On the other hand, the CTCF motif CTRL2, downstream of the LAT enhancer, was ∼3-fold less enriched by 6 h relative to latency but remained significantly more enriched than the negative control, gC (Fig. 4D). Collectively, these data show that binding of CTCF is significantly diminished at early times in a reactivation episode in HSV-1. Further, our data indicate that CTCF occupancy of CTCF clusters in HSV-1 may be regulated individually, due to the fact that CTCF binding is not completely abolished at the CTRL2 site, as it is in clusters upstream of the IE promoters ICP0 and ICP4.
Fig 4.
CTCF occupation of CTCF binding clusters in HSV-1 following reactivation with NaB in mouse TG. Mice latently infected with HSV-1 17Syn+ were subjected to i.p. treatment with NaB, and the TG were harvested at 1 h, 2 h, 3 h, 4 h, and 6 h. ChIP assays using anti-CTCF were combined with real-time PCR analyses to compare TG from NaB-treated mice versus latently infected mouse TG. All ratios reported are normalized to the B/I ratios of the host control Tsix imprinting/choice center CTCF site A for that experiment. The time following NaB treatment is represented on the x axis. The 0-h time point samples are latently infected mice not subjected to NaB treatment. (A) CTCF occupation on the CTRL2 domain (downstream of the LAT enhancer) following NaB treatment. There was a significant decrease in CTCF occupation of this site by 2 h post-NaB treatment (>3-fold; P < 0.01). (B) CTCF occupation on the CTa′m domain (upstream of the ICP0 promoter) following NaB treatment. A significant decrease in CTCF occupation was found at 2 h post-NaB treatment (one-way ANOVA; P < 0.04), and it was essentially abolished by 6 h post-NaB treatment. (C) CTCF occupation of the CTRS3 region (upstream of the ICP4 promoter) following NaB significantly decreased by 2 h (>5-fold; P < 0.03) and was essentially abolished at 6 h post-NaB treatment. (D) CTCF occupation of the CTRL2 binding motif in HSV-1 was significantly diminished relative to latency (A) but not completely abolished by 6 h post-NaB treatment at the site, as determined by one-way ANOVA of the normalized B/I ratios for CTRL2 at 6 h compared to the negative viral control, gC (P < 0.05).
Loss of CTCF binding to HSV-1 is not due to CTCF degradation following the application of NaB.
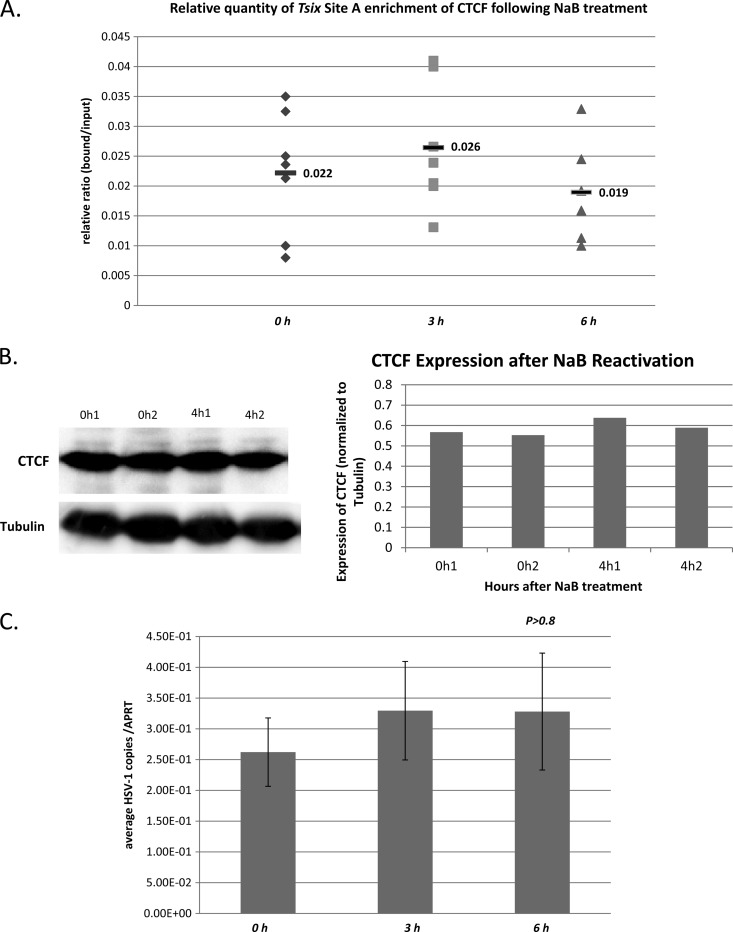
To further confirm that the loss of CTCF enrichment at these sites was not due to an overall loss or degradation of the CTCF protein, we compared the enrichments of CTCF bound to the cellular control, Tsix imprinting/choice center CTCF site A, at 0 h, 3 h, and 6 h post-NaB treatment and performed Western blot (WB) analyses using anti-CTCF with two separate antibodies (Millipore and Abcam) at 4 h post-NaB treatment. We found no significant change in the enrichment of CTCF bound to Tsix imprinting/choice center CTCF site A over any of these time points relative to latency, and the B/I ratios were consistent among experiments at all time points analyzed (Fig. 5A). One-way ANOVA was done by comparing the relative B/I ratios determined for Tsix imprinting/choice center CTCF site A during latency and the B/I ratios for Tsix imprinting/choice center CTCF site A at 3 h or 6 h (P > 0.3 and P > 0.5, respectively). Western blot analysis of total CTCF found in the TG during latency and 4 h post-NaB treatment showed no significant loss or degradation in total CTCF protein following NaB treatment (Fig. 5B). Finally, to confirm that the changes observed in CTCF occupation were not a result of differences in genome copy numbers between experiments, input samples from each ChIP assay were used to determine the HSV-1 genome copy numbers during latency and following NaB-induced reactivation by determining the numbers of copies of HSV-1 DNA polymerase in each sample. These values were normalized to the quantities of mouse APRT in each sample, determined from real-time PCR of each input at 0, 3, and 6 h post-NaB treatment (Fig. 5C). One-way ANOVA was done by comparing the number of normalized relative genome copies during latency to the number of normalized genome copies at 3 h or 6 h post-NaB treatment. We found no significant difference in the numbers of genomes present from each assay, regardless of the time point analyzed following NaB treatment (P > 0.8). Therefore, the loss of CTCF binding to the HSV-1 genome is not a consequence of global CTCF loss or CTCF degradation following NaB treatment.
Fig 5.
Loss of CTCF occupation on the HSV-1 genome is not a consequence of global degradation of CTCF following NaB treatment. To confirm that the loss of CTCF occupation on HSV-1 was not due to an overall loss or degradation of the CTCF protein, we compared the enrichments of CTCF bound to the cellular control, Tsix imprinting/choice center CTCF site A, at 0 h, 3 h, and 6 h post-NaB treatment and performed Western blot analyses using anti-CTCF with two separate antibodies (Millipore and Abcam) at 4 h post-NaB treatment. (A) Relative B/I ratios following ChIP for the cellular control Tsix site A. The ratios obtained from ChIP assays done on latent TG were compared to the B/I ratios of the host control Tsix site A at 3 and 6 h post-NaB treatment. One-way ANOVA comparisons between the B/I ratios for latent versus B/I ratios at 3 h and then at 6 h showed no significant difference in the CTCF occupation of the site following NaB treatment (P > 0.5). (B) Western blot analysis of global CTCF expression in latent mouse TG and in latent mice subjected to NaB at 4 h showed no change or degradation in cellular CTCF. A traditional WB protocol was used in which the membrane was incubated in anti-CTCF antibody (Abcam number ab70303) at a 1:5,000 dilution overnight at 4°C. After exposure, the blots were analyzed with ImageQuant TL software to determine the average intensity of both CTCF and tubulin bands. The CTCF band was normalized to tubulin to control for variations in loading, and the normalized values are plotted for two individual experiments, each containing two TG. (C) The number of HSV-1 genomes per ChIP sample was determined using real-time PCR. The relative numbers of copies of HSV-1 DNA polymerase and mouse APRT were determined by PCR for each input fraction from ChIP assays at 0 (latent), 3, and 6 h post-NaB treatment and are presented as a ratio of HSV-1 DNA pol to APRT. One-way ANOVA of the individual ratios of DNA pol to APRT showed no significant change in HSV-1 copies following NaB treatment through 6 h. The error bars indicate standard deviations from the mean.
