Abstract
Rift Valley fever virus (RVFV) is a zoonotic pathogen capable of causing serious morbidity and mortality in both humans and livestock. The lack of efficient countermeasure strategies, the potential for dispersion into new regions, and the pathogenesis in humans and livestock make RVFV a serious public health concern. The receptors, cellular factors, and entry pathways used by RVFV and other members of the family Bunyaviridae remain largely uncharacterized. Here we provide evidence that RVFV strain MP-12 uses dynamin-dependent caveola-mediated endocytosis for cell entry. Caveolae are lipid raft domains composed of caveolin (the main structural component), cholesterol, and sphingolipids. Caveola-mediated endocytosis is responsible for the uptake of a wide variety of host ligands, as well as bacteria, bacterial toxins, and a number of viruses. To determine the cellular entry mechanism of RVFV, we used small-molecule inhibitors, RNA interference (RNAi), and dominant negative (DN) protein expression to inhibit the major mammalian cell endocytic pathways. Inhibitors and RNAi specific for macropinocytosis and clathrin-mediated endocytosis had no effect on RVFV infection. In contrast, inhibitors of caveola-mediated endocytosis, and RNAi targeted to caveolin-1 and dynamin, drastically reduced RVFV infection in multiple cell lines. Expression of DN caveolin-1 also reduced RVFV infection significantly, while expression of DN EPS15, a protein required for the assembly of clathrin-coated pits, and DN PAK-1, an obligate mediator of macropinocytosis, had no significant impact on RVFV infection. These results together suggest that the primary mechanism of RVFV MP-12 uptake is dynamin-dependent, caveolin-1-mediated endocytosis.
INTRODUCTION
Rift Valley fever virus (RVFV) is a mosquito-borne zoonotic pathogen within the family Bunyaviridae, genus Phlebovirus. Large outbreaks of RVFV infection have occurred predominantly in sub-Saharan Africa. However, recent outbreaks outside of the African continent, in the Arabian Peninsula, have raised concerns about the potential intercontinental spread of this virus to Europe, Asia, and the Americas (4, 10, 20, 82). In livestock, RVFV infection causes frequent abortions and a newborn mortality rate as high as 95%, which leads to significant economic loss during an outbreak of RVFV. Human infection with RVFV is typically associated with an acute febrile illness but can lead to more-severe symptoms, such as retinal vasculitis, encephalitis, and fatal hepatitis with hemorrhagic fever (10). RVFV is considered a select agent with bioterrorism and agroterrorism potential. Currently there are no specific therapies for use against this pathogen.
RVFV is a spherical enveloped virus with a diameter of 90 to 100 nm and a tripartite, single-stranded RNA genome in a negative and ambisense orientation. The virion surface displays two membrane-anchored glycoproteins that are arranged into pentameric and hexameric units to form a unique T=12 icosahedral structure (37). The envelope proteins, Gn and Gc, are derived from cotranslational cleavage of a precursor protein (GPC), and the Gc protein is predicted to employ a class II fusion mechanism, like the alphavirus E1 and flavivirus E proteins (37, 87). Like that of other bunyaviruses, RVFV entry is thought to occur via endocytosis, since membrane fusion and infection are induced by a low pH (35, 36). Entry into host cells is the first step in virus infection and represents an attractive target for therapeutic intervention. In addition, the mechanisms underlying RVFV entry are critically linked to tropism and pathogenesis. However, understanding of the RVFV entry pathways and of the cellular factors involved prior to the release of the viral genome into the cytosol is limited.
Some of the major endocytotic pathways used by viruses to enter host cells include clathrin-mediated endocytosis (CME), caveola-mediated endocytosis (CavME), and macropinocytosis (67). The best-studied and most common method of virus entry is CME, a rapid process that is dependent on clathrin and multiple other adaptor molecules (i.e., dynamin II, EPS15) that coat vesicles and induce internalization (25, 39, 67). The formation of clathrin-coated pits is a constitutive process but can also be actively initiated by its cargo. After clathrin-coated pits have assembled at the cell membrane, the large GTPase dynamin II causes scission of the vesicles from the membrane, and these mature clathrin-coated vesicles, with a diameter of ∼120 nm, then transfer their cargo to endosomes (25, 27, 39, 67). CME is involved in the uptake of hormones and nutrients that are essential to the cell, such as iron-laden transferrin, cholesterol-laden low-density lipoproteins, and growth factors (39). Viruses that have been shown to enter cells by CME include influenza viruses (98, 117), hepatitis C virus (11), vesicular stomatitis virus (VSV) (51, 107), African swine fever virus (45), and the bunyavirus family member Hantaan virus (27, 50). CavME is a ligand-triggered process with kinetics slower than those of CME and, like CME, requires dynamin II for the scission of vesicles from the membrane. Caveolae are highly structured lipid raft microdomains enriched in caveolin-1 (CAV-1), cholesterol, and sphingolipids, with a flask-shaped morphology (77). Caveolins (including CAV-1) are scaffolding proteins, expressed on the cytoplasmic side of the plasma membrane, that interact with gangliosides, integrins, and cholesterol and are the major structural proteins of caveolae (19, 43, 78). CavME is responsible for mediating numerous transduction signals, for transcytosis, and for the uptake of extracellular ligands, bacterial toxins, and membrane components (39). Some viruses that are known to enter cells via CavME include nonenveloped viruses, such as simian virus 40 (SV40) (79, 80), echovirus 1 (61), BK polyomavirus (BKV) (32), human papillomavirus type 31 (104), and avian reovirus (46), and enveloped viruses, such as hepatitis B virus (60), amphotropic murine leukemia virus (5), respiratory syncytial virus (118), coronavirus 229E (73), and Newcastle disease virus (14). Macropinocytosis is used by the cell to internalize extracellular fluid and is largely dependent on actin remolding. Macropinosomes, vesicles with a diameter up to 10 μm, are formed when membrane ruffles fold back onto the plasma membrane to form fluid-filled cavities that close by membrane fusion (39, 65, 93). Macropinocytosis involves multiple signaling events resulting in ruffle formation, mediated by phosphinositide 3-kinase (PI3K), Rho GTPase Rac-1, and p21-activated kinase 1 (PAK-1), and macropinosome closure, facilitated by PI3K and PAK-1 phosphorylation of C-terminal binding protein 1 (CtBP-1) (53, 65, 108, 114). Macropinocytosis is unique in that it is not dependent on dynamin II for separation from the plasma membrane (67). Viruses that have been shown to use macropinocytosis for entry include vaccinia virus (VacV) (64, 66), Ebola virus (71, 88), African swine fever virus (90), and Nipah virus (83).
Recent studies with Uukuniemi virus (UUKV), a phlebovirus related to RVFV, have revealed that UUKV entry is mainly clathrin independent; the virus is transported from early endosomes to late endosomes; and infection is dependent on normal maturation of late endosomes (58). These studies indicate a potential for other bunyaviruses to utilize a similar non-clathrin-mediated entry mechanism. It has also been suggested, on the basis of the role of protein kinase C (PKC) epsilon in RVFV infection, that macropinocytosis may contribute to RVFV entry (35). However, to date, the pathway involved in RVFV entry has not been thoroughly and systematically investigated. To elucidate the cellular factors involved in RVFV entry, we used an attenuated strain of RVFV, RVFV MP-12. Wild-type (WT) RVFV is a select agent, and experiments using WT RVFV have to be performed in high-level biosafety containment labs, whereas RVFV MP-12 can be handled at biosafety level 2. RVFV strain MP-12 was derived from the wild-type Egyptian strain ZH548 by propagation for 12 serial tissue culture passages in the presence of the mutagenic agent 5-fluorouracil (15). Strain MP-12, originally reported to differ by 4 amino acids (115), differs in only 3 amino acids in the mature glycoproteins (Gn/Gc) from wild-type ZH548 according to recent sequencing data (9), and the in vitro tropism of RVFV MP-12 seems not to be altered from that of WT RVFV, making it likely that cellular factors required for RVFV MP-12 entry are also necessary for WT RVFV entry (12, 15). In this study, we use small-molecule inhibitors, RNA interference (RNAi), and dominant negative (DN) protein expression to investigate the entry of RVFV in multiple cell types (HeLa, HepG2, and 293T cells). In addition, viruses with known endocytosis entry pathways were used as specificity controls in the panoply of assays. By combining all of these approaches, we provide an accurate and detailed description of the RVFV MP-12 uptake mechanism. Our data clearly indicate that RVFV MP-12 enters and infects multiple cell lines independently of CME and macropinocytosis. Instead, the primary mechanism of virus uptake appears to be dynamin II-dependent, caveolin-1-mediated endocytosis.
MATERIALS AND METHODS
Cells and culture conditions.
All cell lines were maintained in culture medium supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin (Invitrogen) at 37°C under 5% CO2. HEK293T (human embryonic kidney), HeLa (human cervix carcinoma), BSC40 (African green monkey kidney), and HepG2 (hepatocellular carcinoma) cells were cultured in Dulbecco's modified Eagle's medium (DMEM), while Vero (African green monkey kidney) cells were maintained in minimum essential medium alpha.
Viruses.
The recombinant RVFV vaccine strain MP-12 generated to carry a green fluorescent protein (GFP) gene (RVFV-MP-12-GFP) in place of the NSs gene has been described previously (47). Authentic nonrecombinant RVFV strain MP-12 was obtained from C. J. Peters (University of Texas Medical Branch). The recombinant VSV expressing GFP (VSV-GFP) was a kind gift from Adolfo Garcia-Sastre (Mount Sinai School of Medicine) and was derived from the Indiana serotype 1 strain (33). The recombinant VacV expressing GFP (VacV-GFP) was obtained through the NIH Biodefense and Emerging Infections Research Resources Repository, NIAID (NR-624) and was derived from the Western Reserve strain (30). RVFV MP-12, RVFV-MP-12-GFP, and VSV-GFP were propagated in Vero cells, while VacV-GFP was grown in BSC-40 cells. The concentrations of PFU for RVFV MP-12, RVFV-MP-12-GFP, and VSV-GFP were quantified in Vero cells by using a standard plaque assay consisting of an agarose overlay with crystal violet staining. VacV-GFP PFU were obtained by dilution of the virus and infection of BSC-40 cells without an agarose overlay. For all infection experiments, cells were first incubated with each virus at a multiplicity of infection (MOI) of 1 (diluted in complete medium) for 3 h and were then washed with phosphate-buffered saline (PBS) and incubated in fresh complete medium for 14 to 16 h at 37°C.
Inhibitor treatment.
HeLa or HepG2 cells were plated overnight in complete medium in a collagen-coated 96-well dish at 1 × 104 cells per well or in a collagen-coated 384-well dish at 3 × 103 cells per well. Unless otherwise indicated, all inhibitors were purchased from EMD Millipore. The following inhibitors were initially resuspended in dimethyl sulfoxide (DMSO) and were then diluted in complete medium to obtain the final concentrations indicated: p21-activated kinase peptide inhibitor (PAK18) (20 μM), the inactive control peptide PAK18-R192A (PAK-NC) (20 μM), p21-activated kinase inhibitor III (IPA-3) (10 μM), a Rac1 inhibitor (Rac-I) (100 μM), the PI3K inhibitor Ly294002 (50 μM), the cell-permeant 17β-hydroxy analog of wortmannin (HWT), a PI3K inhibitor (0.5 μM), cytochalasin D (CD) (1 μM), latrunculin A (LA) (2 μM), jasplakinolide (JP) (1 μM), the vacuolar H+-ATPase inhibitor bafilomycin A (BAF) (100 nM), cholesterol-disrupting nystatin (Nys) (30 μM) and progesterone (Prog; Sigma)(10 μM), and the dynamin II inhibitor dynasore (DYN) (80 μM) or phorbol 12-myristate 13-acetate (PMA) (10 μM). As a control, the cells were also incubated with 50 μM DMSO, representing the highest concentration of DMSO to which the cells were exposed with the inhibitors listed above. The Na+/H+ exchange inhibitor 5-(N-ethyl-N-isopropyl)amiloride (EIPA; Sigma) was resuspended in methanol at 33 mM prior to dilution in medium to obtain a final concentration of 10 μM, 25 μM, or 50 μM. The rest of the inhibitors used were first resuspended in H2O and then diluted in complete medium. These inhibitors include the lysosomotropic agent ammonium chloride (NH4Cl; 50 mM), the serine/threonine phosphatase inhibitor okadaic acid (OA pre; 100 nM), and the cationic amphiphilic compound chlorpromazine (CPZ; 6.5 μg/ml; Sigma). The concentration of CPZ used in this study has been shown to specifically block CME by preventing clathrin-coated pit assembly at the cell surface (11, 48, 66, 107, 109). Inhibitors were used at previously published concentrations, and concentration curves were performed with all of the inhibitors to determine the concentration that resulted in the maximum decrease in the expected target without altering cell viability (2, 5, 11, 13, 17, 21, 26, 28, 31, 32, 38, 40–42, 44–46, 48–51, 53, 59, 61, 64, 66, 71, 72, 74, 76, 77, 81, 85, 88, 90, 94, 102, 107, 109, 114, 120, 123–126). The endocytosis pathway targeted by each inhibitor is outlined in Table 1. Cells were incubated with medium alone, DMSO, or one of the indicated inhibitors (see Fig. 2 and 3) for 1 h prior to and during incubation with RVFV-MP-12-GFP, VacV-GFP, or VSV-GFP at an MOI of 1 for 3 h at 37°C. Alternatively, untreated cells were incubated with the indicated virus (see Fig. 2 and 3) for 2 h, washed with PBS, and then incubated with complete medium alone or with 100 nM OA for 1 h (OA post). Cells were then washed with PBS and were incubated in complete medium at 37°C for 14 to 16 h. alamarBlue reagent (Invitrogen) was added to cells in complete medium to make a 1× final concentration, and cells were incubated for 45 min at 37°C. Fluorescence was measured at an excitation wavelength of 570 nm and an emission wavelength of 590 nm (EX570/EM590); then cells were washed with PBS, and 100 μl (96-well plate) or 50 μl (384-well plate) of Thermo Scientific radioimmunoprecipitation assay (RIPA) buffer with 1× Halt protease inhibitors (Thermo Scientific) was added per well. GFP fluorescence was measured at EX488/EM510 after freeze-thawing. To account for differences in cell number from well to well, GFP values were normalized to cell viability readings based on alamarBlue fluorescence, and the background (uninfected cells) was subtracted. The percentage was determined by dividing the average of the treated and infected samples by the average of untreated or DMSO-treated and infected samples. These experiments were performed in triplicate for each cell type 3 or more times, and the average (± standard deviation) of three independent experiments is shown.
