Abstract
Growing evidence from mammals suggests that host microRNAs (miRNAs) play important roles in the antiviral immune response. However, the roles of invertebrate miRNAs in response to virus infection remain to be investigated. Based on our previous studies, the shrimp miR-7 was found to be upregulated in response to white spot syndrome virus (WSSV) infection. In this study, the results showed that shrimp miR-7 could target the 3′-untranslated region (3′UTR) of the WSSV early gene wsv477, implying that miR-7 was involved in viral DNA replication. In insect High Five cells, the synthesized miR-7 significantly decreased the expression level of the fluorescent construct bearing the 3′UTR of wsv477 compared with the expression of the control constructs. When the activity of transfected miR-7 was blocked by locked-nucleic-acid (LNA)-modified anti-miR-7 oligonucleotide (AMO-miR-7), the repression of luciferase gene expression by miR-7 was relieved. In vivo, when the synthesized miR-7 was injected into shrimp, the numbers of WSSV genome copies/mg gills were 1,000-fold lower than those of WSSV only at 72 and 96 h postinfection. The results indicated that the blocking of endogenous miR-7 by AMO-miR-7 led to about a 10-fold increase of WSSV genome copies/mg gills in WSSV-infected shrimp compared with the control WSSV only. Further, it was revealed that the host Dicer1 was an important component for the biogenesis of miR-7, which had a large effect on virus infection. Therefore, our study revealed a novel regulatory function for an invertebrate miRNA in host-virus interactions by targeting the viral early gene.
INTRODUCTION
MicroRNAs (miRNAs) are an extensive category of small noncoding RNAs, 18 to 26 nucleotides (nt) in length, that have emerged as important regulators in many biological processes (1, 2, 5, 6, 9, 26). Two decades ago, the first miRNA, lin-4, was identified in Caenorhabditis elegans (20). Now, about 21,643 miRNAs from plants, animals, and viruses (miRBase, release 18.0; http://www.mirbase.org) have been discovered. A miRNA is transcribed by RNA polymerase II or III as primary microRNA (pri-miRNA), which is further cleaved by the microprocessor complex comprising Drosha (RNase III) and DGCR8/Pasha (RNA binding protein) into a 60- to 80-nucleotide precursor miRNA (pre-miRNA) with a stem-loop hairpin structure (5). The pre-miRNA is then exported to the cytoplasm by exportin-5 and GTP-binding cofactor Ran and further processed by cytoplasmic RNase III enzyme Dicer into approximately 22-nt double-stranded miRNA:miRNA* duplexes (5). One strand of the duplex is preferentially selected, depending on the relative thermodynamic stability of its 5′ end, and incorporated into the RNA-induced silencing complex (RISC) (24, 27). Typically, mature miRNAs bind to target sites in the 3′UTRs of target mRNAs, which leads to translation repression and/or mRNA degradation (3, 21).
Recent findings have shown that the expression patterns of host miRNAs can be altered by virus infection, such as hepatitis C virus (HCV), HIV-1, human cytomegalovirus, and Epstein-Barr virus (EBV) (8, 14, 33). These changes reflect that the host miRNAs may play important roles in host-virus interactions. In mammals, some host miRNAs may inhibit virus invasion by targeting viral mRNAs or host transcripts beneficial to the virus. RNA interference (RNAi) experiments reveal that the knockdown of Drosha and Dicer participating in mammalian miRNA biogenesis can result in a decreased production of mature miRNAs and thus increase the host sensitivity to viral infection (23, 31). It is demonstrated that the administration of synthetic miRNA mimics of miR-196, miR-296, miR-351, miR-431, and miR-448 significantly reduces the accumulation of HCV RNA, because the transfected miRNA mimics match sites in the viral genome (26). The host miR-32 has been shown to inhibit the replication of the retrovirus primate foamy virus (PFV) in infected HeLa cells (19). When miR-32 is blocked using locked-nucleic-acid (LNA)-modified antisense oligonucleotides, virus production is enhanced (19). Recently, it was reported that a host miR-24 is differentially expressed following Heliothis virescens ascovirus (HvAV-3e, Ascoviridae) infection in the Helicoverpa zea fat body cell, which leads to a negative impact on viral replication because of targeting of the viral DNA-dependent RNA polymerase and its β subunit transcript (16).
To date, the existing studies on the roles of miRNAs in host-virus interactions have been carried out predominantly in vertebrates. However, information about the involvement of host miRNAs in invertebrate-virus interactions is still very limited (4). In our previous study, the small RNA sequencing of the white spot syndrome virus (WSSV)-infected shrimp revealed that 31 host miRNAs were involved in virus infection (15). Among them, the shrimp miR-7 was found to be upregulated in response to WSSV challenge. In the present study, the shrimp miR-7 was characterized. The results showed that miR-7 played crucial roles in the virus-host interactions by targeting the 3′untranslated region (3′UTR) of the viral early gene.
MATERIALS AND METHODS
Shrimp culture and shrimp infection by WSSV.
Shrimp (Marsupenaeus japonicus) with an average body weight of 15 g were cultured in groups of 20 individuals in the same tank filled with seawater. First, the shrimp were cultured temporarily for 2 to 3 days. Three shrimp were randomly selected for the PCR detection of WSSV with WSSV-specific primers (see Table S1 in the supplemental material) to ensure that the shrimp were WSSV free before the experiments. Then the virus-free shrimp were infected with 0.1 ml of WSSV virus solution at 105 genome copies/ml by intramuscular injection using a syringe with a 29-gauge needle. The WSSV solution was obtained from virus-infected shrimp (34). The tissues of WSSV-infected shrimp were homogenized in TN buffer (20 mM Tris-HCl, 400 mM NaCl, pH 7.4) at 0.1 g/ml. After centrifugation at 2,000 × g for 10 min, the supernatant was diluted to 1:100 with 0.9% NaCl and filtered through a 0.45-μm-pore-size filter. At different postinfection times, the WSSV-infected shrimp were collected for later use.
RNA extraction.