CTCF binding to CTRL2, CTa′m, and CTRS3 is disrupted following explant-induced reactivation.
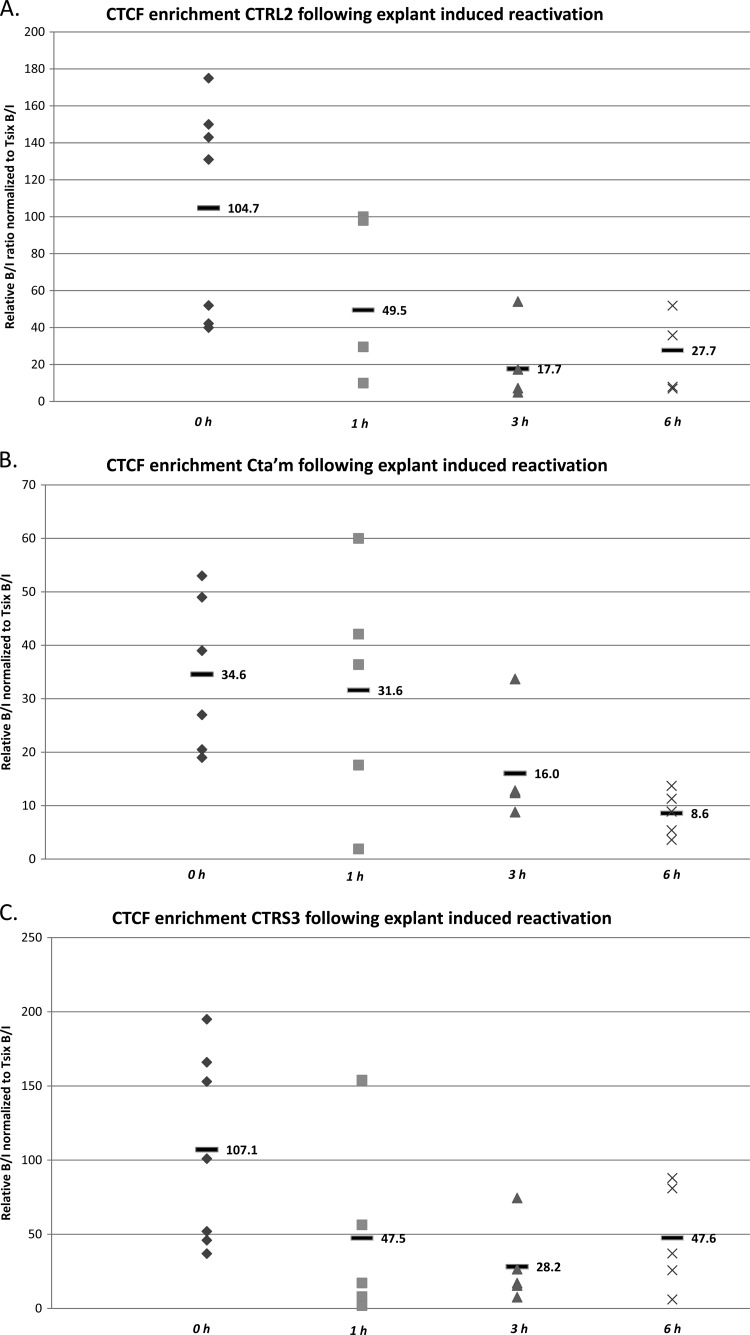
While the use of NaB as a reactivation stimulus has been well documented with a number of herpesviruses, we still acknowledged that NaB has pleiotropic effects in vivo. Therefore, to ensure the changes being observed with respect to decreased CTCF occupation of these binding sites were lost as a consequence of reactivation and not as a direct result of NaB treatment, we confirmed our results using an alternative method of HSV-1 reactivation in the mouse. Here, we employed the explant-induced HSV-1 reactivation model, an ex vivo model that has been consistently and reliably used to study HSV-1 reactivation in latently infected mice (1, 17). We harvested mouse TG latently infected with wild-type 17Syn+, placed them in supplemented medium, and incubated them at 37°C with 5% CO2 for 1, 3, or 6 h, as previously reported (1). ChIP assays were performed as described for NaB experiments. The results of our explant-induced reactivation ChIP experiments confirmed there was a significant loss of CTCF occupation at the CTRL2 (>5-fold; P < 0.009) and CTRS3 (∼4-fold; P < 0.02) binding domains by 3 h postexplant and a significant (>2-fold; P < 0.04) loss of CTCF binding at the CTa′m site by 3 h postexplant (Fig. 6A, B, and C). Further, this was consistent with our observations showing a significant loss of CTCF binding following NaB treatment (see Fig. 4A, B, and C). One notable difference between the two models was the CTCF binding at the CTRS3 domain in HSV-1. While the fold changes in CTCF occupation at each site were consistent between the two reactivation models by 3 h postreactivation, CTCF binding was not abolished at the CTRS3 motif, as was observed in the NaB-treated mice. We noted that at the CTRS3 domain, CTCF occupation significantly decreased by 3 h postexplant, consistent with NaB-treated mice at this time point. Based on our data from ChIP experiments following explant-induced reactivation, CTCF binding at the CTRS3 motif appeared to be increasing at 6 h postexplant. This finding is not surprising and is consistent with other transient epigenetic changes to the HSV-1 genome following reactivation. Specifically, epigenetic changes to euchromatin in the lytic regions of HSV-1 during reactivation are transient in nature, and latency and reactivation are reversible states; therefore, it is logical that changes to CTCF occupation are also transient. We suspect that we may be observing the reoccupation of CTRS3 by 6 h postexplant, which was not observed in the NaB-treated mice due to differences in reactivation kinetics between the two models.
Fig 6.
CTCF occupation following explant-induced reactivation of HSV-1 in mouse TG. Mice latently infected with HSV-1 17Syn+ were subjected to explant-induced HSV-1 reactivation for 1 h, 3 h, and 6 h by placing TG in supplemented medium and incubating each sample at 37°C with 5% CO2 for either 1, 3, or 6 h. ChIP assays using anti-CTCF were combined with real-time PCR analyses to compare explanted TG to TG from latently infected mice. All ChIP assays contained 3 mice (6 TG) pooled, and ChIPs postexplant were repeated 5 times (n = 5). All ratios reported are normalized to the B/I ratios of the host control Tsix imprinting/choice center CTCF site A for that experiment. The time following explant is represented on the x axis. The horizontal bars in the figures represent the averages of the individual normalized ratios from ChIP for each time point. (A) CTCF occupation of the CTRL2 domain following explant is significantly lower by 3 h postexplant than during latency (>5-fold decrease; P < 0.009). (B) CTCF enrichment of the CTa′m domain following explant is ∼2-fold lower by 3 h postexplant (P < 0.04) and is essentially abolished by 6 h postexplant. (C) CTCF enrichment of the CTRS3 domain following explant is significantly lower by 3 h postexplant (∼4-fold; P < 0.02). These data are similar to what was observed over 6 h following NaB-induced HSV-1 reactivation of latently infected mice.
CTa′m and CTRS3 motifs in HSV-1 display enhancer-blocking activity.