Table 1.
Pharmacological inhibitors, their abbreviations, and their specificities
| Reagent | Tag | Endocytosis pathway blocked | Specificity | Reference(s) |
|---|---|---|---|---|
| PAK-1 inhibitor peptide | PAK-18 | Macropinocytosis | Autoinhibitory domain of PAK-1 | 88, 124, 125 |
| Inactive control peptide (PAK18-R192A) | PAK-NC | Control for PAK-18 | Inactive peptide that controls for nonspecific effects | |
| RacGEF inhibitor (NSC23766) | RAC-I | Macropinocytosis | Inhibits Rac-1 GDP/GTP exchange | 26, 85, 90 |
| p21-activated kinase inhibitor III | IPA-3 | Macropinocytosis | Prevents PAK-1 activation (irreversible) | 26, 66, 90 |
| 5-(N-Ethyl-N-isopropyl)amiloride | EIPA | Macropinocytosis | Inhibits Na+/H+ exchange | 38, 53, 88 |
| Cell-permeant 17β-hydroxy analog of wortmannin | HWT | Macropinocytosis | Irreversible PI3K inhibitor | 64, 66 |
| LY294002 | Macropinocytosis | Reversible PI3K inhibitor | 44, 71, 90 | |
| Cytochalasin D | CD | Macropinocytosis | Actin-depolymerizing agent | 40, 64, 94 |
| Latrunculin A | LA | Macropinocytosis and clathrin | Sequesters actin monomers | 31, 40, 64, 120 |
| Jasplakinolide | JP | Macropinocytosis and caveolin | Stabilizes actin polymers | 31, 64, 94 |
| Ammonium chloride | NH4Cl | All pH dependent | Blocks acidification of endosomes | 74 |
| Bafilomycin A1 | BAF | All pH dependent | Inhibits vacuolar H+-ATPase (irreversible) | 123 |
| Chlorpromazine | CPZ | Clathrin | Prevents clathrin-coated pit assembly at the cell surface | 11, 49, 72, 107 |
| Dynasore | DYN | Caveolin and clathrin | Blocks dynamin II | 40–42, 46, 51 |
| Okadaic acid | OA pre | Caveolin and clathrin | Removes cell surface caveolae and blocks CME | 5, 13, 59, 76, 77, 126 |
| Okadaic acid; cells treated for 1 h, postinfection | OA post | Enhances caveolin | Promotes CavME of bound cargo | 5, 76, 77, 126 |
| Phorbol 12-myristate 13-acetate | PMA | Caveolin | Decreases the level of caveolae at the cell surface | 2, 14, 102 |
| Nystatin + progesterone | Nys+Prog | Caveolin | Sequesters and blocks cholesterol | 2, 17, 21, 114 |
Fig 2.
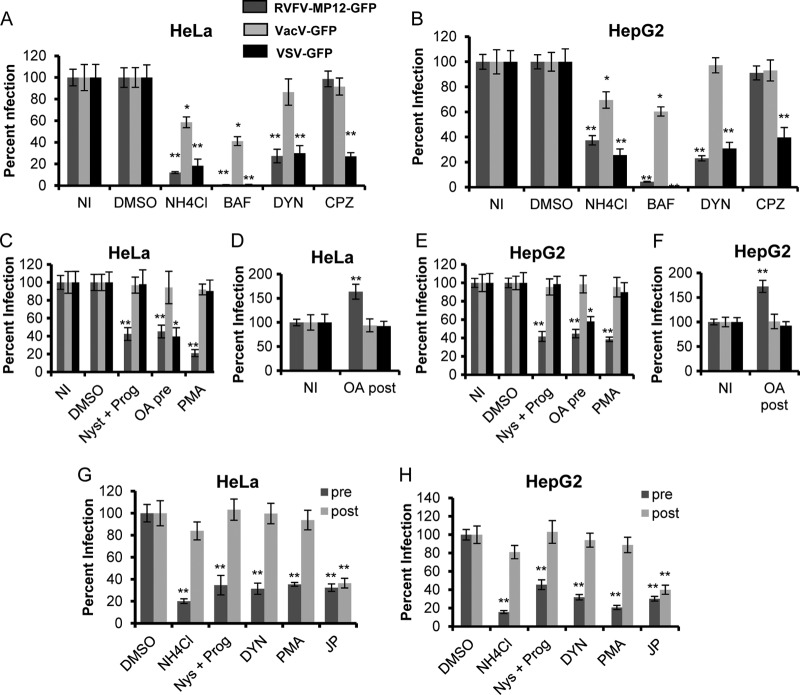
Inhibitors of caveolar but not clathrin-mediated endocytosis block RVFV infection. Average levels of infection detected by GFP fluorescence (± standard deviations), compared to those for untreated or DMSO-treated controls, are shown. (A and B) HeLa (A) or HepG2 (B) cells were either left untreated (NI) or pretreated with DMSO, 50 mM NH4Cl, 100 nM BAF, 6.5 μg/ml CPZ, or 80 μM DYN for 1 h. The pretreated cells were incubated with the indicated virus at an MOI of 1 for 3 h in the presence of inhibitors. (C and E) HeLa (C) or HepG2 (E) cells were either left untreated or pretreated with DMSO, 30 μM Nys plus 10 μM Prog, 100 nM OA (pre), or 10 μM PMA for 1 h. Inhibitors were also present during 3 h of incubation with RVFV-MP-12-GFP, VacV-GFP, or VSV-GFP at an MOI of 1. (D and F) HeLa (D) or HepG2 (F) cells were infected with the indicated virus at an MOI of 1 for 2 h, washed, and then treated with 100 nM OA for 1 h (post). (G and H) HeLa (G) or HepG2 (H) cells were either left untreated or pretreated (pre) with DMSO, 50 mM NH4Cl, 30 μM Nys plus 10 μM Prog, 80 μM DYN, 10 μM PMA, or 1 μM JP for 1 h. The inhibitors were also present during the 3 h of incubation with RVFV-MP-12-GFP at an MOI of 1. Alternatively, HeLa (G) or HepG2 (H) cells were incubated for 1 h with RVFV-MP-12-GFP at an MOI of 1. Cells were washed to remove unbound virus and were then incubated with the indicated inhibitors (post) for 4 h. GFP expression was normalized to cell titers measured by alamarBlue fluorescence. The percentage of infection was determined by taking untreated or DMSO-treated and infected samples as 100% infected. Shown are the means for three independent experiments performed in triplicate (**, P < 0.01; *, P < 0.05).
Fig 3.
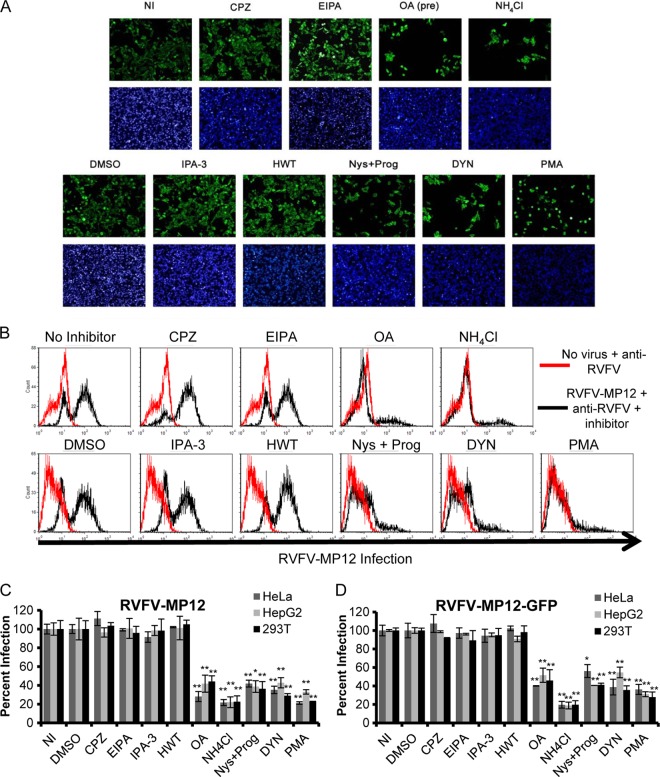
Inhibitors of caveola-mediated endocytosis, but not inhibitors of CME or macropinocytosis, reduce the percentage of cells infected with authentic RVFV MP-12 or recombinant RVFV-MP-12-GFP. HeLa cells (A, C, and D), 293T cells (B, C, and D), or HepG2 cells (C and D) were either left untreated (NI), pretreated with 6.5 μg/ml CPZ, 25 μM EIPA, 100 nM OA, or 50 mM NH4Cl for 1 h (A and B, top), or treated with DMSO, 10 μM IPA-3, 0.5 μM HWT, 30 μM Nys plus 10 μM Prog, 80 μM DYN, or 10 μM PMA for 1 h (A and B, bottom). The inhibitors were also present during the 3 h of incubation with authentic RVFV MP-12 (A, B, and C) or RVFV-MP-12-GFP (D) at an MOI of 1. (A) Infection of HeLa cells was detected by immunofluorescence using anti-RVFV polyclonal antibodies (green) and the nuclear dye DAPI (blue). This assay was performed in triplicate three or more times with similar results. Images shown are representative of 10 images/well of a 96-well plate. (B) Infection of 293T cells was detected by flow cytometry using anti-RVFV. The data shown are representative of results from three similar experiments performed in duplicate. Red histograms represent uninfected cells that were left untreated (top) or were treated with DMSO (bottom); black histograms represent RVFV-MP-12-infected cells treated with the indicated inhibitors. (C and D) Infection was detected by flow cytometry using an anti-RVFV antibody (C) or GFP expression (D). The quantity of infected cells relative to that of untreated or DMSO-treated controls is given as the percentage of infection. Shown are the means (± standard deviations) for three independent experiments performed in duplicate for each cell type (**, P < 0.01; *, P < 0.05).
Time-of-addition experiments.
HeLa or HepG2 cells were plated overnight in complete medium in a collagen-coated 384-well dish at 3 × 103 cells per well. Small-molecule inhibitors were added at the concentrations given above for 1 h prior to and during infection for pretreatment samples (pre). Alternatively, untreated cells were incubated with RVFV-MP-12-GFP (MOI, 1) for 1 h, washed with PBS to remove unbound virus, and then incubated with small-molecule inhibitors in complete medium for 4 h (post). Cells were then washed with PBS and were incubated in complete medium at 37°C overnight. Following 16 h of incubation, alamarBlue reagent was added to cells in complete medium to make a 1× final concentration, and cells were incubated for 45 min at 37°C. Fluorescence was measured at EX570/EM590; then cells were washed with PBS, and 50 μl of Thermo Scientific RIPA buffer with 1× Halt protease inhibitors (Thermo Scientific) was added per well. GFP fluorescence was measured at EX488/EM 510 after freeze-thawing. GFP values were normalized to cell viability readings, and the background (uninfected cells) was subtracted. The percentage was determined by dividing the average of the treated and infected samples by the average of untreated or DMSO-treated and infected samples. These experiments were performed in triplicate for each cell type 3 or more times, and the average (± standard deviation) of three independent experiments is shown.
Inhibitor treatment and infection with RVFV MP-12 and RVFV-MP-12-GFP analyzed by flow cytometry and imaging.
HeLa, HepG2, or 293T cells were plated overnight in complete medium at 2 × 105 per well on collagen-coated 12-well plates for flow cytometry, and HeLa cells were plated at 1 × 104 per well on collagen-coated 96-well plates for image analysis. Cells were incubated with either no inhibitor, the DMSO control, 6.5 μg/ml CPZ, 25 μM EIPA, 100 nM OA, 50 mM NH4Cl, 10 μM IPA-3, 0.5 μM HWT, 30 μM Nys plus 10 μM Prog, 80 μM DYN, or 10 μM PMA for 1 h prior to and during infection with RVFV-MP-12-GFP or authentic RVFV MP-12 (MOI, 1). Cells were then washed with PBS and incubated in complete medium at 37°C for 16 h. For imaging, HeLa cells were first fixed with 4% paraformaldehyde for 30 min at room temperature (RT) and then washed once with Tris-buffered saline (TBS). Samples infected with RVFV-MP-12-GFP were incubated with 300 nM 4′,6-diamidino-2-phenylindole (DAPI) in PBS for 2 min at RT, washed twice with PBS, and left in PBS for imaging.
Samples infected with authentic RVFV MP-12 were fixed, permeabilized with 90% methanol for 10 min, washed with PBS, put in a block (BD bovine serum albumin [BSA] stain buffer plus 3% normal goat serum plus 0.5% Triton) for 1 h, and then incubated for 1 h at RT with an anti-RVFV mouse polyclonal antibody at 1:500 (kindly provided by Robert Tesh, University of Texas Medical Branch) in a block. After two washes with PBS, Alexa Fluor 488-conjugated goat anti-mouse IgG (Invitrogen) was added in a block at 1:750 for 30 min at RT. These samples were washed twice with PBS, incubated with 300 nM DAPI in PBS for 2 min at RT, washed twice with PBS, imaged using an Olympus IX70 inverted microscope, and indexed using ImagePro software (version 6.3). For each well, a series of 10 green images was taken through the narrow blue filter (EX470 to 490/EM515) to capture infection; to visualize the nuclei, images were taken with a DAPI filter (EX330 to 385/EM420). Experiments for image analysis were performed in triplicate with HeLa cells three or more times; data are representative of results from three similar experiments.