Total RNAs were extracted from the fresh tissues of shrimp using a mirVanaPTMP miRNA isolation kit according to the manufacturer's instructions (Ambion, USA). Potentially contaminating DNA was removed using RNase-free DNase I (TaKaRa, Japan) at 37°C for 30 min. The concentration of the extracted RNA was determined using a NanoDrop ND-100 spectrophotometer (NanoDrop Technologies, Wilmington, DE), and the extracted RNA was immediately stored at −80°C until use.
Preparation of siRNAs and RNAi assays in shrimp.
Based on the gene sequences of shrimp Dicer1 (GenBank accession number GU265733.1), Dicer2 (GenBank accession number JQ349041), and viral wsv477 (GenBank accession number DQ121373), the small interfering RNAs (siRNAs) specifically targeting these genes were separately synthesized according to the design rule for RNAi (34). As controls, one nucleotide was mutated at random in the siRNA sequences of Dicer1, Dicer2, and wsv477, yielding the corresponding mutation-siRNAs. The sequences of Dicer1-siRNA, Dicer1-mutation-siRNA, Dicer2-siRNA, Dicer2-mutation-siRNA, wsv477-siRNA, and wsv477-mutation-siRNA are listed in Table S1 in the supplemental material.
All the siRNAs were synthesized using the in vitro transcription T7 kit for siRNA synthesis (TaKaRa, Japan) according to the manufacturer's instructions. The synthesized siRNAs were dissolved in siRNA solution (50 mM Tris-HCl, 100 mM NaCl, pH 7.5) and quantified by spectrophotometry. The siRNAs (Dicer1-siRNA, Dicer1-mutation-siRNA, Dicer2-siRNA, Dicer2-mutation-siRNA, wsv477-siRNA, and wsv477-mutation-siRNA) were injected into the lateral area of the fourth abdominal segment at 30 μg/shrimp using a syringe with a 29-gauge needle. As a negative control, phosphate-buffered saline (PBS) (0.1 M, pH 7.4) was used in the injections instead of the siRNAs. Three shrimp from each treatment, selected at random, were collected at different times after the siRNA injection and subjected to Northern blotting and real-time PCR. All assays were repeated three times.
Quantitative real-time PCR.
To analyze the expressions of shrimp Dicer1 and Dicer2 genes, quantitative real-time PCR (qRT-PCR) was conducted using the gene-specific primers and TaqMan fluorogenic probes. Shrimp β-actin was used as the control. The primers and TaqMan probes for Dicer1, Dicer2, and β-actin are provided in Table S1 in the supplemental material. The reaction mixtures were prepared in a total volume of 25 μl containing 12.5 μl of Premix Ex Taq (TaKaRa, Japan), 100 ng of cDNA template, 0.5 μl of 10 μM forward and reverse primers, and 0.5 μl of 10 μM TaqMan fluorogenic probes to a final concentration of 0.2 μM. The qRT-PCR conditions were 95°C for 1 min and 40 cycles of 95°C for 15 s and 57°C for 45 s.
To monitor the viral replication in shrimp, qRT-PCR was performed using WSSV forward and reverse primers and a TaqMan fluorogenic probe (see Table S1 in the supplemental material). The primers were used to amplify a region from position 260075 to position 260138 of the WSSV genome (GenBank accession number AF332093.1) (22). A linearized plasmid containing a 1,400-bp DNA fragment from the WSSV genome was used as the internal standard for real-time PCR (22). The internal standard plasmid was serially diluted 10-fold for the generation of a standard curve in real-time PCR. About 20 mg of shrimp gills was collected, and the genomic DNA extraction was performed using a SQ tissue DNA kit (Omega Bio-tek, Norcross, GA) according to the manufacturer's instruction. The extracted DNA template and the internal standard plasmid were subjected to real-time PCR. The PCR mixture (25 μl) contained 12.5 μl Premix Ex Taq (TaKaRa, Japan), 1 μl DNA template, 0.5 μl of 10 μM primers, and 0.5 μl of 10 μM TaqMan fluorogenic probe to a final concentration of 0.2 μM. The real-time PCR conditions were 95°C for 1 min, 45 cycles of 30 s at 95°C and 30 s at 52°C, and 30 s at 72°C.
Northern blotting.
Small RNAs were separated on a denaturing 15% polyacrylamide gel containing 7 M urea and transferred to a Hybond-N+ nylon membrane (Amersham Biosciences, Buckinghamshire, United Kingdom). After UV cross-linking (Ultra-Violet Products Ltd., USA), the membrane was prehybridized in DIG Easy Hyb granule buffer (Roche, Basel, Switzerland) for 0.5 h, followed by hybridization with digoxigenin (DIG)-labeled miR-7 or U6 probes (see Table S1 in the supplemental material) at 45°C overnight. Detection was performed with the DIG High Prime DNA labeling and detection starter kit II (Roche, Basel, Switzerland) following the manufacturer's instructions.
For detection of the wsv477, wsv076, and vp28 transcripts, the shrimp gill tissues were collected. The extracted RNAs were detected with the DIG-labeled wsv477 probe, wsv076 probe, and vp28 probe and the shrimp β-actin probe (see Table S1 in the supplemental material).
Western blotting.
The protein samples were analyzed in a 12% SDS-PAGE gel and transferred onto a nitrocellulose membrane (Bio-Rad, USA). The membrane was immersed in blocking buffer (3% bovine serum albumin [BSA] in PBS, pH 7.2) at 4°C overnight. Subsequently, the membrane was incubated with a specific antibody (anti-Dicer1 IgG, anti-Dicer2 IgG, anti-β-actin IgG, or anti-wsv477 IgG), followed by incubation with alkaline phosphatase (AP)-conjugated goat anti-mouse IgG (Sigma) for 1 h and detected with nitroblue tetrazolium (NBT) and 5-bromo-4-chloro-3-indolylphosphate (BCIP) solutions (BBI, Canada).
Plasmid construction.
