Abstract
HIV-1 replicates poorly in macaque cells, and this had hindered the advancement of relevant nonhuman primate model systems for HIV-1 infection and pathogenesis. Several host restriction factors have been identified that contribute to this species-specific restriction to HIV-1 replication, but these do not fully explain the poor replication of most strains of HIV-1 in macaque cells. Only select HIV-1 envelope variants, typically those derived from viruses that have been adapted in cell culture, result in infectious chimeric SIVs encoding HIV-1 envelope (SHIVs). Here we demonstrate that most circulating HIV-1 variants obtained directly from infected individuals soon after virus acquisition do not efficiently mediate entry using the macaque CD4 receptor. The infectivity of these viruses is ca. 20- to 50-fold lower with the rhesus and pig-tailed macaque versus the human CD4 receptor. In contrast, culture-derived HIV-1 envelope variants that facilitate efficient replication in macaques showed similar infectivity with macaque and human CD4 receptors (within ∼2-fold). The ability of an envelope to mediate entry using macaque CD4 correlated with its ability to mediate entry of cells expressing low levels of the human CD4 receptor and with soluble CD4 sensitivity. Species-specific differences in the functional capacity of the CD4 receptor to mediate entry mapped to a single amino acid difference at position 39 that is under strong positive selection, suggesting that the evolution of CD4 may have been influenced by its function as a viral receptor. These results also suggest that N39 in human CD4 may be a critical residue for interaction of transmitted HIV-1 variants. These studies provide important insights into virus-host cell interactions that have hindered the development of relevant nonhuman primate models for HIV-1 infection and provide possible markers, such as sCD4 sensitivity, to identify potential HIV-1 variants that could be exploited for development of better SHIV/macaque model systems.
INTRODUCTION
Rhesus (rh) and pig-tailed (pt) macaques are commonly used as model systems to study HIV-1 infection. However, there are multiple restrictions to HIV-1 replication in macaques that are effectively antagonized by SIV proteins but not by their HIV-1 counterparts. Chimeric SIV/HIV-1 proviruses (SHIVs) that encode the relevant SIV antagonists replicate in macaque cells, and such SHIVs are important models of HIV-1 infection and pathogenesis and are used to evaluate vaccine approaches. However, developing SHIVs based on specific HIV-1 sequences has been a hit-or-miss proposition due to considerable variation in replication of the resulting viruses in macaque cells. The sequences encoding the envelope protein (Env) are a particularly important component of SHIVs because Env is a highly antigenic protein that facilitates entry, and thus it is a critical target for vaccine and prevention approaches that focus on inhibiting virus entry. However, the current SHIVs poorly represent the envelopes characteristic of viruses that are spreading in the population, including the dominant HIV-1 subtypes that are fueling the HIV-1 pandemic.
The initial SHIVs were constructed using variants from chronic or later stages of HIV-1 infection, most of which were lab-adapted variants that used the CXCR4 coreceptor (27, 30, 36, 49, 53, 54). Because most HIV-1 variants use CCR5 as a coreceptor for entry, particularly those that establish a new HIV-1 infection, more recent SHIVs have been constructed using CCR5-tropic (R5) HIV-1 envelope sequences (7, 20, 23, 24, 36, 42, 43, 45, 55, 56). The R5 SHIVs were also constructed from viruses isolated in culture from chronically infected individuals, with the exception of two SHIVs that encode envelope variants derived from recently infected infants (55, 56). There are no SHIVs derived from HIV-1 variants obtained directly from individuals soon after sexual infection, even though sexual transmission is the major mode of HIV-1 spread. This limits the utility of this model system for studies of transmission and prevention.
Another limitation of most of existing SHIVs is that they encode subtype B HIV-1 envelope sequences, which account for only ∼10% of worldwide infections (21). Given the extensive sequence and antigenic diversity between subtypes, it is unclear whether the results obtained with SHIV-Bs will be generalizable across subtypes (28). Subtypes A, C, and D are most prevalent in sub-Saharan Africa, which carries the highest burden of new HIV-1 infections and HIV-1 related deaths (21). There are SHIVs that encode subtype C envelope sequences (7, 32, 34, 42, 55, 56), as well as an SHIV encoding envelope sequences from a circulating recombinant CRF_AE, virus, a common HIV-1 subtype in Southeast Asia (23). However, SHIVs that include subtype A and D determinants have not been successfully generated.
Initial attempts to generate subtype A-based SHIVs (SHIV-As) that replicated in macaque cells were unsuccessful (22). Subsequent studies showed that several subtype A variants were unable to mediate entry using the pt macaque CD4 (ptCD4) receptor. The culturing of virus encoding subtype A envelope in pt macaque cells resulted in evolution of two adapted viruses with single amino acid changes in the extracellular envelope protein, gp120, i.e., mutations A204E and G312V, that increased the efficiency of entry into cells expressing macaque CD4 (26).
The basis for the functional difference in macaque and human CD4 receptors is unknown. The N-terminal immunoglobulin-like domain (D1) of human CD4 is the primary determinant of HIV-1 Env-CD4 interactions (reviewed in reference 6). The early mutagenesis and binding studies implicating D1 of CD4 were supported by later structural analyses, which identified F43 and R59 as particularly important HIV-1 contact residues (29). The critical residues for HIV-1 envelope binding to macaque CD4 have not specifically been defined. There are 12 amino acid differences in the D1 domain between macaque and human CD4, including an R-to-K change at position 59 that could impact HIV-1 envelope interaction.
Early studies suggested that the macaque CD4 protein was a functional HIV-1 receptor, including both ptCD4 (17) and rhCD4 (8). However, these studies all focused on envelope variants from subtype B-infected individuals at chronic stages of infection derived after virus amplification culture (HIV LAI [17] and HIV-1 ADA and JR-F [8]). In contrast, the studies suggesting that macaque CD4 was a suboptimal HIV-1 receptor focused on several subtype A envelope sequences obtained without culture amplification, four of five of which were from individuals in acute/early infection (26). Thus, the basis for differences observed in macaque CD4-mediated entry by HIV-1 Env used in various studies could be due to biological differences between viruses of different subtypes and/or from different stages of infection, both of which are important variables for designing relevant SHIV models. Here, we examined the ability of subtype A, B, C, and D envelope variants obtained early after HIV-1 acquisition to mediate infection of cells using macaque CD4. We found that most of these Envs showed limited ability to facilitate infection using the macaque CD4 receptor. The CD4 determinant responsible for differences in the ability of the macaque and human CD4 to function as an HIV-1 receptor was identified.
MATERIALS AND METHODS
Cells.