For flow cytometry, HeLa, HepG2, and 293T cells were washed once with PBS; then 293T cells were removed from the plate with 500 μl PBS, and HeLa and HepG2 cells were detached by incubation in 5 mM EDTA (Invitrogen) for 5 min at 37°C. These cells were transferred to 1.5-ml centrifuge tubes and were spun down at 5,000 × g for 10 min. RVFV-MP-12-GFP-infected samples were incubated with 4% paraformaldehyde for 30 min prior to resuspension in PBS for flow analysis using a FACScan flow cytometer with CellQuest software (BD Biosciences). The cells infected with RVFV MP-12 were first incubated in 4% paraformaldehyde in PBS for 30 min, then resuspended in PBS containing 100 mM glycine, and incubated in the dark at 4°C overnight. Cells were incubated with an anti-RVFV mouse polyclonal antibody in permeabilization buffer (PBS containing 0.1% saponin, 20 mM EDTA, 0.02% sodium azide, and 2% FBS) for 1 h with rotation at 4°C. Cells were washed 3 times with PBS then incubated with Alexa Fluor 488 goat anti-mouse IgG (Invitrogen), at 1:750 in permeabilization buffer for 30 min rotating at 4°C. Cells were washed three times with PBS and were then resuspended in PBS and analyzed using a FACScan flow cytometer with CellQuest software (BD Biosciences). Flow cytometry experiments were performed in duplicate for each cell type two or more times, and the average (± standard deviation) of two independent experiments is shown. Cells were counted as infected if their FL-1 fluorescence was greater than that of the untreated or DMSO treated, uninfected cells (Fig. 3B, red histograms). The quantity of infected cells relative to that of untreated or DMSO-treated controls is given as the percentage of infection. For experiments with RVFV MP-12, uninfected cells were similarly probed with anti-RVFV and an appropriate secondary antibody to control for any nonspecific binding.
siRNA treatment.
All small interfering RNAs (siRNAs) were purchased from Qiagen. The siRNAs used in this study include the scrambled control siRNA (AllStars negative-control siRNA), the GFP-22 siRNA, vATPV1A siRNAs (FlexiTube GeneSolution for ATP6V1A and Hs_ATP6V1A_5), caveolin-1 siRNAs (FlexiTube GeneSolution for CAV-1), dynamin 2 (DMN2) siRNAs (FlexiTube GeneSolution for DMN2), clathrin heavy-chain (CHC) siRNAs (FlexiTube GeneSolution GS1213 for CLTC), epidermal growth factor receptor pathway substrate 15 (EPS15) siRNAs (FlexiTube GeneSolution for EPS15), PAK-1 siRNAs (FlexiTube GeneSolution for PAK-1) and phosphoinositide-3-kinase (PI3K) siRNAs (FlexiTube GeneSolution GS5295 for PIK3R1). All lyophilized siRNA constructs were resuspended in RNase- and DNase-free H2O to obtain a stock concentration of 20 μM. The FlexiTube GeneSolution siRNAs were four different siRNA constructs that were resuspended at 20 μM. To optimize the time of infection posttransfection and the siRNAs used for target knockdown, the individual FlexiTube siRNAs were mixed together in 1 tube at a 1:1:1:1 ratio and were transfected into HeLa cells or HepG2 cells at a final concentration of 50 nM using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's instructions. The samples were harvested at 48 h, 60 h, and 72 h posttransfection, and the efficiency of target downregulation was assessed by Western blot analysis. Once optimal target knockdown was measured by Western blotting, individual siRNAs were transfected at a final concentration of 50 nM, and the siRNAs that resulted in the specific maximum decrease in the expected target without altering cell viability were used for additional studies.
For the experiments shown, individual siRNA constructs indicated in Fig. 4 were transfected into HeLa or HepG2 cells at a final concentration of 50 nM using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's instructions. Twenty-four hours posttransfection, fresh medium was added. Forty-eight hours posttransfection, cells were plated overnight in complete medium in a collagen-coated 384-well plate at 3 × 103 cells per well. Sixty hours posttransfection, cells were incubated either with no virus or with RVFV-MP-12-GFP, VacV-GFP, or VSV-GFP (MOI, 1) for 3 h at 37°C; then the cells were washed with PBS and were incubated in complete medium overnight. Following 16 h of incubation, alamarBlue reagent was added to the cells in complete medium to make a 1× final concentration, and the cells were incubated for 1 h at 37°C. Fluorescence was measured at EX570/EM590; then cells were washed with PBS, and 50 μl of Thermo Scientific RIPA buffer with 1× Halt protease inhibitors (Thermo Scientific) was added per well. GFP fluorescence was measured at EX488/EM510 after freeze-thawing. GFP values were normalized to cell viability readings, and the background (uninfected cells) was subtracted. The percentage was determined by dividing the average of the transfected and infected samples by the average of infected samples transfected with scrambled control siRNA. These experiments were performed in triplicate for each cell type three or more times, and the average (± standard deviation) of three independent experiments is shown.
Fig 4.
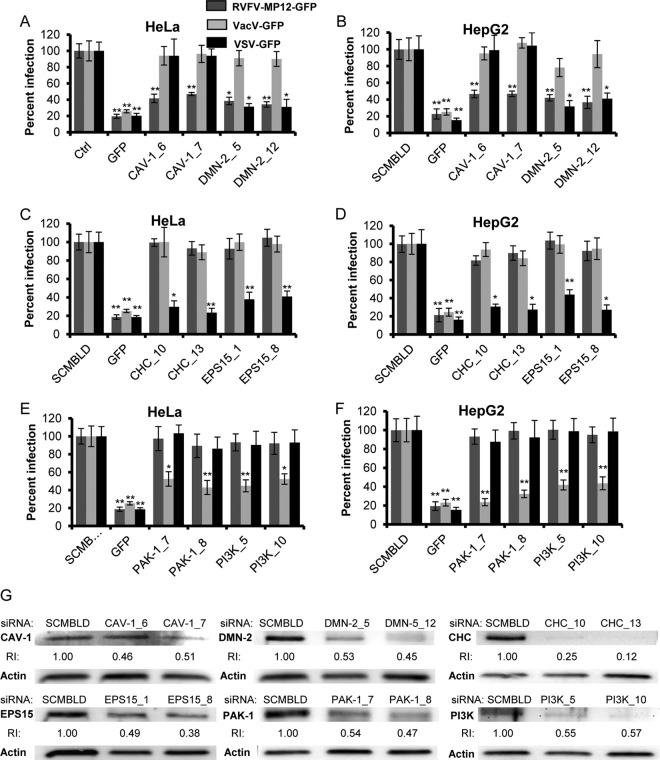
Dynamin 2 and caveolin-1 downregulation with siRNA reduces RVFV infection. Average levels of infection detected by GFP fluorescence (± standard deviations), compared to those for scrambled siRNA controls, are shown. (A through F) HeLa (A, C, and E) or HepG2 (B, D, and F) cells were transfected with 50 nM siRNA: either scrambled siRNA (SCMBLD), siRNA targeted against GFP, or two independent siRNAs (each) directed against CAV-1 or DMN2 (A and B), CHC or EPS15 (C and D), or PAK-1 or PI3K (E and F). Sixty hours posttransfection, cells were incubated either without virus or with RVFV-MP-12-GFP, VacV-GFP, or VSV-GFP at an MOI of 1 for 3 h. The percentage of infection was determined by taking cells transfected with SCMBLD siRNA as 100% infected. GFP expression was normalized to cell titers measured by alamarBlue fluorescence. Shown are the means for three independent experiments performed in triplicate (**, P < 0.01; *, P < 0.05). (G) Transfected HeLa cells were washed and lysed at 60 h posttransfection, and whole-cell lysates were analyzed by Western blotting. The relative reduction index (RI) was calculated as the quotient of the densitometry signal for the target protein band divided by that for actin, which was then normalized by the ratio obtained with SCMBLD siRNA (considered to be 1). The data represent one of three experiments with similar results.
Western blot analysis.
The efficiency of target downregulation with individual siRNAs was assessed by Western blot analysis. Samples were harvested at 60 h posttransfection. Transfected cells were washed with PBS, trypsinized, resuspended in PBS, spun down, and then resuspended in Thermo Scientific RIPA buffer with 1× Halt protease inhibitors. Twenty-five to 50 μg of lysate was separated on Mini-Protean TGX precast gels (Bio-Rad), transferred to polyvinylidene difluoride (PVDF) membranes, blocked with 0.01% Tween 20 and 5% dry milk in TBS for 1 h to overnight, and probed with mouse anti-clathrin heavy chain (dilution, 1:500; Thermo Scientific), rabbit anti-dynamin 2 (1:750; Abcam), rabbit anti-CAV-1 (1:200), rabbit anti-EPS15 (1:200), rabbit anti-PAK-1 (1:200), rabbit anti-PI3K p85α (Santa Cruz Biotechnology [SCBT]), or rabbit anti-V-ATPase subunit A (1:500; GeneTex) for 1 h to overnight in a block. After five washes with 0.01% Tween 20 in TBS (TBS-T), a Pierce horseradish peroxidase (HRP)-conjugated polyclonal goat anti-mouse or goat anti-rabbit secondary antibody (Thermo Scientific) was added at 1:2,500 for 30 min at room temperature in a block. Proteins were visualized with an Alpha Innotech (San Leandro, CA) imager using a SuperSignal West Femto sensitivity substrate (Thermo Scientific). The blots were cut after transfer for development with more than one antibody. The cut strips with 30- to 50-kDa proteins were probed for 1 h with an HRP-conjugated rabbit anti-actin polyclonal antibody (Novus) at 1:1,000 and were then washed and visualized with an Alpha Innotech (San Leandro, CA) imager using a SuperSignal West Femto sensitivity substrate (Thermo Scientific).
Cloning of wild-type PAK-1, EPS15, and CAV-1, controls, and DN mutants.
The human CAV-1 DNA plasmid was purchased from Origene, and the full-length, wild-type CAV-1 open reading frame was cloned into a pIRES vector (Clontech). The internal ribosome entry site (IRES)-containing bicistronic vectors allow the simultaneous expression of two proteins separately from the same RNA transcript. WT-CAV-1 was cloned upstream of an mCherry reporter gene. The DN CAV-1 construct was assembled similarly to WT-CAV-1; however, the sequence coding the first 81 amino acids was removed and replaced with a start codon (CavΔ1-81). Similar caveolin constructs have been described previously (60). The control EPS15D3Δ2 construct (control EPS15) (7) and the DN EPS15Δ95/295 mutant construct (DN-EPS15) (6) were provided by Alice Daurty-Varsat and Nathalie Sauvonnet (Institut Pasteur, Paris, France). To visualize EPS15 and EPS15 expression under a red fluorescence channel, the GFP versions of these constructs were used as cloning templates. The GFP genes from the pEGFP-C1 (Clontech) vector templates were removed and replaced with the mCherry reporter to create an in-frame fusion with the EPS15 proteins. Wild-type and DN PAK-1 plasmids were purchased from Addgene (Cambridge, MA). Wild-type full-length PAK-1 is Myc tagged on the N-terminal end, while the DN PAK-1 construct comprising the autoinhibitory domain was N-terminally tagged with glutathione S-transferase (GST).
Transfection and transient expression.
293T or HeLa cells were plated at 1 × 105 per well on a collagen-coated glass-bottom 24-well plate for imaging studies, or at 4 × 105 per well on a collagen-coated 6-well plate for flow cytometry. The next day, 293T cells were transfected with 0.5 μg/well (24-well plate) or 2.5 μg/well (6-well plate) of WT CAV-1-mCherry (WT-CAV-1), DN CAV-1-mCherry (DN-CAV-1), control EPS15-mCherry (clone D3D2; control EPS15), DN EPS15 mCherry (EPS15Δ95/295; DN-EPS15), WT-PAK-1-myc, or DN-PAK-1-GST using TransIT-LT1 (Mirus) according to the manufacturer's instructions. For transfection of HeLa cells, the medium was changed to serum-free Opti-MEM (Invitrogen), and cells were transfected with 0.75 μg/well (24-well plate) or 2.5 μg/well (6-well plate) of the constructs given above by using 1.5 μl/well (24-well plate) or 5 μl/well (6-well plate) Lipofectamine 2000 (Invitrogen). After 4 to 6 h of incubation with DNA plus Lipofectamine 2000, the serum-free medium was removed and was replaced with complete medium for overnight incubation at 37°C.
Infection studies with DN mutants and analysis by imaging and flow cytometry.
Sixteen to 24 h posttransfection, 293T and HeLa cells were incubated with either RVFV-MP-12-GFP, VacV-GFP, or VSV-GFP (MOI, 1) for 3 h in complete DMEM. After incubation with the virus, cells were washed once with PBS and were then incubated overnight in complete medium at 37°C. After 16 h of incubation, cells were fixed with 4% paraformaldehyde for 30 min at RT. Alternatively, 24 h posttransfection, 293T cells were serum starved for 30 min prior to treatment with either no ligand, 4 μg/ml cholera toxin subunit B (CTX-B) conjugated to Alexa Fluor 488 (Invitrogen), or 50 μg/ml human transferrin (hTfn) conjugated to Alexa Fluor 488 (Invitrogen) in complete DMEM buffered with 20 mM HEPES (pH 7.2) for 30 min at 4°C; then the plate was transferred to 37°C for 2 h. After incubation with labels, cells were washed once with 0.1 M glycine–0.1 M NaCl in H2O (pH 3.0) to remove any uninternalized label and were then fixed with 4% paraformaldehyde for 30 min at RT. For imaging, cells transfected with WT-PAK-Myc or DN-PAK-GST were permeabilized by the addition of 90% methanol for 10 min. These cells were washed twice with PBS, put in a block for 1 h, and then incubated with mouse anti-c-Myc (dilution, 1:200) or rabbit anti-GST (1:100) (SCBT) in a block for 1 h. After two washes with PBS, an Alexa Fluor 555-conjugated F(ab′)2 fragment of goat anti-mouse IgG or an Alexa Fluor 555-conjugated F(ab′)2 fragment of goat anti-rabbit IgG (Invitrogen) was added at 1:750 for 30 min at RT in a block. These cells were then washed twice with PBS; then all transfected cells were incubated with 300 nM DAPI in PBS for 2 min at RT, washed twice with PBS, imaged at a magnification of ×40 using an Olympus IX70 microscope, and analyzed with ImagePro software 6.3 (version 6.3). Experiments for image analysis were performed with HeLa and 293T cells, three or more times; images are representative of results from three similar experiments.