To confirm the preliminary observation that the shrimp miR-7 could target the viral wsv477 gene, the wsv477 3′UTR and enhanced green fluorescent protein (EGFP) gene were cloned into a pIZ/V5-His vector (Invitrogen, USA). The EGFP gene was amplified from the pEGFP vector (BD Biosciences, USA) using EGFP forward and reverse primers (see Table S1 in the supplemental material) and cloned into the pIZ vector to produce a control EGFP construct. Subsequently, the wsv477 3′UTR (see Table S1) was cloned into the pIZ vector downstream of EGFP using the XbaI and SacII restriction sites to generate the EGFP-wsv477 construct. As a control, the wsv477 3′UTR sequence (GTCTTCC) complementary to the miR-7 seed sequence was mutated to ACTCCTT to yield the EGFP-Δwsv477 construct (see Table S1). All the recombinant plasmids were confirmed by sequencing.
Cell culture, transfection, and fluorescence assays.
Insect High Five cells (Invitrogen) were cultured at 27°C in Express Five serum-free medium (SFM) (Invitrogen) containing l-glutamine (Invitrogen) as a monolayer. When the cells were at about 70% confluence, they were transfected with 2 μg of EGFP, EGFP-wsv477, or EGFP-Δwsv477. At the same time, the cells were transfected with 100 pmol of either synthesized miR-7 (see Table S1 in the supplemental material) or a synthesized control miRNA (see Table S1). All the miRNAs were synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China). To inhibit the activity of miR-7, the plasmid (EGFP, EGFP-wsv477, or EGFP-Δwsv477)-transfected cells were simultaneously transfected with miR-7 (100 pmol) and LNA-modified anti-miR-7 oligonucleotide (AMO-miR-7) (100 pmol) or with miR-7 (100 pmol) and LNA-modified anti-control miRNA oligonucleotide (AMO-control miRNA) (100 pmol) (see Table S1). The AMOs were obtained from Exiqon A/S (Vedbaek, Denmark). For the LNA-modified anti-miR-7 oligonucleotide (AMO-miR-7), the sequence complementary to the miR-7 sequence was modified at the third, sixth, and tenth nucleotides with LNA, at the thirteenth and sixteenth nucleotides with 2′-O-methoxyethyl (MOE), and at the remaining nucleotides with phosphorothioate. For the LNA-modified anti-control miRNA oligonucleotide (AMO-control miRNA), the sequence complementary to the control miRNA sequence was modified as described above. All transfections were carried out in triplicate with Cellfectin transfection reagent (Invitrogen) according to the manufacturer's protocol. After being cultured for 12 h, the transfected cells were seeded to 96-well plates at concentrations of 2.0 × 104 cells per well. At 48 h after transfection, the EGFP fluorescence of the cells was monitored with a Flex Station II microplate reader (Molecular Devices, USA) at 490/510 nm of excitation/emission (Ex/Em). The fluorescence values were corrected by subtracting the autofluorescence of cells not expressing EGFP. All the experiments were repeated biologically three times.
In vivo analysis.
The shrimp were simultaneously injected with WSSV virions (104 genome copies/shrimp) and 30 μg of one of the siRNAs (Dicer1-siRNA, Dicer1-mutation-siRNA, wsv477-siRNA, or wsv477-mutation-siRNA), with WSSV virions (104 genome copies/shrimp) and 30 μg of a synthesized miRNA (miR-7 or control miRNA), or with WSSV virions (104 genome copies/shrimp) and 15 μg of a LNA-modified anti-miRNA oligonucleotide (AMO-miR-7 or AMO-control miRNA). WSSV only (104 genome copies/shrimp) and PBS solution only were included in the injections as controls. To remedy the loss of injected oligonucleotides that might have been degraded in vivo, at 48 h after the first injection, 30 μg of siRNAs, 30 μg of synthesized miRNAs, or 15 μg of LNA-modified anti-miRNA oligonucleotides was injected separately into the same shrimp again. For each treatment, a group of 20 individual shrimps were used and gill tissues were collected at different times after the first injection (0, 12, 24, 48, 72, and 96 h). Three shrimp specimens from each treatment, selected at random, were subjected to real-time PCR to quantify the WSSV genome copies or to Northern blot analysis.
Statistical analysis.
The data from three independent experiments were analyzed by one-way analysis of variance (ANOVA) to calculate the means and standard deviations (SD) of the triplicate assays.
RESULTS
Interaction between host miR-7 and viral wsv477 gene.
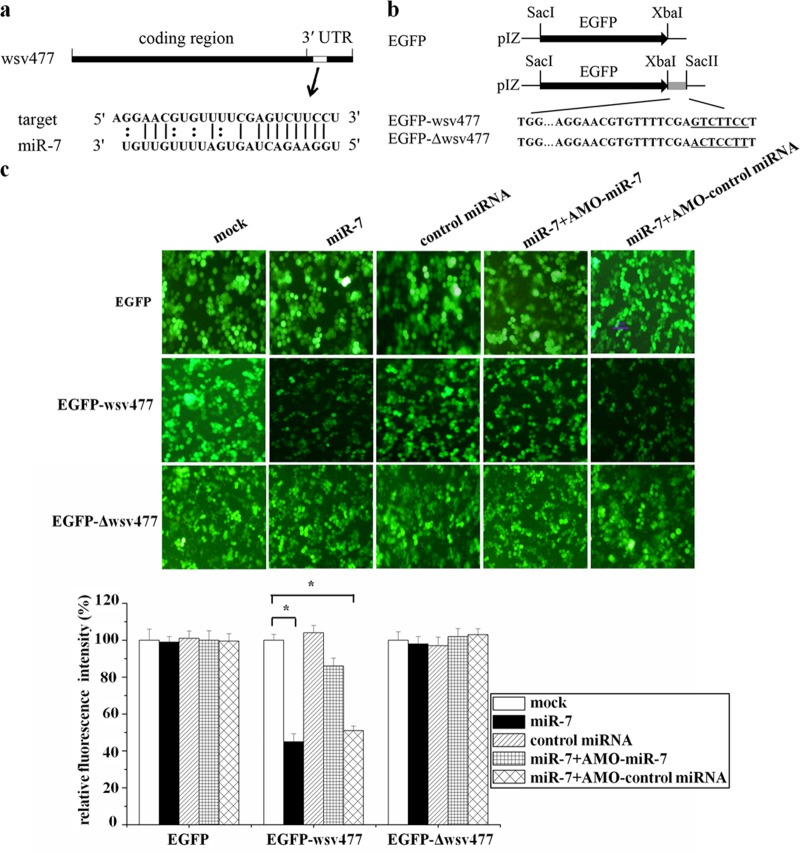
Growing evidence has indicated that host miRNAs play important roles in host-virus interactions in vertebrates, thus providing a novel and exciting dimension to study similar interactions in invertebrate hosts. In our previous study (15), 31 shrimp miRNAs were differentially expressed in response to WSSV infection, suggesting that these miRNAs were involved in virus-shrimp interactions. Among the 31 miRNAs, the prediction analysis of miRNA targets showed that the shrimp miR-7 could target the viral wsv477, a WSSV early gene involved in viral DNA replication (Fig. 1a), indicating that the host miR-7 might play important roles in virus infection.
