Abstract
Assembly of the herpesvirus tegument is poorly understood but is believed to involve interactions between outer tegument proteins and the cytoplasmic domains of envelope glycoproteins. Here, we present the detailed characterization of a multicomponent glycoprotein-tegument complex found in herpes simplex virus 1 (HSV-1)-infected cells. We demonstrate that the tegument protein VP22 bridges a complex between glycoprotein E (gE) and glycoprotein M (gM). Glycoprotein I (gI), the known binding partner of gE, is also recruited into this gE-VP22-gM complex but is not required for its formation. Exclusion of the glycoproteins gB and gD and VP22's major binding partner VP16 demonstrates that recruitment of virion components into this complex is highly selective. The immediate-early protein ICP0, which requires VP22 for packaging into the virion, is also assembled into this gE-VP22-gM-gI complex in a VP22-dependent fashion. Although subcomplexes containing VP22 and ICP0 can be formed when either gE or gM are absent, optimal complex formation requires both glycoproteins. Furthermore, and in line with complex formation, neither of these glycoproteins is individually required for VP22 or ICP0 packaging into the virion, but deletion of gE and gM greatly reduces assembly of both VP22 and ICP0. Double deletion of gE and gM also results in small plaque size, reduced virus yield, and defective secondary envelopment, similar to the phenotype previously shown for pseudorabies virus. Hence, we suggest that optimal gE-VP22-gM-gI-ICP0 complex formation correlates with efficient virus morphogenesis and spread. These data give novel insights into the poorly understood process of tegument acquisition.
INTRODUCTION
The herpesvirus virion is a complex particle consisting of a DNA genome-containing capsid surrounded by a compartment termed the tegument and an outer, host cell-derived lipid envelope that contains virus-encoded glycoproteins (29). The prototypic alphaherpesvirus herpes simplex virus 1 (HSV-1) packages at least 26 tegument proteins and up to 20 envelope proteins (35). Because of this complexity and previously recognized redundancy among the structural proteins, the molecular mechanisms involved in the assembly of this virus remain poorly understood. An attractive and widely accepted model proposes that viral glycoproteins embedded within membranes of the late secretory pathway recruit components of the outer tegument, through which viral capsids, surrounded by an inner tegument, bud to produce mature virions (25, 33). In spite of this favored model, there are few examples in the literature of well-defined tegument-glycoprotein interactions shown to be involved in virus assembly. For example, the major tegument components VP16 and VP22 have been reported to interact with gH and gD, respectively, but the contribution these interactions make to tegument assembly and virus morphogenesis have not yet been proven (5, 17, 20).
The nonessential glycoprotein gE is the only envelope protein thus far convincingly shown to interact directly with and be required for the recruitment of individual tegument proteins into the alphaherpesvirus virion. In HSV-1, gE has recently been shown to be required for the packaging of the membrane-bound tegument protein UL11 via an interaction between the cytoplasmic tail of gE and the C terminus of UL11, thereby representing the only published example of a single glycoprotein being required to package a tegument protein (17, 22). The cytoplasmic tail of gE has also been shown to interact with the tegument protein VP22, a protein that is also not essential for virus assembly (11). This was first demonstrated in pseudorabies virus (PRV), where yeast two-hybrid assays indicated such an interaction (19). More recently, a similar interaction has been identified between HSV-1 VP22 and the cytoplasmic tail of gE of HSV-1 (17, 43, 49). The relationship between the HSV-1 proteins has been explored in detail and has been shown using pulldown of VP22 on a GST-gE cytoplasmic tail fusion protein, relocalization of VP22 to Golgi-localized gE in cotransfected cells, and coimmunoprecipitations from infected cells (43, 49). A conserved C-terminal domain of VP22 was shown to coprecipitate gE in infected cells, and importantly, deletion of a 12-residue region in this domain abrogated both interaction with gE and assembly of VP22 into the virion (21, 43, 49).
Nonetheless, unlike the UL11-gE interaction, a paradox exists with the gE/VP22 complex in PRV as a virus lacking gE, and its partner gI in PRV is still able to package full-length VP22 to wild-type levels (19). In the case of HSV-1, it has been reported that virus lacking the cytoplasmic tail of gE (22) also packages VP22 to wild-type levels. Hence, these results suggest an alternative assembly pathway for VP22 that utilizes the same essential C-terminal region. VP22 is known to interact with a second major tegument protein, VP16, but this interaction is not required to package VP22 into the virion (12, 21, 43). A second candidate for an alternative packaging route is the cytoplasmic tail of the glycoprotein gM. Like gE and VP16, HSV-1 gM has been shown to coimmunoprecipitate with VP22 in infected cells (49). Direct binding studies between VP22 and gM from PRV and HSV-1 have also indicated the potential for these two proteins to interact directly, but in HSV-1 at least this interaction was much less convincing than the aforementioned gE-VP22 interaction (49). Furthermore, unlike the case for HSV-1 VP22 and gE, coexpression of HSV-1 VP22 and gM by cotransfection of expressing plasmids revealed no ability of gM to relocalize VP22 to the Golgi region, leaving the significance of the VP22-gM interaction unclear (49). Nonetheless, simultaneous deletion of gE, its known binding partner gI, and gM in PRV was shown to completely abrogate VP22 assembly into the virion (19). As single deletions of gM in both PRV and HSV-1 have been shown to be dispensable for VP22 incorporation into the virion (19, 46), it would seem that it is the combination of the gE/gI and gM deletions that abolishes VP22 assembly in PRV virions.
Taken together, this combination of data hints at the presence of an as-yet poorly characterized network of protein-protein interactions involving VP22, which is required for VP22 virion incorporation. Furthermore, as VP22 is required for virion assembly of the immediate-early protein ICP0 (11, 38), it is likely that any interaction that impinges on the recruitment of VP22 to the virus particle will indirectly affect ICP0 assembly. In this paper, we demonstrate that gE forms a specific complex with gM in the infected cell that is dependent on the presence of VP22. This complex also contains the gE binding partner gI and the VP22 partner ICP0 but does not recruit gD or gB or the well-characterized VP22-binding protein VP16. We show that gE, gM, and gI are individually dispensable for VP22 virion incorporation in HSV-1 but that a virus lacking both gE and gM in combination packages greatly reduced levels of both VP22 and ICP0. The phenotype of this double glycoprotein mutant virus—small plaque size, reduced virus yield, and defective secondary envelopment as determined by electron microscopy—suggests that formation of this multicomponent complex is important for efficient virus production. This study represents the first detailed characterization of a higher-order glycoprotein-tegument complex in infected cells involved in the recruitment of several tegument proteins into the herpesvirus particle. Our data therefore provide valuable new insights into the molecular mechanisms that underpin the most widely accepted model for herpesvirus morphogenesis.
MATERIALS AND METHODS
Cells and viruses.
Vero cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% newborn calf serum and antibiotics. HFFF cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and antibiotics.
The wild-type HSV-1 strain sc16 and the ΔgE, ΔgEgM, and ΔgI mutants were kindly provided by Helena Browne (2, 4), and the ΔgM virus was kindly donated by Colin Crump (46) (both from the University of Cambridge, Cambridge, United Kingdom). Our Δ22 virus expressing green fluorescent protein (GFP) in place of VP22 (169v) (11) and our GFP-22 mutant expressing GFP-tagged full-length VP22 (166v) (14) have been described previously. Our ΔgEbind virus, which expresses GFP-tagged VP22 with a small lesion within the gE-binding domain of VP22, is also described elsewhere (49). Viruses expressing GFP tagged to residues 108 to 301, 160 to 301, and 212 to 301 of VP22 and GFP tagged to variants of VP22 expressing mutated phosphorylation sites have been described before (21, 45). Extracellular virions were purified on Ficoll gradients from the infected cell medium of between 3 × 108 and 1 × 109 Vero cells, as described previously (14). Viruses were routinely titrated on Vero cells.
Antibodies.
The gD (LP14), VP16 (LP1), gB (R69), and gM (Ab980) antibodies were kindly provided by Tony Minson and Helena Browne (University of Cambridge). Antibodies specific for gE (3114), gE in the context of the gE-gI complex (3063), and gI (3104) were kindly donated by David Johnson (Oregon Health and Science University, Portland, OR). The gE antibody 3114 was used for Western blotting only; for immunoprecipitations, we used a commercially available gE antibody (ab6510) (AbCam, Cambridge, United Kingdom). The VP5 (3B6) and ICP0 (11060) antibodies are also commercially available from Virusys (Taneytown, MD) and Santa Cruz (Santa Cruz, CA), respectively. Our VP22-specific antibodies AGV600 and AGV031 are described elsewhere (13) and were used for immunoprecipitations and Western blotting, respectively. Horseradish peroxidase-conjugated secondary antibodies were purchased from Bio-Rad Laboratories (Hercules, CA). For gM and VP22 Western blots of immunoprecipitation samples, the TrueBlot horseradish peroxidase-conjugated rabbit secondary antibody (eBioscience, San Diego, CA), which does not recognize denatured immunoglobulin, was used.