For flow cytometry, after incubation with virus or labels, HeLa and 293T cells were washed once with PBS; then 293T cells were removed from the plate with 500 μl PBS, and HeLa cells were detached by incubation in 5 mM EDTA (Invitrogen) for 5 min at 37°C. These cells were transferred to 1.5-ml centrifuge tubes and were spun down at 5,000 × g for 10 min. All samples were incubated with 4% paraformaldehyde for 30 min prior to resuspension in PBS for flow analysis using a FACScan flow cytometer with CellQuest software (BD Biosciences). For analysis of cells transfected with WT-PAK-1-Myc or DN-PAK-1-GST, untransfected and transfected cells were incubated with mouse anti-c-myc (dilution, 1:200) or rabbit anti-GST (1:100) (SCBT) in permeabilization buffer for 1 h. After two washes with PBS, an Alexa Fluor 555-conjugated F(ab′)2 fragment of goat anti-mouse IgG, or an Alexa Fluor 555-conjugated F(ab′)2 fragment of goat anti-rabbit IgG (Invitrogen) was added at 1:750 for 30 min at RT in permeabilization buffer. Cells were washed three times with PBS and were then resuspended in PBS and analyzed using a FACScan flow cytometer with CellQuest software (BD Biosciences). For analysis of flow cytometry data, gates were set using untransfected and uninfected controls. For every flow experiment, samples included untransfected cells infected with each GFP-expressing virus to compensate for GFP, and uninfected cells transfected with each plasmid to compensate for mCherry or the Alexa Fluor 555-conjugated secondary antibody. For experiments with WT-PAK-1-Myc or DN-PAK-1-GST, untransfected cells were similarly probed with anti-c-Myc, anti-GST, and an appropriate secondary antibody to control for any nonspecific binding. The quadrants were set based on the FL-1 and FL-2 values of untransfected/uninfected controls (with staining for Fig. 7). For each cell type, the gates were set based on the level of expression of each plasmid (FL-2) in the absence of infection.
Fig 7.
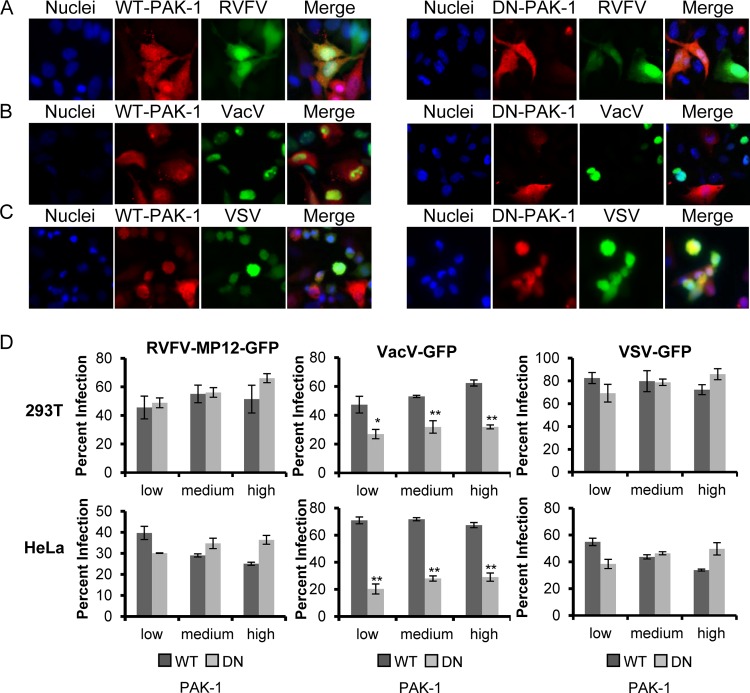
Expression of dominant negative PAK-1 blocks VacV-GFP infection but has no effect on infection with RVFV-MP-12-GFP or VSV-GFP. (A to C) HeLa cells transfected with WT-PAK-1-Myc (left) or DN-PAK-1-GST (right) (both red) were incubated with RVFV-MP-12-GFP (A), VacV-GFP (B), or VSV-GFP (C) (all green) at an MOI of 1 for 3 h. After cells were fixed with 4% paraformaldehyde, they were probed with anti-c-Myc or anti-GST antibodies, followed by an Alexa Fluor 555-conjugated secondary antibody (red), and were then treated with DAPI to visualize the nuclei (blue). Infection of PAK-1-expressing cells with a recombinant virus expressing GFP was observed using an Olympus IX70 microscope. Images were cropped, but relative cell sizes were maintained. The images shown are representative of results from three independent experiments with similar results. (D) 293T (top) or HeLa (bottom) cells transfected with WT-PAK-1 or DN-PAK-1 were incubated with RVFV-MP-12-GFP (left), VacV-GFP (center), or VSV-GFP (right) at an MOI of 1 for 3 h. Infection in relation to the level of overexpressed PAK-1 was scored by flow cytometry (data not shown). The quantity of infected cells expressing low, medium, or high levels of WT-PAK-1 or DN-PAK-1, relative to that of all cells expressing low, medium, or high levels of WT-PAK-1 or DN-PAK-1, is given as the percentage of infection. Shown are the means (± standard deviations) for three independent experiments for each cell type (**, P < 0.01; *, P < 0.05).
Statistical analysis.
Raw data for infection assays measured by GFP fluorescence were compared by using a two-tailed t test for each individual experiment. Values obtained with inhibitors suspended in DMSO were compared to those for DMSO-treated samples, and values obtained with inhibitors suspended in H2O were compared to those for untreated infected samples. For siRNA-treated samples, values obtained with each individual siRNA construct were compared to values obtained with wells transfected with scrambled siRNA on the same plate. For plate assays, untreated and DMSO-treated controls, or scrambled siRNA and GFP siRNA, were included on every plate, with no virus, RVFV-MP-12-GFP, VacV-GFP, and VSV-GFP. Inhibitor-treated, siRNA-transfected, and DN construct-expressing cells that were infected with RVFV-MP-12-GFP were compared to control samples infected with RVFV-MP-12-GFP, as was the case for VacV-GFP and VSV-GFP. P values were considered significant when they were <0.05 (single asterisk) and very significant when they were <0.01 (double asterisk). The P values shown in the figures and the text were based on the highest P values obtained from three independent experiments.
RESULTS
Role of macropinocytosis and actin in RVFV entry.
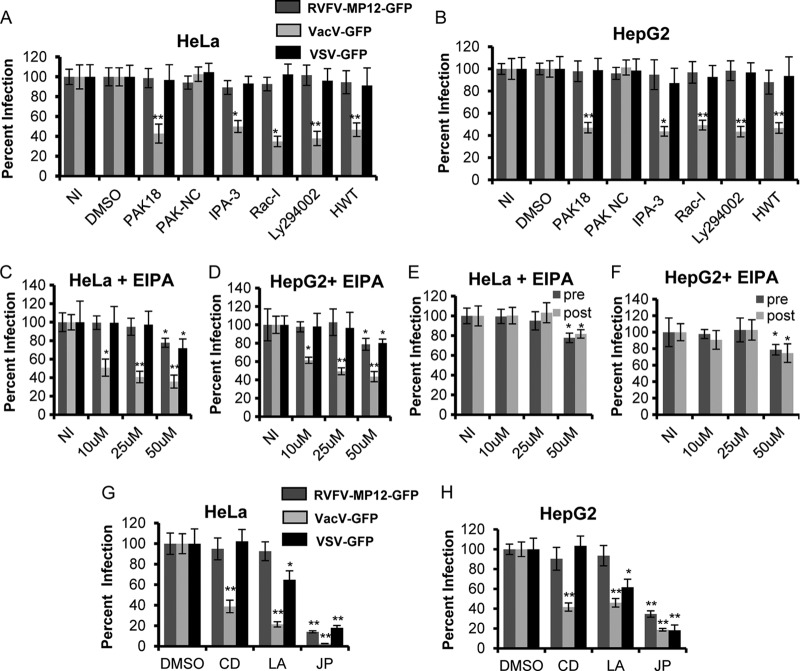
A recent review by Mercer and Helenius outlined the minimal set of requirements to satisfy the definition of virus macropinocytosis (65). These criteria include a block of entry with inhibitors of actin dynamics, Rho GTPases, Na+/H+ exchangers, and kinases such as PAK-1 and PI3K. To test the role of macropinocytosis in RVFV entry, HeLa and HepG2 cells were pretreated with an inhibitor of Rac (RAC-I), PAK-1 inhibitors PAK-1 inhibitor peptide (PAK18) and PAK-1 inhibitor III (IPA-3), and the PI3K inhibitors LY-294002 and 17β-hydroxy wortmannin (HWT). The pretreated cells were then infected at an MOI of 1 with the attenuated RVFV strain MP-12 expressing GFP (RVFV-MP-12-GFP). As a control, pretreated cells were infected at an MOI of 1 with either VacV-GFP (a large DNA virus that has been shown to enter cells by macropinocytosis) or VSV-GFP (an acid-activated, enveloped RNA virus that is known to employ CME for entry into host cells) to test the specificity and efficacy of the inhibitors (64, 67). Each virus used in these experiments is a recombinant strain that expresses GFP upon entry and replication. Following infection, the cells were incubated with alamarBlue, a reagent that enables measurement of cellular metabolic activity. Inhibitors were used at concentrations that have been shown previously to block target activity and that exhibited no cytotoxic or pleiotropic effects, as determined by alamarBlue staining and infection with a control virus(es). Pretreatment with the inhibitors at concentrations that are known to specifically block macropinocytosis (26, 62, 64, 66, 71, 88, 90) significantly decreased VacV-GFP infection of HeLa and HepG2 cells but had little or no effect on VSV-GFP or RVFV-MP-12-GFP infection (Fig. 1A and B).
Fig 1.
RVFV does not enter cells via macropinocytosis. Average levels of infection detected by GFP fluorescence (± standard deviations), compared to those for untreated or DMSO-treated controls, are shown. (A and B) HeLa (A) or HepG2 (B) cells were either left untreated (NI) or pretreated with 50 μM DMSO, 20 μM PAK18, 20 μM PAK-NC, 10 μM IPA-3, 100 μM Rac-I, 50 μM Ly294002, or 0.5 μM HWT for 1 h. (C and D) HeLa (C) or HepG2 (D) cells were either left untreated or pretreated with 10, 25, or 50 μM 5-(N-ethyl-N-isopropyl)amiloride (EIPA) for 1 h. The inhibitors were also present during the 3 h of incubation with RVFV-MP-12-GFP, VacV-GFP, or VSV-GFP at an MOI of 1. (E and F) HeLa (E) or HepG2 (F) cells were either left untreated or pretreated (pre) with 10, 25, or 50 μM EIPA for 1 h. The inhibitors were also present during the 3 h of incubation with RVFV-MP-12-GFP at an MOI of 1 at 37°C. Alternatively, HeLa (E) or HepG2 (F) cells were incubated for 1 h with RVFV-MP-12-GFP at an MOI of 1; then cells were washed to remove unbound virus and were subsequently incubated with 10, 25, or 50 μM EIPA (post) for 4 h. (G and H) HeLa cells (G) or HepG2 cells (H) were pretreated with either DMSO, 1 μM CD, 2 μM LA, or 1 μM JP for 1 h. The inhibitors were also present during the 3 h of incubation with RVFV-MP-12-GFP, VacV-GFP, or VSV-GFP at an MOI of 1. GFP expression was normalized to cell titers measured by alamarBlue fluorescence. The percentage of infection was determined by taking untreated or DMSO-treated and infected samples as 100% infected. Shown are the means for three independent experiments performed in triplicate (**, P < 0.01; *, P < 0.05).
Another common method for analyzing the role of macropinocytosis is to treat cells with the Na+/H+ exchange inhibitor ethyl-N-isopropyl amiloride (EIPA) prior to and during infection (26, 40, 53, 64, 71, 88). Na+/H+ exchange is required to maintain the pH at the plasma membrane necessary for Rac activation (53). HeLa and HepG2 cells were treated with 10, 25, or 50 μM EIPA for 1 h prior to infection and during 3 h of incubation with RVFV-MP-12-GFP, VacV-GFP, or VSV-GFP at an MOI of 1. Higher EIPA concentrations resulted in a decrease in cell viability. Infection of HeLa cells with VacV-GFP was reduced by 60% ± 6.3% with 25 μM EIPA and by 65% ± 7.0% with 50 μM EIPA, with similar results in HepG2 cells (Fig. 1C and D). Infection of HeLa and HepG2 cells with RVFV-MP-12-GFP was not affected by treatment with 10 or 25 μM EIPA but was mildly affected with the highest concentration (50 μM) of EIPA; however, this effect was also observed with VSV-GFP infection (Fig. 1C and D). To determine if the effect observed with EIPA was specific to the entry of RVFV-MP-12-GFP, EIPA was added 1 h after infection with RVFV-MP-12-GFP at an MOI of 1. It is thought that RVFV-MP-12 binding and entry occur within 1 h of infection, because blocking of endosomal acidification 1 h postinfection has no effect on infection (35). Treatment of HeLa (Fig. 1E) and HepG2 (Fig. 1F) cells with 50 μM EIPA 1 h postinfection reduced infection to the same extent as pretreatment, suggesting that the slight effect observed with 50 μM EIPA (Fig. 1C and D) is not specific to entry. These results show that under conditions in which macropinocytosis is inhibited, RVFV infection is unaffected, strongly suggesting that RVFV does not use macropinocytosis for cell entry during infection.
Macropinocytosis relies largely on actin arrangements, such that viruses that enter via macropinocytosis are not internalized by cells that have been pretreated with inhibitors that disrupt actin dynamics (65). Infection of cells with VacV or Ebola virus, both known to enter cells via macropinocytosis, is blocked by treatment with the actin filament production inhibitor cytochalasin D (CD), the actin monomer-sequestering drug latrunculin A (LA), and the actin-stabilizing drug jasplakinolide (JP) (63–66, 71, 110, 121). Treatment of cells with LA has also been shown to inhibit VSV endocytosis, suggesting that actin assembly is also important for CME (120). The actin-stabilizing drug JP has been shown to disrupt the dynamic rearrangement of microfilaments and to significantly inhibit infection with BKV, a virus shown to enter via CavME (31, 32). To determine the sensitivity of RVFV infection to these actin-modulating drugs, cells were treated with each inhibitor prior to and during incubation with RVFV-MP-12-GFP, VacV-GFP, or VSV-GFP at an MOI of 1. As expected, the inhibitors dramatically reduced infection with VacV-GFP (Fig. 1G and H). Infection with VSV-GFP was significantly decreased with LA and very significantly decreased with the actin-stabilizing drug JP (Fig. 1G and H). RVFV-MP-12-GFP infection was unaffected by CD and LA, indicating that RVFV internalization is independent of an intact actin cytoskeleton (Fig. 1G and H). However, RVFV-MP-12-GFP infection was reduced by 76% ± 1.1% in HeLa cells and by 65% ± 3.3% in HepG2 cells treated with JP.01). Although RVFV infection is dependent on microfilament assembly and disassembly, as indicated by inhibition with JP, its insensitivity to other inhibitors of actin dynamics sets RVFV apart from viruses that enter via macropinocytosis or CME, implying that RVFV enters via an alternative route.
Roles of clathrin-mediated endocytosis and caveolin-mediated endocytosis in RVFV entry.