HEK 293T cells, Cf2Th/syn CCR5 cells, JC.24 cells, and RC.49 cells were maintained in Dulbecco modified Eagle medium (Invitrogen) supplemented with 10% heat-inactivated fetal calf serum (FCS) and 2 mM l-glutamine. Cf2Th/syn CCR5 cells, which are dog thymocytes engineered to express high levels of codon optimized human CCR5 (38), were further supplemented with 400 μg of Geneticin (Gibco)/ml to maintain CCR5 expression. The JC.24 and RC.49 cell lines, which are HeLa-derived cells lines that express comparable levels of CCR5 but different amounts (∼40-fold) of CD4 (47), are referred to throughout as CD4HIGH and CD4LOW cells.
Envelope clones and mutagenesis.
The envelope clones used in the present study included the following: 12 subtype B HIV-1 env clones from early infection (RHPA4259.7, REJO4541.67, SC422661.8, WITO4160.33, TRO.11, CAAN5342.A2, QH0692.42, AC10.0.29, THRO4156.18, TRJO4551.58, PVO.4, and 6535.3 [31]), 13 subtype C HIV-1 env clones from early infection (ZM249M.PL1, Du156.12, ZM135M.PL10a, ZM214M.PL15, ZM53M.PB12, ZM233M.PB6, CAP45.2.00.G3, CAP210.2.00.E8, Du172.17, Du422.1, ZM109F.PB4, and ZM197M.PB7 [31, 32] and QC406.70 M.ENV.F3 [3]), 10 subtype A HIV-1 env clones from early infection (QB726.70 M.ENV.B3, QF495.23 M.ENV.A3, QG984.21 M.ENV.A3, QH343.21 M.ENV.A10, and QH359.21 M.ENV.C1 [3] and Q259.d2.17, Q168b23, Q461e2, Q769h5, and Q842d16 [35]), 4 subtype D HIV-1 env clones from early infection (QA013.70I.ENV.H1, QA465.59 M.ENV.A1, QB857.110I.ENV.B3, and QD435.100 M.ENV.B5 [3]), and 7 env clones representing variants that are known to infect macaque cells (BaL.01 [34], YU-2 [33], SF162 [9], SF162P3 [24], and 89.6 [10], SIV MneCL8 [44, 46], and Q23-17 A204E and Q23-17 G312V [26]).
Nucleotide changes encoding the A204E and G312V mutations were introduced into the following env's: QC406.70 M.ENV.F3 (called QC406F3 here), QA013.70I.ENV.H1 (QA013H1), QB857.110I.ENV.B3 (QB857B3), and QD435.100 M.ENV.B5 (QD435B5) using standard methods similar to those described previously (26). The subsequent envelope mutants were sequenced through the entirety of the env open reading frame to verify that no undesired nucleotide changes had occurred.
Preparation of green fluorescent protein (GFP) reporter pseudoviruses.
GFP reporter pseudoviruses were generated in 293T cells. At 24 h prior to transfection, 293T cells were plated at 5 × 105 cells/well in a six-well dish. The cells were cotransfected with 667 ng of Q23Δenv-GFP (26) and 333 ng of the Env clone of interest using Fugene 6 (Roche) at a ratio of 3 μl of Fugene to 1 μg of DNA according to the manufacturer's instructions. Pseudoviruses were harvested 72 h after transfection, centrifuged at 1,300 rpm for 5 min to clear cellular debris, divided into aliquots, and frozen at −80°C until use. To estimate the virus titer, 2 to 10 μl of thawed viral supernatant was used to infect Cf2Th/syn CCR5 huCD4 cells, and the cells were analyzed for GFP expression as described below. The percentage of GFP-positive cells was used to estimate the infecting MOI (e.g., 10% GFP positive Cf2Th/syn CCR5 huCD4 cells corresponds to a multiplicity of infection (MOI) of 0.1, and the MOI was used to estimate the virus titer based on the volume applied to the cells.
Generation of stable cells expressing human, rhesus, and pig-tailed macaque CD4 variants.
The expression plasmids encoding CD4 receptors have been described previously: human CD4 (huCD4), pig-tailed macaque CD4 (ptCD4) (26), and rhesus macaque CD4 (rhCD4) (48). These plasmids were used as templates to amplify the CD4 open reading frame using forward primer 5′-GATGTCGACATGAACCGGGGAGTCCC-3′ with reverse primer 5′-GGTCTCGAGTCAAATGGGGCTACATG-3′ for huCD4 and forward primer 5′- GATGTCGACATGAACCGGGGAATCCC-3′ with the same reverse primer for ptCD4 and rhCD4 using TaqPlus Precision using methods similar to those described previously (26). The resulting PCR products were digested with SalI and XhoI (the restriction sites are underlined in the primers), cloned into pLXSH (an MLV-based retroviral vector encoding a hygromycin resistance gene) (37), and verified by sequencing.
Virus-like particles (VLPs) were generated by cotransfecting the pLXSH vector encoding the CD4 of interest, an MLV-based retroviral packaging vector (pJK3 [2]) and a plasmid expressing the vesicular stomatitis virus G protein under the cytomegalovirus promoter (pMD.G [41]) at a ratio of 1:1:0.1. Supernatants were harvested at 48 h posttransfection, filtered through a 0.22-μm-pore-size filter, and used immediately to infect CF2Th/syn CCR5 cells that had been plated 24 h prior in a T75 flask at 105 cells. The supernatant with VLP (ca. 4 to 5 ml) was added in the presence of 10 μg of DEAE dextran/ml at 37°C, followed by incubation for 3 h before bringing the medium volume up to 10 ml. This procedure was repeated on the same cells the next day with the VLP supernatant harvested from transfected cells at 72 h. The following day, cells were split 1:2 into new flasks and allowed to recover in the absence of drug selection for 24 h. The medium was then supplemented with 400 μg of Geneticin/ml (to maintain CCR5 expression) and 300 μg of hygromycin B (Invitrogen)/ml to select for cells expressing CD4. The cultures were maintained in these conditions, changing the medium every 3 to 4 days until cultures that had not been exposed to VLPs were completely clear of cells (typically 10 to 14 days). The cells that survived drug selection were maintained thereafter with 150 μg of hygromycin B/ml.
