Abstract
Hypoviruses Cryphonectria hypovirus 1 (CHV-1)/EP713, CHV-1/Euro7, and CHV-1/EP721, which infect the chestnut blight fungus Cryphonectria parasitica, differ in their degrees of virulence attenuation (hypovirulence), symptom expression, and viral RNA accumulation, even though they share between 90% and 99% amino acid sequence identity. In this report we examine whether this variability is influenced by interactions with the C. parasitica Dicer gene dcl2-dependent RNA-silencing antiviral defense response. The mild symptoms exhibited by strains infected with CHV-1/Euro7 and CHV-1/EP721 relative to those with severe hypovirus CHV-1/EP713 did not correlate with a higher induction of the RNA-silencing pathway. Rather, dcl2 transcripts accumulated to a higher level (∼8-fold) following infection by CHV-1/EP713 than following infection by CHV-1/Euro7 (1.2-fold) or CHV-1/EP721 (1.4-fold). The differences in dcl2 transcript accumulation in response to CHV-1/EP713 and CHV-1/EP721 were unrelated to the suppressor of RNA silencing, p29, encoded by the two viruses. Moreover, the coding strand viral RNA levels increased by 33-, 32-, and 16-fold for CHV-1/EP713, CHV-1/Euro7, and CHV-1/EP721, respectively, in Δdcl2 mutant strains. This indicates that a very robust antiviral RNA-silencing response was induced against all three viruses, even though significant differences in the levels of dcl2 transcript accumulation were observed. Unexpectedly, the severe debilitation previously reported for CHV-1/EP713-infected Δdcl2 mutant strains, and observed here for the CHV-1/Euro7-infected Δdcl2 mutant strains, was not observed with infection by CHV-1/EP721. By constructing chimeric viruses containing portions of CHV-1/EP713 and CHV-1/EP721, it was possible to map the region that is associated with the severe debilitation of the Δdcl2 mutant hosts to a 4.1-kb coding domain located in the central part of the CHV-1/EP713 genome.
INTRODUCTION
Field strains of the chestnut blight fungus Cryphonectria parasitica harboring virulence-attenuating RNA mycoviruses in the family Hypoviridae exhibit considerable variabilities in virulence levels, the magnitude of associated phenotypic traits such as reduced sporulation, and viral RNA accumulation (1, 7, 15, 16, 17, 25). Hypoviruses associated with these natural populations correspondingly exhibit significant levels of nucleotide sequence diversity (1, 7). In this regard, variabilities in virulence levels, symptom expression, and viral RNA accumulation are exhibited by three closely related hypoviruses for which infectious cDNA clones have been developed. C. parasitica strains infected with the prototypic hypovirus Cryphonectria hypovirus 1 (CHV-1)/EP713 are severely compromised in their ability to form cankers and sporulate on chestnut trees (6). They also exhibit reduced growth rates on culture media. In contrast, C. parasitica strains infected with hypovirus CHV-1/Euro7 aggressively colonize chestnut tree tissue upon inoculation, forming cankers 3- to 4-fold larger than those produced by CHV-1/EP713-infected strains before halting expansion and produce significant levels of asexual spores (5). CHV-1/Euro7-infected strains actually grow faster than the corresponding uninfected strains on artificial media. These two hypovirus isolates, which share greater than 90% identity at the amino acid level, have been referred to as severe and mild hypovirus isolates, respectively, by analogy with descriptive terms used for plant viruses. Hypovirus CHV-1/EP721 causes phenotypic changes that are indistinguishable from those caused by CHV-1/Euro7, with which it shares 99% identity at the amino acid level, but accumulates to only 20% the level of CHV-1/Euro7 and 10% the level of CHV-1/EP713, based on the level of viral double-stranded replicative-form RNA. Domain-swapping studies have identified a 2.5-kb coding region as being responsible for the low CHV-1/EP721 viral RNA accumulation (23).
We recently demonstrated that an inducible RNA-silencing pathway consisting of a single Dicer protein, DCL2, and a single Argonaute protein, AGL2, serves as an effective antiviral defense response in C. parasitica (27, 29). Transcript levels for dcl2 were found to significantly increase 10- and 15-fold following infection by hypovirus CHV-1/EP713 or Mycoreovirus 1 (MyRV1)/Cp9B21 by a mechanism that was repressed by the hypovirus suppressor of RNA silencing, p29 (26, 39). Inactivation of the C. parasitica RNA-silencing pathway by the disruption of dcl2 significantly increased susceptibility to mycovirus infection, as evidenced by a highly debilitating growth phenotype (27). Surprisingly, both dcl2 and agl2 were also found to contribute to viral RNA recombination (29, 38). The frequent production of hypovirus-derived defective interfering RNAs (DI RNAs) observed in infected C. parasitica was absent in infected Δdcl2 or Δagl2 deletion strains, and the stability of nonviral sequences in hypovirus-based viral vectors was dramatically increased in strains in which the RNA-silencing pathway was inactivated.
The extent and nature of plant virus symptom expression, movement, and accumulation are governed in large part by interactions with the plant host RNA-silencing pathways (reviewed in references 9, 13, and 14). Considerable evidence implicates virus-encoded suppressors of RNA silencing in enhancing virus replication and movement and in eliciting symptoms by virtue of the impairment of micro-RNA-regulated host developmental pathways (2, 12, 19). Here we examine whether variabilities in hypovirus-mediated symptom expression and hypovirus RNA accumulation are related to differential interactions with the host RNA-silencing antiviral defense response.
MATERIALS AND METHODS
Fungal strains and growth conditions.