It has been reported that the entry of RVFV and VSV requires endosomal acidification and that VacV-GFP is partially sensitive to inhibitors of acidification (35, 36, 57, 58, 63, 65, 66, 119). Ammonium chloride (NH4Cl) and bafilomycin A (BAF), inhibitors that block the acidification of endosomes, were used as positive controls for the inhibition of endocytosis (74, 123). To investigate the role of CME in RVFV internalization, cells were treated with the dynamin II inhibitor dynasore (DYN) and the CME-specific inhibitor chlorpromazine (CPZ) for 1 h prior to infection and during infection with the viruses indicated in Fig. 2 at an MOI of 1 (11, 48, 66, 67, 107, 109). As expected, infection of HeLa or HepG2 cells with RVFV-MP-12-GFP or VSV-GFP was very significantly reduced by treatment with NH4Cl or BAF (Fig. 2A and B). VacV-GFP infection was decreased by 41% ± 5.0% in NH4Cl-treated HeLa cells and by 59% ± 4.1% in BAF-treated HeLa cells, with similar results in HepG2 cells. Treatment of cells with DYN decreased RVFV-MP-12-GFP infection by 73% ± 6.3% in HeLa cells and by 77% ± 2.0% in HepG2 cells and decreased VSV-GFP infection by 70% ± 6.9% in HeLa cells and by 69% ± 4.8% in HepG2 cells but had no effect on VacV-GFP infection (Fig. 2A and B). These results indicate that RVFV infection requires dynamin II GTPase activity, suggesting internalization by CME or CavME. When cells were treated with the CME-specific inhibitor CPZ, VSV-GFP infection was reduced by 73% ± 3.4% in HeLa cells and by 60.3% ± 7.9% in CPZ-treated HepG2 cells (Fig. 2A and B), while RVFV-MP-12-GFP and VacV-GFP infection of HeLa and HepG2 cells was unaffected (Fig. 2A and B). These results propose that RVFV entry is dependent on dynamin II but independent of CME.
Next, we tested the effects of several CavME inhibitors on RVFV infection. CavME is a dynamic process that is regulated by the presence of caveolae at the plasma membrane and by the relative activity of multiple kinases and phosphatases (55, 70, 77). Cholesterol is a major factor of caveolae, and depletion of cholesterol from the plasma membrane reduces the number of invaginated caveolae (16, 43, 55). To address the role of CavME in RVFV infection, cells were treated with nystatin and progesterone (Nys plus Prog) and the PKC activator phorbol 12-myristate 13 acetate (PMA) prior to and during incubation with the viruses indicated in Fig. 2 (MOI, 1). Treatment of cells with a combination of Nys and Prog impairs lipid raft formation (17, 55, 94), and treatment with PMA increases the degradation of caveolin-1, thereby decreasing the number of caveolae at the cell surface (14, 42, 102, 118). Treatment with Nys plus Prog reduced the level of RVFV-MP-12-GFP infection by an average of 65% ± 8.9% in HeLa cells and by 64% ± 5.4% in HepG2 cells (Fig. 2C and E) but had no significant effect on infection with VacV-GFP or VSV-GFP. A greater decrease in RVFV-MP-12-GFP infection was observed with higher concentrations of Nys and/or Prog; however, these concentrations also significantly decreased infection with VacV-GFP and VSV-GFP, or reduced cell viability (data not shown). PMA treatment reduced the level of RVFV-MP-12-GFP infection of HeLa cells by 79% ± 0.8% and of HepG2 cells by 61% ± 2.5% (Fig. 2C and E). In contrast, treatment of cells with PMA had no significant effect on infection with VacV-GFP or VSV-GFP.
To further address the role of CavME in RVFV infection, cells were treated with okadaic acid (OA), a serine/threonine phosphatase inhibitor that specifically blocks phosphatases 1 and 2A (PP1 and PP2A) (5, 76, 77, 126). Treatment with OA prior to the addition of CavME-utilizing viruses results in a decrease in infection, whereas the addition of OA 1 to 2 h after virus addition promotes CavME of bound cargo and causes an increase in infection (5, 76, 77, 126). Pretreatment of cells with OA has also been shown to decrease the early steps of VSV infection by blocking penetration and uncoating; however, incubation with OA 1 h after virus addition has no effect on VSV infection (5, 28, 76, 77, 122). Treatment of HeLa cells with 100 nM OA prior to and during incubation with the virus decreased the level of RVFV-MP-12-GFP infection by an average of 55% ± 8.0% and that of VSV-GFP infection by an average of 60% ± 8.6%, with similar results in HepG2 cells (Fig. 2C and E). Treatment with OA after virus binding enhanced RVFV-MP-12-GFP infection of HeLa cells by 173% ± 19%, and that of HepG2 cells by an average of 165% ± 13.7% (Fig. 2D and F), indicating that RVFV internalization is significantly increased upon stimulation of CavME. Consistent with previous results, this postentry treatment did not change VSV-GFP infection of HeLa and HepG2 cells (5). VacV-GFP infection was unaffected by treatment with OA before, during, or after incubation with virus, suggesting that macropinocytosis is not regulated by PP1 or PP2A. These results together indicate that RVFV entry is distinct from VacV or VSV entry, and they strongly suggest that RVFV is endocytosed via caveolae.
To determine whether the inhibitors shown to decrease RVFV-MP-12-GFP infection were blocking an early or a late step in the RVFV life cycle, we performed time-of-addition experiments. Treatment with NH4Cl 40 min to 1 h postinfection had no effect on infection with UUKV or RVFV-MP-12, suggesting that the binding and entry of these viruses occur within the first hour (35, 58). Therefore, agents that specifically affect entry should have no effect on infection when added 1 h postinfection. The inhibitors known to block CavME, DYN, Nys plus Prog, and PMA, very significantly blocked RVFV-MP-12-GFP infection of HeLa and HepG2 cells when added 1 h prior to and during incubation with virus at an MOI of 1 (Fig. 2G and H) but had no effect on infection when added postentry (Fig. 2G and H). In contrast, the actin-stabilizing drug JP was inhibitory when added either prior to infection or postentry, suggesting that JP blocks late steps in the life cycle of RVFV (Fig. 2G and H). The results demonstrate that the blockade in RVFV infection with CavME inhibitors is at the virus entry step.
Inhibitors of CavME decrease the percentage of cells infected with RVFV-MP-12-GFP and authentic RVFV-MP-12.
The recombinant RVFV vaccine strain MP-12 lacking the NSs gene and expressing GFP (RVFV-MP-12-GFP) is an extremely useful tool for rapid and sensitive screening of inhibitors of RVFV (47). By using a sensitive plate reader assay that measures GFP fluorescence and cell viability, we were able to control for the specificity and efficacy of the small-molecule inhibitors by including VacV-GFP and VSV-GFP infection to exclude nonspecific effects. To validate the results obtained with the plate reader and RVFV-MP-12-GFP, we used a 96-well immunofluorescence assay (IFA) to quantitatively analyze the percentage of cells infected with authentic nonrecombinant RVFV MP-12. HeLa cells were treated with the inhibitors indicated in Fig. 3 for 1 h prior to infection and during infection with RVFV-MP-12 at an MOI of 1. At 16 h postinfection, cells were fixed, and infection was visualized by IFA. The images shown in Fig. 3A are representative of results from three similar experiments. Cells treated with inhibitors resuspended in H2O were compared to untreated cells (Fig. 3A, top row), and cells treated with inhibitors resuspended in DMSO were compared to DMSO-treated cells (Fig. 3A, bottom row). As with previous assays, treatment of cells with inhibitors specific for macropinocytosis (EIPA, IPA-3, HWT) or CME (CPZ) had no effect on the percentage of cells infected with RVFV-MP-12, but there was a noticeable decrease in infection with recombinant RVFV-MP-12-GFP (data not shown) and nonrecombinant RVFV-MP-12 in cells pretreated with DYN, OA, Nys plus Prog, or PMA (Fig. 3A).
To quantify the results obtained with the IFA discussed above, flow cytometry was used to measure the percentage of inhibitor-treated cells infected with authentic RVFV MP-12 or recombinant RVFV-MP-12-GFP at an MOI of 1. HeLa, HepG2, and 293T cells were treated with the inhibitors indicated in Fig. 3B for 1 h prior to infection and during infection with RVFV MP-12 or RVFV-MP-12-GFP. To measure infection with RVFV MP-12, uninfected and RVFV MP-12-infected cells were fixed and stained with anti-RVFV antibodies. RVFV MP-12-infected cells were compared to uninfected cells probed with the same antibodies. Treatment of 293T cells with the CME inhibitor CPZ had no effect on the percentage of cells infected with RVFV MP-12 (Fig. 3B, top, and C). Similarly, treatment with the macropinocytosis inhibitor EIPA, IPA-3, or HWT did not reduce the percentage of 293T cells, HeLa cells, or HepG2 cells infected with RVFV MP-12 (Fig. 3B and C). Treatment of 293T cells, HeLa cells, and HepG2 cells with CPZ and macropinocytosis inhibitors also had no effect on the percentage of cells infected with RVFV-MP-12-GFP (Fig. 3D). As expected, treatment of cells with NH4Cl drastically reduced the percentage of cells infected with RVFV MP-12 (Fig. 3C) or RVFV-MP-12-GFP (Fig. 3D). The percentage of 293T cells infected with RVFV MP-12 was reduced by an average of 71% ± 2.2% with DYN, by 56% ± 6.0% with OA, by 64% ± 7.7% with Nys plus Prog, and by 77% ± 0.1% with PMA (Fig. 3B and C), with similar results for HeLa and HepG2 cells (Fig. 3C). The percentage of cells infected with RVFV MP-12 or RVFV-MP-12-GFP in the presence of these inhibitors was reduced to levels similar to those measured by IFA, flow cytometry, and GFP fluorescence, indicating that these virus strains use the same mechanism of entry (Fig. 2 and 3C and D).
Downregulation of dynamin 2 and caveolin-1 reduces RVFV infection.
To further characterize the means by which RVFV enters host cells, we downregulated endogenous proteins that are required for CavME, CME, and macropinocytosis with small interfering RNA (siRNA) and assessed the effects on RVFV infection. To block CME and CavME, but not macropinocytosis, we used siRNA to dynamin II (DMN2). To specifically block CME, we used siRNA targeted to clathrin heavy chain (CHC) or to epidermal growth factor receptor pathway substrate clone 15 (EPS15), an accessory molecule required for the assembly of clathrin-coated pits (6). To specifically block CavME, we used siRNA targeted to caveolin-1 (CAV-1), and to specifically block macropinocytosis, we used siRNA targeted to PAK-1 and to PI3K. We also used siRNA targeted to vacuolar H+-ATPase (ATP6V1A) to block vacuolar acidification (data not shown) and siRNA targeted to GFP as a positive control for the downregulation of virus-derived GFP fluorescence. Protein expression in siRNA-transfected cells was assessed by immunoblotting 48 h, 60 h, and 72 h posttransfection to determine when protein levels were most efficiently reduced (data not shown). We found that expression of all target proteins was most consistently reduced between 60 h and 72 h posttransfection; thus, in subsequent experiments, siRNA-transfected cells were infected with the virus at an MOI of 1at 60 h posttransfection, and two independent siRNA constructs were used per target. Infection with RVFV-MP-12-GFP was reduced by an average of 58% ± 5.2% in HeLa cells expressing CAV-1_6 siRNAs and by an average of 53% ± 1.9% in HeLa cells expressing CAV-1_7 siRNAs; similar results were observed with HepG2 cells (Fig. 4A and B). The decrease in RVFV-MP-12-GFP infection correlated well with the decreased steady-state levels of CAV-1 at 60 h posttransfection, as detected by immunoblotting (Fig. 4G). VacV-GFP and VSV-GFP infection of HeLa and HepG2 cells was unaffected by CAV-1 downregulation. Downregulation of DMN2, a protein required for CavME and CME, caused a substantial decrease in RVFV-GFP and VSV-GFP infection of HeLa and HepG2 cells (Fig. 4A and B). As expected, downregulation of the CME mediators CHC and EPS15 had no effect on RVFV-MP-12-GFP or VacV-GFP infection but reduced VSV-GFP infection of HeLa cells by averages of 73% ± 5.6% and 61% ± 6.8%, respectively, with similar results in HepG2 cells (Fig. 4C and D). Downregulation of the macropinocytosis markers PAK-1 and PI3K had no effect on RVFV-MP-12-GFP infection or VSV-GFP infection, despite a 46% to 53% decrease in PAK-1 protein expression and a 43% to 45% decrease in PI3K protein expression at 60 h posttransfection (Fig. 4G). Downregulation of PAK-1 and PI3K decreased VacV-GFP infection of HeLa cells by averages of 52% ± 8.0% and 51% ± 6.3%, respectively, with similar results in HepG2 cells (Fig. 4E and F), which correlates with the decreased steady-state levels of these proteins. These data support the role of CAV-1 and DMN2 in RVFV entry and infection.
Expression of dominant negative caveolin-1 blocks RVFV infection.