Cells that had high levels of CD4 were obtained by sorting the surviving transduced cells on a FACSAria II cell sorter (BD Biosciences) using a CD4 monoclonal antibody that cross-reacts with macaque and human CD4 (BD BioSciences [unpublished data]). Briefly, 1 × 106 to 3 × 106 cells were treated with 5 mM EDTA to remove them from the tissue culture flask and washed in phosphate-buffered saline (PBS) containing 2% fetal bovine serum (FBS). The cells were resuspended in 200 μl of PBS–2% FBS, followed by incubation with 4 μl of allophycocyanin-conjugated mouse anti-human CD4 antibody (BD Biosciences) at room temperature for 30 min. The cells were then washed again in PBS–2% FBS, resuspended to a concentration of 106 cells/ml in PBS–1% FBS–1 mM EDTA, and filtered through a 35-μm-pore-size nylon mesh cap (BD Falcon) before sorting. The cells with the highest levels of CD4 expression, representing the top 20 to 30% of CD4-expressing cells, were retained and used for infection assays.
Infection and analysis of Cf2Th/syn CCR5-based cells and CD4HIGH/LOW cells.
Cf2Th/syn CCR5 cells stably expressing CD4 were plated 24 h prior to infection at 2.5 × 104 cells/well in 24-well tissue culture plates in 500 μl of drug-free medium. The cells were infected in duplicate wells in the presence of 10 μg of DEAE-dextran/ml by spinoculation for 60 to 90 min at 1,200 × g at an estimated MOI of 0.1 in 100 μl of medium. After 48 h, the cells were washed in cold PBS, incubated with 100 μl of 5 mM EDTA until they lost adherence to the well, and fixed in 500 μl of 1% paraformaldehyde. The fixed cells were transferred to flow cytometry tubes (BD Falcon) that were centrifuged at 800 × g for 5 min. The excess fix was decanted, and the cells were analyzed for GFP expression on a BD FACSCalibur flow cytometer. The data from ∼104 live cells, as determined by side-scatter and forward-scatter profiles, were analyzed by using FlowJo version 9.4.6. Cells whose GFP expression was >200 fluorescence units—a gating that captured only 0.01 to 0.02% of uninfected cells—were gated as GFP positive.
Infections and analysis of the CD4HIGH and CD4LOW cells were performed as described for the Cf2Th/syn CCR5 cells except that 4 × 104 cells were plated on the day prior to infection, 25 mM EDTA was used to remove the cells from the tissue culture well, and only ∼5 × 103 live cells were analyzed for GFP expression.
Construction of chimeric CD4 and CD4 mutants.
NheI restriction digest sites, which are present within the vector and within the CD4 ORF at codon 176 in the mature protein, were used to generate chimeric CD4 molecules encoding D1 and D2 of the huCD4 receptor in the context of the ptCD4 receptor (huD1D2). The resulting clone was verified by sequence analysis to ensure that the fragment had been cloned in the proper orientation.
Chimeric ptCD4 sequences containing the huCD4 D1, N-terminal D1 and C-terminal D1 domains were constructed using a two-step overlap extension PCR approach described previously (11) with some changes. In the first round, the huCD4 fragment of interest, as well as regions of ptCD4 overlapping with the huCD4 fragment and comprising the remainder of the ptCD4 ORF, was amplified by PCR from 25 ng of plasmid encoding huCD4 or ptCD4 (26) for 25 cycles. Up to 4 μl of each gel-purified product was pooled together for use as a template in the second round. Similarly, the second-round amplification was performed for 25 cycles with the forward primer 5′- GATGGATCCATGAACCGGGGAATCCC-3′ and the reverse primer 5′-GGTGTCGACTCAAATGGGGCTACATG-3′, except in the case of the chimeric ptCD4 expressing the C terminus of the huCD4 D1 domain, in which 5′-GATGGATCCATGAACCGGGGAGTCCC-3′ was used as a forward primer. The amplicons were purified using a QIAquick PCR purification kit, digested with BamHI and SalI (restriction sites are underlined in the primers), gel purified, and ligated into the pBabe-Puro expression vector (40).
Site-directed mutagenesis was performed in huCD4 and ptCD4 using the overlap extension protocol described above. All primers used to make and CD4 mutants are available upon request.
Infection of cells transiently expressing CD4 and CCR5.
Infection of cells that transiently expressed the CD4 and CCR5 were performed as described previously (26). Briefly, 293T cells were transfected with CD4 and CCR5 expression plasmids in six-well dishes using Fugene 6 transfection reagent. After 40 h, the cells were removed from the dish using 5 mM EDTA and plated at 80,000 cells/well in 500 μl in a 24-well dish, with 2 × 105 to 3 × 105 cells kept for analysis of CD4 and CCR5 expression by flow cytometry. At 4 to 5 h after the cells were plated, infections were performed in duplicate by spinoculation in the presence of 10 μg of DEAE-dextran/ml with dilutions of GFP pseudovirus. The cells were fixed with cold PBS–1% formaldehyde–0.2% glutaraldehyde 72 h after infection, and the GFP-positive cells were counted by eye to determine the number of infected cells.
Data presentation and analysis.
All data were plotted, and Mann-Whitney U tests, Wilcoxon signed-rank tests, and Spearman correlations were performed where indicated using Prism version 5.0a (GraphPad Software).
RESULTS
Infection of huCD4, ptCD4, and rhCD4 cell lines with diverse SHIV, SIV, and HIV-1 Env variants.
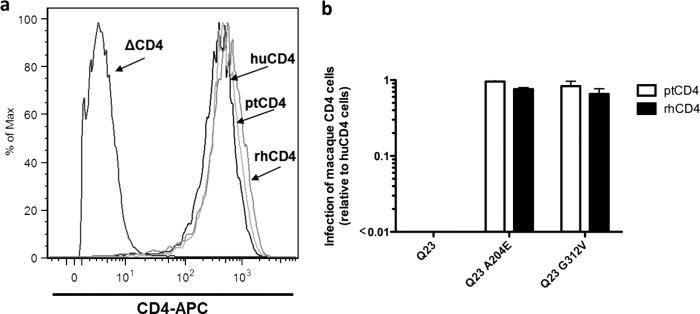
To examine the ability of different primate CD4s to mediate HIV entry, Cf2Th/syn CCR5 cells (38) expressing human (hu), pig-tailed (pt), or rhesus (rh) CD4 were infected with GFP-reporter viruses derived from the subtype A Q23-17 envelope with known differences in ptCD4 receptor usage (26). In a prior study, we showed that the inability of HIV-1 subtype A envelope variants to infect cells expressing macaque CD4 was associated with poor replication of the corresponding virus in pig-tailed macaque lymphocytes (26). CD4 surface expression was comparable for the three Cf2Th/syn CCR5 cell lines expressing the different CD4 receptors (Fig. 1a). The wild-type Q23-17 pseudovirus showed ∼100-fold reduction in infectivity in cells expressing ptCD4 compared to huCD4 (Fig. 1b), which in consistent with results of our previous study (26). Pseudoviruses generated with variants of Q23-17 that encoded A204E or G312V infected cells expressing ptCD4 with high efficiency comparable to the levels seen in cells expressing huCD4 (0.95- and 0.83-fold infection for ptmac versus huCD4 for A204E and G312V, respectively, Fig. 1b). Similar results were observed for the rhCD4 receptor (Fig. 1b), which was not tested in the prior study (26), demonstrating that the rhCD4 is also a suboptimal receptor for the prototype primary subtype A variant.