SDS-PAGE and Western blotting.
Protein samples were analyzed on 10% polyacrylamide gels and subjected to electrophoresis in Tris-glycine buffer. All samples were boiled for 3 min prior to electrophoresis except for those used for gM blots, which were heated to 42°C for 20 min. Gels were then either stained in Coomassie blue or transferred to nitrocellulose membrane for Western blot analysis using antibodies as indicated. Western blots were developed using SuperSignal West Pico chemiluminescent substrate (Thermo Fisher Scientific).
Immunoprecipitation assay.
Vero cells grown in 6-cm dishes were infected with the relevant viruses at a multiplicity of 1. After 24 h, the cells were washed twice with phosphate-buffered saline (PBS), solubilized in 1 ml radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 0.1% SDS, 1% Na deoxycholate, 1% NP-40, protease inhibitors) and incubated on ice for 20 min. The cells were then centrifuged at 12 K for 30 min at 4°C, and the supernatant was collected. The supernatant was precleared with protein A Sepharose beads at a sample-bead ratio of 10:1 for 1 h at 4°C with rotation. The precleared lysate was then incubated with antibody at a sample-antibody ratio of 100:1 (monoclonal) or 40:1 (polyclonal) at 4°C for 3 h with rotation. Protein A Sepharose beads were added at a sample-bead ratio of 10:1 and incubated for 1 h at 4°C with rotation, and the resulting protein A-antibody complexes were washed three times in a buffer consisting of 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, and 1% (vol/vol) NP-40. The immunoprecipitated proteins were then analyzed by SDS-PAGE followed by Western blotting.
Expression and purification of GST-tagged proteins.
Plasmid expressing GST fused to the C-terminal 126 residues of glycoprotein M was kindly provided by Colin Crump (University of Cambridge). GST-gM and GST alone were expressed in Escherichia coli strain BL21 by inducing a 250-ml culture with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) followed by a further 3-h incubation. The cells were then pelleted, resuspended in 10 ml PBS containing protease inhibitors and 1 mg/ml lysozyme, and left on ice for 30 min. Following sonication, Triton X-100 was added to 1%, and the extracts were incubated for a further 30 min of rotating at 4°C and centrifuged at 4 K for 30 min at 4°C. The soluble supernatant was added to 200 μl of a 50:50 suspension of glutathione Sepharose beads and rotated for 1 h at 4°C, and the unbound protein was washed off the beads by 3 washes with PBS.
Pulldown of infected cell extracts on GST-tagged proteins.
Vero cells in 6-cm dishes were infected with the relevant viruses, and 16 h later cells were washed with PBS and harvested in 1 ml lysis buffer (50 mM Tris [pH 7.5], 200 mM NaCl, 2 mM MgCl, 1% NP-40, and protease inhibitors). The samples were left on ice for 20 min and centrifuged at 12 K for 30 min at 4°C. A total of 400 μl of supernatant was mixed with the relevant GST fusion protein already bound to glutathione Sepharose beads. Following 2 to 3 h of rotating at 4°C, the beads were washed 3 times with lysis buffer, and samples of each were analyzed by SDS-PAGE and Western blotting.
Transmission electron microscopy.
Cells for electron microscopy (EM) were fixed in 0.5% glutaraldehyde in 200 mM sodium cacodylate buffer for 30 min, washed in buffer, and secondarily fixed in reduced 1% osmium tetroxide and 1.5% potassium ferricyanide for 60 min. The samples were washed in distilled water and stained overnight at 4°C in 0.5% magnesium uranyl acetate, washed in distilled water, and dehydrated in graded ethanol. The samples were then embedded flat in the dish in Epon resin. Resin-filled stubs were placed on embedded cell monolayers and polymerized. Ultrathin sections (typically 50 to 70 nm) were cut parallel to the dish and examined in an FEI Tecnai electron microscope with charge-coupled-device (CCD) camera image acquisition.
RESULTS
Characterization of HSV-1 lacking gE and/or gM.
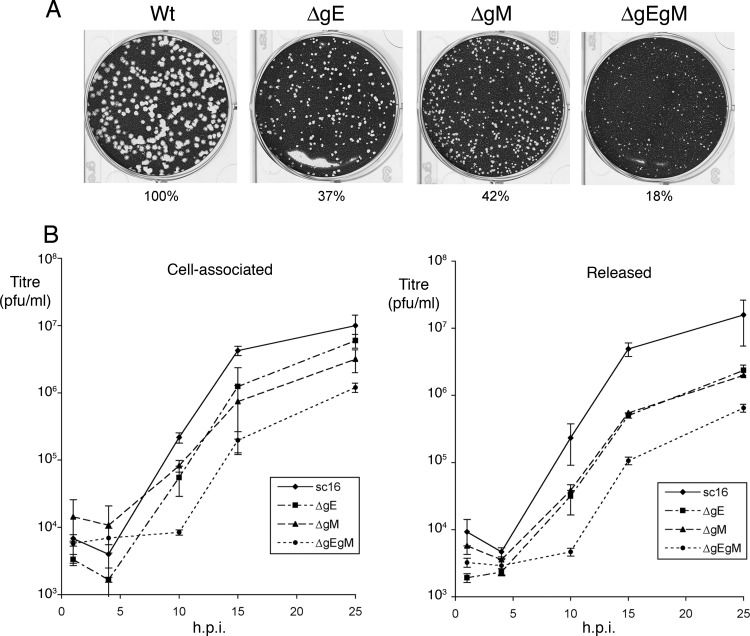
The starting point for this work was our existing data that both gE and gM coimmunoprecipitate with VP22 in infected cell lysates (49). Such a result could indicate either two different VP22-glycoprotein complexes or a single complex comprising all three proteins. To further examine the relationship between VP22, gE, and gM, we made use of viruses lacking expression of gE (ΔgE), gM (ΔgM), or both gE and gM (ΔgEgM) based on the wild-type (WT) strain sc16, kindly provided by Helena Browne and Colin Crump (University of Cambridge). In PRV, it has been shown that simultaneous deletion of gE, gI, and gM results in a 1,000-fold drop in virus production (3) but in HSV-1 double deletion of gE and gM has been characterized as having limited but variable effects on virus replication depending on cell type (4). Before using the HSV-1 mutants in our studies for protein-protein interactions, we first characterized their growth in Vero cells. Plaque analysis indicated that, as expected from previous studies (4), all three mutant viruses formed plaques on Vero cells, but they exhibited reduced plaque size compared to that of the WT, with the plaques size of the double-deletion mutant being over 5-fold less than that of the WT (Fig. 1A). In the case of the ΔgM mutant, its plaque size was similar to that published before (46). Furthermore, one-step growth curves showed that the relative plaque size of the virus mutants correlated with their ability to replicate in a single cycle, with the single mutants producing and releasing around 8-fold less virus while the double mutant produced up to 50-fold less virus than the WT virus (Fig. 1B). Although the defects in virus replication that we observed were not as extensive as those observed in the PRV study, it should be noted firstly that, in the PRV study, gI had also been deleted and secondly that the differences we observed in the replication of the ΔgEgM double-deletion mutant in HSV was more substantial than described previously (4).
Fig 1.
Growth characteristics of HSV-1 lacking gE and/or gM. (A) Vero cells in 6-well plates were infected with approximately 100 PFU of WT (sc16), ΔgE, ΔgM, or ΔgEgM viruses and fixed and stained 4 days later. The area of 20 representative plaques for each virus was measured using ImageJ software, and the average relative size was calculated as a percentage of WT plaques. (B) Single-step growth curves of WT, ΔgE, ΔgM, or ΔgEgM viruses were carried out by infecting Vero cells at a multiplicity of 5 and harvested at the indicated times for cell-associated or released virus. All growth curves were carried out in triplicate.
Coimmunoprecipitation of gM and gE with VP22.