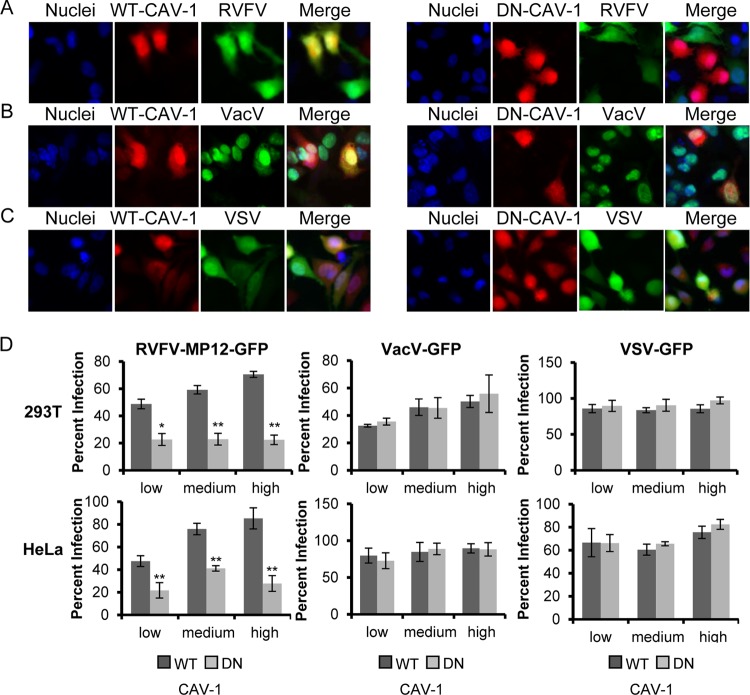
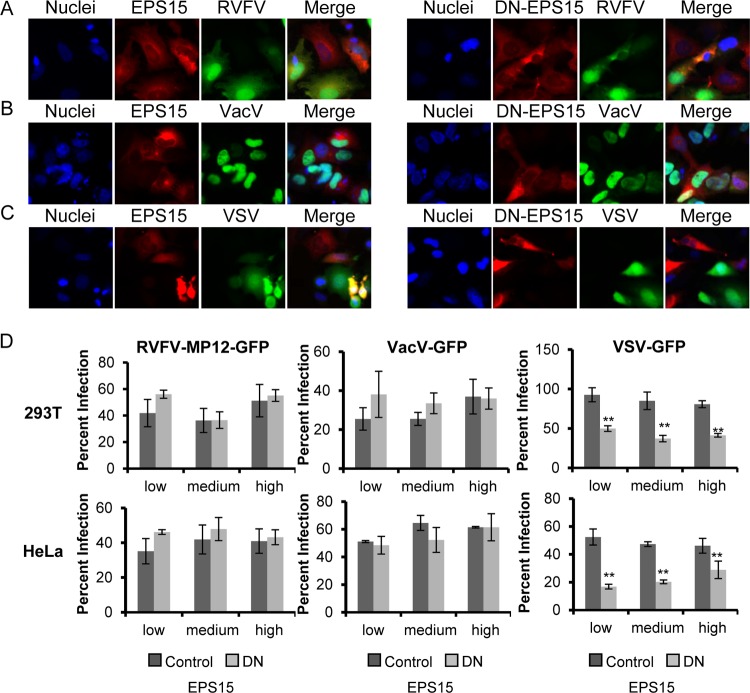
Another method used to analyze the function of endocytosis pathways in virus entry is to express DN mutant versions of cellular proteins in target cells. Overexpression of DN mutants overwhelms the wild-type proteins and prevents their function. This approach has been used extensively to characterize the entry pathways of multiple viruses, including bluetongue virus 1 (40), Ebola virus (88), tiger frog virus (41), VacV (66), and hepatitis B virus (60). We took a similar approach, using a deletion mutant of CAV-1 (DN-CAV-1), which acts as a DN inhibitor of CavME and blocks the uptake of the marker cholera toxin subunit B (CTX-B) (60); a deletion mutant of EPS15 (DN-EPS15), which acts as a DN inhibitor of CME and blocks the uptake of human transferrin (hTfn) (86); and a mutant of PAK-1 comprising the autoinhibitory domain (DN-PAK-1), which has been shown previously to inhibit VacV and Ebola virus infection (64, 88). HeLa and 293T cells were transiently transfected to express WT-CAV-1, DN-CAV-1, WT-PAK-1, DN-PAK-1, DN-EPS15, or a form of EPS15 that does not inhibit CME (D3Δ2; referred to as “control EPS15”) (86). At 24 h posttransfection, cells were incubated with RVFV-MP-12-GFP, VacV-GFP, or VSV-GFP at an MOI of 1 for 3 h at 37°C; then the cells were washed and were incubated overnight in complete medium. The cells were then fixed and processed for fluorescence microscopy or flow cytometry. Untransfected cells (no red) and cells expressing WT-CAV-1 were readily infected with RVFV-MP-12-GFP (Fig. 5A, left), whereas infection was reduced in cells expressing DN-CAV-1, and RVFV-MP-12-GFP preferentially infected neighboring untransfected cells (Fig. 5A, right). The inhibitory effect of DN-CAV-1 expression on RVFV-MP-12-GFP infection was specific, as evidenced by the fact that the levels of infection with VacV-GFP and VSV-GFP were the same for cells expressing DN-CAV-1 and WT-CAV-1 (Fig. 5B and C). To quantify the effect of the expression of DN mutants on infection with RVFV-MP-12-GFP, VacV-GFP, or VSV-GFP, flow cytometry was used. To account for the fact that the dominant effect of mutant proteins can sometimes depend on the level of expression, we grouped the data into those for cells expressing low, medium, and high levels of exogenous protein (data not shown). In cells expressing DN-CAV-1, RVFV-MP-12-GFP infection was decreased by an average of 61% ± 6.3% in HeLa cells and by 52% ± 3.8% in 293T cells (Fig. 5D, left), whereas VacV-GFP infection and VSV-GFP infection were unaffected by DN-CAV-1 expression (Fig. 5D, center and right). Expression of DN-EPS15 had no effect on RVFV-MP-12-GFP or VacV-GFP infection (Fig. 6A and B and Fig. 6D, left and center) but decreased VSV-GFP infection by an average of 57% ± 5.0% in HeLa cells and by 46% ± 4.0% in 293T cells (Fig. 6C and Fig. 6D, right). Similarly, the levels of RVFV-MP-12-GFP and VSV-GFP infection were the same in cells expressing DN-PAK-1 and those expressing WT-PAK-1 (Fig. 7A, C, and D, left and right), whereas expression of DN-PAK-1 decreased VacV infection by an average of 63% ± 0.6% in HeLa cells and by 40% ± 2.8% in 293T cells (Fig. 7B and D, center). These results confirm the specificity of the DN mutants and demonstrate that CavME is required for RVFV entry, whereas CME and macropinocytosis are not, in agreement with the results reported in this study for inhibitors and siRNA.
Fig 5.
Expression of dominant negative caveolin-1 blocks RVFV infection. (A to C) HeLa cells transfected with WT-CAV-1-mCherry (left) or DN-CAV-1-mCherry (right) (both red) were incubated with RVFV-MP-12-GFP (A), VacV-GFP (B), or VSV-GFP (C) (all green) at an MOI of 1 for 3 h. After cells were fixed with 4% paraformaldehyde, they were treated with DAPI to visualize the nuclei (blue). Infection of mCherry-expressing cells with a recombinant virus expressing GFP was observed using a 40× objective on an Olympus IX70 microscope. Images were cropped, but relative cell sizes were maintained. The images shown are representative of results from three independent experiments with similar results. (D) 293T cells (top) or HeLa cells (bottom) transfected with WT-CAV-1 or DN-CAV-1 were incubated with RVFV-MP-12-GFP (left), VacV-GFP (center), or VSV-GFP (right) at an MOI of 1 for 3 h. Infection in relation to the level of overexpressed CAV-1 was scored by flow cytometry (data not shown). The quantity of infected cells expressing low, medium, or high levels of WT-CAV-1-mCherry or DN-CAV-1-mCherry relative to that of all cells expressing low, medium, or high levels of WT-CAV-1-mCherry or DN-CAV-1-mCherry is given as the percentage of infection. Shown are the means (± standard deviations) for three independent experiments for each cell type (**, P < 0.01; *, P < 0.05).
Fig 6.
Expression of dominant negative EPS15 decreases the level of infection with VSV-GFP but not with RVFV-MP-12-GFP or VacV-GFP. (A to C) HeLa cells transfected with control EPS15-mCherry (left) or DN-EPS15-mCherry (right) (both red) were incubated with RVFV-MP-12-GFP (A), VacV-GFP (B), or VSV-GFP (C) (all green) at an MOI of 1 for 3 h. After cells were fixed with 4% paraformaldehyde, they were treated with DAPI to visualize the nuclei (blue). Infection of mCherry-expressing cells with a recombinant virus expressing GFP was observed using an Olympus IX70 microscope. Images were cropped, but relative cell sizes were maintained. The images shown are representative of results from three independent experiments with similar results. (D) 293T (top) or HeLa (bottom) cells transfected with control EPS15 or DN-EPS15 were incubated with RVFV-MP-12-GFP (left), VacV-GFP (center), or VSV-GFP (right) at an MOI of 1 for 3 h. Infection in relation to the level of overexpressed EPS15 was scored by flow cytometry (data not shown). The quantity of infected cells expressing low, medium, or high levels of control-EPS15-mCherry or DN-EPS15-mCherry relative to that of all cells expressing low, medium, or high levels of control-EPS15-mCherry or DN-EPS15-mCherry is given as the percentage of infection. Shown are the means (± standard deviations) for three independent experiments for each cell type (**, P < 0.01; *, P < 0.05).
DISCUSSION
Viruses are obligate parasites that have developed multiple ways to exploit host cell processes so as to support infection. Different viruses have evolved distinct methods for taking advantage of host cell functions to support their specific needs. For example, viruses have been shown to target different endocytic pathways to deliver their genetic material to sites optimal for replication and to avoid immune regulation (24, 40). While it has been shown that RVFV enters cells through pH-dependent endocytosis, the specific endocytic pathway has not been clearly defined (35, 36). To clearly demonstrate the entry mechanism employed by RVFV MP-12, multiple independent assays were performed; results were confirmed in multiple cell types; specificity and efficiency were evaluated using viruses with known entry mechanisms; and time-of-addition experiments were used to distinguish a block in virus entry from a block at a later stage in the virus life cycle. Cellular perturbations using small-molecule inhibitors, siRNA, and mutant protein overexpression provide evidence that CavME serves as the primary endocytic pathway for RVFV MP-12 uptake in mammalian cell lines.
CavME is a slow, ligand-triggered process characterized by a dependency on cholesterol, lipid rafts, dynamin II, and a complex signaling pathway that is regulated by tyrosine kinases and phosphatases (67). Cholesterol is an important component of lipid rafts and is required for the formation of caveolae and for CavME (43). Treatment with Nys plus Prog significantly reduced the level of RVFV infection, suggesting that cholesterol rich domains are required for entry (70). Inhibition of dynamin II activity and treatment with the PP2A inhibitor OA block both CavME and CME. Upon further examination of RVFV internalization, we found that pretreatment with the dynamin II inhibitor dynasore or OA blocked VSV-GFP infection and RVFV infection. However, treatment with OA after virus exposure increased the level of RVFV entry, as expected for a caveola-dependent endocytic pathway, whereas there was no effect on infection with VSV-GFP (5, 79, 101). The mechanism for OA enhancement of CavME is unknown, but inhibition of PP2A prevents endosome fusion with the lysosome, which has a positive effect on SV40 infection (5, 126). VSV penetration occurs in early endosomes 6 to 30 min after binding, so it is unaffected by OA treatment 1 h postinfection (5, 51). Previous studies have shown that treatment of cells with the PKC activator PMA specifically blocks CavME by preventing caveola formation and invagination (2, 14, 102). In this study, we observed a significant decrease in RVFV infection in cells treated with PMA, while VacV-GFP and VSV-GFP infection were unaffected. Time-of-addition experiments demonstrated that the inhibitors of CavME (DYN, Nys plus Prog, OA, and PMA) specifically block RVFV entry and have no effect when added after virus binding and entry. These inhibitors were able to efficiently block infection with a recombinant RVFV expressing GFP in place of NSs, as well as infection with authentic RVFV MP-12. Similar results were obtained regardless of whether infection was measured by GFP fluorescence using a plate reader, by immunofluorescence microscopy, or by flow cytometry. In addition, the results obtained by immunofluorescence microscopy and flow cytometry with RVFV-MP-12-GFP were identical to those obtained with nonrecombinant RVFV, suggesting that the expression of GFP in place of NSs has no effect on viral entry. We also found that suppression of CAV-1 activity by siRNA or expression of DN-CAV-1 reduced RVFV infection but had no effect on CME or macropinocytosis. These results clearly demonstrate that RVFV entry is dependent on mediators of CavME.
In contrast to previous studies, we found no evidence for the involvement of macropinocytosis in RVFV entry and infection (35). Cells were treated with multiple inhibitors of macropinocytosis (PAK18, IPA-3, Rac-I, Ly294002, HWT, and EIPA), and despite a significant decrease in VacV-GFP infection with each inhibitor, none of the inhibitors blocked RVFV infection. The entry of viruses dependent on macropinocytosis and infection by those viruses require multiple actin cytoskeletal rearrangements and are sensitive to inhibitors of actin dynamics. Disruption of actin polymers with CD and LA had a significant effect on VacV infection but did not reduce RVFV infection. We also measured the effect of siRNA-mediated suppression of endogenous PAK-1 and PI3K, key regulators of macropinocytosis, and found that infection with VacV was significantly reduced, whereas infection with RVFV or VSV was unaffected. Finally, expression of DN-PAK-1 had no impact on infection with RVFV, despite a decrease in VacV infection observed by imaging and flow cytometry. These results strongly suggest that RVFV entry is not dependent on macropinocytosis.
Recent studies using CPZ, siRNA, and microscopy indicate a role for CME in the entry of bunyaviruses Hantaan virus, Oropouche virus, and Crimean-Congo hemorrhagic fever virus (CCHFV) (50, 92, 99), and it has been suggested that RVFV may enter cells via CME (95). In this study, we found no evidence for a role of CME in RVFV infection. Treatment of cells with the CME inhibitor CPZ, transfection of siRNA targeted to CHC and EPS15, and expression of DN-EPS15 significantly reduced VSV-GFP infection. However, these treatments had no effect on RVFV infection, suggesting that RVFV entry is independent of CME. The use of different entry pathways by members of the same virus family has been well documented for polyomaviruses, hepadnaviruses, and papillomaviruses (2, 60, 84, 104); therefore, the discrepancy between Hantaan virus, Oropouche virus, CCHFV, and RVFV may be a consequence of different uptake strategies used within the family Bunyaviradae. At the level of the whole Bunyaviradae family, there is great variance in size, shape, and the structure of the glycoprotein lattices between members (75, 116), and it is likely that the overall viral particle properties, including size (90 to 100 nm) and glycoprotein structure, influence the endocytic pathway.
Unlike the rest of the Bunyaviradae family, the members of the genus Phlebovirus have similar surface morphologies and a constant size and are only mildly pleiomorphic (75). Recent cryoelectron tomography of UUKV and RVFV MP-12 revealed that both Phlebovirus members have the same T=12 icosahedral symmetry, a structure that has been reported only for these two viruses (37, 75). Moreover, a recent study reported that the entry mechanism for UUKV is mainly clathrin independent, and electron microscopic (EM) studies revealed that the majority of UUKV particles were located in noncoated, small cytoplasmic vesicles (58), consistent with small caveolar vesicles. Here we confirm a clathrin-independent role for RVFV and propose that members of the Phelobovirus genus use similar endocytic viral entry mechanisms. These similarities between UUKV and RVFV, despite the vast difference in their glycoprotein sequences, also suggest that the three amino acid differences between MP-12 and wild-type RVFV strain ZH548 are unlikely to change the entry mechanism of the virus.