Fig 1.
Infection of Cf2Th/syn CCR5 cells that stably express hu-, pt-, and rhCD4 with GFP reporter pseudoviruses expressing subtype A Q23-17 Env variants. (a) Flow cytometric analysis. Expression of hu-, pt-, and rhCD4 (indicated by arrows) on the surface of stably transduced CF2Th/syn CCR5. ΔCD4 refers to the parental Cf2Th/syn CCR5 cell line, which does not express CD4. (b) Infection of ptCD4 cells (white bars) and rhCD4 cells (black bars) relative to huCD4 cells with GFP pseudoviruses expressing the Q23-17, Q23-17 A204E and Q23-17 G312V Env variants. The virus tested is denoted on the x axis. The y axis shows the ratio of GFP-positive cells expressing ptCD4 or rhCD4 relative to GFP-positive cells expressing huCD4. A ratio of 1 indicates equal infection of cells expressing the macaque and human CD4 receptors with the indicated virus. Error bars represent the standard deviation of the mean from two independent experiments.
SHIV and SIV Envs from viruses that have previously been shown to replicate in macaque lymphocytes—HIV 89.6, BaL, SF162, SF162p3, and YU2 (19, 36, 45, 49, 59) and SIVMneCl8 (44)—were tested in the CF2Th/syn CD4 cell panel. The ability of the SIV/SHIV Envs to mediate infection of macaque CD4 cells ranged from 0.30-fold (for 89.6) to 0.81-fold (for YU-2) in cells expressing ptCD4 versus huCD4 and from 0.36-fold (for SIVMneCl8) to 0.89-fold (for SF162p3) in rhCD4 cells (Fig. 2a). The median relative infection among SIV/SHIV strains was 0.61-fold for cells expressing ptCD4 cells and 0.71-fold for cells expressing rhCD4 compared to cells expressing huCD4.
Fig 2.
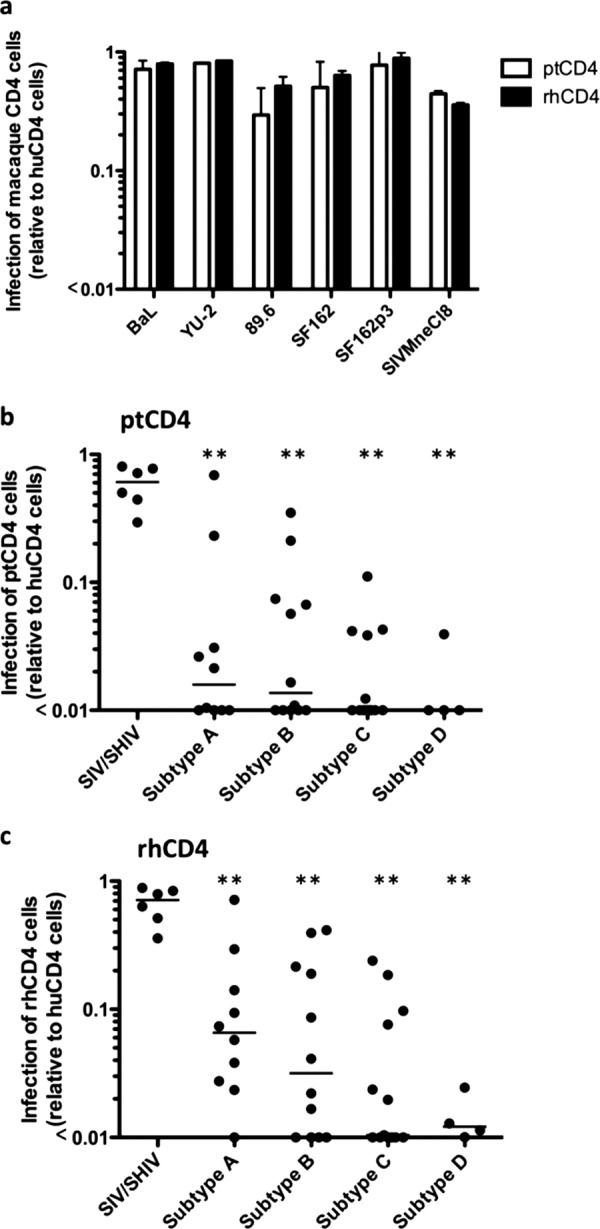
Infection of cells expressing the macaque CD4 receptor with GFP reporter pseudoviruses expressing Envs from diverse HIV-1 and SIV strains. Infection of ptCD4 cells and rhCD4 cells are shown relative to huCD4 cells on the y axis in all three display panels. (a) Infection of cells expressing ptCD4 (white bars) and rhCD4 cells (black bars) with GFP pseudoviruses expressing Envs from SHIV/SIV envelope variants denoted on the x axis. Error bars represent the standard deviation of the mean from two independent experiments. B and C) Acute/early virus infection of cells expressing ptCD4 (b) and rhCD4 (c) relative to cells expressing huCD4. The subtype of the envelope of the GFP reporter pseudoviruses tested is indicated on the x axis. Each point on the plot represents the mean infectivity of a given virus obtained from duplicate independent experiments, and the line represents the median measurement for each subtype. The SHIV/SIV Envs are the same as shown in panel A, shown here for reference. **, P < 0.01 compared to SIV/SHIV Envs (Mann-Whitney U test).
To define the potential of HIV-1 variants more relevant to the global pandemic, viruses expressing Envs from HIV-1 sequences obtained directly from infected individuals during the acute/early stage of their infection were examined for their ability to infect cells expressing ptCD4 and rhCD4. Of the 39 acute/early-stage Envs tested, 34 (>87%) showed >10-fold lower infectivity in cells expressing ptCD4 cells relative to cells expressing huCD4 (Fig. 2b). The median relative infection of ptCD4 cells for each of the four subtypes was significantly lower than SIV/SHIV Envs (P < 0.01 for all subtypes [Mann-Whitney U test]). Among the 13% that demonstrated the highest infectivity with ptCD4, two were subtype A, two were subtype B, and one was subtype C. One subtype A virus (Q842d16) infected cells expressing ptCD4 at levels comparable to those seen with HIV envelopes from infectious SHIV proviruses.