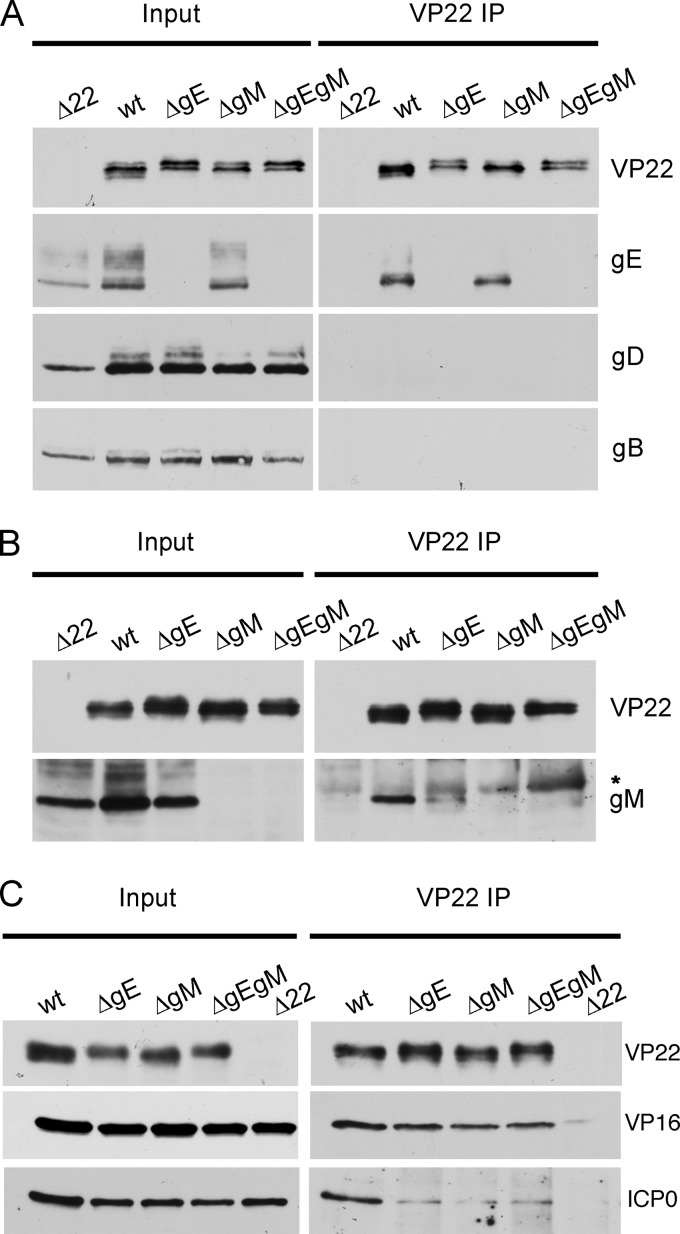
The requirement for gE and gM in each other's interaction with VP22 was examined by immunoprecipitating VP22 from Vero cells infected with each of the above-described viruses, with our previously described VP22-null (Δ22) virus acting as a negative control (11). Western blotting of the input extracts indicated that the proteins being examined were expressed to similar levels in all infections with the exception of the Δ22 infection (Fig. 2A, input), which we have shown previously to express reduced levels of virus proteins (11). Coimmunoprecipitation of gE with VP22 was clearly detected in cells infected with WT virus and was unaffected by the absence of gM, indicating that gM is not required to form the gE-VP22 complex in infected cells (Fig. 2A, VP22 IP). Importantly, neither gD nor gB precipitated with VP22 in WT-infected cells, confirming that VP22 does not indiscriminately bind all glycoproteins (Fig. 2A). In contrast, although gM coimmunoprecipitated with VP22 from WT-infected cells, only trace amounts precipitated with VP22 in the absence of gE (Fig. 2B). Interestingly, VP22 expressed in the absence of gE migrated more slowly on the gel compared to VP22 expressed in the presence of gE, suggesting that gE is in some way required for the appropriate modification of VP22 during infection (Fig. 2A). As this slow-migrating form has previously been shown to represent phosphorylated VP22, with the faster-migrating form representing nonphosphorylated VP22 (15, 16), it seems plausible that gE is somehow required for the expression of VP22 lacking phosphorylation.
Fig 2.
Immunoprecipitation of VP22-specific complexes from cells infected with viruses lacking gE and/or gM. (A to C) Whole-cell lysates harvested 24 h after infection from Vero cells infected with Δ22, sc16 (wt), ΔgE, ΔgM, or ΔgEgM viruses were subjected to immunoprecipitation with an anti-VP22 antibody and analyzed by Western blotting for the presence of gE, gD, or gB (A), gM (B), and VP16 or ICP0 (C). *, immunoglobulin heavy chain; note that the immunoglobulin heavy chain is visible in this gM blot, and no others, because the ordinary rabbit secondary antibody, rather than the TrueBlot secondary antibody, was used here.
To determine if the interaction of VP22 with its other known tegument binding partners was affected by the absence of gE and/or gM, VP22 coimmunoprecipitations were also tested for the presence of VP16 or ICP0 (Fig. 2C). Interestingly, VP16 coimmunoprecipitated efficiently with VP22 in all infected cell lysates (Fig. 2C, VP16), indicating that neither gE nor gM is required in the formation of the VP22-VP16 complex. However, the presence of ICP0 in the VP22-specific complex was greatly reduced when either of the glycoproteins was absent (Fig. 2C, ICP0), suggesting that the interaction of ICP0 with VP22 may be facilitated by both glycoproteins.
gE forms a VP22-dependent complex with gM.
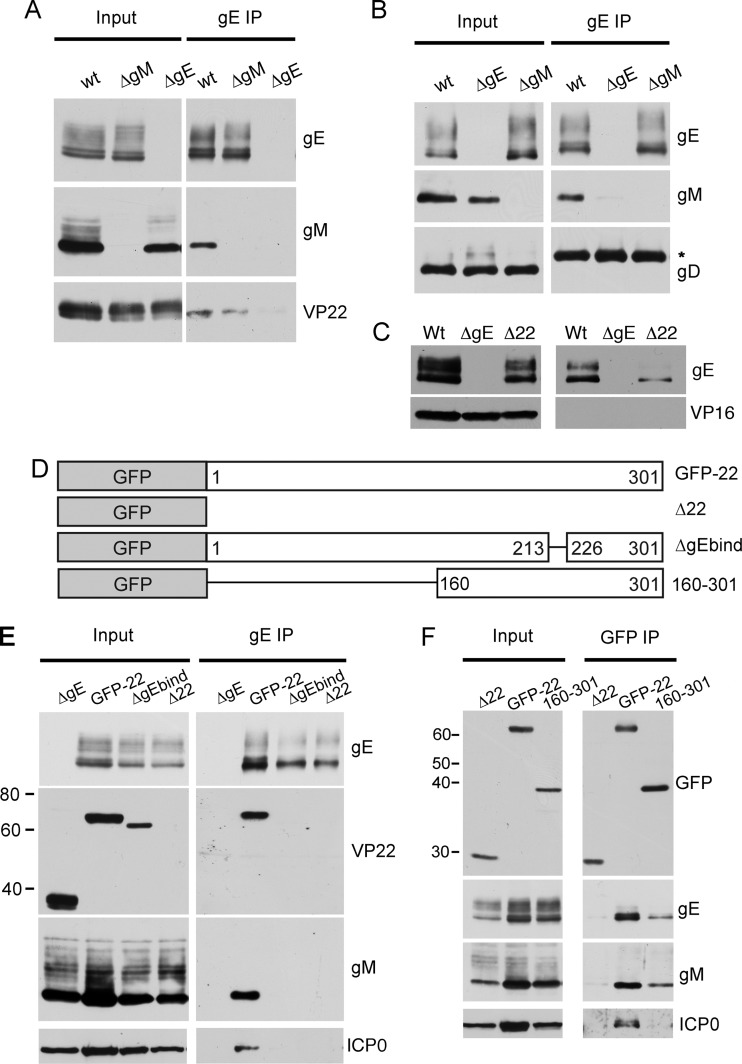
To further examine the nature of these complexes, we next immunoprecipitated gE from infected cells. Here, the ΔgE mutant served as a negative control. As expected from the reciprocal VP22 immunoprecipitations, VP22 bound to gE from WT-infected cells (Fig. 3A, wt). Furthermore, VP22 interacted with gE in the absence of gM (Fig. 3A, ΔgM). Interestingly, we were also able to routinely coprecipitate gM with gE from WT-infected cells (Fig. 3A and B, wt), demonstrating that gE and gM exist in a complex in the infected cell. Importantly, the lack of gD pulldown in the gE immunoprecipitation shows that gE does not indiscriminately bind all viral glycoproteins (Fig. 3B). In addition, VP16 did not precipitate with gE, confirming that the VP22-VP16 complex is distinct from the gE-VP22 complex (Fig. 3C).
Fig 3.
VP22 bridges gE and gM to form a multicomponent complex in infected cells. (A to C) Vero cells were infected with WT HSV-1 or the ΔgE or ΔgM viruses, and gE was immunoprecipitated from whole-cell lysates harvested 24 h using antibody ab6510. Samples were subjected to Western blot analysis using antibodies specific for gE, gM, VP22, gD, or VP16. *, immunoglobulin heavy chain. (D) Schematic illustrating our previously described VP22 mutant viruses expressing GFP-tagged full-length VP22 (GFP-22), GFP in place of VP22 (Δ22), GFP-tagged VP22 with a deletion between residues 213 to 226 (ΔgEbind), or GFP tagged to the C-terminal half of VP22 (160 to 301). (E and F) As for panel A, except Vero cells were infected with the viruses described in panel D or the ΔgE mutant and samples analyzed by Western blotting using antibodies specific for gE, gM, VP22, GFP, or ICP0. Molecular weight marker sizes (kDa) are shown on the left.