Reassortment experiments between MP-12 and virulent RVFV strains indicate that attenuating mutations exist in each of the viral genome segments (89). Compared to ZH548, the MP-12 M segment contains changes in the untranslated regions and the preglycoprotein, in addition to the three residue changes in the mature glycoproteins (Gn/Gc) (9). Due to the similarities between UUKV and RVFV described above, added to the fact that there is only one amino acid change in the predicted cell attachment protein Gn (87), it is highly unlikely that MP-12 attenuation in the M segment is a result of endocytosis pathway changes. MP-12 M segment attenuation is more likely due to a combination of defects that alter efficiency in genome segment replication, glycoprotein processing, virus attachment, and/or membrane fusion.
To our knowledge, RVFV is the first documented arbovirus that is internalized by CavME, and although this endocytic pathway has been analyzed in mammalian cells, there are no studies of CavME as an entry pathway for viruses in insect cells (1, 8, 22, 23, 54, 91, 103, 105, 113). Caveolins are evolutionarily conserved from humans to nematodes, and caveolin function has been shown in multiple nonmammalian systems, including the moth cell line Sf9 derived from Spodoptera frugiperda (3), the zebra fish Danio rerio, the frog species Xenopus laevis, and the honeybee Apis mellifera (52). It is possible that RVFV uses a similar, caveola-mediated internalization route in mammalian and insect cells. In addition, understanding of the entry mechanism used by RVFV could provide new insights for the discovery of its receptor(s). CavME is a regulated process that can be induced or stimulated by its cargo. Multiple studies have shown that the receptors for ligands internalized by CavME, including receptors for bacteria and viruses, localize to caveolae (18, 19, 29, 32, 68, 69, 96, 97, 100, 111, 112). Therefore, it is likely that the putative receptor for RVFV either is a component of caveolae or moves into caveolae once RVFV interacts with the host cell (19). Identification of the endocytic entry pathway for RVFV should expedite the process of identifying its receptor(s) and could contribute to the development of countermeasures against this virus.
ACKNOWLEDGMENTS
We thank Steve Branda for critical reading of the manuscript.
This work was supported by a Laboratory Directed Research and Development grant given to O.A.N. at Sandia National Laboratories (SNL). SNL is a multiprogram laboratory managed and operated by Sandia Corporation, a wholly owned subsidiary of Lockheed Martin Corporation, for the U.S. Department of Energy's National Nuclear Security Administration under contract DE-AC04-94AL85000.
Footnotes
Published ahead of print 19 September 2012
REFERENCES
- 1. Acosta EG, Castilla V, Damonte EB. 2011. Infectious dengue-1 virus entry into mosquito C6/36 cells. Virus Res. 160:173–179 [DOI] [PubMed] [Google Scholar]
- 2. Anderson HA, Chen Y, Norkin LC. 1996. Bound simian virus 40 translocates to caveolin-enriched membrane domains, and its entry is inhibited by drugs that selectively disrupt caveolae. Mol. Biol. Cell 7:1825–1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Avisar D, Segal M, Sneh B, Zilberstein A. 2005. Cell-cycle-dependent resistance to Bacillus thuringiensis Cry1C toxin in Sf9 cells. J. Cell Sci. 118:3163–3171 [DOI] [PubMed] [Google Scholar]
- 4. Balkhy HH, Memish ZA. 2003. Rift Valley fever: an uninvited zoonosis in the Arabian peninsula. Int. J. Antimicrob. Agents 21:153–157 [DOI] [PubMed] [Google Scholar]
- 5. Beer C, Andersen DS, Rojek A, Pedersen L. 2005. Caveola-dependent endocytic entry of amphotropic murine leukemia virus. J. Virol. 79:10776–10787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Benmerah A, Bayrou M, Cerf-Bensussan N, Dautry-Varsat A. 1999. Inhibition of clathrin-coated pit assembly by an Eps15 mutant. J. Cell Sci. 112(Pt 9):1303–1311 [DOI] [PubMed] [Google Scholar]
- 7. Benmerah A, et al. 1998. AP-2/Eps15 interaction is required for receptor-mediated endocytosis. J. Cell Biol. 140:1055–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bernard E, et al. 2010. Endocytosis of Chikungunya virus into mammalian cells: role of clathrin and early endosomal compartments. PLoS One 5:e11479 doi:10.1371/journal.pone.0011479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bird BH, Khristova ML, Rollin PE, Ksiazek TG, Nichol ST. 2007. Complete genome analysis of 33 ecologically and biologically diverse Rift Valley fever virus strains reveals widespread virus movement and low genetic diversity due to recent common ancestry. J. Virol. 81:2805–2816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bird BH, Ksiazek TG, Nichol ST, Maclachlan NJ. 2009. Rift Valley fever virus. J. Am. Vet. Med. Assoc. 234:883–893 [DOI] [PubMed] [Google Scholar]
- 11. Blanchard E, et al. 2006. Hepatitis C virus entry depends on clathrin-mediated endocytosis. J. Virol. 80:6964–6972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bouloy M, et al. 2001. Genetic evidence for an interferon-antagonistic function of Rift Valley fever virus nonstructural protein NSs. J. Virol. 75:1371–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cammisotto PG, et al. 2010. Receptor-mediated transcytosis of leptin through human intestinal cells in vitro. Int. J. Cell Biol. 2010:928169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cantín C, Holguera J, Ferreira L, Villar E, Munoz-Barroso I. 2007. Newcastle disease virus may enter cells by caveola-mediated endocytosis. J. Gen. Virol. 88:559–569 [DOI] [PubMed] [Google Scholar]
- 15. Caplen H, Peters CJ, Bishop DH. 1985. Mutagen-directed attenuation of Rift Valley fever virus as a method for vaccine development. J. Gen. Virol. 66(Pt 10):2271–2277 [DOI] [PubMed] [Google Scholar]
- 16. Chang WJ, et al. 1994. Purification and characterization of smooth muscle cell caveolae. J. Cell Biol. 126:127–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen J, Chen J-K, Harris RC. 2012. Angiotensin II induces epithelial-to-mesenchymal transition in renal epithelial cells through reactive oxygen species/Src/caveolin-mediated activation of an epidermal growth factor receptor–extracellular signal-regulated kinase signaling pathway. Mol. Cell. Biol. 32:981–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen X, Shank S, Davis PB, Ziady AG. 2011. Nucleolin-mediated cellular trafficking of DNA nanoparticle is lipid raft and microtubule dependent and can be modulated by glucocorticoid. Mol. Ther. 19:93–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cheng ZJ, Singh RD, Marks DL, Pagano RE. 2006. Membrane microdomains, caveolae, and caveolar endocytosis of sphingolipids. Mol. Membr. Biol. 23:101–110 [DOI] [PubMed] [Google Scholar]
- 20. Chevalier V, Pepin M, Plee L, Lancelot R. 2010. Rift Valley fever—a threat for Europe? Euro Surveill. 15:19506. [PubMed] [Google Scholar]
- 21. Chi F, Wang L, Zheng X, Jong A, Huang SH. 2011. Recruitment of α7 nicotinic acetylcholine receptor to caveolin-1-enriched lipid rafts is required for nicotine-enhanced Escherichia coli K1 entry into brain endothelial cells. Future Microbiol. 6:953–966 [DOI] [PubMed] [Google Scholar]
- 22. Chu JJ, Leong PW, Ng ML. 2006. Analysis of the endocytic pathway mediating the infectious entry of mosquito-borne flavivirus West Nile into Aedes albopictus mosquito (C6/36) cells. Virology 349:463–475 [DOI] [PubMed] [Google Scholar]
- 23. Chu JJ, Ng ML. 2004. Infectious entry of West Nile virus occurs through a clathrin-mediated endocytic pathway. J. Virol. 78:10543–10555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cosset FL, Lavillette D. 2011. Cell entry of enveloped viruses. Adv. Genet. 73:121–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. DeTulleo L, Kirchhausen T. 1998. The clathrin endocytic pathway in viral infection. EMBO J. 17:4585–4593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. de Vries E, et al. 2011. Dissection of the influenza A virus endocytic routes reveals macropinocytosis as an alternative entry pathway. PLoS Pathog. 7:e1001329 doi:10.1371/journal.ppat.1001329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Doherty GJ, McMahon HT. 2009. Mechanisms of endocytosis. Annu. Rev. Biochem. 78:857–902 [DOI] [PubMed] [Google Scholar]
- 28. Du G, Huang P, Liang BT, Frohman MA. 2004. Phospholipase D2 localizes to the plasma membrane and regulates angiotensin II receptor endocytosis. Mol. Biol. Cell 15:1024–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dugan AS, Eash S, Atwood WJ. 2005. An N-linked glycoprotein with α(2,3)-linked sialic acid is a receptor for BK virus. J. Virol. 79:14442–14445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Earl PL, Americo JL, Moss B. 2003. Development and use of a vaccinia virus neutralization assay based on flow cytometric detection of green fluorescent protein. J. Virol. 77:10684–10688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Eash S, Atwood WJ. 2005. Involvement of cytoskeletal components in BK virus infectious entry. J. Virol. 79:11734–11741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Eash S, Querbes W, Atwood WJ. 2004. Infection of Vero cells by BK virus is dependent on caveolae. J. Virol. 78:11583–11590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ebert O, et al. 2003. Oncolytic vesicular stomatitis virus for treatment of orthotopic hepatocellular carcinoma in immune-competent rats. Cancer Res. 63:3605–3611 [PubMed] [Google Scholar]
- 34. Eiblmaier M, et al. 2008. Correlating EGFR expression with receptor-binding properties and internalization of 64Cu-DOTA-cetuximab in 5 cervical cancer cell lines. J. Nucl. Med. 49:1472–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Filone CM, et al. 2010. Rift Valley fever virus infection of human cells and insect hosts is promoted by protein kinase C epsilon. PLoS One 5:e15483 doi:10.1371/journal.pone.0015483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Filone CM, Heise M, Doms RW, Bertolotti-Ciarlet A. 2006. Development and characterization of a Rift Valley fever virus cell-cell fusion assay using alphavirus replicon vectors. Virology 356:155–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Freiberg AN, Sherman MB, Morais MC, Holbrook MR, Watowich SJ. 2008. Three-dimensional organization of Rift Valley fever virus revealed by cryoelectron tomography. J. Virol. 82:10341–10348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fretz M, et al. 2006. Effects of Na+/H+ exchanger inhibitors on subcellular localisation of endocytic organelles and intracellular dynamics of protein transduction domains HIV-TAT peptide and octaarginine. J. Control Release 116:247–254 [DOI] [PubMed] [Google Scholar]
- 39. Ghigo E. 2010. A dilemma for viruses and giant viruses: which endocytic pathway to use to enter cells? Intervirology 53:274–283 [DOI] [PubMed] [Google Scholar]
- 40. Gold S, Monaghan P, Mertens P, Jackson T. 2010. A clathrin independent macropinocytosis-like entry mechanism used by bluetongue virus-1 during infection of BHK cells. PLoS One 5:e11360 doi:10.1371/journal.pone.0011360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Guo CJ, et al. 2011. Entry of tiger frog virus (an iridovirus) into HepG2 cells via a pH-dependent, atypical, caveola-mediated endocytosis pathway. J. Virol. 85:6416–6426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Guo CJ, et al. 2012. Infectious spleen and kidney necrosis virus (a fish iridovirus) enters Mandarin fish fry cells via caveola-dependent endocytosis. J. Virol. 86:2621–2631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hansen CG, Nichols BJ. 2009. Molecular mechanisms of clathrin-independent endocytosis. J. Cell Sci. 122:1713–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hartmann W, et al. 2009. Activation of phosphatidylinositol-3′-kinase/AKT signaling is essential in hepatoblastoma survival. Clin. Cancer Res. 15:4538–4545 [DOI] [PubMed] [Google Scholar]
- 45. Hernaez B, Alonso C. 2010. Dynamin- and clathrin-dependent endocytosis in African swine fever virus entry. J. Virol. 84:2100–2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Huang WR, et al. 2011. Cell entry of avian reovirus follows a caveolin-1-mediated and dynamin-2-dependent endocytic pathway that requires activation of p38 mitogen-activated protein kinase (MAPK) and Src signaling pathways as well as microtubules and small GTPase Rab5 protein. J. Biol. Chem. 286:30780–30794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ikegami T, Won S, Peters CJ, Makino S. 2006. Rescue of infectious Rift Valley fever virus entirely from cDNA, analysis of virus lacking the NSs gene, and expression of a foreign gene. J. Virol. 80:2933–2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ivanov AI. 2008. Pharmacological inhibition of endocytic pathways: is it specific enough to be useful? Methods Mol. Biol. 440:15–33 [DOI] [PubMed] [Google Scholar]
- 49. Ivanov AI, Nusrat A, Parkos CA. 2004. Endocytosis of epithelial apical junctional proteins by a clathrin-mediated pathway into a unique storage compartment. Mol. Biol. Cell 15:176–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jin M, et al. 2002. Hantaan virus enters cells by clathrin-dependent receptor-mediated endocytosis. Virology 294:60–69 [DOI] [PubMed] [Google Scholar]
- 51. Johannsdottir HK, Mancini R, Kartenbeck J, Amato L, Helenius A. 2009. Host cell factors and functions involved in vesicular stomatitis virus entry. J. Virol. 83:440–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kirkham M, et al. 2008. Evolutionary analysis and molecular dissection of caveola biogenesis. J. Cell Sci. 121:2075–2086 [DOI] [PubMed] [Google Scholar]
- 53. Koivusalo M, et al. 2010. Amiloride inhibits macropinocytosis by lowering submembranous pH and preventing Rac1 and Cdc42 signaling. J. Cell Biol. 188:547–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kolokoltsov AA, Fleming EH, Davey RA. 2006. Venezuelan equine encephalitis virus entry mechanism requires late endosome formation and resists cell membrane cholesterol depletion. Virology 347:333–342 [DOI] [PubMed] [Google Scholar]
- 55. Lajoie P, Nabi IR. 2010. Lipid rafts, caveolae, and their endocytosis. Int. Rev. Cell Mol. Biol. 282:135–163 [DOI] [PubMed] [Google Scholar]