Viruses expressing acute/early Envs from all subtypes also showed decreased infectivity of rhCD4 cells compared to the SIV/SHIV strains (Fig. 2c, P < 0.01 for all subtypes [Mann-Whitney U test]), with 29 of the 39 acute/early Envs tested (>74%) having a >10-fold decrease in their ability to infect rhCD4 cells compared to huCD4 cells. In general, viruses expressing acute/early Env variants had higher infectivity in cells expressing rhCD4 compared to cells expressing ptCD4 (median, 0.049-fold versus 0.018-fold relative to huCD4, P = 0.0003 by Wilcoxon signed-rank test). Despite the statistically significant differences in infection between the rhCD4 and ptCD4 receptors, the magnitude of the difference was small, and neither macaque receptor was optimal as an entry receptor compared to huCD4.
Overall, there were few or no differences in macaque CD4-mediated entry among the subtypes. There were no significant differences in ptCD4-mediated entry between subtypes (P > 0.23 for each comparison [Mann-Whitney U test]). The median infection of ptCD4 cells relative to huCD4 among subtypes ranged from <0.01-fold for subtypes C and D to 0.02-fold for subtype A. In the case of rhCD4-mediated infection, the median ratio of infection for rhCD4/huCD4 for each subtype ranged from 0.01-fold for subtype C to 0.07-fold for subtype A strains. The difference in median infection between subtypes A and D was statistically significant (P = 0.0337 [Mann-Whitney U test]); however, all other differences between subtypes were not statistically significant (P > 0.08 for other comparisons [Mann-Whitney U test]).
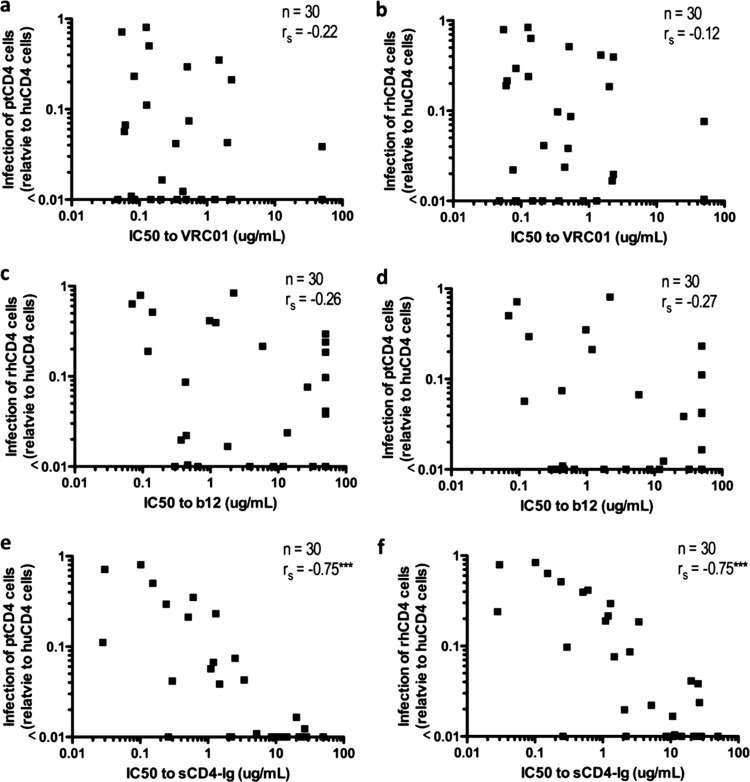
Association between entry using macaque CD4 and sensitivity to b12, VRC01, and soluble CD4.
An increase in sensitivity to soluble CD4 (sCD4) was a characteristic of Env variants that were derived by adapting subtype A HIV-1 to replicate in macaque cells (26). To determine whether sensitivity to sCD4 (human) predicts whether an envelope variant can mediate entry using the macaque CD4 receptors, we took advantage of an extensive collection of neutralization data published by Wu et al. (60) using viruses that overlapped the virus panels used in the present study. Fifty percent inhibitory concentrations (IC50s) were available for sCD4 and for two monoclonal antibodies that target the CD4 binding site—b12 (4, 51) and VRC01 (60)—for 30 of the 45 Envs that were examined as part of the present study, including both the SHIV Envs and the acute/early HIV Envs. When the IC50 values were compared to infectivity data from Fig. 2 for the 30 overlapping viruses, there was no significant association between neutralization by b12 or VRC01 and the relative infection of ptCD4 cells or rhCD4 cells (Fig. 3a to d). However, there was a significant association between sensitivity to sCD4-Ig and the relative infection of both ptCD4 cells (Fig. 3e, P < 0.0001) and rhCD4 cells (Fig. 3f, P < 0.0001), suggesting that sensitivity to human sCD4 is a marker of the ability of a variant to use the macaque CD4 receptor.
Fig 3.
Relationship between infection of cells expressing macaque CD4 and sensitivity to neutralization by CD4 binding site-directed monoclonal antibodies and sCD4. The association with the infection of ptCD4 cells (a, c, and e) and the infection of rhCD4 cells (b, d, and f) was determined using infectivity data from Fig. 2 and neutralization IC50 values, as described previously (60). Each dot represents a variant tested for infection of cells expressing macaque CD4 in Fig. 2. rs, Spearman rank correlation coefficient. ***, P < 0.0001 (Spearman correlation).
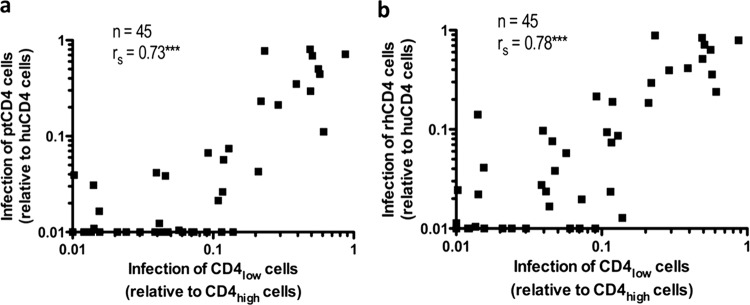
Association between macaque CD4-mediated entry and the ability to infect cells expressing low levels of human CD4.