C. parasitica strains were maintained on potato dextrose agar (PDA) (Difco, Detroit, MI) at 22 to 24°C on the laboratory bench top. Strains used for the preparation of RNA for real-time reverse transcriptase PCR (RT-PCR) analysis were grown for 7 days under the same conditions on PDA overlaid with a cellophane membrane. The origins and sources of wild-type C. parasitica strain EP155 (ATCC 38755), the isogenic hypovirus CHV-1/EP713-containing strain EP713 (ATCC 52571), and CHV-1/Euro7-containing strain Euro7 (ATCC 66021) have been described by Chen and Nuss (5). Hypovirus CHV-1/EP721 was identified in the Italian hypovirulent strain HI2 and then transferred by anastomosis to North American strain EP60 (ATCC 38765) isolated from Michigan (W. MacDonald, personal communication), resulting in a hypovirulent strain, EP721 (ATCC 66024). The genome sequence of CHV-1EP721 was determined, and an infectious cDNA clone, p721, was constructed by Lin et al. (23). CHV-1/EP721-containing strain EP155 (CHV-1/EP721), used for symptom comparison and dcl2 induction studies, was made by transfecting EP155 with in vitro-synthesized viral transcripts of CHV-1/EP721 cDNA clone p721, as described by Chen et al. (3). Null mutants of C. parasitica strain EP155 containing disruption of Dicer genes dcl1 (Δdcl1 strain) and dcl2 (Δdcl2 strain) have been described by Segers et al. (27). All virus-infected strains used for symptom comparison were generated by the transfection of EP155 or the Δdcl2 strain with synthetic infectious transcripts of each virus, respectively.
Construction of chimeric viruses.
Chimeras of hypoviruses CHV-1/EP713 and CHV-1/EP721 were made by swapping the domains of two cDNA clones, pRFL4 and p721AMX. Plasmid pRFL4 is an infectious cDNA clone made by Mark Craven in 1993 that contains the full-length CHV-1/EP713 cDNA preceded by the T7 polymerase promoter and flanked at each end by NotI sites, based on a modified pTZ19 vector (U.S. Biochemicals, Cleveland, OH). The full-length CHV-1/EP713 12.7-kb cDNA can be released by NotI digestion. Plasmid p721AMX is identical to the infectious cDNA clone p721 described by Lin et al. (23) for CHV-1/EP721 except that an XbaI site was introduced by point mutation at genome position 12,391 (the 3′ noncoding region). This new restriction site serves as a molecular marker for identification. Swapping of the different domains was dependent on the unique restriction enzyme sites of CHV-1/EP713 (AscI, 40 bp upstream of the viral sequence; NarI, genome coordinate 5,312; XbaI, genome coordinate 9,437) and CHV-1/EP721 (NotI, 31 bp upstream of the viral sequence; NarI, genome coordinate 5,311; BsmI, genome coordinate 9,384) and fusion PCR to rebuild the chimeric junction sequences. Detailed descriptions of the plasmid construction are available upon request.
Deletion of p29 from CHV-1/Euro7 and CHV-1/EP721.
Infectious cDNA clone pE7T-N of CHV-1/Euro7 was constructed by Chen and Nuss (5). CHV-1/Euro7 and CHV-1/EP721 mutants lacking p29 were made by PCR. The majority of the p29 coding region from nucleotide (nt) 567 to nt 1223 was deleted, but the remaining portion of p29 was still in frame, and the p29/p40 autocatalytic cleavage site remained intact. The rebuilt fragments between the NotI and NheI sites were inserted back into pE7T-N or p721AMX, resulting in the deletion mutants Euro7Δp29 and EP721Δp29.
RNA isolation and semiquantitative RT-PCR.
Total RNA was prepared from fungal cells as previously described by Deng and Nuss (11). Semiquantitative real-time RT-PCR was performed on an Applied Biosystems 7300 fast real-time PCR system using the protocol described by Suzuki and Nuss (30). Primers specific for the positive-strand viral RNA present as single-strand RNA and in replicative-form double-stranded RNA were used for first-strand cDNA synthesis. Primer-probe sets were selected to avoid amplification of viral defective interfering (DI) RNAs (38) and corresponded to genome coordinates 9049 to 9121 for CHV-1/EP713, nt 9021 to 9094 for CHV-1/Euro7, and nt 9022 to 9094 for CHV-1/EP721. Calculation of the transcript accumulation values was performed with the comparative threshold cycle method using 18S rRNA values to normalize for variations in the template concentrations.
RESULTS
Differential response of Dicer gene dcl2 expression induction to infection by different hypoviruses.
We recently reported 10- and 15-fold inductions in transcript accumulation for the RNA-silencing antiviral Dicer gene dcl2 following infection by the severe hypovirus CHV-1/EP713 and mycoreovirus MyRV/Cp9B21 (39). Given the milder symptoms and lower levels of viral RNA accumulation reported for strains infected with hypoviruses CHV-1/Euro7 and CHV-1/EP721 (5, 23), we considered the possibility that the milder symptoms and reduced viral RNA accumulation were due to a stronger induction of the RNA-silencing antiviral defense response. To test this possibility we initiated infections of C. parasitica strain EP155 with infectious transcripts of the three hypoviruses. This resulted in the relative changes in mycelial growth and colony morphology previously described (5, 23) and shown in Fig. 1A, C, E, and G.
Fig 1.