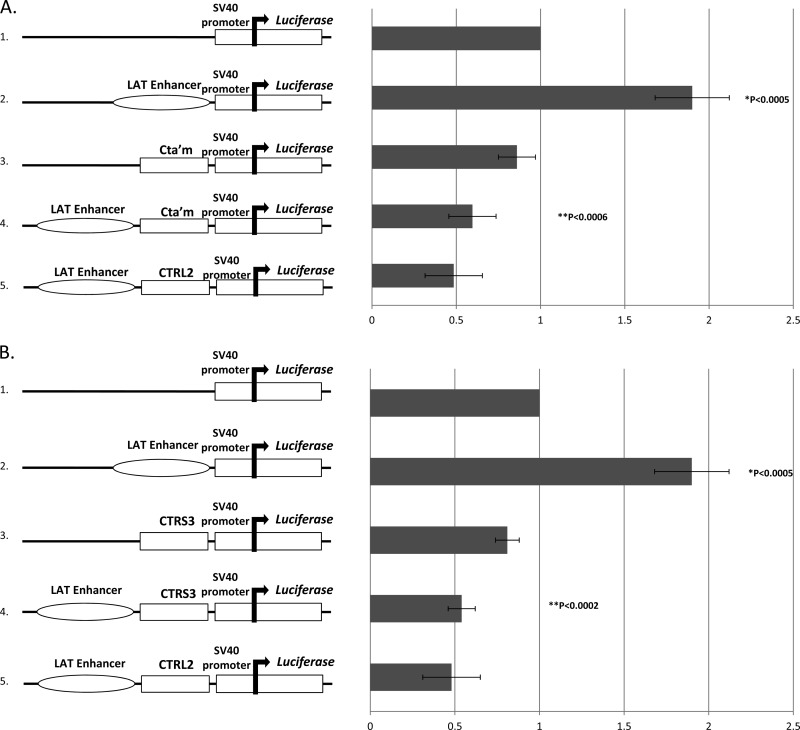
Considering that the CTRL2 region has already been characterized as an insulator with enhancer-blocking and silencing properties (2), we anticipated that we would observe a loss of CTCF binding to the region following reactivation. Loss of CTCF binding to CTRL2 could result in loss of insulator activity, which could, in turn, allow the LAT enhancer to activate the IE regions of HSV-1. This change could be responsible for the subsequent cascade of IE gene activation at early times in a reactivation event. However, it was more intriguing that a significant loss of CTCF binding to both the CTa′m and CTRS3 domains was also observed in parallel following the application of reactivation stimuli. Each of these CTCF clusters lies upstream of IE promoters. Our data further indicated that each CTCF binding domain identified in HSV-1 might be a functional insulator that is independently regulated. In order to determine whether these motifs were functional insulators, we set out to determine if the CTa′m and/or the CTRS3 region was able to act as an enhancer blocker of the LAT enhancer. To do so, we generated luciferase reporter constructs to test whether each of these CTCF binding motifs possessed enhancer-blocking ability. A number of reporter constructs were generated (see Materials and Methods), including a construct containing a 589-bp fragment corresponding to the LAT enhancer element. Additionally, a 1.3-kb fragment containing sequences flanking the previously identified core 217-bp CTa′m CTCF cluster and a 1.6-kb fragment containing sequences flanking the previously identified 111-bp core CTRS3 CTCF cluster were also constructed and tested for insulator activity. Constructs were generated to include ∼1,000 bp flanking each motif due to the fact that functional properties of eukaryotic insulator elements are located 5′ and 3′ to the core CTCF binding motifs (46). Enhancer elements have been routinely characterized in cellular literature as distance-independent elements having the capability of routinely activating gene promoters at distances of >2 kb; therefore, the addition of 1,000-bp sequences in our plasmid constructs should not have an effect on the functions of the core CTCF motifs, as related to distance (5, 22). An additional luciferase construct identical to the previously reported and characterized CTRL2 construct (containing a fragment of 2.1 kb) was also generated as a positive control for the enhancer-blocking assays. Transient-transfection assays were done using each construct in triplicate on rabbit skin cells to assess for enhancer-blocking activity. We found that the construct containing the LAT enhancer (pGL3-control/LTE) (Fig. 7A, construct 2) was capable of enhancing transcription from a heterologous promoter (the SV40 promoter) by ∼2-fold (P < 0.0005). Conversely, the 1.3-kb CTa′m construct (pGL3-control/CTa′m) (Fig. 7A, construct 3) alone slightly reduced the effect of the SV40 promoter-driven luciferase expression (enhanced by the SV40 enhancer present in the commercial construct) (Fig. 7A, construct 1). However, when the CTa′m fragment was positioned between the LAT enhancer and the SV40 promoter in the construct (pGL3-control/LTE+CTa′m) (Fig. 7A, construct 4), it essentially blocked the effects of the LAT enhancer, as measured by the >3-fold reduction in luciferase expression relative to the LAT enhancer construct (P < 0.0006). These findings are comparable to what was found with the previously characterized CTRL2 motif (2). Similarly, when the 1.6-kb CTRS3 fragment was directionally cloned into the reporter constructs, the CTRS3 fragment itself induced a slight reduction in SV40 promoter-driven luciferase expression, and when positioned between the LAT enhancer and the SV40 promoter (pGL3-control/LTE+CTRS3) (Fig. 7B, construct 4), the motif was able to block the enhancer activity of the LAT enhancer, as measured by the >3.5-fold reduction in luciferase expression relative to the LAT enhancer construct (Fig. 7B, construct 2). This demonstrates that both of the CTCF clusters individually possess classic enhancer-blocking activity and can function as insulators for the LAT enhancer of HSV-1.
Fig 7.
CTa′m and CTRS3 binding motifs in HSV-1 display insulator activity as enhancer blockers. Luciferase reporter constructs were generated to test the ability of the CTCF binding motif to block the LAT enhancer. The enhancer-blocking activity of the CTa′m and CTRS3 motifs of HSV-1 were assayed in transient-transfection assays using rabbit skin cells. All transfections were done in triplicate wells of rabbit skin cells. All transfection assays were repeated 5 times (n = 5). *, P values representing unpaired one-tailed Student's t tests in pairwise comparisons to the pGL3-control construct. **, P values representing comparison to the pGL3-control/LTE construct. The data were collected in relative luciferase units and normalized to an internal Renilla luciferase control. All values are expressed relative to the pGL3-control vector, which is normalized to a value of 1. The error bars represent standard deviations from the mean. (A) (1) pGL3-control vector from Promega (containing an SV40 promoter) was used as a base for all reporter constructs generated. (2) Reporter construct containing the 589-bp fragment LAT enhancer (pGL3-control/LTE). (3) A 1.3-kb reporter construct encompassing CTa′m (pGL3-control/CTa′m). (4) Reporter construct with the CTa′m fragment positioned between the SV40 promoter and the LAT enhancer (pGL3-control/LTE+CTa′m). (5) CTRL2 construct used as a positive control (2). The CTa′m fragment was able to significantly block LAT enhancer activity on the SV40 promoter in transient transfections. (B) (1) pGL3-control vector from Promega (containing an SV40 promoter) was used as a base for reporter constructs generated. (2) Reporter construct containing the 589-bp fragment LAT enhancer (pGL3-control/LTE). (3) A 1.6-kb reporter construct encompassing CTRS3 (pGL3-control/CTRS3). (4) Reporter construct with the CTRS3 fragment positioned between the SV40 promoter and the LAT enhancer (pGL3-control/LTE+CTRS3). (5) CTRL2 construct used as a positive control (2). The CTRS3 fragment was able to block LAT enhancer activity on the SV40 promoter. The CTa′m and CTRS3 sites of HSV-1 displayed enhancer-blocking activity comparable to that of the previously reported CTRL2 enhancer blocker of HSV-1 (2).
The 17Δ348 and KD6 mutant viruses lack ICP0 and ICP4 transcripts during latency and following the application of reactivation stimuli in mouse TG.
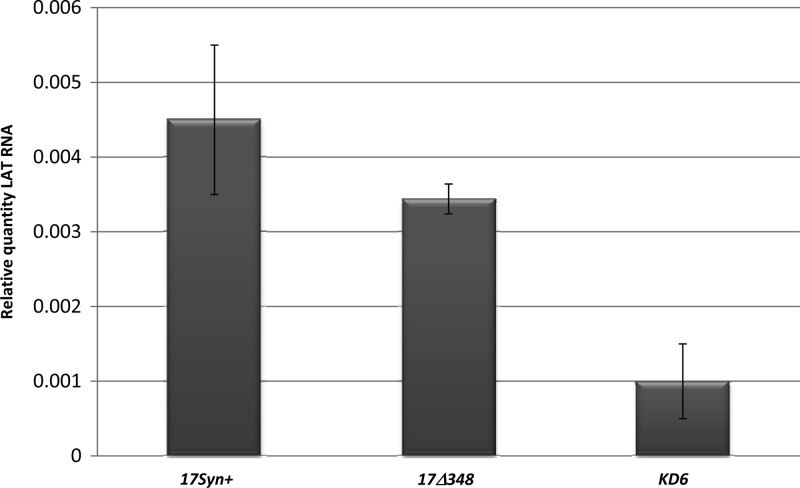
It has been reported that that by 28 days postinfection, mice latently infected with KD6 have no detectable IE transcripts (ICP0, ICP4, or ICP27) present in ganglia as measured by in situ hybridization, even though the virus maintains levels of LAT equivalent to those in the wild-type virus (37). It has also been reported that the LAT enhancer mutant 17Δ348 expresses LAT equivalent to that of the wild-type virus (8), yet both mutants display severely attenuated reactivation phenotypes in vivo (4, 30). We utilized each of these mutants to determine the effects that IE may have on loss of CTCF binding to the HSV-1 genome following reactivation in the presence of LAT. qRT-PCR was performed on the TG of mice latently infected with either 17Δ348 or KD6 to quantitatively measure transcript levels of LAT, ICP0, and ICP4. In both recombinant viruses, we were unable to detect any measurable IE transcripts during latency. In comparison, baseline levels of ICP0 and ICP4 were detected in 17Syn+ (references 1 and 31 and data not shown). This was not an unexpected finding, particularly for KD6, as previous reports had indicated that the virus lacked any lytic transcription beyond 14 days postinfection (37). However, when we quantitated the LAT transcript abundance for each of the mutant viruses, we found a measurable reduction in the amount of LAT present (∼3-fold) in latent KD6 TG relative to 17Δ348 and wild-type virus (Fig. 8). This finding was somewhat surprising, because previous reports showed LAT levels to be equivalent to those of the wild-type virus during latency in KD6 ganglia. However, it must be noted that the KD6 mutant virus was derived from the parent KOS, and not 17Syn+, which could account for the differences in LAT RNA abundance between the viruses. The implications of this finding and how it could relate to CTCF binding are further discussed below (see Discussion). Nonetheless, the fact that each mutant virus lacked measurable IE expression in the presence of LAT allowed us to evaluate only the effects of altered IE transcription (relative to the wild type).