We next repeated these gE immunoprecipitations with the Δ22 virus and our previously described HSV-1 mutants expressing GFP-tagged full-length VP22 (GFP-22) (14) or GFP-tagged VP22 with a small lesion within the conserved gE-binding domain of VP22 (49) (Fig. 3D, ΔgEbind). Similarly to WT HSV-1, gE coimmunoprecipitated VP22 and gM from cells infected with the GFP-22 virus (Fig. 3E). As shown previously by VP22 immunoprecipitation (49), VP22 did not interact with gE in cells infected with the ΔgEbind mutant. Interestingly, gM did not bind gE in cells infected with the ΔgEbind or VP22-null viruses, demonstrating that an interaction between gE and VP22 is required for formation of the gE-gM complex. Moreover, ICP0 was also present in the gE-specific complex in WT-infected cells but was absent from gE-specific complexes lacking VP22 (Fig. 3E, ICP0), confirming that ICP0 is recruited into the gE-VP22-gM complex in a VP22-dependent fashion. As our previous studies had shown that the C-terminal half of VP22 (160 to 301 in Fig. 3D) was sufficient for assembly of VP22 into the virion (21), we next tested the ability of this region of VP22 to interact with gE and/or gM during infection. Coimmunoprecipitation of VP22 from cells infected with HSV-1 expressing GFP 160 to 301 revealed that both gE and gM were able to complex with the C-terminal half of VP22 (Fig. 3F). In contrast, ICP0 was not present in this multicomponent complex, indicating that the N-terminal half of VP22 is likely to be involved in bringing ICP0 into this complex (Fig. 3F, ICP0). This confirms our previous data showing a requirement for the N terminus of VP22 in the assembly of ICP0 into the virion (38). Collectively, our data show that VP22, ICP0, gE, and gM are present within the same complex in infected cells and that, although VP22 is not essential for virus assembly, it is fundamental to the formation of this multicomponent complex.
VP22 expressed in the absence of gE can bind to the cytoplasmic domain of gM.
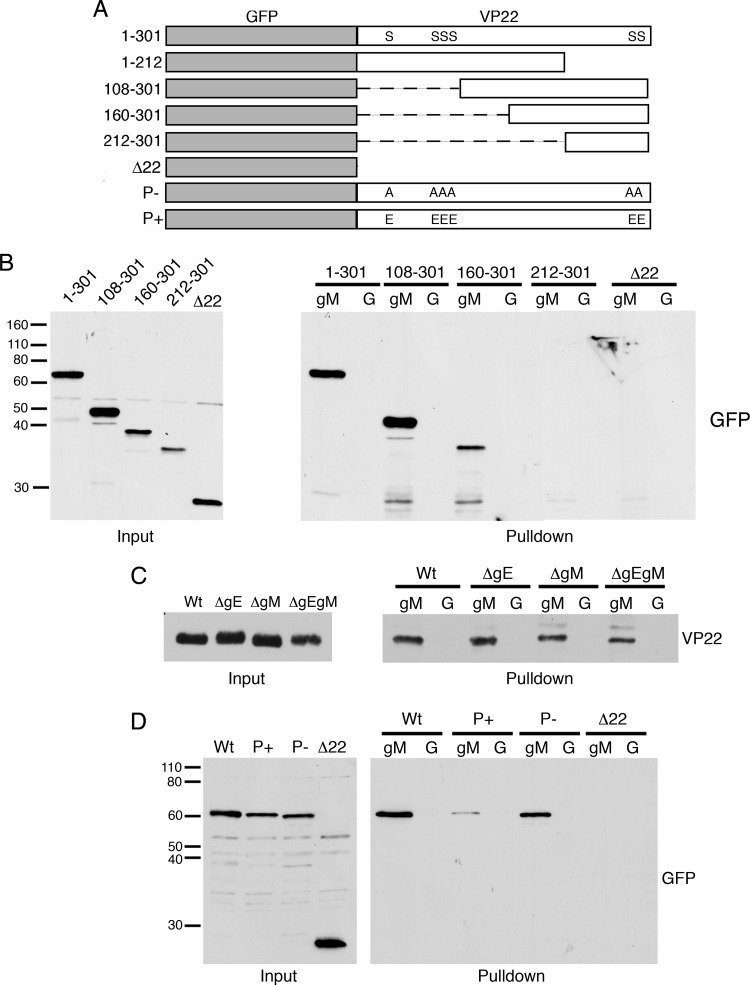
The above-described data demonstrate that gM requires VP22 to form a complex with gE. As it was difficult to detect a complex between gM and VP22 in the absence of gE (Fig. 2B), it is possible that gE is somehow required for formation of a VP22-gM complex. We were unable to investigate complex formation by immunoprecipitation of gM because the antibody available to us is a rabbit polyclonal antibody that in our hands routinely nonspecifically coprecipitated the gE-gI Fc receptor complex (data not shown). Hence, to determine if VP22 expressed in ΔgE-infected cells was capable of binding gM, we carried out pulldown assays of infected cell extracts using a GST-gM (cytoplasmic tail) fusion protein bound to glutathione Sepharose beads, as described previously (49). The resulting complexes bound to gM were then analyzed by Western blotting for the presence of VP22. We first confirmed that the pulldown from infected cells reproduced our results from VP22 coimmunoprecipitations, by carrying out pulldowns with extracts from cells infected with viruses expressing truncated versions of VP22 (Fig. 4A). These confirmed that region 160 to 301 of VP22, the region that coimmunoprecipitates gE and gM and is packaged into virions (21), was able to bind to the cytoplasmic tail of gM in vitro (Fig. 4B). We next tested the pulldown of VP22 expressed in cells infected with the gE and gM deletion mutants. Interestingly, these showed that, in spite of not efficiently coimmunoprecipitating gM, the VP22 population present in ΔgE-infected cells had the ability to interact with GST-gM in the absence of gE (Fig. 4C).
Fig 4.
VP22 interaction with the cytoplasmic tail of gM. (A) Schematic of VP22 variants expressed by recombinant viruses used in panels B and D. (B to D) GST-gM (gM) or GST alone (G) bound to glutathione Sepharose beads was incubated with lysates from Vero cells infected with viruses as indicated and analyzed by SDS-PAGE followed by Western blotting for GFP (B and D) or VP22 (C). Molecular weight marker sizes (kDa) are shown on the left.
Data presented in Fig. 1B suggest that there may a correlation between posttranslational modification of VP22 and its ability to bind gM. We therefore tested binding of gM to two other variants of VP22 in which the previously identified phosphorylated residues had been mutated to alanines to mimic nonphosphorylated VP22 (P−) or to glutamic acids to mimic the phosphorylated form of VP22 (P+) (45). Using cell extracts from infections with HSV-1 expressing either of these two variants, we found that VP22 with a constitutively high negative charge at its phosphorylation sites (P+) consistently bound to gM with less efficiency than the noncharged version (Fig. 4D). Taken together, these results suggest that VP22 from infected cells has the ability to bind to gM in the absence of gE but that the prevalence of modified/negatively charged VP22 may limit the efficiency or stability of this interaction as detected by coimmunoprecipitations.
The gE-VP22-gM complex also incorporates gI.
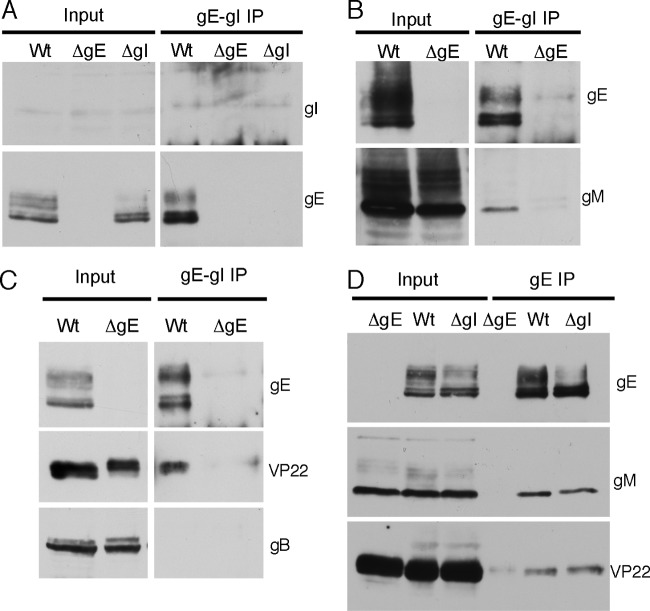
In infected cells, gE interacts with gI to form the HSV-1 Fc receptor (26). We therefore sought to determine whether gI is also recruited into the gE-VP22-gM complex or whether a separate gE-gI complex exists. Initial attempts to determine if gI coprecipitated with VP22 were hampered by our inability to detect gI in infected cell lysates with the monoclonal antibody available to us (data not shown). To overcome this issue, we made use of an alternative monoclonal antibody (3063) that reportedly recognizes gE only in the context of gI (6). We validated the specificity of this antibody by using it to immunoprecipitate the gE-gI complex from cells infected with WT HSV-1, the ΔgE mutant, or the ΔgI mutant. Once again, no gI was observed in the input cell lysate or following gE-gI immunoprecipitation (Fig. 5A). However, the fact that this antibody immunoprecipitated gE from cells infected with the WT virus but not the ΔgI virus, despite equivalent levels of gE expression in both viruses (Fig. 5A, input), confirms that this antibody recognizes gE only when incorporated into the gE-gI complex. When we repeated this experiment, we found that gM consistently coimmunoprecipitated with gE using this antibody (Fig. 5B). In addition, VP22 precipitation with gE was also observed with this antibody (Fig. 5C). These data demonstrate that at least a proportion of the gE recruited into the VP22 packaging complex is also bound to gI. The fact that gB was not coimmunoprecipitated with the gE-gI complex (Fig. 5C) reconfirms that the VP22 packaging complex does not indiscriminately incorporate all viral glycoproteins.