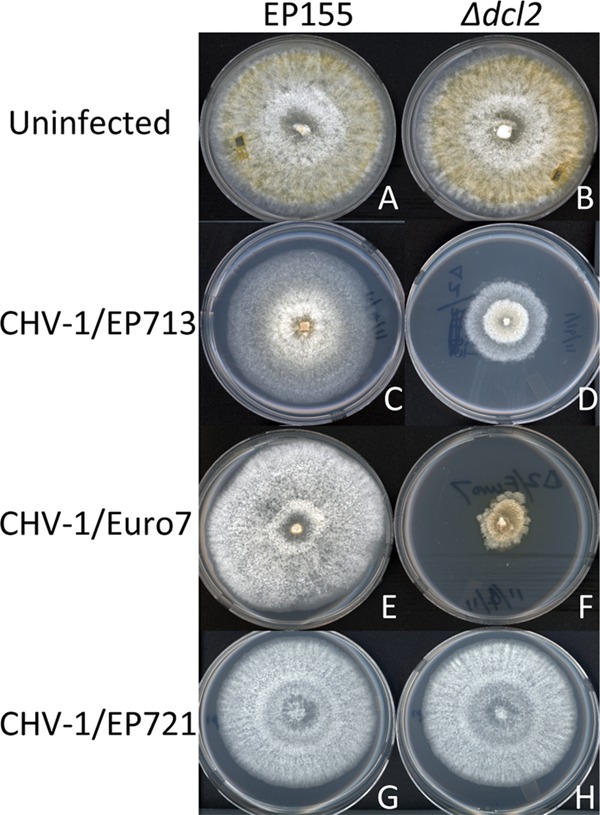
Colony morphologies of wild-type Cryphonectria parasitica strain EP155 (left column) and mutant strain Δdcl2 (right column) following infection by hypoviruses CHV-1/EP713, CHV-1/Euro7, and CHV-1/EP721. The colony morphologies for the uninfected EP155 and Δdcl2 strains are shown at the top. Infections were initiated by transfection with the transcripts of the corresponding hypoviruses (3), and infected colonies were transferred to potato dextrose agar. The photographs were taken on day 7 of the experiment.
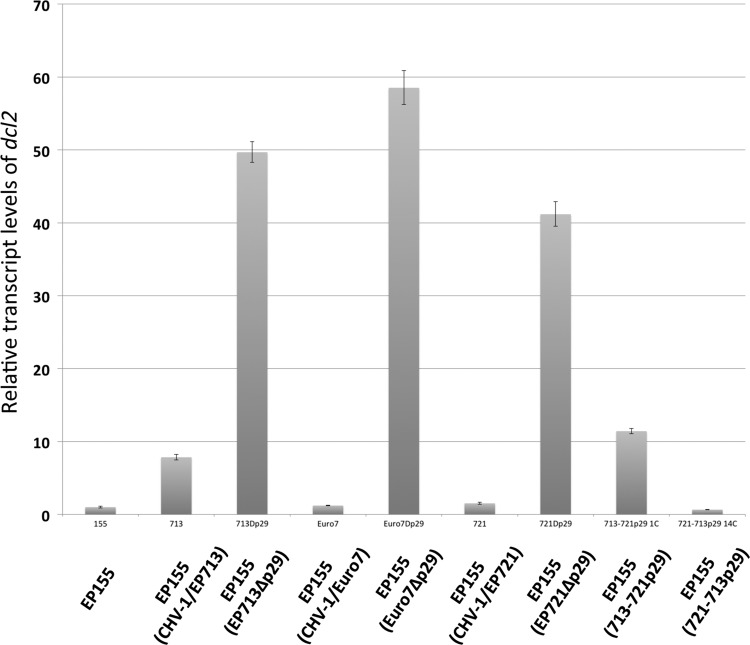
Semiquantitative real-time RT-PCR measurement of dcl2 transcript levels showed that, rather than inducing a higher level of dcl2 transcript accumulation, CHV-1/Euro7 and CHV-1/EP721 infections resulted in lower dcl2 transcript levels of 1.2- and 1.4-fold, respectively, than the >8-fold increase observed for CHV-1/EP713 infection (Fig. 2).
Fig 2.
Accumulation of dcl2 transcripts in response to different hypoviruses. The relative levels of dcl2 transcripts were measured by semiquantitative real-time RT-PCR for uninfected strain EP155 and strain EP155 infected by CHV-1/EP713, CHV-1/Euro7, CHV-1/EP721, their corresponding Δp29 mutants, and the chimeric CHV-1/EP713 and CHV-1EP721 viruses containing the swapped p29 coding domains 713/721p29 and 721/713p29, as indicated at the bottom. The values on the y axis were normalized to the dcl2 transcript level in uninfected strain EP155, set to a value of 1.
We recently reported that infection by a mutant CHV-1/EP713 virus that lacked the p29 suppressor of RNA silencing resulted in a >30-fold superinduction of dcl2 transcript accumulation (39), leading to the conclusion that p29 functions directly or indirectly to repress dcl2 transcript induction. As shown in Fig. 2, the p29 deletion mutants of CHV-1/Euro7 (Euro7Δp29) and CHV-1/EP721 (EP721Δp29) also caused a superinduction of dcl2 transcript accumulation following infection, indicating that the corresponding p29 proteins also repress dcl2 transcript accumulation. This raised the possibility that the low levels of dcl2 transcript accumulation observed for CHV-1/Euro7- and CHV-1/EP721-infected EP155 may be due to more effective suppression of dcl2 transcript accumulation by the CHV-1/Euro7- and CHV-1/EP721-encoded p29s.
To test this possibility, we constructed chimeric infectious cDNA clones in which the p29 coding regions of CHV-1/EP713 and CHV-1/EP721 were swapped. That is, we replaced the p29 coding region in CHV-1/EP713 with the CHV-1/EP721 p29 sequence to make the 713/721p29 chimeric virus and did the reverse to make the 721/713p29 chimeric virus. As shown in Fig. 2, the 713/721p29 virus caused an increase in dcl2 transcripts of ∼11-fold, similar to the increase in response to CHV-1/EP713 infection, while the 721/713p29 chimeric virus caused no increase in dcl2 transcript accumulation, similar to the observations for CHV-1/EP721. Thus, the differences in dcl2 transcript accumulation in response to CHV-1/EP713 and CHV-1/EP721 infections appear to be independent of the p29 proteins encoded by the two viruses.
Viral RNA accumulation and differential virus-mediated phenotypic changes in RNA-silencing-deficient Δdcl2 host strain.
The observation that dcl2 transcript levels increase by ∼10-fold in response to CHV-1/EP713 and an unrelated mycoreovirus (39) while showing little to no increase in response to two hypoviruses that shared greater than 90% nucleotide sequence similarity with CHV-1/EP713 was surprising and raised questions about the role of the dcl2-dependent RNA-silencing pathway as an antiviral defense against these viruses. To investigate this more closely, we introduced the three viruses into a dcl2 deletion mutant strain.