Fig 1.
Interaction between host miR-7 and viral wsv477 gene. (a) The region of the wsv477 3′UTR sequence targeted by miR-7 predicted with TargetScan. The seed sequence of miR-7 is underlined. (b) Constructs of the wild-type and mutated 3′UTRs of the viral wsv477 gene. The sequences complementary to the seed region of miR-7 are underlined. (c) Interaction between host miR-7 and the viral wsv477 gene in insect cells. The insect High Five cells were cotransfected with miR-7 and different plasmids (EGFP, EGFP-wsv477, or EGFP-Δwsv477). To act as a control, the control miRNA was included in the transfections. The constructs EGFP-wsv477 and EGFP-Δwsv477 contained the wild-type and mutant wsv477, respectively. To inhibit the activity of miR-7, the plasmid (EGFP, EGFP-wsv477, or EGFP-Δwsv477)-transfected cells were transfected simultaneously with miR-7, AMO-miR-7, or AMO-control miRNA. At 48 h after transfections, fluorescent images were obtained (upper panel). Lane headings indicate the miRNAs and LNA-modified anti-miRNA oligonucleotides (AMOs) that were used. The plasmids that were used for transfections are indicated on the left. The effects of the miRNAs and/or AMOs on gene expressions of the different plasmids (EGFP, EGFP-wsv477, or EGFP-Δwsv477) were determined by comparing fluorescence intensities in the absence of miRNAs and/or AMOs with that of the mock transfection (lower panel). *, statistically significant differences (P < 0.05).
To characterize the interaction between the host miR-7 and the viral wsv477 gene, the synthesized miR-7 and the plasmid EGFP-wsv477 consisting of EGFP and the wsv477 3′UTR were cotransfected into insect High Five cells because a shrimp cell line was not available (Fig. 1b). The results showed that the fluorescence intensity in the cotransfected cells was significantly decreased compared with the intensity in the EGFP-wsv477-only cells, indicating that miR-7 inhibited the expression of the wsv477 gene by targeting its 3′UTR (Fig. 1c). The control miRNA had no effect on the expression of EGFP-wsv477. When the activity of synthesized miR-7 was inhibited by the LNA-modified AMO-miR-7, the fluorescence intensity in the synthesized miR-7, AMO-miR-7, and EGFP-wsv477-cotransfected cells was recovered (Fig. 1c). To investigate the specificity of the interaction between miR-7 and wsv477 3′UTR, the wsv477 3′UTR sequence complementary to the miR-7 seed sequence was mutated, yielding the pIZ/EGFP-Δwsv477 construct. It was revealed that the synthesized miR-7 had no effect on the expression of EGFP in the synthesized miR-7 and EGFP-Δwsv477 cotransfected cells (Fig. 1c), showing that the miR-7 seed sequence was required to target the wsv477 3′UTR. The above results indicated that the shrimp miR-7 could target the viral wsv477 3′UTR.
To further investigate the in vivo interaction between host miR-7 and the viral wsv477 gene in shrimp, the synthesized miR-7 was injected into WSSV-infected shrimp. The results indicated that miR-7 was overexpressed in shrimp injected with the synthesized miR-7 at 12, 24, and 48 h after miRNA injection compared with that in shrimp injected with the control of WSSV only (Fig. 2a). In this study, Northern blotting was used to detect simultaneously the amount and size of the wsv477 transcript. It was found that the overexpression of miR-7 led to a significant decrease in the transcript levels of the wsv477 gene at 12 h postinfection (Fig. 2b). However, miR-7 had little effect on the expressions of the other viral genes (an early gene and a late gene of WSSV) (Fig. 2b). To inhibit the expression of endogenous miR-7, AMO-miR-7 was injected into WSSV-infected shrimp. The results showed that the shrimp miR-7 expression was inhibited by AMO-miR-7 (Fig. 2a). In this case, the wsv477 gene was transcribed normally (Fig. 2b). The Western blots that were performed at the same time yielded results that were similar to those obtained from the Northern blots (Fig. 2c). These results showed that changes in miR-7 levels could alter the expression of viral gene wsv477 in shrimp in vivo.
Fig 2.