Fig 8.
LAT RNA abundance following latency in 17Syn+, 17Δ348, and KD6 viruses. LAT transcript abundance was determined following infection at 28 days. RNA was isolated and analyzed by RT-PCR using primers and probes specific for the LAT intron. The relative quantities were determined for LAT and then normalized to the relative quantities of mouse APRT to account for variations between samples. The average values for each virus are presented, along with standard deviations from the mean. All samples were done in triplicate with 5 mice (10 TG).
CTCF occupation of the CTRL2 domain appears to be LAT dependent during latency.
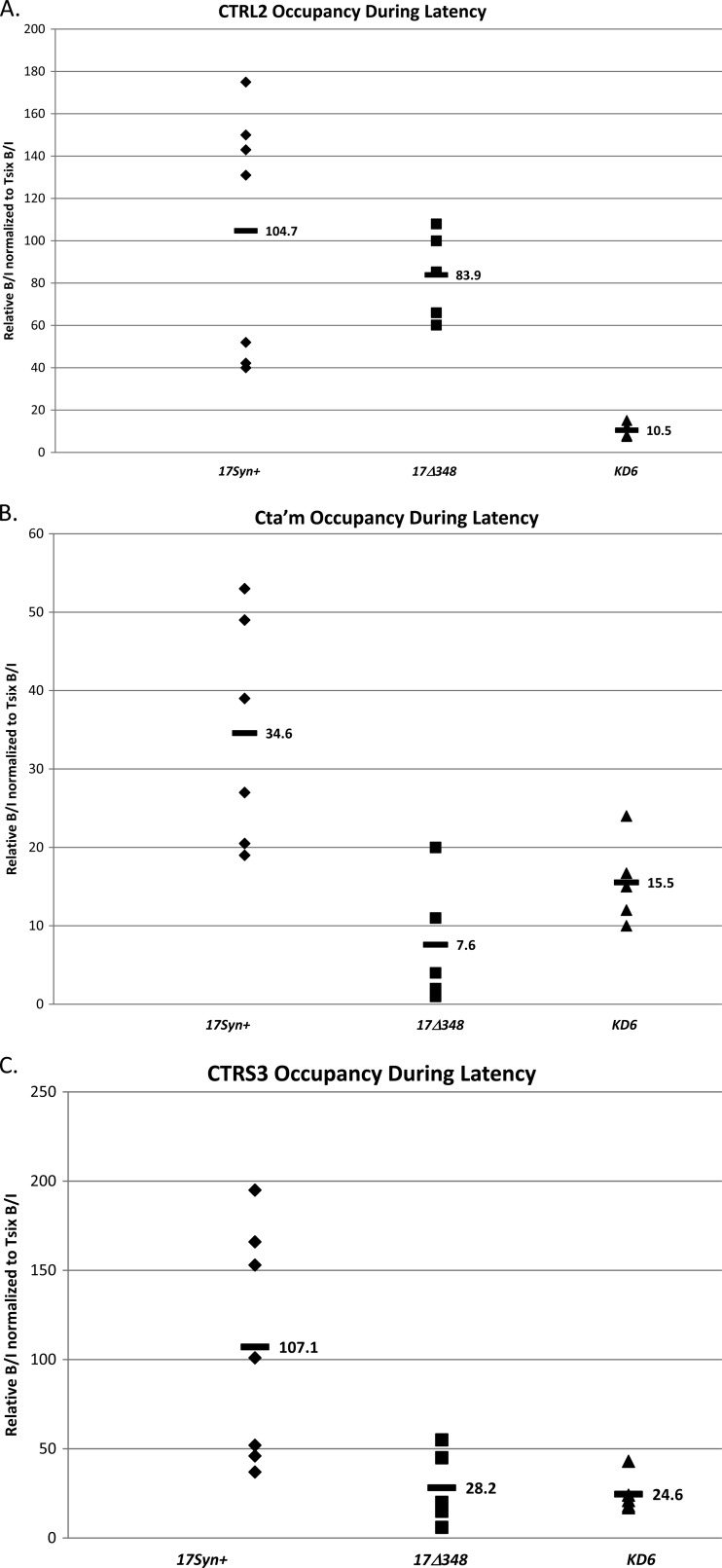
We have shown that the CTRL2, CTa′m, and CTRS3 binding motifs of HSV-1 are enriched in bound CTCF during latency (Fig. 3). We have further demonstrated that CTCF binding to the HSV-1 genome may contribute significantly to the maintenance of the latent infection in neurons through its insulator activity as an enhancer blocker of the LAT enhancer. Conversely, the occupation of CTCF on HSV-1 sites is disrupted at early times during reactivation, leading to the possibility that a loss of insulator activity (which could result in the LAT enhancer activating distal IE promoters) is fundamental to reactivation. To explore the elements required, not only for the deposition of CTCF on the HSV-1 genome during latency, but to tease out mechanisms by which CTCF binding may be disrupted during a reactivation event, as well, we analyzed the patterns of CTCF binding to the two recombinant viruses 17Δ348 and KD6. Data have shown that by 28 days postinfection, mice latently infected with KD6 have no detectable IE transcripts (ICP0, ICP4, or ICP27) present in ganglia (37). We subsequently confirmed these data using qRT-PCR. Additionally, we have determined that mice infected with 17Δ348 also lack detectable ICP0 and ICP4 transcripts during latency in the TG. Finally, both KD6 and 17Δ348 have severely restricted reactivation phenotypes in vivo. The first notable difference between these three viruses was with respect to the CTCF enrichment of the CTRL2 region during latency (Fig. 9A). Relative to the wild type, the IE transcript-null KD6 had essentially no CTCF bound to this domain, while the LAT enhancer mutant 17Δ348 remained enriched in CTCF at the CTRL2 motif. This was an interesting finding, because when we quantified the abundance of LAT transcripts during latency, we found a 3-fold reduction in LAT abundance in KD6 relative to the wild type and 17Δ348 (Fig. 8). These data suggest that abundant LAT transcription may significantly contribute to occupation of CTCF in the CTRL2 domain during latency. Conversely, the presence of the LAT enhancer element does not contribute to CTCF occupation of the site during latency, as the KD6 recombinant maintains the intact enhancer element, yet CTCF binding is significantly attenuated in the KD6 mutant at CTRL2.
Fig 9.
CTCF occupation of CTRL2, CTa′m, and CTRS3 during latency in viruses. ChIP data are presented for each site during latency for comparison of the three viruses used. All ChIPs were validated as described in Materials and Methods. Each data point represents a normalized B/I ratio for 3 mice (6 TG pooled). Each time point contains a minimum of 5 data points (n = 5). All relative B/I ratios from each gene region were normalized to the B/I ratio for the cellular control Tsix site A B/I for that ChIP assay. The horizontal bars represent the average normalized B/I ratios for a given gene region from all ChIP assays at that time point. Statistical data were determined by one-way analysis of variance by comparing the normalized B/I ratio of each gene region to the normalized B/I ratio for 17Syn+. (A) CTRL2 occupation during latency. No statistical difference was found between CTCF occupation of 17Δ348 and 17Syn+ (P > 0.1). (B) CTa′m occupation during latency. There was a 2-fold reduction in CTCF bound to the site in KD6 and a 4-fold reduction in 17Δ348 during latency compared to the wild type (P < 0.02 and P < 0.005, respectively). (C) CTRS3 occupation during latency. Both recombinants have ∼4-fold lower CTCF occupation at this site during latency than the wild type (P < 0.05).