- 56. Li Z, et al. 2007. Erlotinib effectively inhibits JAK2V617F activity and polycythemia vera cell growth. J. Biol. Chem. 282:3428–3432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lozach P-Y, et al. 2011. DC-SIGN as a receptor for phleboviruses. Cell Host Microbe 10:75–88 [DOI] [PubMed] [Google Scholar]
- 58. Lozach PY, et al. 2010. Entry of bunyaviruses into mammalian cells. Cell Host Microbe 7:488–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lucocq J, Warren G, Pryde J. 1991. Okadaic acid induces Golgi apparatus fragmentation and arrest of intracellular transport. J. Cell Sci. 100:753–759 [DOI] [PubMed] [Google Scholar]
- 60. Macovei A, et al. 2010. Hepatitis B virus requires intact caveolin-1 function for productive infection in HepaRG cells. J. Virol. 84:243–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Marjomäki V, et al. 2002. Internalization of echovirus 1 in caveolae. J. Virol. 76:1856–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Meier O, et al. 2002. Adenovirus triggers macropinocytosis and endosomal leakage together with its clathrin-mediated uptake. J. Cell Biol. 158:1119–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mercer J, Helenius A. 2010. Apoptotic mimicry: phosphatidylserine-mediated macropinocytosis of vaccinia virus. Ann. N. Y. Acad. Sci. 1209:49–55 [DOI] [PubMed] [Google Scholar]
- 64. Mercer J, Helenius A. 2008. Vaccinia virus uses macropinocytosis and apoptotic mimicry to enter host cells. Science 320:531–535 [DOI] [PubMed] [Google Scholar]
- 65. Mercer J, Helenius A. 2009. Virus entry by macropinocytosis. Nat. Cell Biol. 11:510–520 [DOI] [PubMed] [Google Scholar]
- 66. Mercer J, et al. 2010. Vaccinia virus strains use distinct forms of macropinocytosis for host-cell entry. Proc. Natl. Acad. Sci. U. S. A. 107:9346–9351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Mercer J, Schelhaas M, Helenius A. 2010. Virus entry by endocytosis. Annu. Rev. Biochem. 79:803–833 [DOI] [PubMed] [Google Scholar]
- 68. Moriyama T, Marquez JP, Wakatsuki T, Sorokin A. 2007. Caveolar endocytosis is critical for BK virus infection of human renal proximal tubular epithelial cells. J. Virol. 81:8552–8562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Mudhakir D, Harashima H. 2009. Learning from the viral journey: how to enter cells and how to overcome intracellular barriers to reach the nucleus. AAPS J. 11:65–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Nabi IR, Le PU. 2003. Caveolae/raft-dependent endocytosis. J. Cell Biol. 161:673–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Nanbo A, et al. 2010. Ebolavirus is internalized into host cells via macropinocytosis in a viral glycoprotein-dependent manner. PLoS Pathog. 6:e1001121 doi:10.1371/journal.ppat.1001121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Nawa M, Takasaki T, Yamada K, Kurane I, Akatsuka T. 2003. Interference in Japanese encephalitis virus infection of Vero cells by a cationic amphiphilic drug, chlorpromazine. J. Gen. Virol. 84:1737–1741 [DOI] [PubMed] [Google Scholar]
- 73. Nomura R, et al. 2004. Human coronavirus 229E binds to CD13 in rafts and enters the cell through caveolae. J. Virol. 78:8701–8708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ohkuma S, Poole B. 1978. Fluorescence probe measurement of the intralysosomal pH in living cells and the perturbation of pH by various agents. Proc. Natl. Acad. Sci. U. S. A. 75:3327–3331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Overby AK, Pettersson RF, Grunewald K, Huiskonen JT. 2008. Insights into bunyavirus architecture from electron cryotomography of Uukuniemi virus. Proc. Natl. Acad. Sci. U. S. A. 105:2375–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Parton RG, Joggerst B, Simons K. 1994. Regulated internalization of caveolae. J. Cell Biol. 127:1199–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Parton RG, Simons K. 2007. The multiple faces of caveolae. Nat. Rev. Mol. Cell Biol. 8:185–194 [DOI] [PubMed] [Google Scholar]
- 78. Pelkmans L. 2005. Secrets of caveolae- and lipid raft-mediated endocytosis revealed by mammalian viruses. Biochim. Biophys. Acta 1746:295–304 [DOI] [PubMed] [Google Scholar]
- 79. Pelkmans L, Kartenbeck J, Helenius A. 2001. Caveolar endocytosis of simian virus 40 reveals a new two-step vesicular-transport pathway to the ER. Nat. Cell Biol. 3:473–483 [DOI] [PubMed] [Google Scholar]
- 80. Pelkmans L, Puntener D, Helenius A. 2002. Local actin polymerization and dynamin recruitment in SV40-induced internalization of caveolae. Science 296:535–539 [DOI] [PubMed] [Google Scholar]
- 81. Penzes P, et al. 2003. Rapid induction of dendritic spine morphogenesis by trans-synaptic ephrinB-EphB receptor activation of the Rho-GEF kalirin. Neuron 37:263–274 [DOI] [PubMed] [Google Scholar]
- 82. Pepin M, Bouloy M, Bird BH, Kemp A, Paweska J. 2010. Rift Valley fever virus(Bunyaviridae: Phlebovirus): an update on pathogenesis, molecular epidemiology, vectors, diagnostics and prevention. Vet. Res. 41:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Pernet O, Pohl C, Ainouze M, Kweder H, Buckland R. 2009. Nipah virus entry can occur by macropinocytosis. Virology 395:298–311 [DOI] [PubMed] [Google Scholar]
- 84. Pho MT, Ashok A, Atwood WJ. 2000. JC virus enters human glial cells by clathrin-dependent receptor-mediated endocytosis. J. Virol. 74:2288–2292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Pontow S, Harmon B, Campbell N, Ratner L. 2007. Antiviral activity of a Rac GEF inhibitor characterized with a sensitive HIV/SIV fusion assay. Virology 368:1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Querbes W, Benmerah A, Tosoni D, Di Fiore PP, Atwood WJ. 2004. A JC virus-induced signal is required for infection of glial cells by a clathrin- and eps15-dependent pathway. J. Virol. 78:250–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Rusu M, et al. 2012. An assembly model of Rift Valley fever virus. Front. Microbiol. 3:254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Saeed MF, Kolokoltsov AA, Albrecht T, Davey RA. 2010. Cellular entry of Ebola virus involves uptake by a macropinocytosis-like mechanism and subsequent trafficking through early and late endosomes. PLoS Pathog. 6:e1001110 doi:10.1371/journal.ppat.1001110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Saluzzo JF, Smith JF. 1990. Use of reassortant viruses to map attenuating and temperature-sensitive mutations of the Rift Valley fever virus MP-12 vaccine. Vaccine 8:369–375 [DOI] [PubMed] [Google Scholar]
- 90. Sánchez EG, et al. 2012. African swine fever virus uses macropinocytosis to enter host cells. PLoS Pathog. 8:e1002754 doi:10.1371/journal.ppat.1002754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Sánchez-San Martín C, Liu CY, Kielian M. 2009. Dealing with low pH: entry and exit of alphaviruses and flaviviruses. Trends Microbiol. 17:514–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Santos RI, et al. 2008. Oropouche virus entry into HeLa cells involves clathrin and requires endosomal acidification. Virus Res. 138:139–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Schelhaas M. 2010. Come in and take your coat off—how host cells provide endocytosis for virus entry. Cell. Microbiol. 12:1378–1388 [DOI] [PubMed] [Google Scholar]
- 94. Schelhaas M, et al. 2012. Entry of human papillomavirus type 16 by actin-dependent, clathrin- and lipid raft-independent endocytosis. PLoS Pathog. 8:e1002657 doi:10.1371/journal.ppat.1002657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Schmaljohn CS, Nichol ST. 2007. Bunyaviridae, p 1741–1789 In Knipe DM, et al. (ed), Fields virology, 5th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 96. Shin JS, Abraham SN. 2001. Co-option of endocytic functions of cellular caveolae by pathogens. Immunology 102:2–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Shin JS, Gao Z, Abraham SN. 2000. Involvement of cellular caveolae in bacterial entry into mast cells. Science 289:785–788 [DOI] [PubMed] [Google Scholar]
- 98. Sieczkarski SB, Whittaker GR. 2002. Influenza virus can enter and infect cells in the absence of clathrin-mediated endocytosis. J. Virol. 76:10455–10464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Simon M, Johansson C, Mirazimi A. 2009. Crimean-Congo hemorrhagic fever virus entry and replication is clathrin-, pH- and cholesterol-dependent. J. Gen. Virol. 90:210–215 [DOI] [PubMed] [Google Scholar]
- 100. Sinclair JF, O'Brien AD. 2002. Cell surface-localized nucleolin is a eukaryotic receptor for the adhesin intimin-gamma of enterohemorrhagic Escherichia coli O157:H7. J. Biol. Chem. 277:2876–2885 [DOI] [PubMed] [Google Scholar]
- 101. Singh RD, et al. 2006. Caveolar endocytosis and microdomain association of a glycosphingolipid analog is dependent on its sphingosine stereochemistry. J. Biol. Chem. 281:30660–30668 [DOI] [PubMed] [Google Scholar]
- 102. Smart EJ, Ying YS, Conrad PA, Anderson RG. 1994. Caveolin moves from caveolae to the Golgi apparatus in response to cholesterol oxidation. J. Cell Biol. 127:1185–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Smit JM, Moesker B, Rodenhuis-Zybert I, Wilschut J. 2011. Flavivirus cell entry and membrane fusion. Viruses 3:160–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Smith JL, Campos SK, Ozbun MA. 2007. Human papillomavirus type 31 uses a caveolin 1- and dynamin 2-mediated entry pathway for infection of human keratinocytes. J. Virol. 81:9922–9931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Stiles KM, Kielian M. 2011. Alphavirus entry: NRAMP leads the way. Cell Host Microbe 10:92–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Sun L, Hemgard GV, Susanto SA, Wirth M. 2010. Caveolin-1 influences human influenza A virus (H1N1) multiplication in cell culture. Virol. J. 7:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Sun X, Yau VK, Briggs BJ, Whittaker GR. 2005. Role of clathrin-mediated endocytosis during vesicular stomatitis virus entry into host cells. Virology 338:53–60 [DOI] [PubMed] [Google Scholar]
- 108. Swanson JA. 2008. Shaping cups into phagosomes and macropinosomes. Nat. Rev. Mol. Cell Biol. 9:639–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Taylor GM, Sanders DA. 2003. Structural criteria for regulation of membrane fusion and virion incorporation by the murine leukemia virus TM cytoplasmic domain. Virology 312:295–305 [DOI] [PubMed] [Google Scholar]
- 110. Taylor MP, Koyuncu OO, Enquist LW. 2011. Subversion of the actin cytoskeleton during viral infection. Nat. Rev. Microbiol. 9:427–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Tayyari F, et al. 2011. Identification of nucleolin as a cellular receptor for human respiratory syncytial virus. Nat. Med. 17:1132–1135 [DOI] [PubMed] [Google Scholar]
- 112. Upla P, et al. 2004. Clustering induces a lateral redistribution of α2β1 integrin from membrane rafts to caveolae and subsequent protein kinase C-dependent internalization. Mol. Biol. Cell 15:625–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. van der Schaar HM, et al. 2008. Dissecting the cell entry pathway of dengue virus by single-particle tracking in living cells. PLoS Pathog. 4:e1000244 doi:10.1371/journal.ppat.1000244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Vela EM, Zhang L, Colpitts TM, Davey RA, Aronson JF. 2007. Arenavirus entry occurs through a cholesterol-dependent, non-caveolar, clathrin-mediated endocytic mechanism. Virology 369:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Vialat P, Muller R, Vu TH, Prehaud C, Bouloy M. 1997. Mapping of the mutations present in the genome of the Rift Valley fever virus attenuated MP-12 strain and their putative role in attenuation. Virus Res. 52:43–50 [DOI] [PubMed] [Google Scholar]
- 116. Walter CT, Barr JN. 2011. Recent advances in the molecular and cellular biology of bunyaviruses. J. Gen. Virol. 92:2467–2484 [DOI] [PubMed] [Google Scholar]
- 117. Wang H, Jiang C. 2009. Influenza A virus H5N1 entry into host cells is through clathrin-dependent endocytosis. Sci. China C Life Sci. 52:464–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Werling D, et al. 1999. Involvement of caveolae in the uptake of respiratory syncytial virus antigen by dendritic cells. J. Leukoc. Biol. 66:50–58 [DOI] [PubMed] [Google Scholar]
- 119. Whitbeck JC, Foo CH, Ponce de Leon M, Eisenberg RJ, Cohen GH. 2009. Vaccinia virus exhibits cell-type-dependent entry characteristics. Virology 385:383–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Yarar D, Waterman-Storer CM, Schmid SL. 2005. A dynamic actin cytoskeleton functions at multiple stages of clathrin-mediated endocytosis. Mol. Biol. Cell 16:964–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Yonezawa A, Cavrois M, Greene WC. 2005. Studies of Ebola virus glycoprotein-mediated entry and fusion by using pseudotyped human immunodeficiency virus type 1 virions: involvement of cytoskeletal proteins and enhancement by tumor necrosis factor alpha. J. Virol. 79:918–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Yoon MJ, et al. 2003. Localization of Tie2 and phospholipase D in endothelial caveolae is involved in angiopoietin-1-induced MEK/ERK phosphorylation and migration in endothelial cells. Biochem. Biophys. Res. Commun. 308:101–105 [DOI] [PubMed] [Google Scholar]
- 123. Yoshimori T, Yamamoto A, Moriyama Y, Futai M, Tashiro Y. 1991. Bafilomycin A1, a specific inhibitor of vacuolar-type H+-ATPase, inhibits acidification and protein degradation in lysosomes of cultured cells. J. Biol. Chem. 266:17707–17712 [PubMed] [Google Scholar]
- 124. Zhao L, et al. 2006. Role of p21-activated kinase pathway defects in the cognitive deficits of Alzheimer disease. Nat. Neurosci. 9:234–242 [DOI] [PubMed] [Google Scholar]
- 125. Zhao Z, et al. 2009. p21-activated kinase mediates rapid estradiol-negative feedback actions in the reproductive axis. Proc. Natl. Acad. Sci. U. S. A. 106:7221–7226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Zhu D, Xiong WC, Mei L. 2006. Lipid rafts serve as a signaling platform for nicotinic acetylcholine receptor clustering. J. Neurosci. 26:4841–4851 [DOI] [PMC free article] [PubMed] [Google Scholar]