Given that macaque CD4 usage was correlated with sensitivity to sCD4, we also examined whether it was correlated with the ability to utilize low levels of cell surface human CD4, another property that is attributed to increased CD4 binding site exposure (12–15, 18, 52, 58). For this purpose, the SIV/SHIV Envs and acute/early Envs were examined for their ability to mediate the infection of cells expressing low levels of CD4 relative to cells expressing high levels of CD4 (CD4LOW and CD4HIGH cells). There was a strong association between both ptCD4 and rhCD4 usage (defined as a ratio of infectivity for cells expressing the macaque versus human CD4 receptor [Fig. 2]) and the ability to use low levels of huCD4 (P < 0.0001 [Spearman correlation]; Fig. 4). Viruses that effectively utilized macaque CD4 for entry (i.e., had infectivity ptCD4/huCD4 or rhCD4/huCD4 ratios of close to 1) were also able to infect CD4lLOW cells at levels that were similar to CD4HIGH cells (i.e., had CD4LOW/CD4HIGH ratios close to 1).
Fig 4.
Relationship between infection of cells expressing macaque CD4 and infection of cells expressing low levels of human CD4. The association between infection of cells expressing ptCD4 (a) or rhCD4 (b) and cells expressing low levels of human CD4 was examined. The CD4LOW/CD4HIGH infectivity, which represents the ratio of infection of a given virus from cells expressing low levels of huCD4 versus cells expressing high levels of huCD4 (47), was compared to the data from Fig. 2 for each virus, and rs represents the Spearman rank correlation coefficient. ***, P < 0.0001 (Spearman correlation).
Effect of amino acid changes within and outside of the CD4 binding site on macaque CD4-mediated entry.
A number of naturally occurring polymorphisms in Env that flank the CD4 binding site, including D279 (58), N283 (14), and N362 (57), have previously been shown to be associated with the ability to use low levels of CD4 to mediate infection. Given the strong correlation between infection of CD4LOW cells and entry using macaque CD4, the acute/early Envs were examined for an association between the presence of these polymorphisms and their ability to infect cells expressing the macaque versus human CD4 receptors. The presence of N283 was significantly associated with increased infection of rhCD4 cells (P = 0.0439 [Mann-Whitney U test]) but not ptCD4 cells (Table 1). The presence of D279 and N362 were not associated with the ability of a virus to mediate entry by macaque CD4, as measured by the number of infected cells expressing the macaque receptor compared to cells expressing the human receptor for the viruses tested (Table 1).
Table 1.
Association of D279, N362, and N283 polymorphisms in Env with the relative efficiency of entry using macaque versus human CD4 receptors
| Presence of polymorphisma | nb | Infectionc |
|||
|---|---|---|---|---|---|
| ptCD4 |
rhCD4 |
||||
| Median (range) | P | Median (range) | P | ||
| D279 | 0.7188 | 0.3749 | |||
| + | 12 | 0.03 (0.01–0.11) | 0.02 (0.01–0.24) | ||
| – | 31 | 0.01 (0.01–0.69) | 0.01 (0.01–0.71) | ||
| N283 | 0.2391 | 0.0439 | |||
| + | 10 | 0.03 (0.01–0.59) | 0.12 (0.01–0.57) | ||
| – | 33 | 0.01 (0.01–0.69) | 0.02 (0.01–0.71) | ||
| N362 | 0.3162 | 0.7902 | |||
| + | 17 | 0.01 (0.01–0.35) | 0.05 (0.01–0.41) | ||
| – | 26 | 0.01 (0.01–0.69) | 0.02 (0.01–0.71) | ||
Indicates the presence (+) or absence (–) of the indicated polymorphism.
n, Number of Envs with or without the indicated polymorphism.
Infection of cells expressing macaque CD4 relative to cells expressing huCD4. P values were calculated using a Mann-Whitney U test comparing the medians.
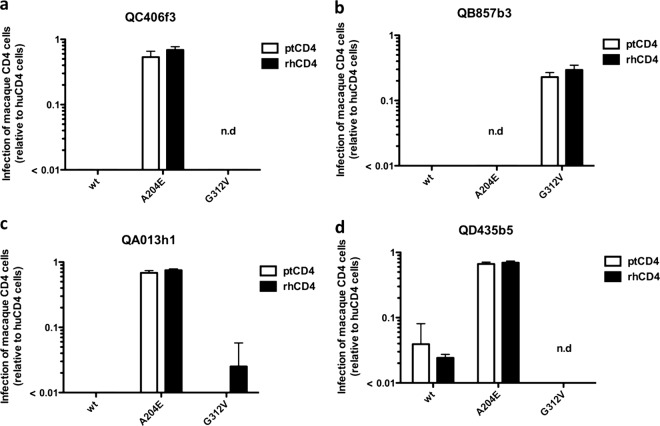
Changes in envelope at positions A204E and G312V in subtype A HIV-1 increase the level of infection in cells expressing the ptCD4 receptor to levels similar to cells expressing the huCD4 receptor (26). To determine whether the A204E and G312V amino acid changes conferred the same phenotype to envelopes from the other major global subtypes, we introduced these two changes individually into 3 subtype D Envs (QB857b3, QA013h1, and QD435b5) and one subtype C Env (Q406f3). The introduction of the A204E and G312V changes to the subtype C and D Envs had variable effects, including leading to decreased infectivity of cells expressing the huCD4 receptor for the QC406f3 G312V, QB857b3 A204E, and QD435b5 G312V variants, which were not tested further. For the Env variants that retained their infectivity using the huCD4 receptor, there was an increase in infectivity for cells expressing macaque CD4. The effect was most pronounced for subtype D QA013h1 A204E variant, where there was a >80-fold increase relative to wild-type virus in rhCD4 cells, with slightly lower increases for the other two viruses tested (Fig. 5). The G312V change had variable effects; in the case of subtype D QB857b3, introduction of the G312V change increased infection in ptCD4 and rhCD4 cells by approximately 20- and 30-fold, respectively. However, the G312V change had very little impact on the ability of another subtype D Env, QA013h1, to use either macaque CD4 receptor (<2-fold).
Fig 5.
Infection of cells expressing macaque CD4 with pseudoviruses expressing subtype C and D A204E and G312V Env variants. The infection of ptCD4 cells (white bars) and rhCD4 cells (black bars) relative to huCD4 cells with GFP pseudoviruses was evaluated. The envelope variant tested, which includes wild-type (wt) and the corresponding A204E and G312V Env variants, is indicated on the x axis for four different viral envelopes as follows: subtype C, QC406f3 (a); subtype D, Q857b3 (b); subtype D variant, QA013h1 (c); and subtype D variant, QD435b5 (d). Error bars represent the standard deviation of the mean from two independent experiments. n.d., not determined due to low infectivity of the mutant virus.
Species-specific determinants of CD4-mediated entry by HIV-1.