Fig 5.
The gE-VP22-gM complex incorporates gI. (A to C) Vero cells infected with WT HSV-1, the ΔgE virus, or the ΔgI virus (panel A only) were harvested 24 h after infection and the gE-gI complex was immunoprecipitated using antibody 3063, specific for gE in the context of the gE-gI complex. Samples were analyzed by Western blotting using antibodies as indicated. (D) Vero cells infected with WT HSV-1 or the ΔgE or ΔgI viruses were harvested for 24 h, and gE was immunoprecipitated using antibody 6510, specific for free and gI-complexed gE. Samples were analyzed by Western blotting using antibodies specific for gE, gM, or VP22.
We next evaluated the impact gI deletion has on the formation of the VP22 complex itself. Glycoprotein E was immunoprecipitated from Vero cells infected with the WT and ΔgI viruses; the ΔgE mutant served as a negative control. Note that the gE antibody (ab6510) used here and in experiments depicted in Fig. 2 recognizes free gE as well as gE in the context of gI. As before, both gM and VP22 coimmunoprecipitated with gE from cells infected with WT virus (Fig. 5D, Wt). Furthermore, comparable levels of gM and VP22 also coprecipitated with gE in the absence of gI (Fig. 5D, ΔgI). Hence, although gI is recruited into this multicomponent packaging complex, we find no evidence to suggest that gI contributes to its formation.
VP22 fails to package into virions lacking both gE and gM.
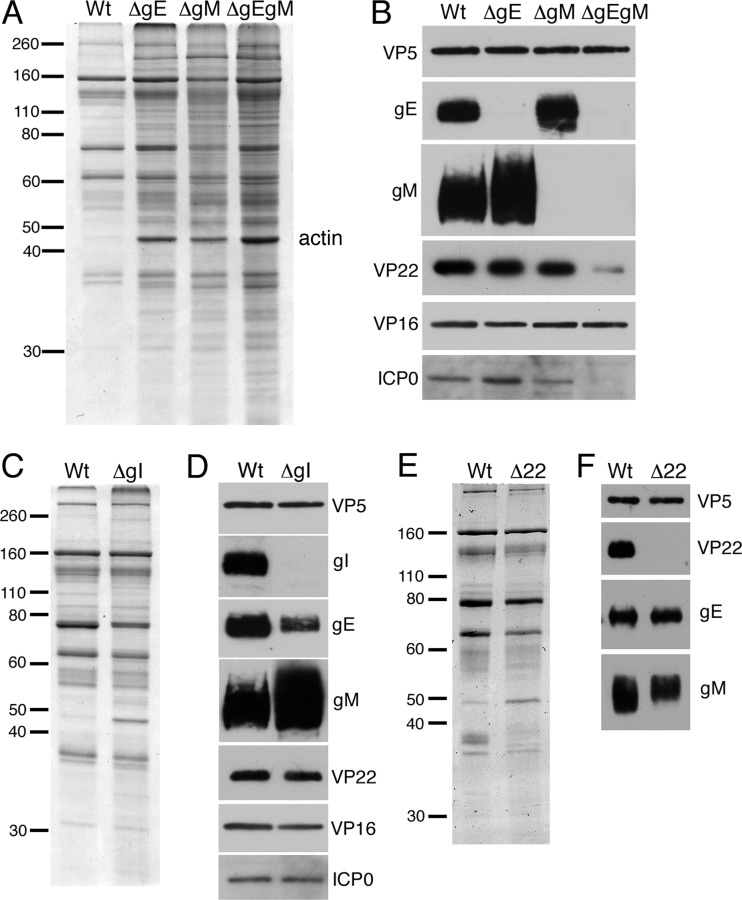
The above-described results suggest that although optimal complex formation requires both gE and gM, infected cell VP22 can interact with the cytoplasmic tail of both glycoproteins in the absence of each other. To evaluate the roles of gE and gM in VP22 assembly into HSV-1 virions, extracellular particles were gradient purified from Vero cells infected with WT, ΔgE, ΔgM, or ΔgEgM viruses and analyzed by SDS-PAGE followed by staining with Coomassie blue or Western blotting. In several purifications, the ΔgM and ΔgEgM viruses both proved difficult to purify in large amounts; however, small but discrete bands were formed during gradient purification. Coomassie staining of the particles revealed that although the glycoprotein mutant virions contained a higher background of overall protein content than the WT virions, the major components appeared similar, with the exception of the actin band at 43 kDa, which was greatly enriched in all mutant virions (Fig. 6A). Western blotting for various components indicated that assembly of gE into the virion is independent of gM, and vice versa (Fig. 6B), as has been shown previously for PRV (19).
Fig 6.
Relative assembly of VP22 into HSV-1 virions isolated from Vero cells infected with glycoprotein mutant viruses. (A and B) Gradient purified extracellular sc16 (WT), ΔgE, ΔgM, or ΔgEgM virions were analyzed by Coomassie blue staining (A) or by Western blotting using antibodies as indicated (B). Molecular weight marker sizes (kDa) are shown on the left. (C and D) Gradient-purified extracellular sc16 (WT) or ΔgI virions were analyzed by SDS-PAGE followed by Coomassie blue staining (C) or Western blotting with antibodies as indicated (D). Molecular weight marker sizes (kDa) are shown on the left. (E and F) Extracellular WT (s17) or Δ22 virions purified from BHK cells were analyzed by Coomassie blue staining (E) or Western blotting using antibodies as indicated (F). Molecular weight marker sizes (kDa) are shown on the left.
However, although VP22 was packaged into ΔgE and ΔgM virions to the level of the WT virus, its assembly was greatly reduced in ΔgEgM virions (Fig. 6B, VP22). Blotting for another tegument protein, VP16, showed that this protein was packaged at equivalent levels in all viruses, and hence the reduction of VP22 in the absence of both gE and gM was specific (Fig. 6B, VP16). In contrast, ICP0 packaging directly correlated with the levels of its binding partner VP22 and as such was diminished only in the ΔgEgM virions (Fig. 6D, ICP0). We therefore conclude that, as previously published (38), VP22 is the major viral determinant for ICP0 virion incorporation and that the requirement for gE and gM does not extend beyond their role in facilitating VP22 packaging.
To confirm that gI is not required for VP22 assembly in HSV-1, extracellular particles from Vero cells infected with WT or ΔgI viruses were gradient purified and analyzed by SDS-PAGE followed by staining with Coomassie blue or Western blotting. Virions were equalized according to the major capsid protein VP5 (Fig. 6C), and subsequent blotting showed that both VP22 and ICP0 were efficiently packaged to WT levels in the absence of gI (Fig. 6D). Interestingly, the ΔgI virions appeared to assemble less gE but more gM than WT virions, indicating that the assembly of these glycoproteins may be linked to gI (Fig. 6D).
Finally, to determine if VP22 is involved in packaging either gE or gM into the virion, we purified virions from our Δ22 virus and its parent s17 and analyzed equivalent amounts as judged by VP5 loading (Fig. 6E). Blotting for VP22 showed that as expected, VP22 was absent from the Δ22 virions (Fig. 6F). However, the levels of gE and gM were similar in both sets of virions, confirming that the absence of the tegument protein VP22 has no effect on the recruitment of these glycoproteins to the virus, as is the case in PRV (19).
Defective secondary envelopment by HSV-1 in the absence of both gE and gM.