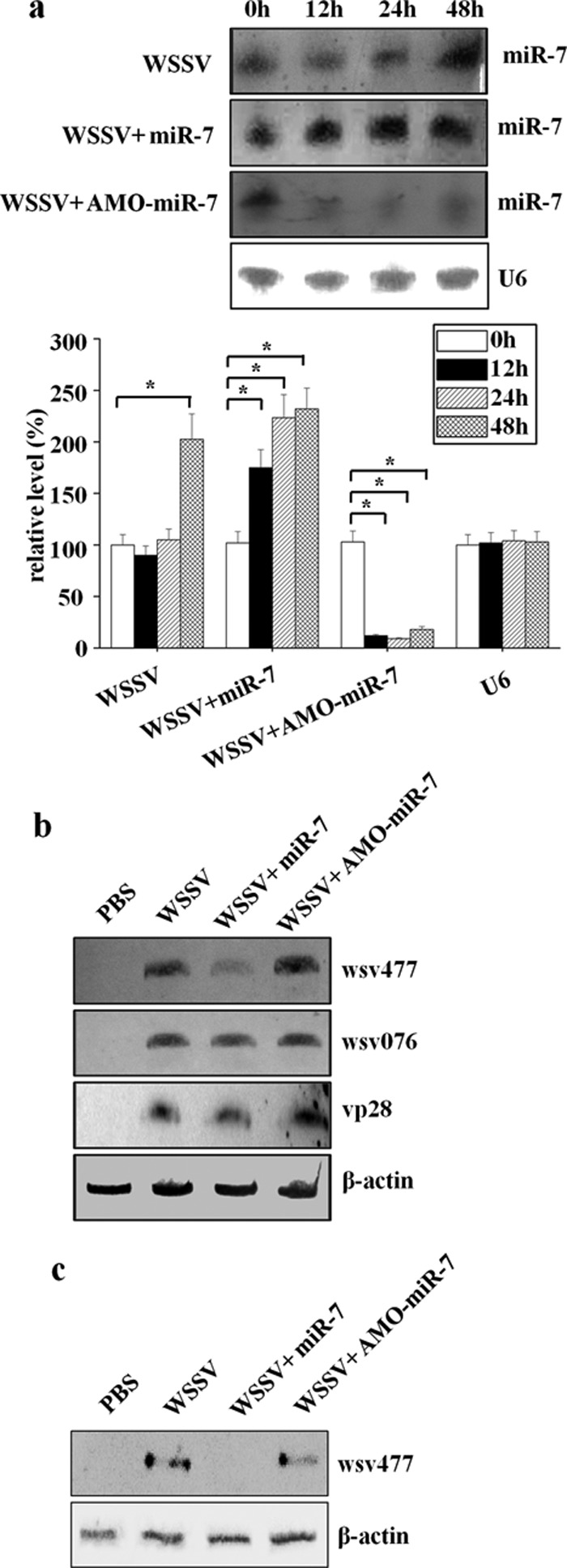
In vivo interaction between host miR-7 and viral wsv477 gene in shrimp. (a) Expression of miR-7 in shrimp. Either miR-7 or the LNA-modified anti-miR-7 oligonucleotide (AMO-miR-7) was injected into WSSV-infected shrimp. At different times (0, 12, 24, and 48 h) after injection, the shrimp gills were collected and subjected to Northern blotting with the miR-7-specific probe (upper panel). Shrimp U6 was used as a control. The bar chart in the lower panel represents the densitometric measurement of miR-7 expression in the Northern blot. The statistically significant differences between treatments are indicated with asterisks (*, P < 0.05; **, P < 0.01). (b) Effects of miR-7 on the expression of the viral wsv477 gene in Northern blotting. To detect the expression of wsv477, the shrimp specimens collected at 12 h after the injection of WSSV plus miR-7 or WSSV plus AMO-miR-7 were characterized by Northern blotting using the wsv477-specific probe. PBS and WSSV alone were used as controls. To evaluate the effects of miR-7 on the expressions of other viral genes, the mRNAs of wsv076 (an early gene of WSSV) and vp28 (a late gene of WSSV) were probed with gene-specific probes. The shrimp β-actin was used as a control. Lane headings indicate the solutions that were injected. The probes that were used are indicated on the right. (c) Effects of miR-7 on wsv477 expression in Western blotting. Western blot analyses were conducted with AP-conjugated goat anti-mouse IgGs using shrimp gills collected at 12 h after the injection of either PBS, WSSV, WSSV plus miR-7, or WSSV plus AMO-miR-7. The shrimp β-actin was used as a control. Lane headings indicate the solutions that were injected. The antibodies that were used are shown on the right.
Requirement of host Dicer1 for the biogenesis of miRNA.
To investigate the biogenesis pathway of miRNA, the full-length Dicer1 and Dicer2 genes of shrimp were cloned according to the reported sequences (10, 29, 35). The sequence analyses indicated that Dicer1 and Dicer2 shared high sequence similarities with Dicer homologs in other animals (see Fig. S1 in the supplemental material).
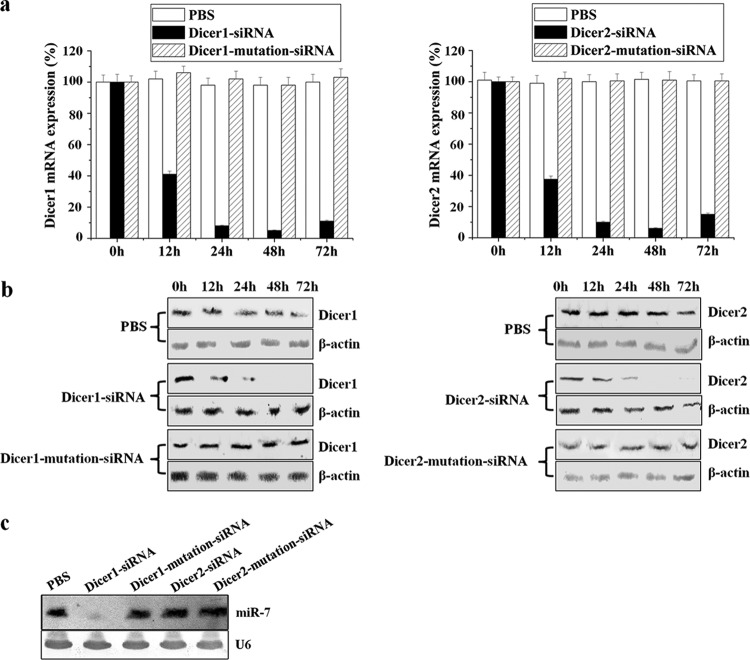
To determine whether Dicer1 or Dicer2 was implicated in the miRNA biogenesis, the shrimp Dicer1 and Dicer2 were separately silenced with sequence-specific siRNA followed by the detection of miR-7. The qRT-PCR and Western blot results showed that the expressions of Dicer1 and Dicer2 were repressed by the gene-specific siRNAs from 48 to 72 h after siRNA injection (Fig. 3a and b). The mutation-siRNAs of Dicer1 and Dicer2 had no effect on the expressions of Dicer1 and Dicer2 (Fig. 3a and b). The data indicated that the siRNAs were highly sequence specific, which could silence the Dicer1 or Dicer2 genes in shrimp in vivo.