As we previously reported, the Δdcl2 strain is more susceptible to CHV-1/EP713 infection, as evidenced by the very debilitating growth phenotype shown in Fig. 1D. A similar phenotype was observed for the CHV-1/Euro7-infected Δdcl2 colony (Fig. 1F). Surprisingly, the CHV-1/EP721-infected Δdcl2 strain (Fig. 1H) exhibited a phenotype indistinguishable from that of CHV-1/EP721-infected wild-type strain EP155 (Fig. 1G). That is, CHV-1/EP721 failed to elicit the debilitating growth phenotype in the absence of an intact RNA-silencing pathway.
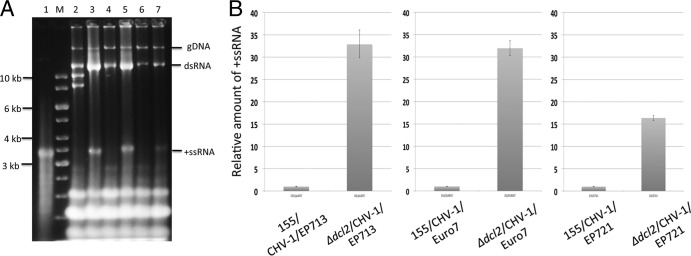
Agarose gel analysis of the viral RNA levels in the infected Δdcl2 strain showed a significant increase for CHV-1/EP713 and CHV-1/Euro7, particularly in the form of a prominent band between the 3-kb and 4-kb DNA markers that comigrates with the full-length, 12,734-nt synthetic CHV-1/EP713 coding strand transcript (Fig. 3A). Nuclease digestion and strand-specific RT-PCR (data not shown) confirmed this band to be single-stranded positive-sense viral RNA. A lower increase in the level of viral RNA accumulation was generally observed for CHV-1/EP721-infected Δdcl2 strains (Fig. 3A, lanes 6 and 7). Note that the magnitude of increases in positive-sense single-stranded RNA (ssRNA) for all three viruses in the Δdcl2 strain were underestimated by gel analysis due to the lower binding efficiency of ethidium bromide to ssRNA than to double-stranded RNA (dsRNA).
Fig 3.
Hypovirus RNA accumulation in the C. parasitica Δdcl2 mutant strain. (A). Gel analysis of RNA isolated from hypovirus-infected wild-type C. parasitica strain EP155 and the Δdcl2 mutant strain. Approximately 10 μg of total nucleic acids isolated from each infected strain was loaded and subjected to electrophoresis in 1% agarose gels set at 15 V overnight. Lane 1 contains 0.5 μg of the in vitro-synthesized 12,734-nt transcript from infectious CHV-1/EP713 cDNA clone pRFL4 (see Materials and Methods). Lane M contains DNA size markers. Lanes 2 through 7 contain nucleic acids from strain EP155 infected with CHV-1/EP713, the Δdcl2 mutant infected with CHV-1/EP713, EP155 infected with CHV-1/Euro7, the Δdcl2 mutant infected with CHV-1/Euro7, EP155 infected with CHV-1/EP721, and the Δdcl2 mutant infected with CHV-1EP721, respectively. The migration positions of genomic DNA (gDNA) and the viral double-stranded replicative-form RNA (dsRNA) and positive-sense single-stranded viral RNA (+ssRNA) are indicated on the right. The bands migrating near the 8- to 10-kb markers in lane 2 (CHV-1/EP713-infected strain EP155) are defective viral RNAs commonly observed in infected wild-type C. parasitica strains but not observed in the RNA-silencing mutant Δdcl2 strain (38). Note the significant increase in accumulation of viral positive-sense ssRNA. (B). Relative accumulation of positive-sense ssRNA hypovirus RNA in infected wild-type C. parasitica strain EP155 and Δdcl2 mutant strain determined by real-time RT-PCR. The values on the y axis were normalized to the viral RNA levels in the infected wild-type strain EP155. The relative levels of positive-sense ssRNA for CHV-1/EP713, CHV-1/Euro7, and CHV-1/EP721 increased 33-, 32-, and 16-fold in the Δdcl2 strain relative to the levels in strain EP155, respectively.
To gain a more accurate estimate of the relative increases in viral RNA in the Δdcl2 strain, semiquantitative real-time RT-PCR measurements were performed using primers designed to amplify an internal portion (73 bp for CHV-1/EP713 and CHV-1/EP721 and 74 bp for CHV-1/Euro7) of the positive-strand RNAs present in replicative-form dsRNA and as ssRNA transcripts but absent in the DI RNAs that accumulate in CHV-1/EP713-infected strain EP155. The results, presented in Fig. 3B, indicate relative increases of positive-strand RNAs in the Δdcl2 strain of 33-, 32-, and 16-fold for CHV-1/EP713, CHV-1/Euro7, and CHV-1/EP721, respectively. These increases indicate that all three hypoviruses are subject to robust DCL2-directed RNA silencing in strain EP155 and that an effective antiviral RNA-silencing response does not require a significant increase in dcl2 transcript accumulation. While the fold increase in CHV-1/EP721 RNA accumulation in the Δdcl2 mutant was generally lower than the increases for CHV-1/EP713 and CHV-1/Euro7, we did observe that the CHV-1/EP721-infected Δdcl2 strains accumulated higher levels of viral RNA after multiple passages to fresh media (data not shown) while retaining the nondebilitating phenotype. Thus, while it is conceivable that these lower levels of viral RNA accumulation could possibly contribute to the failure of CHV-1/EP721 to elicit the Δdcl2 strain-debilitating growth phenotype, a strict correlation between viral RNA accumulation and phenotype has not been observed.
Mapping of the virus-mediated Δdcl2 strain-debilitating growth phenotype to a 4.1-kb portion of the CHV-1/EP713 central coding domain.