There are 21 amino acid differences between the D1 and D2 domains of human and macaque CD4, 12 within D1 (Fig. 6), and the ptCD4 and rhCD4 amino acid sequences are identical in these regions. Of note, a commonly used reference sequence for rhCD4 (GenBank accession no. NM_001042662) differed from the sequence of the rhCD4 clone we used here (16). We therefore sequenced the third and fourth exons, which encode the entire D1 and D2 domains, from 15 unrelated Indian rhesus macaques and 15 unrelated Chinese rhesus macaques. All 30 animals had identical sequences that matched the sequence of the clone used here (shown in Fig. 6a), demonstrating that the rhesus CD4 used in the functional studies represents the dominant allele in the population.
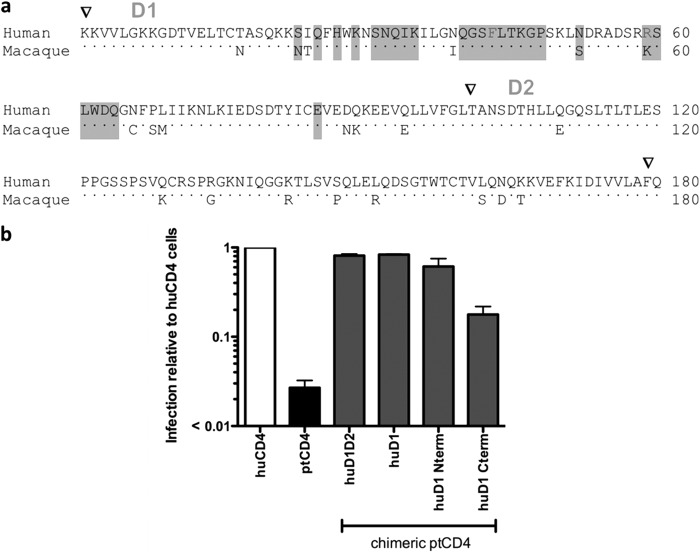
Fig 6.
Identification of subdomains in huCD4 sufficient for infection mediated by HIV-1. (a) Amino acid alignment of the D1 and D2 domains of human and macaque CD4. Arrowheads above the alignment denote the beginning and end of the D1 and D2 domains. Dots indicate conserved residues, and positions where macaque and human sequence differ are shown as letters. The residues that are highlighted in gray have previously been implicated in gp120 binding (1) and include F43 and R59 (underlined) that are predicted to form hydrogen bonds with HxBc2 gp120 (29). The black arrow indicates the approximate boundary (the overlap PCR covered a conserved region corresponding to amino acids 43 to 52) in the huD1 Nterm and huD1 Cterm chimeras. (b) Results of infection of 293T cells transiently expressing huCCR5 and huCD4, ptCD4, or chimeric ptCD4 variants with GFP reporter pseudoviruses. The prototype subtype A Q23-17 Env was used for these studies. The y axis shows the ratio of GFP-positive cells expressing ptCD4 or rhCD4 relative to GFP-positive cells expressing huCD4. A ratio of 1 indicates equal infection of cells expressing the chimeric and huCD4 receptors with the indicated virus. Error bars represent the standard deviation of the mean from duplicate independent experiments.
To investigate which of the amino acid differences between huCD4 and ptCD4 were most critical for mediating infection by circulating HIV-1 strains, chimeric CD4s were constructed by introducing huCD4-specific residues to ptCD4, and these were examined for gain of function using a prototype subtype A Q23-17 Env, which shows limited infectivity with ptCD4 (Fig. 6b). Chimeric CD4s containing the human D1-D2 region (huD1D2) and D1 region (huD1) mediated infection by a pseudovirus expressing the Q23-17 Env, as well as the huCD4 (Fig. 6b). This finding suggests that the determinants for differences in the function of the macaque and human CD4 receptors are within D1.
Additional chimeric CD4s encoding ptCD4 and huCD4 residues in the N-terminal of D1 or the C-terminal of D1 were generated. A chimeric ptCD4 encoding the N terminus of huD1 (huD1Nterm; Fig. 6b) mediated HIV-1 entry nearly as well as huCD4 and ∼50-fold higher than ptCD4. The ptCD4 encoding the C terminus of huD1 also was a better HIV-1 receptor than ptCD4- by ∼ 10-fold. However, entry was still ∼ 6-fold lower than for huCD4 (Fig. 6b). These findings indicate that the residues most critical for entry by HIV-1 Q23-17 reside in the N terminus of the D1 domain of CD4 (which includes T17, S23, I24, and N39 in the human receptor); however, there also may be some influence of residues in the C terminus of the D1 domain on HIV-1 entry.
To further investigate which of the amino acid changes in the N terminus of D1 was sufficient to allow infection by HIV-1, ptCD4 mutants encoding the individual T17, S23, I24, and N39 changes were generated and tested. The N17, S23, and I24 changes to ptCD4 did not increase the infectivity of HIV-1 relative to ptCD4. However, the introduction of an N at position 39 in the context of the ptCD4 receptor increased infection to levels that were comparable to huCD4 (Fig. 7a). Thus, the I39N change in ptCD4 was sufficient to allow infection by virus expressing the prototype subtype A Env.
Fig 7.