CTCF occupation of the CTa′m and CTRS3 domains of HSV-1 is dependent on IE gene transcription during latency.
We also analyzed the CTCF occupation of the CTCF binding domains upstream of the IE promoters (ICP0 and ICP4) of KD6 and 17Δ348 during latency. ChIP analyses revealed a 2-fold reduction in CTCF bound to the CTa′m site of KD6 and a 4-fold reduction in CTCF bound to CTa′m of 17Δ348 relative to the wild type (P < 0.02 and P < 0.005, respectively) (Fig. 9B). Similarly, when we analyzed the CTRS3 motif, upstream of the ICP4 promoter, again we found an ∼4-fold reduction of CTCF bound to the site in both KD6 and 17Δ348 recombinants relative to the wild type (Fig. 9C). Considering that both viruses have undetectable IE transcripts during latency as measured by qRT-PCR but the 17Δ348 virus has no deletion of ICP4 or any other IE gene region, it is reasonable to think that the presence of IE transcripts, even baseline levels reported in the wild-type virus, are likely a critical component in the CTCF occupation of these insulator elements during latency. Furthermore, collectively, these data also indicate that each CTCF binding domain present in HSV-1 may in fact be independently regulated during latency in a site-specific manner, either by the presence of abundant LAT transcription or by IE transcripts.
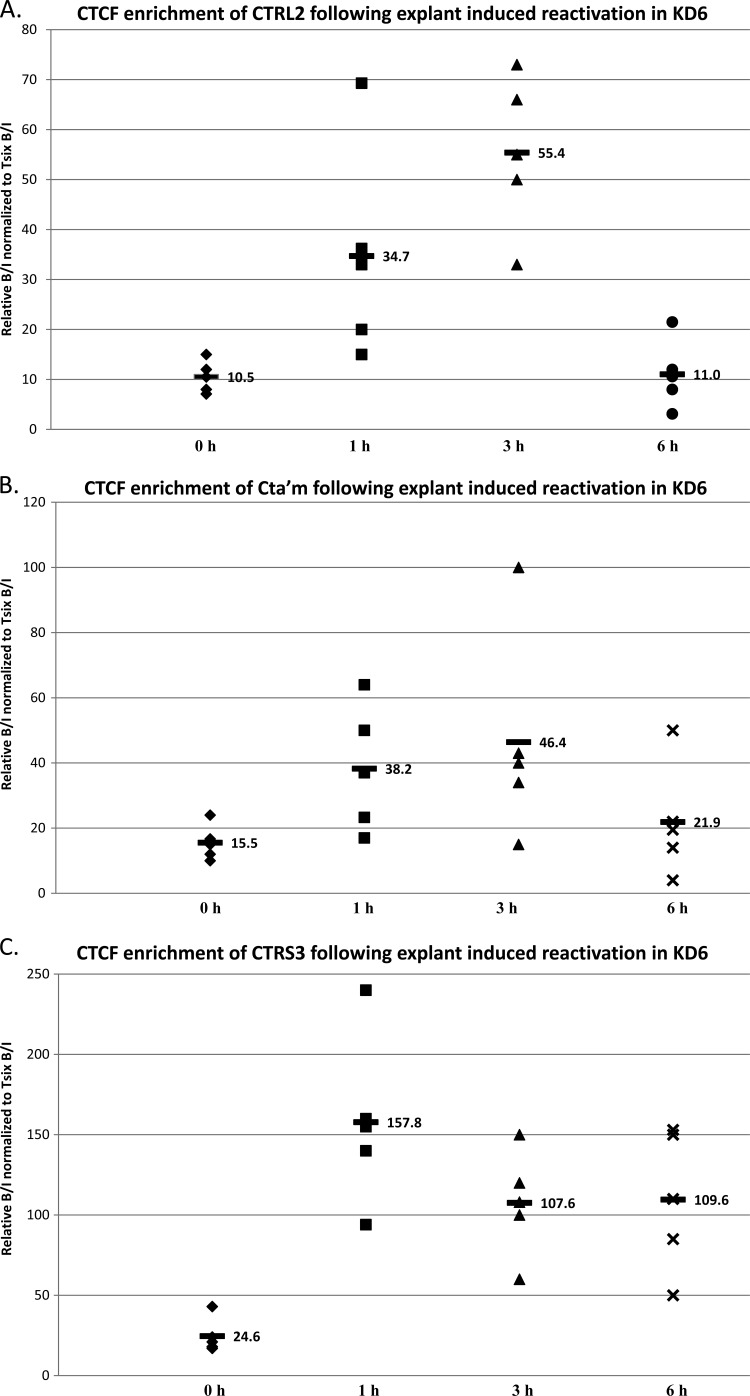
CTCF occupation of the CTRL2 and CTRS3 domains in KD6 significantly increases following the application of a reactivation stimulus.
Considering the multifaceted role of CTCF in the context of both eukaryotes and gamma herpesviruses, we hypothesized that disruption of CTCF binding to the HSV-1 genome might be a fundamental step in the process of reactivation, perhaps facilitated by an initial increase in IE gene transcription following the application of a reactivation stimulus. This is a somewhat difficult possibility to assess, particularly in the mouse model. We and others have shown that LAT transcript abundance rapidly decreases (likely by LAT degradation) by 4 h postexplant or post-NaB reactivation in parallel with the changes in chromatin at these times (1, 31). However, while IE transcripts were detected, the studies were unable to detect significant increases in IE gene transcription in parallel with chromatin changes to ICP0 and ICP4 postreactivation in the mouse (through 4 h), indicating that only small increases in the production of IE transcripts might be occurring at early times following the application of a reactivation stimulus (1, 31). Therefore, to tease out mechanisms by which IE transcription might contribute to the displacement of CTCF at early times postreactivation, we sought to determine the effects that the absence of IE gene transcription would have on CTCF occupation of these insulators at very early times following a reactivation stimulus in the IE-null recombinant. In wild-type 17Syn+, the CTCF occupation of the CTCF binding motifs CTRL2, CTa′m, and CTRS3 decrease significantly following both NaB- and explant-induced reactivation. To determine whether IE gene transcription is required for the disruption of CTCF binding to each of these sites, we performed additional experiments in which the TG of mice latently infected with KD6 were subjected to explant-induced reactivation for 1, 3, or 6 h. Subsequent ChIP analyses and real-time PCR were done (see Materials and Methods). Analyses of the CTRL2 (downstream of the LAT enhancer) and the CTRS3 (upstream of ICP4) revealed that CTCF occupation at both sites significantly increased following explant: there was an average 3-fold increase in CTCF at the CTRL2 motif at 3 h postexplant (P < 0.05) (Fig. 10A) and an average 5-fold increase in CTCF at the CTRS3 motif by 1 h postexplant (Fig. 10C) in the ICP4-null recombinant KD6. In contrast, we detected no significant change in CTCF bound to the CTa′m site upstream of the ICP0 promoter following explant of KD6 TG at any time through 6 h postexplant (Fig. 10B). These data, taken together, are very intriguing and indirectly suggest that changes in IE gene expression following a reactivation stressor in wild- type HSV-1 might be a major contributing factor in the loss of CTCF binding to the CTCF insulator sites of HSV-1.
Fig 10.
CTCF occupation following explant-induced reactivation of HSV-1 in mouse TG latently infected with KD6. Mice latently infected with HSV-1 KD6 were subjected to explant-induced HSV-1 reactivation for 1 h, 3 h, and 6 h by placing TG in supplemented medium and incubating each sample at 37°C with 5% CO2 for either 1, 3, or 6 h. ChIP assays using anti-CTCF were combined with real-time PCR analyses to compare explanted TG to TG from latently infected mice. All ChIP assays contained 3 mice (6 TG) pooled, and ChIPs postexplant were repeated 5 times (n = 5). All ratios reported are normalized to the B/I ratios of the host control Tsix imprinting/choice center CTCF site A for that experiment. The time following explant is represented on the x axis. The horizontal bars represent the average of the individual normalized ratios from ChIP for each time point. (A) CTCF occupation of the CTRL2 domain following explant is >5-fold more enriched in CTCF by 3 h postexplant than during latency. (B) CTCF occupation of the CTa′m domain following explant is ∼2.5-fold more enriched in CTCF by 3 h postexplant. (C) CTCF occupation of the CTRS3 domain following explant is >5-fold more enriched in CTCF by 1 h postexplant.