To further investigate the phenotypic response of the Δdcl2 strain to hypovirus infection, we tested whether specific viral domains contributed to the absence of the Δdcl2 strain-debilitating growth phenotype for CHV-1/EP721 by constructing chimeric viruses from the CHV-1/EP713 and CHV-1/EP721 infectious cDNA clones. As shown in Fig. 4A through D, the CHV-1/EP713–CHV-1/EP721 chimeric viruses constructed by swapping the corresponding p29 coding regions, 713/721p29 and 721/713p29, caused phenotypic changes for the Δdcl2 strain that were indistinguishable from the parental viruses. Thus, the p29 coding region does not contribute to the difference in the Δdcl2 strain-debilitating phenotypes elicited by CHV-1/EP713 and CHV-1/EP721.
Fig 4.
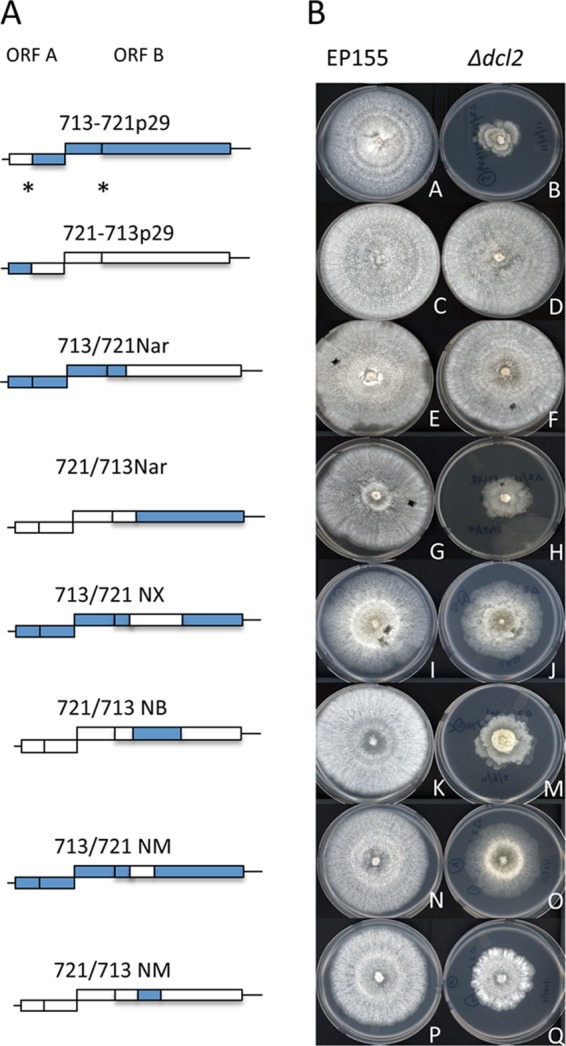
Colony morphologies conferred by CHV-1/EP713–CHV-1/EP721 chimeric viruses in wild-type strain EP155 and the Δdcl2 mutant strain. (A) Schematic diagrams of chimeric viruses. The genomes of CHV-1/EP713 and CHV-1/EP721 contain two open reading frames designated ORF A and ORF B, as indicated at the top. The coding regions of individual chimeric viruses derived from the CHV-1/EP713 genome are indicated as blue boxes, and the CHV-1/EP721 genome-derived coding regions are indicated as white boxes. The autocatalytic cleavage sites for p29 and p48 are indicated by asterisks. The 5′ and 3′ noncoding terminal regions are indicated as lines. (B) Colony morphologies conferred by CHV-1/EP713–CHV-1/EP721 chimeric viruses in wild-type strain EP155 (left column) and the Δdcl2 mutant strain (right column). Infections were initiated by transfection with transcripts of the corresponding chimeric virus (3), and infected colonies were transferred to potato dextrose agar. The photographs were taken on day 7 of the experiment.
Advantage was taken of a common NarI restriction site near genome coordinate 5300 in the infectious CHV-1/EP713 and CHV-1/EP721 cDNA clones to swap the 5′-terminal and 3′-terminal portions of the two viruses. As shown in Fig. 4F and H, chimeric virus 721/713Nar, which contains the 3′-terminal half from CHV-1/EP713, causes the debilitating growth phenotype in the Δdcl2 strain, while chimeric virus 713/721Nar, which contains the 3′-terminal half from CHV-1/EP721, does not. These results clearly demonstrate that a viral determinant is responsible for the difference in Δdcl2 strain-debilitating growth phenotypes and resides in the 3′-terminal region of the viral genome.
The next pair of chimeras, 713/721NX and 721/713NB, were constructed by exchanging a 4.1-kb fragment that extended from the NarI site near genomic coordinate 5300 to an XbaI site in CHV-1/EP721 or a BsmI site in CHV-1/EP713 near genomic coordinate 9400 (a second BsmI site near coordinate 9200 in CHV-1/EP721 was eliminated by point mutagenesis to facilitate chimeric virus construction). As shown in Fig. 4M, chimeric virus 721/713NB caused a debilitating phenotype in the Δdcl2 strain similar to that produced by chimeric virus 721/713Nar. However, chimeric virus 713/721NX caused a reduced growth phenotype compared to that caused by virus 713/721Nar but certainly not to the level exhibited by the 721/713NB-infected Δdcl2 strain (Fig. 4J). These results place the Δdcl2 strain-debilitating determinant between coordinates 5300 and 9400 in CHV-1/EP713.
Chimeric viruses 713/721NM and 721/713NM were constructed by exchanging an ∼2.5-kb fragment that extended from the NarI site (∼5300) to an MluI site near genomic coordinate 7800. Virus 713/721NM reduced Δdcl2 strain growth slightly more than that observed for virus 713/721NX, while virus 721/713NM caused less debilitation of the Δdcl2 strain than was observed for the 721/713NB virus (Fig. 4O and Q), suggesting that this 2.5-kb coding region of CHV-1/EP713 can partially confer the Δdcl2 strain-debilitating growth phenotype. Further dissection of this region failed to provide any additional mapping information.