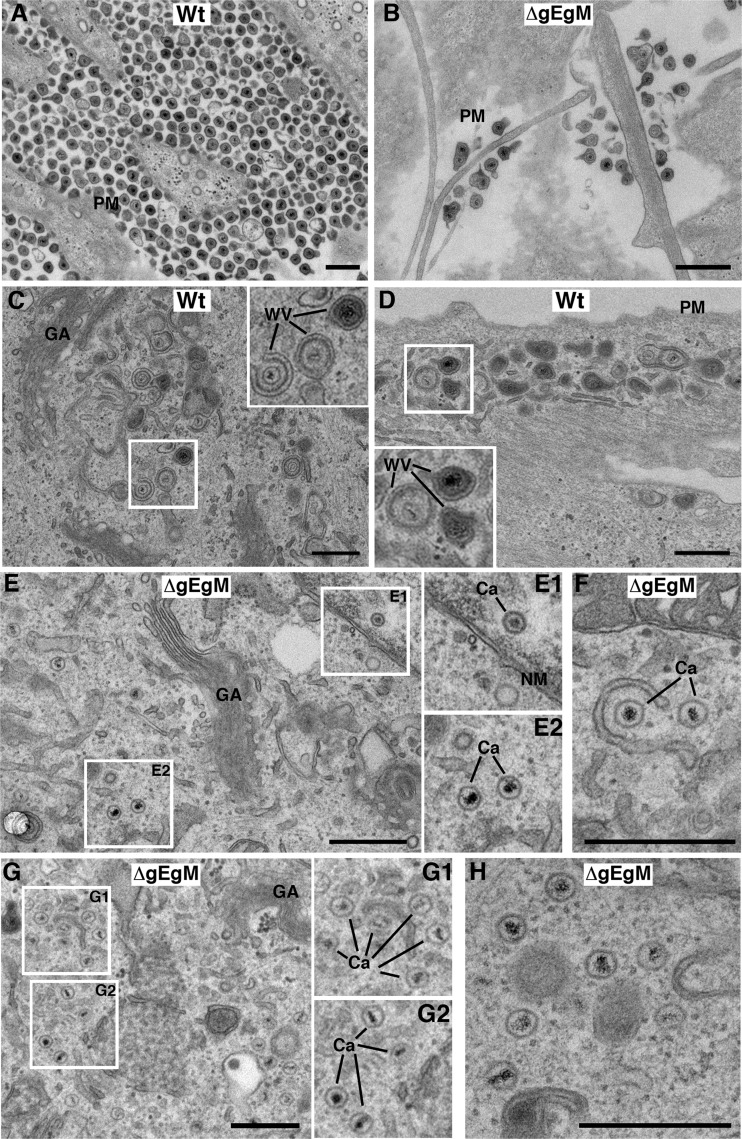
A PRV mutant lacking gE, gI, and gM has been shown to exhibit a profound defect in envelopment, resulting in the accumulation of capsids within the cytoplasm (3). Although the HSV-1 ΔgEgM virus has a less severe phenotype than the aforementioned PRV mutant when measured by single-step growth curve, virus yield and plaque size were nonetheless significantly affected by the absence of these glycoproteins. To compare the phenotype of the HSV-1 mutant to that of PRV, we carried out ultrastructural studies in HFFF cells, a primary human fibroblast cell line that we have recently shown to be a good system for such studies of HSV-1 (24). Plaque assays on HFFF cells indicated that, as in Vero cells, the ΔgEgM mutant of HSV-1 produced small plaques compared to those of the WT virus (data not shown). HFFF cells were infected with WT or ΔgEgM virus at a multiplicity of 2, fixed, and processed for transmission electron microscopy 12 h later. Imaging of the plasma membrane revealed that at this time, large numbers of virions had been released from WT-infected cells (Fig. 7A). In contrast, few released virions were detectable outside ΔgEgM-infected cells (Fig. 7B). Within the cytoplasm of WT-infected cells, areas of active virus wrapping were readily detected both toward the main body of the cell (Fig. 7C) and toward the periphery (Fig. 7D). In ΔgEgM-infected cells, however, while a very few wrapped or wrapping capsids were found (Fig. 7F) and normal numbers of capsids were detected in the nucleus (Fig. 7E and E1), the vast majority of capsids were found as free, unwrapped capsids throughout the cytoplasm (Fig. 7E, E2, G, and H). This indicates that reduced virus yield in ΔgEgM-infected cells can be explained by a drop in secondary envelopment levels and that as previously shown for PRV (3), gE and gM contribute to this fundamental process of HSV-1 morphogenesis. In short, the reduction in VP22 assembly, poor virus yield, and defective secondary envelopment described for PRV lacking gE and gM (3, 19) has now been verified in HSV-1. Hence, although none of the individual components of the multicomponent complex identified here are essential for HSV-1 or PRV, we would suggest that the absence of this complex from infected cells has a deleterious effect on virus maturation.
Fig 7.
HSV-1 lacking glycoproteins E and M is defective in secondary envelopment. HFFF cells infected with the WT or ΔgEgM virus at a multiplicity of 2 were fixed 12 h later and processed for EM. PM, plasma membrane; Ca, capsid; GA, Golgi apparatus; NM, nuclear membrane; WV, wrapped or wrapping virions. Scale bar = 500 nm.
DISCUSSION
The current model for alphaherpesvirus morphogenesis, based on many studies of PRV and HSV-1 in particular, involves assembled capsids translocating from the nucleus to the cytoplasm, where they acquire inner tegument proteins such as UL36 and UL37. At the same time, outer tegument proteins are proposed to assemble onto the cytoplasmic tails of virus glycoproteins embedded in membranes of the exocytic or endocytic pathways. The capsid/inner tegument then buds into these outer tegument-coated membranes to form the mature particle (25, 40). Such a model would be simple to test if in the assembly process a single glycoprotein recruited a single tegument protein to the virion. However, it is clear from studies on both HSV-1 and PRV that the protein-protein interactions involved in virus assembly present a much more complex scenario, as none of the previously described interactions, with the exception of gE-UL11, are requisite for assembly of individual tegument proteins, and there is obvious redundancy among these interactions (39).
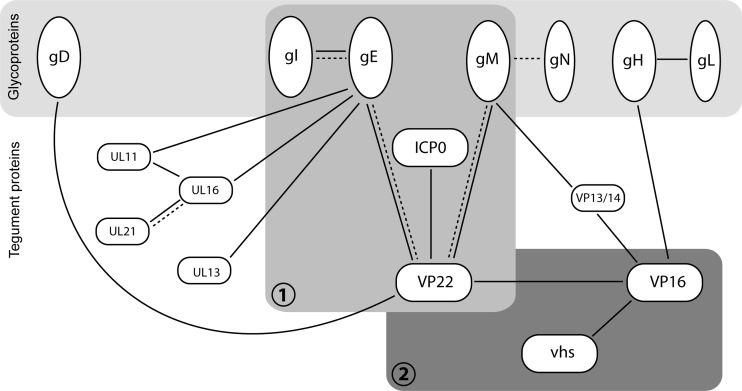
The networks of protein interactions contributing to virus assembly have been largely extrapolated from in vitro studies focused primarily on one or two binding partners, and although large-scale studies have been carried out using yeast two-hybrid analysis of herpesvirus protein interactions, few of the identified networks have been validated in infected cells (18, 52). Here, we have identified a multicomponent glycoprotein-tegument complex present in HSV-1-infected cells that shows individual components of the virus involved in a complicated network of interactions. The complex comprises the glycoproteins gE, gM, and gI and the tegument proteins VP22 and ICP0, and although single interactions between components of this complex have been identified before (gE-gI, VP22-gE, VP22-gM, and VP22-ICP0), such a large multicomponent complex (summarized in Fig. 8) has not been described before. The intricate nature of the interactions identified places VP22 at the center of the complex, with its C terminus binding to the cytoplasmic tails of gE and gM and forming a bridge between them and the N terminus of VP22 recruiting ICP0 into the complex. Our immunoprecipitations indicate that VP22-ICP0 complexes can exist in isolation, as demonstrated recently by yeast two-hybrid studies (18, 54), but it is only when both glycoproteins are present that the complex is efficiently formed and/or stabilized within the cell, suggesting that gE and gM may function to stabilize binding of ICP0 by VP22.
Fig 8.
Network of protein-protein interactions around the HSV-1 tegument protein VP22. Solid lines indicate interactions reported in HSV-1 (5, 8, 12, 20, 22, 23, 26, 34, 38, 42, 48–50, 52, 53). Broken lines indicate interactions shown in PRV (19, 27, 28, 55). The glycoprotein-tegument complex characterized here (complex 1), which is required for VP22 and ICP0 assembly into the virion, is shown in the context of previously reported relevant interactions (19, 38, 49). Whether UL11, UL16, or UL13 are incorporated into the VP22-gE-gM-gI-ICP0 complex via their known interactions with gE remains to be determined (22, 42, 53). Others have observed VP22-gD binding in infected cells (5, 17); however, this interaction was not reproducible in our own hands (49), and gD was not detectable in the VP22-gE-gM-gI-ICP0 complex. The previously characterized VP22-VP16 complex, which also incorporates vhs (12, 48, 50) and which was shown here to be separate from the VP22 assembly complex, is also illustrated (complex 2). For completion, the tegument protein VP13/14 has also been included in the figure. VP13/14 has been shown to interact with the cytoplasmic tail of gM (49) and VP16 (8, 52); however, it is not known which if either of these interactions is involved in VP13/14 assembly.