Fig 3.
Roles of shrimp Dicer1 and Dicer2 in miRNA biogenesis. (a) Detection of Dicer1 and Dicer2 mRNAs by qRT-PCR. The shrimp were injected with sequence-specific Dicer1-siRNA (left panel) or Dicer2-siRNA (right panel). Controls were injected with Dicer1-mutation-siRNA or Dicer2-mutation-siRNA at the same time. PBS only was used as a negative control. At different time points after injection, the shrimp gills were collected and subjected to qRT-PCR to quantify the gene expressions. The numbers indicate the times after injection. Each column represents the mean result of triplicate assays within a 1% standard deviation. (b) Western blots with AP-conjugated goat anti-mouse IgGs using shrimp gills. The siRNAs that were used for injections are indicated on the left. (c) Shrimp were injected with Dicer1-siRNA, Dicer1-mutation-siRNA, Dicer2-siRNA, Dicer2-mutation-siRNA, or PBS. At 48 h after injection, the RNAs were extracted from shrimp gills and subjected to Northern blot analysis with either the miR-7 probe or the U6 probe. The shrimp U6 was used as a control. Lane headings indicate the solutions that were injected. The probes that were used are shown on the right.
When the Dicer1 or Dicer2 genes had been silenced by the siRNAs, the expression of miR-7 in shrimp gill tissues was examined using Northern blotting. The miR-7 gene was not detected in shrimp that had been treated with the Dicer1-siRNA, whereas it was normally expressed in the tissues that had been treated with Dicer1-mutation-siRNA, Dicer2-siRNA, or Dicer2-mutation-siRNA compared with the U6 control (Fig. 3c). This finding clearly demonstrated that the Dicer1 gene was required for the biogenesis of shrimp miRNA.
Roles of miR-7 in shrimp-virus interaction.
All the results described above indicated that the shrimp miR-7 could downregulate the expression of viral early gene wsv477 likely through mRNA degradation/instability and showed that the biogenesis of miR-7 was dependent on shrimp Dicer1. To further examine the effects of miR-7 on virus infection, the expressions of the shrimp Dicer1 and viral wsv477 genes were separately silenced by sequence-specific siRNAs and the endogenous miR-7 was blocked by AMO-miR-7 in WSSV-infected shrimp, followed by examination of virus infection.
After this treatment, the wsv477 mRNA and protein were not detected as early as 12 h after wsv477-siRNA injection (Fig. 4a and b), whereas after the wsv477-mutation-siRNA injection, there was no effect on the expression of the wsv477 gene. As shown in Fig. 4c, the knockdown of the wsv477 gene by wsv477-siRNA led to a statistically significant decrease in the numbers of WSSV genome copies/mg gills at 12 to 96 h postinfection compared with those of WSSV only, while the control wsv477-mutation-siRNA had no effect on virus infection. The results showed that the numbers of WSSV genome copies/mg gills in shrimp treated with wsv477-siRNA were 1,000-fold lower than those of WSSV only at 72 and 96 h postinfection (Fig. 4c). The data indicated that the wsv477 gene played important roles in WSSV replication.
Fig 4.

Roles of host miR-7 in virus-host interactions. (a) Shrimp were injected simultaneously with WSSV and wsv477-siRNA or with WSSV and wsv477-mutation-siRNA, respectively. WSSV only was included in one of the injections. At 12 h postinfection, the shrimp were collected and subjected to Northern blot analysis to detect the wsv477 mRNA. Shrimp actin was used as a control. The probes are indicated on the right. (b) Western blots of shrimp gills with AP-conjugated goat anti-mouse IgGs. Shrimp actin was used as a control. The antibodies are indicated on the right. (c) The shrimp were injected simultaneously with WSSV and siRNA, miRNA, or LNA-modified anti-miRNA oligonucleotides. The solutions that were injected are shown at the top. At different times postinfection, the shrimp gill tissues were collected and subjected to real-time PCR to monitor the WSSV replication. The numbers indicate the time points postinfection. The statistically significant differences between treatments are represented with asterisks (*, P < 0.05; **, P < 0.01). (d) Proposed model for the role of miR-7 in virus-host interactions. Host miR-7 was generated in a Dicer1-dependent manner and decreased the transcript levels of viral early gene wsv477 by directly targeting the 3′UTR of wsv477 mRNA. The Dicer1-miR-7-wsv477 pathway was assumed to be involved in WSSV infection.
To evaluate the effects of miR-7 on WSSV infection, the endogenous miR-7 in WSSV-infected shrimp was inhibited with AMO-miR-7. The results showed that, at 72 and 96 h postinfection, the numbers of WSSV genome copies/mg gills in the AMO-miR-7-treated shrimp significantly increased 10 times compared with those in the control WSSV only, whereas no effect on WSSV replication was observed in the control (AMO-control miRNA) (Fig. 4c). On the other hand, when the synthesized miR-7 was injected into WSSV-infected shrimp, a statistically significant decrease in the numbers of WSSV genome copies/mg gills from 12 to 96 h postinfection, compared with those in WSSV only, was observed (Fig. 4c). At 72 and 96 h postinfection, the numbers of WSSV genome copies/mg gills in shrimp injected with synthesized miR-7 were 1,000-fold lower than those of WSSV only, whereas the control miRNA had no effect on WSSV infection (Fig. 4c). These results indicated that the host miR-7 had an antiviral activity in shrimp to fight WSSV infection.
The Dicer1 gene, required for miR-7 generation, was silenced with Dicer1-siRNA in WSSV-challenged shrimp. The results demonstrated that the knockdown of Dicer1 gene expression resulted in a significant increase in numbers of WSSV genome copies/mg gills at 96 h postinfection compared with those of WSSV only (P < 0.05) (Fig. 4c). The control Dicer1-mutation-siRNA, however, had a negligible effect on virus infection (Fig. 4c). This finding indicated that the host Dicer1 was probably involved in virus-host interactions through its role in the biogenesis of miRNA.