DISCUSSION
A role for RNA silencing as an antiviral defense response in fungi was demonstrated only recently for a limited number of mycoviruses. Suppression of RNA silencing and the production of virus-derived small RNAs were reported for two Aspergillus viruses in Aspergillus nidulans (18). More extensive studies with C. parasitica reported that two very different mycoviruses, hypovirus CHV-1/EP713 and mycoreovirus MyRV1/9B21, caused very similar levels of induction in transcript accumulation for dcl2 (39), the Dicer gene involved in antiviral defense, and debilitating growth phenotypes when introduced into Δdcl2 deletion strains (27). However, the study involving the two additional hypoviruses reported here revealed unexpected variations in the transcriptional activation of the RNA-silencing pathways and in virus-mediated symptom expression in the absence of the RNA-silencing pathway. Robust levels of antiviral RNA silencing of CHV-1/Euro7 and CHV-1/EP721 were inferred in wild-type C. parasitica, as evidenced by the increase in viral RNA accumulation in the Δdcl2 strain in the apparent absence of significant induction of dcl2 transcript accumulation (Fig. 2). The increase in CHV-1/EP721 RNA accumulation in the Δdcl2 strain was not accompanied by the debilitating growth phenotype observed for CHV-1/EP713 and CHV-1/Euro7 infections. Moreover, the difference in the virus-mediated Δdcl2 strain-debilitating phenotype could be mapped to a viral coding domain. These results challenge the previous view that the Δdcl2 strain-debilitating phenotype is due simply to highly elevated levels of viral gene expression in the absence of the RNA-silencing pathway. While providing new insights into the interactions between mycoviruses and host RNA-silencing antiviral defense, the combined results also suggest a higher degree of complexity than previously anticipated.
The initial observations that dcl2 transcript levels increased 10- to 15-fold in response to infection by two very different mycoviruses, a positive single-strand RNA hypovirus and a double-strand RNA mycoreovirus (39), were consistent with the view that a robust antiviral defense response required transcriptional activation of the RNA-silencing pathway. This view is not supported by the results reported here for hypoviruses CHV-1/Euro7 and CHV-1/EP721. Both viruses are subject to RNA silencing, as indicated by the increases in viral RNA accumulation, primarily ssRNA, in the Δdcl2 strain versus wild-type strain EP155. Yet, dcl2 transcript accumulation increased only minimally (1.2- and 1.4-fold) in infected strain EP155 compared to the >8-fold increase with CHV-1/EP713 infection.
Roles for the p29 orthologues of CHV-1/Euro7 and CHV-1/EP721 in suppressing the induction of dcl2 transcript accumulation were confirmed in this study by the observed 40- to 50-fold increase in dcl2 transcript levels in response to infection by the Δp29 mutants of CHV-1/Euro7 and CHV-1/EP721, very similar to the results consistently observed for the CHV-1/EP713 Δp29 mutant (Fig. 2 and reference 39). However, p29 domain-swapping experiments clearly showed that the lower level of dcl2 transcript induction observed for CHV-1/EP721-infected strain EP155 relative to that observed for CHV-1/EP713 infection is not a function of stronger suppression by the CHV-1/EP721-encoded p29 (Fig. 2). It also does not appear to be related to any intrinsic properties of the CHV-1/EP721 and CHV-1/Euro7 RNAs, such as low levels of double-stranded structured regions, since dcl2 transcript superinduction occurs in the absence of the silencing suppressor in response to the Δp29 mutant CHV-1/Euro7 and CHV-1/EP721 viruses (Fig. 2).
In large part, plant virus-encoded suppressors of RNA silencing contribute to symptom expression by interfering with host micro-RNA-directed developmental regulation (22, 35). Minor isolate-specific or mutational amino acid differences can alter suppressor activity and cause differences in associated symptom expression (20, 21, 28, 34, 36, 37, 40). The hypovirus-encoded suppressor p29 has been shown to contribute to virus-mediated suppression of host pigment production and asexual sporulation. The expression of p29 in the absence of virus infection results in reduced pigmentation and sporulation (8, 31) compared to those in the wild-type virus, and replication-competent viruses lacking the p29 coding domain exhibit increased pigmentation and sporulation compared to those in the wild-type virus (8, 31, 33). However, p29-mediated symptom expression is unlikely to involve disruption of the micro-RNA pathways, since evidence for functional micro-RNAs has not been obtained for filamentous fungi (reviewed in reference 10), and disruption of the RNA-silencing pathway is asymptomatic in the absence of virus infection (27). In this regard, a series of studies with chimeric CHV-1 hypoviruses in wild-type and Δdcl2 mutant strains have failed to find contributions of the corresponding p29 silencing suppressor coding domains to hypovirus-specific symptom expression either in the presence (4, 5, 23) or in the absence (Fig. 4A through D) of an intact RNA-silencing pathway.
We recently reported that dcl2 transcript levels increase over 50-fold in response to the production of hairpin dsRNAs (29), which is similar in magnitude to the increase in dcl2 transcript accumulation observed in response to the three hypovirus Δp29 mutant viruses (see Fig. 2 and reference 39). If CHV-1/Euro7 and CHV-1/EP721 RNAs are effectively silenced, as indicated by the differences in viral RNA accumulation in the wild-type and Δdcl2 strains (Fig. 3), with minimal induction of the dcl2 transcripts (Fig. 2), then it is unclear why C. parasitica responds to hairpin RNA or Δp29 mutant viruses with such a large dcl2 transcriptional response. Additional studies are clearly required to fully understand the relationship between the transcriptional induction of the RNA-silencing pathway and the magnitude of the antiviral defense response and the mechanisms that underlie dcl2 transcriptional induction.