The glycoprotein-binding domain of VP22 is conserved among all VP22 homologues (41), suggesting that the gE-VP22-gM complex may be important for alphaherpesvirus replication. Nonetheless, VP22 is not required for replication of all alphaherpesviruses, being dispensable for HSV-1 and PRV but essential for VZV and MDV (9–11, 19, 51). This variable requirement for VP22 may reflect a differential role in assembly of other virus proteins. As illustrated in Fig. 8, there is scope for other proteins to be components of this large complex. Others have shown that UL11, UL16, and UL13 all bind to gE, and hence any of these other tegument proteins may also be incorporated (22, 42, 53). The tegument complex of UL11, UL16, and UL21 is also shown in Fig. 8, as this entire trimeric complex also has the potential to be part of the VP22-gE multicomponent complex (23). Furthermore, in several herpesviruses, gM is known to bind the UL49.5 gene product gN, which in HSV-1 is called UL49A because it is not glycosylated (27, 31, 32, 37). Given that the gE glycoprotein binding partner gI is incorporated into our complex, it is possible that UL49A would also be present by virtue of its binding to gM. However, it is noteworthy that the gM-UL49A interaction has not been confirmed in HSV-1 and that assembly of UL49A itself into the HSV-1 virion remains contentious (1, 35). Importantly, exclusion of gD and VP16 from this glycoprotein-tegument assembly demonstrates that recruitment of virion components into this complex is selective. Although gD has been described as a binding partner for VP22 (5, 17), we have been consistently unable to demonstrate this interaction in our own studies (reference 49 and this study). Hence, any gD-VP22 interaction that occurs during infection would appear to play a limited role. Likewise, VP16 has been shown to interact with VP22 in a well-characterized complex that can be demonstrated in vitro and in infected cells and may also include the host shutoff protein vhs (12, 21, 44, 48, 50). Nonetheless, this complex is not involved in assembly of either VP22 or VP16 into the virion (11, 44) and as such is not recruited into the complex identified here, suggesting that VP16 is recruited to the virion via an additional route. Interestingly, it has been proposed that VP16 is packaged either in the outer tegument by gH (as shown in Fig. 8) or in the inner tegument by VP1/2 (20, 30). The existence of a VP22-VP16 complex that is not involved in assembly of either component also suggests that this complex plays an alternative role in infection, which, as indicated in Fig. 8, could be the regulation of the host shutoff protein vhs activity during infection, as shown by others (47, 50). The tegument protein VP13/14 has also been included in Fig. 8, as it has been shown to interact with both gM and VP16 (8, 49, 52). However, the relationship between VP13/14 and the two complexes shown here remains to be determined.
It is noteworthy that the absence of gE, but not gI or gM, consistently induced a mobility shift in VP22. Others also observed this shift during infection with HSV-1 and PRV with mutations in gE (22, 36). As nonphosphorylated VP22 is preferentially recruited into WT HSV-1 particles (16), it is tempting to speculate that gE might help establish or maintain a nonphosphorylated VP22 subpopulation for assembly into virions. Potential mechanisms for this include gE altering the localization of VP22 so it would no longer be accessible to casein kinase II or UL13, both of which are involved in VP22 phosphorylation (7, 16). Alternatively, UL13 could be sequestered by its known binding to gE (42). Given that VP22-gM binding is also reduced in the absence of gE, the VP22-gM interaction might also be regulated by VP22 phosphorylation. Nonetheless, pulldown of VP22 expressed in the absence of gE on GST-gM indicated that this population of protein was capable of binding to gM in vitro, a result which may indicate that VP22 and/or gM may not localize correctly in the absence of gE to interact efficiently. We previously reported that gE, but not gM, was able to recruit VP22 to secretory pathway membranes in transfected cells, and hence gE may be required for the efficient compartmentalization of VP22 prior to it interacting with gM (49).
Relating complex formation to assembly into the virion is not straightforward. As is the case in PRV (19), the double gE/gM deletion mutant doesn't package VP22 but both single mutants package it to WT levels, suggesting there is redundancy in these glycoproteins and VP22 can bind to the cytoplasmic tails of gE and gM. However, as discussed above, VP22 coprecipitates gM poorly in the absence of gE. VP22-gM association may be dynamic, perhaps becoming stable only in the presence of gE (see above). Moreover, ICP0 is also poorly recruited into the VP22-glycoprotein complex in the absence of either glycoprotein but is assembled to WT levels in the single mutant viruses. It is only when both glycoproteins are absent that VP22 and hence ICP0 fail to package. Since the proportion of expressed VP22 recruited into virus particles remains unknown, an excess of VP22-glycoprotein complexes might form in WT-infected cells compared with the amount incorporated into virions.
Interestingly, although the virions from the glycoprotein mutants appear to package WT levels of most proteins, the ΔgM and ΔgEgM virions were noticeably difficult to isolate. In HSV-1, gM was recently shown to be required for optimal recruitment of the gH-gL glycoprotein complex into the virus particle, a factor which may contribute to the phenotype of these viruses (46). Additionally, PRV lacking gE, gI, and gM has been shown to be severely impaired in the final stages of virion maturation (3, 19). Likewise, we have shown here that HSV-1 lacking gE and gM is also defective in secondary envelopment with unwrapped capsids accumulating in the cytoplasm. Although this phenotype may simply reflect the lack of gE and gM in this virus, it must be considered that it could be explained by a combination of the direct absence of gE and gM and the indirect absence from the virus particle of VP22, ICP0, and any other as-yet-unidentified components of this complex.
It is often proposed that the nature of protein interactions within the tegument confers a degree of order upon this virion compartment, which was previously described as amorphous. To our knowledge, this paper presents the first detailed description of a herpesvirus assembly complex in infected cells that incorporates multiple components. Our data therefore represent an important step toward understanding how higher-order glycoprotein-tegument assemblies form during herpesvirus maturation. In the future, it will be important to expand these studies to gain a better understanding of the wider protein interaction networks leading to assembly of the herpesvirus tegument.
ACKNOWLEDGMENTS
We thank Helena Browne, Colin Crump, David Johnson, and Tony Minson for kindly sharing viruses and antibodies. We also thank Michael Hollinshead for help with electron microscopy.
This work was funded by the Medical Research Council.
Footnotes
Published ahead of print 19 September 2012
REFERENCES
- 1. Adams R, Cunningham C, Davison MD, MacLean CA, Davison AJ. 1998. Characterization of the protein encoded by gene UL49A of herpes simplex virus type 1. J. Gen. Virol. 79:813–823 [DOI] [PubMed] [Google Scholar]
- 2. Balan P, et al. 1994. An analysis of the in vitro and in vivo phenotypes of mutants of herpes simplex virus type 1 lacking glycoproteins gG, gE, gI or the putative gJ. J. Gen. Virol. 75:1245–1258 [DOI] [PubMed] [Google Scholar]
- 3. Brack AR, Dijkstra JM, Granzow H, Klupp BG, Mettenleiter TC. 1999. Inhibition of virion maturation by simultaneous deletion of glycoproteins E, I, and M of pseudorabies virus. J. Virol. 73:5364–5372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Browne H, Bell S, Minson T. 2004. Analysis of the requirement for glycoprotein M in herpes simplex virus type 1 morphogenesis. J. Virol. 78:1039–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chi JH, Harley CA, Mukhopadhyay A, Wilson DW. 2005. The cytoplasmic tail of herpes simplex virus envelope glycoprotein D binds to the tegument protein VP22 and to capsids. J. Gen. Virol. 86:253–261 [DOI] [PubMed] [Google Scholar]
- 6. Collins WJ, Johnson DC. 2003. Herpes simplex virus gE/gI expressed in epithelial cells interferes with cell-to-cell spread. J. Virol. 77:2686–2695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Coulter LJ, Moss HW, Lang J, McGeoch DJ. 1993. A mutant of herpes simplex virus type 1 in which the UL13 protein kinase gene is disrupted. J. Gen. Virol. 74:387–395 [DOI] [PubMed] [Google Scholar]
- 8. Donnelly M, Verhagen J, Elliott G. 2007. RNA binding by the herpes simplex virus type 1 nucleocytoplasmic shuttling protein UL47 is mediated by an N-terminal arginine-rich domain that also functions as its nuclear localization signal. J. Virol. 81:2283–2296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dorange F, Tischer BK, Vautherot JF, Osterrieder N. 2002. Characterization of Marek's disease virus serotype 1 (MDV-1) deletion mutants that lack UL46 to UL49 genes: MDV-1 UL49, encoding VP22, is indispensable for virus growth. J. Virol. 76:1959–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Duffy C, et al. 2006. Characterization of a UL49-null mutant: VP22 of herpes simplex virus type 1 facilitates viral spread in cultured cells and the mouse cornea. J. Virol. 80:8664–8675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Elliott G, Hafezi W, Whiteley A, Bernard E. 2005. Deletion of the herpes simplex virus VP22-encoding gene (UL49) alters the expression, localization, and virion incorporation of ICP0. J. Virol. 79:9735–9745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Elliott G, Mouzakitis G, O'Hare P. 1995. VP16 interacts via its activation domain with VP22, a tegument protein of herpes simplex virus, and is relocated to a novel macromolecular assembly in coexpressing cells. J. Virol. 69:7932–7941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Elliott G, O'Hare P. 1997. Intercellular trafficking and protein delivery by a herpesvirus structural protein. Cell 88:223–233 [DOI] [PubMed] [Google Scholar]