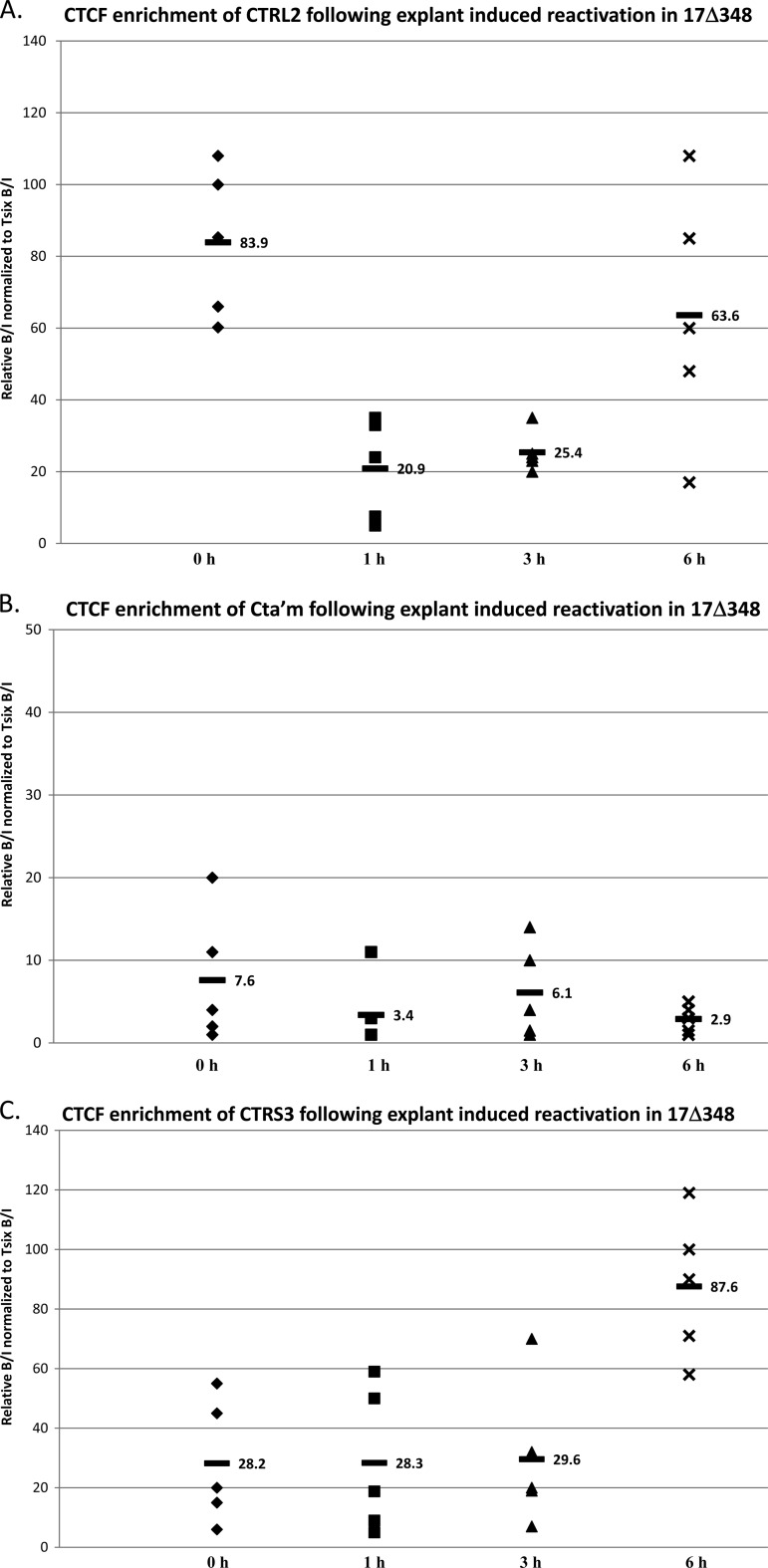
The LAT enhancer element is not required for loss of CTCF occupation at the CTRL2 site.
We hypothesize that the LAT enhancer element of HSV-1 may activate IE promoters to initiate IE transcription following the loss of CTCF in a reactivation event. We have shown in this study that both the CTa′m and CTRS3 sites, upstream of the ICP0 and ICP4 promoters, are functional enhancer-blocking insulators. CTCF occupation of HSV-1 insulator sites is disrupted at very early times following the application of reactivation stimuli in wild-type virus. Our analysis of 17Δ348 by ChIP to determine CTCF occupation of CTRL2 shows that CTCF binding was significantly decreased at the site by 1 h postexplant in a transient manner, similar to what was observed for wild-type virus (Fig. 11A), even in the absence of the LAT enhancer. In contrast, we saw no change in CTCF binding to CTa′m in 17Δ348 (Fig. 11B). Finally, as with the KD6 recombinant, we saw a transient increase in CTCF occupation of the CTRS3 site upstream of the ICP4 promoter in 17Δ348 following explant (Fig. 11C). While our data cannot directly link the LAT enhancer to activating IE genes for transcription, the fact that CTCF occupation of the CTRL2 site decreases in parallel to the wild type in 17Δ348 while no decrease is observed in the CTRS3 site of 17Δ348 following explant does not discredit this as a possibility, and further exploration of the LAT enhancer's ability to activate IE promoters in reactivation is warranted.
Fig 11.
CTCF occupation following explant-induced reactivation of HSV-1 in mouse TG latently infected with 17Δ348. Mice latently infected with HSV-1 17Δ348 were subjected to explant-induced HSV-1 reactivation for 1 h, 3 h, and 6 h by placing TG in supplemented medium and incubating each sample at 37°C with 5% CO2 for either 1, 3, or 6 h. ChIP assays using anti-CTCF were combined with real-time PCR analyses to compare explanted TG to TG from latently infected mice. All ChIP assays contained 3 mice (6 TG) pooled, and ChIPs postexplant were repeated 5 times (n = 5). All ratios reported are normalized to the B/I ratios of the host control Tsix imprinting/choice center CTCF site A for that experiment. The time following explant is represented on the x axis. The horizontal bars in the figures represent the averages of the individual normalized ratios from ChIP for each time point. (A) CTCF occupation of the CTRL2 domain following explant had a 4-fold decrease 1 h postexplant compared to latency. (B) CTCF enrichment of the CTa′m domain following explant did not change. (C) CTCF enrichment of the CTRS3 domain following explant was >3-fold greater by 6 h postexplant.
DISCUSSION
We have shown that the three CTCF binding clusters (CTRL2, CTa′m, and CTRS3) downstream of the LAT enhancer and upstream of the ICP0 and ICP4 promoters, respectively, are highly enriched in CTCF during latency in the mouse TG following ocular inoculation. In addition, each of these three sites possesses characteristics of functional insulators, namely, each has the ability to act as an enhancer blocker of the LAT enhancer. Further, it has been reported that the CTRL2 region downstream of the LAT enhancer also displays a silencer function in transient transfections (2). Considering these findings, it is likely that these CTCF clusters maintain the transcriptional integrity of chromatin domains around the LAT and IE regions in a manner similar to the way in which cellular chromatin domains are regulated by insulators (19, 46). For example, maintenance of chromatin domains in HSV-1 could be facilitated by insulating the LAT enhancer from activating nearby IE promoters, thereby preventing increased IE transcription. Alternatively, these CTCF insulators could also act as corepressive elements with other silencer proteins bound to DNA elements in HSV-1. Both are plausible mechanisms, considering reports in the literature. For example, it has been shown that the LAT 5′ exon region of HSV-1, which contains an enhancer element, is highly enriched in permissive chromatin, while the ICP0 gene (antisense to LAT) (Fig. 1A) is enriched in repressed chromatin. Further, enhancers can routinely facilitate promoter activity in genes over 2 kb away (20). In the context of HSV-1, the LAT enhancer is positioned to activate the ICP0 promoter to increase ICP0 transcription, but during latency, this does not occur. These data, together with previous data showing that the CTRL2 motif is an enhancer-blocking insulator, strongly indicate that the integrity of the LAT and ICP0 chromatin domains is maintained by enhancer-blocking insulators. Alternatively, the cellular literature shows that CTCF can act as a corepressor with other chromatin-modifying proteins, such as YB-1 and sin3, in order to maintain transcriptionally repressive histones on genomic regions (28). Importantly, Amelio et al. have also characterized the CTRL2 cluster as an insulator element capable of a silencer function (2). In the context of HSV-1, lytic promoters are associated with repressive histone modifications, and at least one corepressor protein complex (polycomb-repressive complex 1) has been shown to be recruited to IE regions of HSV-1 to help establish and maintain repressive chromatin marks on IE regions during latency (12, 21, 25). Therefore, the probability that CTCF bound to HSV-1 CTCF clusters may act as a corepressor with silencer or repressive complexes to maintain chromatin domain integrity during latency must be further explored.