Based on the results derived from this study, a model for the role of miR-7 in virus-host interactions was proposed (Fig. 4d). In this model, the biogenesis of miR-7 was assumed to be dependent on host Dicer1, and miR-7 was assumed to function by targeting the 3′UTR of the viral wsv477 mRNA. The wsv477 gene, a viral early gene, was required in WSSV infection, and, therefore, the Dicer1-miR-7-wsv477 pathway was likely to be involved in virus infection.
DISCUSSION
Increasingly, the evidence indicates that the expression profiles of host miRNAs can be altered during virus infection, and some of these changes may represent one aspect of host antiviral responses. In our previous study, it was found that the shrimp miR-7 gene was significantly upregulated in response to WSSV infection, suggesting that miR-7 might be involved in virus infection (15). In the present investigation, a role of miR-7 in virus-host interactions was characterized. It was revealed that miR-7 was lowly expressed in shrimp lymphoid organs (15) and highly expressed in shrimp gills (this study). The results from the in vitro and in vivo assays showed that miR-7 could serve as an antiviral factor by interacting with viral wsv477, an early gene involved in WSSV replication, leading to the inhibition of virus replication. In addition, the observation that silencing of the miRNA processing machinery using siRNA against Dicer1 led to faster replication of WSSV in shrimp provided further compelling evidence in favor of the idea that host miRNAs had antiviral roles. It was reported previously that shrimp Dicer1 and Dicer2 had antiviral effects in shrimp (10, 29, 35). These data indicated that host miRNAs played important roles in shrimp immune defense against virus infection. There is very limited information about the roles of miRNAs in invertebrates, compared with those in vertebrates, in response to virus infection. Our study, therefore, provides the first demonstration that, during virus infection, a crustacean miRNA could have an antiviral role by targeting a viral transcript.
In mammals, data from cultured mammalian cell lines demonstrate that the host miRNAs can be involved in host antiviral responses through direct interactions with viral genes, especially those genes that have roles in viral replication (25, 28). Human miR-24 and miR-93 were shown to interact directly with the RNA-dependent RNA polymerase and phosphoprotein genes of vesicular stomatitis virus (VSV), respectively. These genes encode elements required for polymerase binding and viral genomic replication (25). It was found that human cellular miR-323, miR-491, and miR-654 can bind the influenza PB1-5 gene that encodes a polymerase subunit (28). Recently, an insect (Helicoverpa zea) miRNA (Hz-miR-24) was found to directly target the coding regions of two genes of the Heliothis virescens ascovirus (HvAV-3e, Ascoviridae), the DNA-dependent RNA polymerase gene and its subunit β gene (16). In addition to direct interactions between host miRNAs and virus genes, the host miRNAs can indirectly limit virus replication by targeting host genes that are required for viral replication or by repressing inhibitors of protective innate immune responses. It has been demonstrated that cellular miR-17-5p and miR-20a can inhibit HIV-1 replication by repressing the expression of PCAF, a histone acetyltransferase, that is required for HIV-1 transcriptional elongation (18, 31). It has also been reported that miR-7 is upregulated in human cancer cells in response to mammalian adenovirus infection (30). The upregulation of miR-7 was shown to depend on the activation of the E2F1 protein. Ectopic expression of miR-7 suppresses cell viability and induces autophagy through suppression of oncogenic epidermal growth factor receptor (EGFR) expression (30). Although accumulating evidence indicates that host miRNAs can counteract viral genes, most reported experiments are conducted in vitro, and therefore it remains unclear whether the host miRNAs can function in the physiological contexts in vivo. Our investigations revealed that the administration of miR-7 to shrimp in vivo led to a decrease in the transcript levels of the wsv477 gene, resulting in the inhibition of WSSV replication. Therefore, our study, as well as studies in mammals and insects, indicates that the host miRNAs have major effects on virus infection in animals.
In contrast to the antiviral roles of host miRNAs, the virus can also derive benefits from host miRNA-mediated regulation during virus infection (11, 13, 32). One of the best-established host miRNAs that is known to promote the replication of virus is miR-122, which is specifically and highly expressed in liver tissue (13, 17). miR-122 binds to the 5′UTR of HCV genomic RNA and is essential for the replication of HCV (13, 17). In the present study, the antiviral roles of miR-7 in shrimp have been revealed; however, it is also possible that the virus could derive benefits from host miRNA-mediated regulation. Viruses have flexible genome sequences (28, 32), and, if the miR-7-WSSV interaction was disadvantageous to the virus, the WSSV might have evolved a nucleotide change that would allow it to evade the miRNA-7-mediated regulation. miR-7 is widely conserved in animals and has been shown to be induced in response to infections of mammalian viruses, including murine cytomegalovirus (MCMV), adenovirus, and influenza A virus (7, 12, 30). These findings indicate that miR-7 may have an evolutionarily conserved role that is induced during virus infection. In this context, it is possible that WSSV benefits in some way from the miR-7-mediated regulation. For example, miR-7 regulation might be employed by WSSV to promote a persistent infection in some contexts or to restrict the tissue tropism of the virus to preserve viability of the host during infection.
RNA interference, mediated by siRNAs or miRNAs, is an evolutionarily conserved mechanism that plays an important role in immune responses against virus infection (32). The siRNAs are extensively employed by animal hosts to efficiently defend against RNA virus invasion. However, whether the siRNA pathway in animals is used to counteract DNA viruses is yet to be defined. Increasing evidence indicates that a number of host miRNAs are involved in the antiviral responses, suggesting that miRNAs may be one of the most efficient strategies that animals employ to control DNA virus infections. Unlike siRNAs, the miRNAs require only partial complementarity (seed sequence) to interact with viral transcripts and thus can reduce off-target events better than siRNAs. Therefore, miRNAs are promising candidates for developing novel and efficient therapeutics to control DNA and RNA viruses. In summary, our findings provide novel insights into the molecular events of virus-host interactions directed by miRNAs in crustaceans and, specifically, present an efficient strategy to control WSSV in shrimp.