- 14. Elliott G, O'Hare P. 1999. Live-cell analysis of a green fluorescent protein-tagged herpes simplex virus infection. J. Virol. 73:4110–4119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Elliott G, O'Reilly D, O'Hare P. 1999. Identification of phosphorylation sites within the herpes simplex virus tegument protein VP22. J. Virol. 73:6203–6206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Elliott G, O'Reilly D, O'Hare P. 1996. Phosphorylation of the herpes simplex virus type 1 tegument protein VP22. Virology 226:140–145 [DOI] [PubMed] [Google Scholar]
- 17. Farnsworth A, Wisner TW, Johnson DC. 2007. Cytoplasmic residues of herpes simplex virus glycoprotein gE required for secondary envelopment and binding of tegument proteins VP22 and UL11 to gE and gD. J. Virol. 81:319–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fossum E, et al. 2009. Evolutionarily conserved herpesviral protein interaction networks. PLoS Pathog. 5:e1000570 doi:10.1371/journal.ppat.1000570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fuchs W, et al. 2002. Physical interaction between envelope glycoproteins E and M of pseudorabies virus and the major tegument protein UL49. J. Virol. 76:8208–8217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gross ST, Harley CA, Wilson DW. 2003. The cytoplasmic tail of herpes simplex virus glycoprotein H binds to the tegument protein VP16 in vitro and in vivo. Virology 317:1–12 [DOI] [PubMed] [Google Scholar]
- 21. Hafezi W, Bernard E, Cook R, Elliott G. 2005. Herpes simplex virus tegument protein VP22 contains an internal VP16 interaction domain and a C-terminal domain that are both required for VP22 assembly into the virus particle. J. Virol. 79:13082–13093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Han J, Chadha P, Meckes DG, Jr, Baird NL, Wills JW. 2011. Interaction and interdependent packaging of tegument protein UL11 and glycoprotein E of herpes simplex virus. J. Virol. 85:9437–9446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Harper AL, et al. 2010. Interaction domains of the UL16 and UL21 tegument proteins of herpes simplex virus. J. Virol. 84:2963–2971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hollinshead M, et al. 18 September 2012. Endocytic tubules regulated by Rab GTPases 5 and 11 are used for envelopment of herpes simplex virus. EMBO J. [Epub ahead of print.] doi:10.1038/emboj.2012.262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Johnson DC, Baines JD. 2011. Herpesviruses remodel host membranes for virus egress. Nat. Rev. Microbiol. 9:382–394 [DOI] [PubMed] [Google Scholar]
- 26. Johnson DC, Frame MC, Ligas MW, Cross AM, Stow ND. 1988. Herpes simplex virus immunoglobulin G Fc receptor activity depends on a complex of two viral glycoproteins, gE and gI. J. Virol. 62:1347–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jons A, Dijkstra JM, Mettenleiter TC. 1998. Glycoproteins M and N of pseudorabies virus form a disulfide-linked complex. J. Virol. 72:550–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Klupp BG, Bottcher S, Granzow H, Kopp M, Mettenleiter TC. 2005. Complex formation between the UL16 and UL21 tegument proteins of pseudorabies virus. J. Virol. 79:1510–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Knipe DM, et al. 2007. Fields virology, 5th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 30. Ko DH, Cunningham AL, Diefenbach RJ. 2010. The major determinant for addition of tegument protein pUL48 (VP16) to capsids in herpes simplex virus type 1 is the presence of the major tegument protein pUL36 (VP1/2). J. Virol. 84:1397–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Koyano S, Mar EC, Stamey FR, Inoue N. 2003. Glycoproteins M and N of human herpesvirus 8 form a complex and inhibit cell fusion. J. Gen. Virol. 84:1485–1491 [DOI] [PubMed] [Google Scholar]
- 32. Lake CM, Molesworth SJ, Hutt-Fletcher LM. 1998. The Epstein-Barr virus (EBV) gN homolog BLRF1 encodes a 15-kilodalton glycoprotein that cannot be authentically processed unless it is coexpressed with the EBV gM homolog BBRF3. J. Virol. 72:5559–5564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Leege T, et al. 2009. Effects of simultaneous deletion of pUL11 and glycoprotein M on virion maturation of herpes simplex virus type 1. J. Virol. 83:896–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Loomis JS, Courtney RJ, Wills JW. 2003. Binding partners for the UL11 tegument protein of herpes simplex virus type 1. J. Virol. 77:11417–11424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Loret S, Guay G, Lippe R. 2008. Comprehensive characterization of extracellular herpes simplex virus type 1 virions. J. Virol. 82:8605–8618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lyman MG, Demmin GL, Banfield BW. 2003. The attenuated pseudorabies virus strain Bartha fails to package the tegument proteins Us3 and VP22. J. Virol. 77:1403–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mach M, Kropff B, Dal Monte P, Britt W. 2000. Complex formation by human cytomegalovirus glycoproteins M (gpUL100) and N (gpUL73). J. Virol. 74:11881–11892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Maringer K, Elliott G. 2010. Recruitment of herpes simplex virus type 1 immediate-early protein ICP0 to the virus particle. J. Virol. 84:4682–4696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mettenleiter TC. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mettenleiter TC, Klupp BG, Granzow H. 2009. Herpesvirus assembly: an update. Virus Res. 143:222–234 [DOI] [PubMed] [Google Scholar]
- 41. Mouzakitis G, McLauchlan J, Barreca C, Kueltzo L, O'Hare P. 2005. Characterization of VP22 in herpes simplex virus-infected cells. J. Virol. 79:12185–12198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ng TI, Ogle WO, Roizman B. 1998. UL13 protein kinase of herpes simplex virus 1 complexes with glycoprotein E and mediates the phosphorylation of the viral Fc receptor: glycoproteins E and I. Virology 241:37–48 [DOI] [PubMed] [Google Scholar]
- 43. O'Regan K, Bucks JMA, Murphy MA, Wills JW, Courtney RJ. 2007. A conserved region of the herpes simplex virus type 1 tegument protein VP22 facilitates interaction with the cytoplasmic tail of glycoprotein E (gE). Virology 358:192–200 [DOI] [PubMed] [Google Scholar]
- 44. O'Regan KJ, Murphy MA, Bucks MA, Wills JW, Courtney RJ. 2007. Incorporation of the herpes simplex virus type 1 tegument protein VP22 into the virus particle is independent of interaction with VP16. Virology 369:263–280 [DOI] [PubMed] [Google Scholar]
- 45. Potel C, Elliott G. 2005. Phosphorylation of the herpes simplex virus tegument protein VP22 has no effect on incorporation of VP22 into the virus but is involved in optimal expression and virion packaging of ICP0. J. Virol. 79:14057–14068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ren Y, Bell S, Zenner HL, Lau SY, Crump CM. 2012. Glycoprotein M is important for the efficient incorporation of glycoprotein H-L into herpes simplex virus type 1 particles. J. Gen. Virol. 93:319–329 [DOI] [PubMed] [Google Scholar]
- 47. Sciortino MT, et al. 2007. Replication-competent herpes simplex virus 1 isolates selected from cells transfected with a bacterial artificial chromosome DNA lacking only the UL49 gene vary with respect to the defect in the UL41 gene encoding host shutoff RNase. J. Virol. 81:10924–10932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Smibert CA, Popova B, Xiao P, Capone JP, Smiley JR. 1994. Herpes simplex virus VP16 forms a complex with the virion host shutoff protein vhs. J. Virol. 68:2339–2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Stylianou J, Maringer K, Cook R, Bernard E, Elliott G. 2009. Virion incorporation of the herpes simplex virus type 1 tegument protein VP22 occurs via glycoprotein E-specific recruitment to the late secretory pathway. J. Virol. 83:5204–5218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Taddeo B, Sciortino MT, Zhang W, Roizman B. 2007. Interaction of herpes simplex virus RNase with VP16 and VP22 is required for the accumulation of the protein but not for accumulation of mRNA. Proc. Natl. Acad. Sci. U. S. A. 104:12163–12168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tischer BK, et al. 2007. A self-excisable infectious bacterial artificial chromosome clone of varicella-zoster virus allows analysis of the essential tegument protein encoded by ORF9. J. Virol. 81:13200–13208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Vittone V, et al. 2005. Determination of interactions between tegument proteins of herpes simplex virus type 1. J. Virol. 79:9566–9571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yeh PC, et al. 2011. Direct and specific binding of the UL16 tegument protein of herpes simplex virus to the cytoplasmic tail of glycoprotein E. J. Virol. 85:9425–9436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yu X, et al. 2010. Herpes simplex virus type 1 tegument protein VP22 is capable of modulating the transcription of viral TK and gC genes via interaction with viral ICP0. Biochimie 92:1024–1030 [DOI] [PubMed] [Google Scholar]
- 55. Zuckermann FA, Mettenleiter TC, Schreurs C, Sugg N, Ben-Porat T. 1988. Complex between glycoproteins gI and gp63 of pseudorabies virus: its effect on virus replication. J. Virol. 62:4622–4626 [DOI] [PMC free article] [PubMed] [Google Scholar]