A significant finding of our study was the fact that each of the three CTCF binding clusters in HSV-1 loses CTCF enrichment following the application of NaB or explantation as a reactivation method in the wild-type HSV-1 17Syn+. These findings can be correlated with both changes in LAT transcription and the abundance of permissive histone marks on IE regions of HSV-1 following reactivation in the mouse model and provide an explanation of why the distinct segregation of heterochromatin from euchromatin observed in the latent genome is no longer intact at early times in reactivation. CTCF plays a multifaceted role in both eukaryotic gene regulation and gammaherpesvirus gene regulation. Therefore, it is plausible that a critical component in HSV-1 reactivation would be the loss of CTCF binding to CTCF clusters along the genome. This loss of CTCF binding could (i) facilitate the activation of IE regions by the LAT enhancer of HSV-1 or (ii) result in the spread of heterochromatin associated with the IE regions to the reactivation-critical regions of the LAT. However, perhaps even more intriguing are recent studies done in both eukaryotes and gammaherpesviruses that prove that CTCF regulates gene transcription through the formation of chromatin loop domains. The formation of loop domains during latency is not novel to gammaherpesvirus research, as recent reports have shown CTCF-mediated loop domain formation in latent EBV. The idea that alternative CTCF-mediated loop domains may mitigate latency and reactivation in HSV-1 is an exciting possibility that warrants further exploration.
Finally, our data also suggest that each CTCF domain may be populated and regulated independently during latency. For example, the LAT enhancer recombinant 17Δ348 produces LAT RNA levels comparable to those of wild-type 17Syn+, while the ICP4-null recombinant, KD6 (from the KOS background), has 3-fold less LAT RNA than 17Syn+ and 17Δ348. Even in the presence of LAT, this reduction in LAT levels between the two viruses appears to be responsible for significant differences in the CTCF occupation of the CTRL2 site (downstream of the LAT enhancer). For example, in KD6, there is essentially no CTCF bound to this motif, while 17Δ348 has CTCF occupation at the site equivalent to that of the wild type. This unexpected finding highlights not only the fact that LAT abundance may be dictating CTCF population at the site, but also the fact that there may be very significant differences in the epigenetic regulation of wild-type strains of 17Syn+ and KOS, a highly efficient reactivator and an inefficient reactivator of HSV-1. Further, following explant-induced reactivation of latent 17Δ348 mouse TG, we showed that the CTRL2 domain of 17Δ348 loses CTCF occupation, similar to the wild type, yet this is not the case for the KD6 recombinant (which becomes transiently enriched in CTCF at this site). Our analyses of the RNA abundance of LAT following explantation in both viruses showed that in 17Δ348 there is a rapid degradation of LAT by 3 h postexplant. In contrast, no decreased LAT abundance was observed in KD6 explanted ganglia. These data further support the hypothesis that each CTCF binding domain may be differentially regulated in a site-specific manner by the presence of either LAT or IE transcripts.
Consequently, our ChIP analyses of CTCF occupation show that IE transcription is necessary for the establishment of some insulator domains in HSV-1, namely, around the ICP0 and ICP4 regions of the genome. Our studies found that both 17Δ348 and KD6 were lacking or significantly attenuated in CTCF occupation of the CTa′m and CTRS3 sites upstream of the IE promoters. We found no significant difference in the numbers of viral genomes present in the mouse TG between 17Δ348, KD6, and 17Syn+ latent mice, but we did determine, using qRT-PCR, that both recombinants lacked IE transcripts while wild-type 17Syn+ had detectable IE transcripts during latency. Therefore, based on these data, it seems likely that our findings result from a lack of IE transcription and are not solely based on the presence of LAT during latency. Alternatively, following explant-induced reactivation of KD6 and 17Δ348 latent mouse TG, we found significant increases in CTCF occupancy at both CTRL2 and CTRS3. This is in direct contrast to what was observed for the wild-type virus, where these sites significantly lose CTCF binding in a parallel time frame. These findings were initially perplexing, because we attributed the loss of CTCF directly to IE transcription during reactivation. We also anticipated that without the expression of IE transcription (in the absence of reactivation) we would observe no change in the occupancy of these domains by CTCF following the application of a reactivation stimulus in a nonreactivating IE transcript-null virus. However, recent literature reports focusing on CTCF regulation in eukaryotes have shown that (i) the locations of CTCF binding sites are directly linked to epigenetic markers, particularly H3 methylation (29); (ii) CTCF binding sites are hyperenriched in all three states of H3K4 methylation (3); and (iii) CTCF preferentially binds to regions containing methylation of H3 (33). Taking these reports into consideration, it is apparent that the methylation state of lysine residues of H3 is an integral component in CTCF regulation, and changes in H3 methylation can dramatically alter the CTCF occupation of insulator motifs. The possibility of significant differences existing in H3 methylation profiles between wild-type and recombinant viruses is not without precedent in HSV-1. For example, it has been reported that the IE regions of 17Syn+ are enriched in the facultative heterochromatic marker triMeH3K27 during latency. The same reports have also shown that the LAT promoter deletion virus 17ΔPst is significantly more enriched in this facultative heterochromatic marker in the IE gene regions (2- to 5-fold) than the parent 17Syn+ (25). Therefore, it is plausible that the increased CTCF occupation of CTRL2 and CTRS3 sites in KD6 and 17Δ348 following explantation may result from changes in H3 lysine residue methylation, and this is currently being explored.
In summary, our data show that the CTCF domains positioned within the HSV-1 genome, specifically around the LAT and ICP0 and ICP4 regions of the genome, function as enhancer-blocking insulators during latency. Further, these domains lose CTCF occupancy following the application of reactivation stimuli in wild-type virus. Finally, the CTCF binding domains of HSV-1 are differentially regulated both during latency and at early times following reactivation in the mouse model by the presence of LAT and IE transcripts during latency and following the application of a reactivation stressor.
ACKNOWLEDGMENTS
This work was supported in part by funding from the LSUHSC Department of Pharmacology and Experimental Therapeutics and the Louisiana Lions Eye Foundation and by an unrestricted grant from Research to Prevent Blindness, New York, NY (LSU Department of Ophthalmology-LSUHSC). We also thank David C. Bloom (University of Florida) for providing us with HSV-1 strains 17Syn+, 17Δ348, and KD6.