The observation that CHV-1/EP721 causes indistinguishable symptoms in the presence or absence of an intact RNA–silencing pathway was surprising. Given the large increase in viral coding strand RNA observed for CHV-1/EP713-infected Δdcl2 strains (Fig. 3A), a leading explanation for the associated debilitating phenotype is that concomitant overexpression of viral proteins leads to increased symptom expression. In this regard, we now routinely use the Δdcl2 mutant-infected strain as an experimental tool because of the increased accumulation of CHV-1/EP713 proteins (X. Zhang and D. L. Nuss, unpublished observation). However, two observations suggest that the Δdcl2– strain-debilitating phenotype is not related simply to increased accumulation of viral RNA and general viral protein overexpression but also involves contributions from the expression of specific virus-coding domains. As judged by gel analysis and semiquantitative real-time RT-PCR measurements, CHV-1/EP721 RNA, both the replicative form and the single-stranded RNA, increased in the infected Δdcl2 strain without negatively affecting colony growth relative to that in the infected wild-type strain. This increase occurred in both newly transfected Δdcl2 strains and more so in CHV-1/EP721-infected Δdcl2 strains after multiple passages (data not shown). However, since this increase was less than the increases observed for CHV-1/EP713 and CHV-1/Euro7, the possibility remains that the level of CHV-1/EP721 RNA accumulation was insufficient to elicit the debilitating growth phenotype. More telling is that the construction of the CHV-1/EP713–CHV-1/EP721 chimeric viruses allowed the mapping of the difference in the Δdcl2 strain-debilitating phenotype to a 4.1-kb coding domain extending from CHV-1/EP713 genomic coordinates 5311 to 9437. Interestingly, this region coincides with the portion of open reading frame B (ORF B) implicated in differences in the modulation of host cyclic AMP-dependent signaling by CHV-1/EP713 and CHV-1/Euro7 (24) and overlaps the region (genomic coordinates 5311 to 7843) mapped as contributing to differences in hypovirus RNA accumulation for CHV-1/EP721 and CHV-1/Euro7 (23). The relationship between the multiple response differences elicited by this region of ORF B are intriguing and merit further investigation.
Suzuki et al. (32) demonstrated that hypoviruses could be used as a platform for constructing replication-competent vectors for the expression of foreign genes. However, the utility of these vectors was restricted by the rapid deletion of the foreign gene sequences through RNA recombination. The demonstration by Zhang and Nuss (38) that RNA silencing contributed to viral RNA recombination subsequently provided a solution to the problem of vector instability through the use of Δdcl2 strains for the stable propagation of hypovirus vectors. Unfortunately, the debilitating growth phenotype exhibited by the CHV-1/EP713-based expression vectors limited the potential yields of the expressed foreign proteins. The finding that CHV-1/EP721 infection of Δdcl2 strains fails to cause the debilitating growth phenotype provides the opportunity for using CHV-1/EP721-based vectors for the development of stable high-yield fungal expression systems.
ACKNOWLEDGMENTS
This work was supported in part by Public Health Service grants GM555981 and AI076219 (to D.L.N.).
We thank Baoshan Chen and Haiyan Lin at Guangxi University, China, for providing the infectious CHV1-EP721 cDNA clones.
Footnotes
Published ahead of print 19 September 2012
REFERENCES
- 1. Allemann C, Hoegger P, Heiniger U, Rigling D. 1999. Genetic variation of Cryphonectria hypoviruses (CHV1) in Europe, assessed using restriction fragment length polymorphism (RFLP) markers. Mol. Ecol. 8:843–854 [DOI] [PubMed] [Google Scholar]
- 2. Chapman EJ, Prokhnevsky AI, Gopinath K, Dolja VV, Carrington CJ. 2004. Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step. Genes Dev. 18:1179–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen B, Choi GH, Nuss DL. 1994. Attenuation of fungal virulence by synthetic infectious hypovirus transcripts. Science 264:1762–1764 [DOI] [PubMed] [Google Scholar]
- 4. Chen B, Geletka LM, Nuss DL. 2000. Using chimeric hypoviruses to fine-tune the interaction between a pathogenic fungus and its plant host. J. Virol. 74:7562–7567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen B, Nuss DL. 1999. Infectious cDNA clone of hypovirus CHV1-Euro7: a comparative virology approach to investigate virus-mediated hypovirulence of the chestnut blight fungus Cryphonectria parasitica. J. Virol. 73:985–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Choi GH, Nuss DL. 1992. Hypovirulence of chestnut blight fungus conferred by an infectious viral cDNA. Science 257:800–803 [DOI] [PubMed] [Google Scholar]
- 7. Chung P, Bedker PJ, Hillman BI. 1994. Diversity of Cryphonectria parasitica hypovirulence-associated double-stranded RNAs within a chestnut population in New Jersey. Phytopathology 84:984–990 [Google Scholar]
- 8. Craven MG, Pawlyk DM, Choi GH, Nuss DL. 1993. Papain-like protease p29 as a symptom determinant encoded by a hypovirulence-associated virus of the chestnut blight fungus. J. Virol. 67:6513–6521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Csorba T, Pantaleo V, Burgyan J. 2009. RNA silencing: an antiviral mechanism. Adv. Virus Res. 75:35–71 [DOI] [PubMed] [Google Scholar]
- 10. Dang Y, Qiuying Y, Zhihong X, Liu Y. 2011. RNA interference in fungi: pathways, functions, and applications. Eukaryot. Cell 10:1148–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Deng F, Nuss DL. 2008. Hypovirus papain-like protease p48 is required for initiation but not for maintenance of virus RNA propagation in the chestnut blight fungus Cryphonectria parasitica. J. Virol. 82:6369–6378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Díaz-Pendón JA, Ding SW. 2008. Direct and indirect roles of viral suppressors of RNA silencing in pathogenesis. Annu. Rev. Phytopathol. 46:303–326 [DOI] [PubMed] [Google Scholar]