Supplementary Material
ACKNOWLEDGMENTS
This work was financially supported by the National Natural Science Foundation of China (30830084), the Hi-Tech Research and Development Program of China (863 program of China) (2010AA09Z403), and the Major State Basic Research Development Program (2012CB114403).
Footnotes
Published ahead of print 26 September 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Abrahante JE, et al. 2003. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev. Cell 4:625–637 [DOI] [PubMed] [Google Scholar]
- 2. Ambros V. 2003. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell 113:673–676 [DOI] [PubMed] [Google Scholar]
- 3. Ambros V. 2004. The functions of animal microRNAs. Nature 431:350–355 [DOI] [PubMed] [Google Scholar]
- 4. Asgari S. 2011. Role of microRNAs in insect host-microorganism interactions. Front. Physiol. 2:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bartel DP. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297 [DOI] [PubMed] [Google Scholar]
- 6. Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. 2003. Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113:25–36 [DOI] [PubMed] [Google Scholar]
- 7. Buggele WA, Johnson KE, Horvath CM. 2012. Influenza A virus infection of human respiratory cells induces primary microRNA expression. J. Biol. Chem. doi:10.1074/jbc.M112.387670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cameron JE, et al. 2008. Epstein-Barr virus growth/latency III program alters cellular microRNA expression. Virology 382:257–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Carrington JC, Ambros V. 2003. Role of microRNAs in plant and animal development. Science 301:336–338 [DOI] [PubMed] [Google Scholar]
- 10. Chen YH, et al. 2011. Identification and functional characterization of Dicer2 and five single VWC domain proteins of Litopenaeus vannamei. Dev. Comp. Immunol. 35:661–671 [DOI] [PubMed] [Google Scholar]
- 11. Cullen BR. 2011. Viruses and microRNAs: RISCy interactions with serious consequences. Genes Dev. 25:1881–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dittmer A, Förstemann K. 2012. Murine cytomegalovirus infection of cultured mouse cells induces expression of miR-7a. J. Gen. Virol. 93:1537–1547 [DOI] [PubMed] [Google Scholar]
- 13. Gottwein E, Cullen BR. 2008. Viral and cellular microRNAs as determinants of viral pathogenesis and immunity. Cell Host Microbe 3:375–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Houzet L, et al. 2008. MicroRNA profile changes in human immunodeficiency virus type 1 (HIV-1) seropositive individuals. Retrovirology 5:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huang T, Xu D, Zhang X. 2012. Characterization of host microRNAs that respond to DNA virus infection in a crustacean. BMC Genomics 13:159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hussain M, Asgari S. 2010. Functional analysis of acellular microRNA in insect host-ascovirus interaction. J. Virol. 84:612–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. 2005. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science 309:1577–1581 [DOI] [PubMed] [Google Scholar]
- 18. Kiernan RE, et al. 1999. HIV-1 tat transcriptional activity is regulated by acetylation. EMBO J. 18:6106–6118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lecellier CH, et al. 2005. A cellular microRNA mediates antiviral defense in human cells. Science 308:557–560 [DOI] [PubMed] [Google Scholar]
- 20. Lee RC, Feinbaum RL, Ambros V. 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75:843–854 [DOI] [PubMed] [Google Scholar]
- 21. Lim LP, et al. 2005. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433:769–773 [DOI] [PubMed] [Google Scholar]
- 22. Liu W, Han F, Zhang X. 2009. Ran GTPase regulates hemocytic phagocytosis of shrimp by interaction with myosin. J. Proteome Res. 8:1198–1206 [DOI] [PubMed] [Google Scholar]
- 23. Matskevich AA, Moelling K. 2007. Dicer is involved in protection against influenza A virus infection. J. Gen. Virol. 88:2627–2635 [DOI] [PubMed] [Google Scholar]
- 24. Meister G, Tuschl T. 2004. Mechanisms of gene silencing by double-stranded RNA. Nature 431:343–349 [DOI] [PubMed] [Google Scholar]
- 25. Otsuka M, et al. 2007. Hypersusceptibility to vesicular stomatitis virus infection in Dicer1-deficient mice is due to impaired miR24 and miR93 expression. Immunity 27:123–134 [DOI] [PubMed] [Google Scholar]
- 26. Pedersen IM, et al. 2007. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature 499:919–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schwarz DS, et al. 2003. Asymmetry in the assembly of the RNAi enzyme complex. Cell 115:199–208 [DOI] [PubMed] [Google Scholar]
- 28. Song L, Liu H, Gao S, Jiang W, Huang W. 2010. Cellular microRNAs inhibit replication of the H1N1 influenza A virus in infected cells. J. Virol. 84:8849–8860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Su J, et al. 2008. A key gene of the RNA interference pathway in the black tiger shrimp, Penaeus monodon: identification and functional characterisation of Dicer-1. Fish Shellfish Immunol. 24:223–233 [DOI] [PubMed] [Google Scholar]
- 30. Tazawa H, et al. 2012. Genetically engineered oncolytic adenovirus induces autophagic cell death through an E2F1-microRNA-7-epidermal growth factor receptor axis. Int. J. Cancer doi:10.1002/ijc.27589 [DOI] [PubMed] [Google Scholar]
- 31. Triboulet R, et al. 2007. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science 315:1579–1582 [DOI] [PubMed] [Google Scholar]
- 32. Umbach JL, Cullen BR. 2009. The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Genes Dev. 23:1151–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Varnholt H, et al. 2008. MicroRNA gene expression profile of hepatitis C virus-associated hepatocellular carcinoma. Hepatology 47:1223–1232 [DOI] [PubMed] [Google Scholar]
- 34. Xu J, Han F, Zhang X. 2007. Silencing shrimp white spot syndrome virus (WSSV) genes by siRNA. Antiviral Res. 73:126–131 [DOI] [PubMed] [Google Scholar]
- 35. Yao X, et al. 2010. A Dicer-1 gene from white shrimp Litopenaeus vannamei: expression pattern in the processes of immune response and larval development. Fish Shellfish Immunol. 29:565–570 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.