Footnotes
Published ahead of print 12 September 2012
REFERENCES
- 1. Amelio AL, Giordani NV, Kubat NJ, O'Neil J, Bloom DC. 2006. Deacetylation of the herpes simplex virus type 1 latency-associated transcript (LAT) enhancer and a decrease in LAT abundance precede an increase in ICP0 transcriptional permissiveness at early times postexplant. J. Virol. 80:2063–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Amelio AL, McAnany PK, Bloom DC. 2006. A chromatin insulator-like element in the herpes simplex virus type 1 latency-associated transcript region binds CCCTC-binding factor and displays enhancer-blocking and silencing activities. J. Virol. 80:2358–2368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barski A, et al. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129:823–837 [DOI] [PubMed] [Google Scholar]
- 4. Bhattacharjee PS, et al. 2003. Overlapping subdeletions within a 348-bp in the 5′ exon of the LAT region that facilitates epinephrine-induced reactivation of HSV-1 in the rabbit ocular model do not further define a functional element. Virology 312:151–158 [DOI] [PubMed] [Google Scholar]
- 5. Blackwood EM, Kadonaga JT. 1998. Going the distance: a current view of enhancer action. Science 281:60–63 [DOI] [PubMed] [Google Scholar]
- 6. Bloom DC. 2006. HSV-1 latency and the roles of the LATs, p 325–342 In Sandri-Goldin R. M. (ed), Alpha herpesviruses: molecular and cellular biology, 1st ed Caister Academic Press, Norwich, United Kingdom [Google Scholar]
- 7. Bloom DC, Giordani NV, Kwiatkowski DL. 2010. Epigenetic regulation of latent HSV-1 gene expression. Biochim. Biophys. Acta 1799:246–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bloom DC, et al. 1996. A 348-base-pair region in the latency-associated transcript facilitates herpes simplex virus type 1 reactivation. J. Virol. 70:2449–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Capelson M, Corces VG. 2004. Boundary elements and nuclear organization. Biol. Cell 96:617–629 [DOI] [PubMed] [Google Scholar]
- 10. Chao W, Huynh KD, Spencer RJ, Davidow LS, Lee JT. 2002. CTCF, a candidate trans-acting factor for X-inactivation choice. Science 295:345–347 [DOI] [PubMed] [Google Scholar]
- 11. Chen Q, et al. 2007. CTCF-dependent chromatin boundary element between the latency-associated transcript and ICP0 promoters in the herpes simplex virus type 1 genome. J. Virol. 81:5192–5201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cliffe AR, Garber DA, Knipe DM. 2009. Transcription of the herpes simplex virus latency-associated transcript promotes the formation of facultative heterochromatin on lytic promoters. J. Virol. 83:8182–8190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Creech CC, Neumann DM. 2010. Changes to euchromatin on LAT and ICP4 following reactivation are more prevalent in an efficiently reactivating strain of HSV-1. PLoS One 5:e15416 doi:10.1371/journal.pone.0015416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Daigle D, et al. 2011. Valproic acid antagonizes the capacity of other histone deacetylase inhibitors to activate the Epstein-Barr virus lytic cycle. J. Virol. 85:5628–5643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Danaher RJ, et al. 2005. Histone deacetylase inhibitors induce reactivation of herpes simplex virus type 1 in a latency-associated transcript-independent manner in neuronal cells. J. Neurovirol. 11:306–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Deshmane SL, Fraser NW. 1989. During latency, herpes simplex virus type 1 DNA is associated with nucleosomes in a chromatin structure. J. Virol. 63:943–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Devi-Rao GB, Bloom DC, Stevens JG, Wagner EK. 1994. Herpes simplex virus type 1 DNA replication and gene expression during explant-induced reactivation of latently infected murine sensory ganglia. J. Virol. 68:1271–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dobson AT, Margolis TP, Sedarati F, Stevens JG, Feldman LT. 1990. A latent, nonpathogenic HSV-1-derived vector stably expresses β-galactosidase in mouse neurons. Neuron 5:353–360 [DOI] [PubMed] [Google Scholar]
- 19. Felsenfeld G, et al. 2004. Chromatin boundaries and chromatin domains. Cold Spring Harbor Symp. Quant. Biol. 69:245–250 [DOI] [PubMed] [Google Scholar]
- 20. Giles KE, Gowher H, Ghirlando R, Jin C, Felsenfeld G. 2010. Chromatin boundaries, insulators, and long-range interactions in the nucleus. Cold Spring Harbor. Symp. Quant. Biol. 75:79–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Giordani NV, et al. 2008. During herpes simplex virus type 1 infection of rabbits, the ability to express the latency-associated transcript increases latent-phase transcription of lytic genes. J. Virol. 82:6056–6060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Griffiths AJF, et al. 1999. Modern genetic analysis. W. H. Freeman, New York, NY [Google Scholar]
- 23. Kang H, Wiedmer A, Yuan Y, Robertson E, Lieberman PM. 2011. Coordination of KSHV latent and lytic gene control by CTCF-cohesin mediated chromosome conformation. PLoS Pathog. 7:e1002140 doi:10.1371/journal.ppat.1002140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kubat NJ, Amelio AL, Giordani NV, Bloom DC. 2004. The herpes simplex virus type 1 latency-associated transcript (LAT) enhancer/rcr is hyperacetylated during latency independently of LAT transcription. J. Virol. 78:12508–12518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kwiatkowski DL, Thompson HW, Bloom DC. 2009. The polycomb group protein Bmi1 binds to the herpes simplex virus 1 latent genome and maintains repressive histone marks during latency. J. Virol. 83:8173–8181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liang Y, Vogel JL, Narayanan A, Peng H, Kristie TM. 2009. Inhibition of the histone demethylase LSD1 blocks alpha-herpesvirus lytic replication and reactivation from latency. Nat. Med. 15:1312–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lieberman PM. 2010. Chromatin in viral gene regulation. Preface. Biochim. Biophys. Acta 1799:181. [DOI] [PubMed] [Google Scholar]
- 28. Lutz M, et al. 2000. Transcriptional repression by the insulator protein CTCF involves histone deacetylases. Nucleic Acids Res. 28:1707–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Millau JF, Gaudreau L. 2011. CTCF, cohesin, and histone variants: connecting the genome. Biochem. Cell Biol. 89:505–513 [DOI] [PubMed] [Google Scholar]
- 30. Miller CS, Danaher RJ, Jacob RJ. 2006. ICPO is not required for efficient stress-induced reactivation of herpes simplex virus type 1 from cultured quiescently infected neuronal cells. J. Virol. 80:3360–3368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Neumann DM, Bhattacharjee PS, Giordani NV, Bloom DC, Hill JM. 2007. In vivo changes in the patterns of chromatin structure associated with the latent herpes simplex virus type 1 genome in mouse trigeminal ganglia can be detected at early times after butyrate treatment. J. Virol. 81:13248–13253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Neumann DM, Bhattacharjee PS, Hill JM. 2007. Sodium butyrate: a chemical inducer of in vivo reactivation of herpes simplex virus type 1 in the ocular mouse model. J. Virol. 81:6106–6110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nikolaev LG, Akopov SB, Didych DA, Sverdlov ED. 2009. Vertebrate protein CTCF and its multiple roles in a large-scale regulation of genome activity. Curr. Genomics 10:294–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ohlsson R, Renkawitz R, Lobanenkov V. 2001. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 17:520–527 [DOI] [PubMed] [Google Scholar]
- 35. Phillips JE, Corces VG. 2009. CTCF: master weaver of the genome. Cell 137:1194–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Roizman B. 2011. The checkpoints of viral gene expression in productive and latent infection: the role of the HDAC/CoREST/LSD1/REST repressor complex. J. Virol. 85:7474–7482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sedarati F, Margolis TP, Stevens JG. 1993. Latent infection can be established with drastically restricted transcription and replication of the HSV-1 genome. Virology 192:687–691 [DOI] [PubMed] [Google Scholar]
- 38. Spivack JG, Fraser NW. 1987. Detection of herpes simplex virus type 1 transcripts during latent infection in mice. J. Virol. 61:3841–3847 (Erratum, 62:663, 1988.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Splinter E, et al. 2006. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 20:2349–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stevens JG, Wagner EK, Devi-Rao GB, Cook ML, Feldman LT. 1987. RNA complementary to a herpesvirus α gene mRNA is prominent in latently infected neurons. Science 235:1056–1059 [DOI] [PubMed] [Google Scholar]
- 41. Tempera I, Klichinsky M, Lieberman PM. 2011. EBV latency types adopt alternative chromatin conformations. PLoS Pathog. 7:e1002180 doi:10.1371/journal.ppat.1002180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tempera I, Lieberman PM. 2010. Chromatin organization of gammaherpesvirus latent genomes. Biochim. Biophys. Acta 1799:236–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tempera I, Wiedmer A, Dheekollu J, Lieberman PM. 2010. CTCF prevents the epigenetic drift of EBV latency promoter Qp. PLoS Pathog. 6:e1001048 doi:10.1371/journal.ppat.1001048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wagner EK, Bloom DC. 1997. Experimental investigation of herpes simplex virus latency. Clin. Microbiol. Rev. 10:419–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang QY, et al. 2005. Herpesviral latency-associated transcript gene promotes assembly of heterochromatin on viral lytic-gene promoters in latent infection. Proc. Natl. Acad. Sci. U. S. A. 102:16055–16059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. West AG, Gaszner M, Felsenfeld G. 2002. Insulators: many functions, many mechanisms. Genes Dev. 16:271–288 [DOI] [PubMed] [Google Scholar]
- 47. Zhao H, Dean A. 2004. An insulator blocks spreading of histone acetylation and interferes with RNA polymerase II transfer between an enhancer and gene. Nucleic Acids Res. 32:4903–4919 [DOI] [PMC free article] [PubMed] [Google Scholar]