- 13. Ding SW. 2010. RNA-based antiviral immunity. Nat. Rev. Immunnol. 10:632–644 [DOI] [PubMed] [Google Scholar]
- 14. Ding SW, Voinnet O. 2007. Antiviral immunity directed by small RNAs. Cell 130:413–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Elliston JE. 1978. Pathogenicity and sporulation in normal and diseased strains of Endothia parasitica in American chestnut, p 95–100 In MacDonald WL, Cech FC, Luchok J, Smith C. (ed), Proceedings of the American Chestnut Symposium West Virginia University Press, Morgantown, WV [Google Scholar]
- 16. Elliston JE. 1985. Characterization of dsRNA-free and ds-RNA-containing strains of Endothia parasitica in relation to hypovirulence. Phytopathology 75:151–158 [Google Scholar]
- 17. Enebak SA, MacDonald WL, Hillman BI. 1994. Effect of dsRNA associated with isolates of Cryphonectria parasitica from the central Appalachians and their relatedness to other dsRNAs from North America and Europe. Phytopathology 84:528–534 [Google Scholar]
- 18. Hammond TM, Andrewski MD, Roossinck MJ, Keller NP. 2008. Aspergillus mycoviruses are targets and suppressors of RNA silencing. Eukaryot. Cell 7:350–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kasschau KD, et al. 2003. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev. Cell 4:205–217 [DOI] [PubMed] [Google Scholar]
- 20. Kozlowska-Makulska A, et al. 2010. P0 proteins of European beet-infecting poleroviruses display variable RNA silencing suppression activity. J. Gen. Virol. 91:1082–1091 [DOI] [PubMed] [Google Scholar]
- 21. Lewsey M, Robertson FC, Canto T, Palukaitis P, Carr JP. 2007. Selective targeting of miRNA-regulated plant development by viral countersilencing protein. Plant J. 50:240–252 [DOI] [PubMed] [Google Scholar]
- 22. Li F, Ding SW. 2006. Virus counterdefense: diverse strategies for evading the RNA-silencing immunity. Annu. Rev. Microbiol. 60:503–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lin H, et al. 2007. Genome sequence, full-length infectious cDNA clone, and mapping of viral double-stranded RNA accumulation determinant of hypovirus CHV1-EP721. J. Virol. 81:1813–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Parsley TB, Chen B, Geletka LM, Nuss DL. 2002. Differential modulation of cellular signaling pathways by mild and severe hypovirus strains. Eukaryot. Cell 1:401–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Peever TL, Liu Milgroom Y-CMG. 1998. Incidence and diversity of hypoviruses and other double-stranded RNAs occurring in the chestnut blight fungus Cryphonectria parasitica, in China and Japan. Phytopathology 88:811–817 [DOI] [PubMed] [Google Scholar]
- 26. Segers GC, Van Wezel R, Zhang X, Hong Y, Nuss DL. 2006. Hypovirus papain-like protease p29 suppresses RNA silencing in the natural fungal host and in a heterologous plant system. Eukaryot. Cell 5:896–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Segers GC, Zhang X, Deng F, Sun Q, Nuss DL. 2007. Evidence that RNA silencing as an antiviral defense mechanism in fungi. Proc. Natl. Acad. Sci. U. S. A. 104:12902–12906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Senshu M, et al. 2009. Variability in the level of RNA silencing suppression caused by triple gene block protein 1 (TGBp1) from various potexviruses during infection. J. Gen. Virol. 90:1014–1024 [DOI] [PubMed] [Google Scholar]
- 29. Sun Q, Choi GH, Nuss DL. 2009. A single Argonaute gene is required for induction of RNA silencing antiviral defense and promotes vial recombination. Proc. Natl. Acad. Sci. U. S. A. 106:17927–17932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Suzuki N, Nuss DL. 2002. Contribution of protein p40 to hypovirus-mediated modulation of fungal phenotype and viral RNA accumulation. J. Virol. 76:7747–7759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Suzuki N, Chen B, Nuss DL. 1999. Mapping of a hypovirus p29 protease symptom determinant domain with sequence similarity to potyvirus HC-Pro protease. J. Virol. 73:9478–9484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Suzuki N, Geletka LM, Nuss DL. 2000. Essential and dispensible virus-encoded replication elements revealed by efforts to develop hypoviruses as gene expression vectors. J. Virol. 74:7568–7577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Suzuki N, Maruyama K, Moriyama M, Nuss DL. 2003. Hypovirus papain-like protease p29 functions in trans to enhance viral double-stranded RNA accumulation and vertical transmission. J. Virol. 77:11697–11707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Torres-Barceló C, Martin S, Daros JA, Elena SF. 2008. From hypo- to hypersuppression: effect of amino acid substitutions on the RNA-silencing suppressor activity of the tobacco etch potyvirus HC-Pro. Genetics 180:1039–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Voinnet O. 2005. Induction and suppression of RNA silencing: insights from viral infections. Nat. Rev. Genet. 6:206–220 [DOI] [PubMed] [Google Scholar]
- 36. Voinnet O, Lederer C, Baulcombe DC. 2000. A viral movement protein prevents spread of gene silencing signal in Nicotiana benthamiana. Cell 103:157–167 [DOI] [PubMed] [Google Scholar]
- 37. Wu HW, Lin SS, Chen KC, Yeh SD, Chua NH. 2010. Discriminating mutations of HC-Pro of Zucchini yellow mosaic virus with differential effects on small RNA pathways involved in viral pathogenicity and symptom development. Mol. Plant Microbe Interact. 23:17–28 [DOI] [PubMed] [Google Scholar]
- 38. Zhang X, Nuss DL. 2008. A host dicer is required for defective RNA production and recombinant virus vector RNA instability for a positive sense RNA virus. Proc. Natl. Acad. Sci. U. S. A. 105:16749–16754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang X, Segers GC, Sun Q, Deng F, Nuss DL. 2008. Characterization of hypovirus-derived small RNAs generated in the chestnut blight fungus by an inducible DCL-2-dependent pathway. J. Virol. 82:2613–2619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang X, et al. 2006. Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute cleavage activity to counter plant defense. Genes Dev. 20:3255–3268 [DOI] [PMC free article] [PubMed] [Google Scholar]