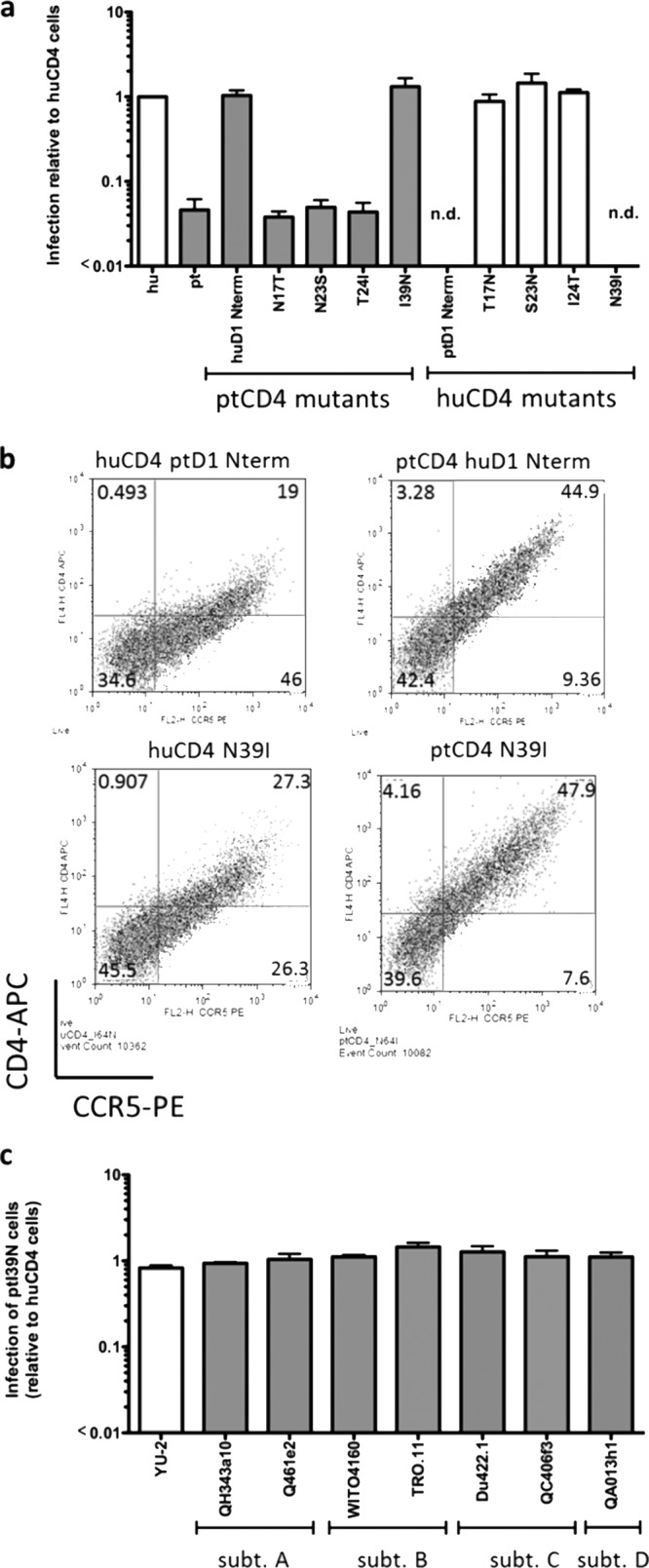
Effect of single residue changes in CD4 on HIV-1 infection. (a) Infection of 293T cells transiently expressing huCCR5 and huCD4, ptCD4, or huCD4 and ptCD4 point mutants with GFP reporter pseudoviruses bearing the Q23-17 Env. The y axis represents viral infection relative to huCD4. Error bars represent the standard deviation of the mean from duplicate independent experiments. n.d., not determined due to insufficient CD4 expression. (b) Flow cytometric analysis of 293T cells transiently expressing huCCR5 and huCD4 or ptCD4 receptors. The y axis represents CD4 expression, and the x axis represents CCR5 expression as determined by flow cytometry. The CD4 receptor that is expressed in the cells is labeled at the top of each plot. These plots are representative of at least three independent experiments. (c) Infection of Cf2Th/syn CCR5 cells stably expressing ptCD4 I39N relative to huCD4-expressing cells with GFP pseudoviruses expressing the YU-2 Env and acute/early Envs from subtypes A to D (as denoted on the x axis). Error bars represent the standard deviation of the mean from two independent experiments.
Reciprocal mutations encoding all four of the amino acid changes described above were introduced into huCD4 to determine whether these residues were necessary to allow infection by HIV-1. The N17T, N23S, and T24I changes all showed activity similar to the huCD4 receptor, further indicating that none of these amino acids plays a significant role as determinants of HIV-1 infection (Fig. 7a). Unfortunately, both huCD4 expressing the N terminus of ptD1 and huCD4 N39I were expressed poorly on the surfaces of cells (Fig. 7b), thus precluding analysis of their receptor function. Thus, while our studies demonstrate that residue N39 is sufficient when introduced into ptCD4 to allow for infection mediated by subtype A HIV-1 Env, its absolute necessity for mediating infection could not be conclusively determined.
To determine whether ptCD4 encoding the I39N change increased infection by other circulating HIV-1 strains, Cf2Th/syn CCR5 cells were engineered to stably express ptCD4 I39N. Infection by viruses carrying subtype A, B, C, and D Envs that showed limited entry using wild-type ptCD4 (<0.01-fold relative infection; Fig. 2) was increased by >90 to 120-fold in ptCD4 I39N cells (Fig. 7c). Thus, the introduction of the I39N amino acid change to ptCD4 is sufficient to allow for increased infection by a wide range of circulating HIV-1 strains from early infection.
DISCUSSION
In this study, we explored the CD4 determinants for HIV-1 infection in the commonly used nonhuman primate models, pig-tailed and rhesus macaques. We found that unlike lab-adapted strains of HIV-1 from chronic infection, most circulating HIV-1 variants derived from recent human infections are impaired in their ability to infect cells using the macaque CD4 receptor. HIV-1 viral envelope variants obtained soon after transmission, including representatives of the major circulating subtypes (A to D) typically showed a 1- to 2-log reduction in entry mediated by macaque CD4 compared to human CD4. A few natural variants were identified that utilized the macaque CD4 receptor with efficiency more comparable to the human CD4 receptor, comparable to envelope variants from existing SHIVs. Our studies suggest that a key predictor of an envelope's ability to engage the macaque CD4 receptor is sensitivity to soluble CD4, thus providing a potential means to screen for HIV-1 envelopes that can be used to construct SHIVs capable of replicating in macaques. Importantly, a single amino acid at position 39 in CD4 was identified that is responsible for species-specific differences in the capacity of the CD4 receptor to facilitate entry of HIV-1. This amino acid is under positive selection (61), suggesting the possibility that the ability of CD4 to function as a viral receptor may be one factor that influenced its evolution.
These studies help explain why it has been so difficult to develop relevant SHIV/macaque models of HIV-1 infection. Many of the current SHIVs were developed using lab-adapted CXCR4-tropic envelope variants and/or envelope variants from viruses obtained from the chronic stages of HIV-1 infection isolated after passage in culture. Here, we show that such SHIVs can enter cells using the macaque CD4 receptor more efficiently than variants derived directly from infected individuals soon after HIV-1 acquisition. This finding provides insight into novel CD4 determinants necessary to support infection by acute/early HIV-1 strains and suggests that lab-adapted and chronic-stage HIV-1 Envs may have significantly altered mechanisms of interaction with CD4 compared to the Envs of recently acquired viruses. Given that early-stage variants from all of the subtypes tested, including subtype B variants, showed poor infectivity with macaque CD4 suggests that differences observed in the function of the macaque CD4 receptor in HIV-1 infection in prior studies (8, 17, 26) does not reflect differences in the viral subtype used to generate the SHIVs but rather differences in either the effect of viral amplification in culture or biological differences between early and later-stage HIV-1 viruses.
The early-stage subtype A, B, C, and D variants examined here represent the types of viruses that are spreading globally and thus the variants that are most critical to study in transmission and prevention models. Importantly, we have identified key characteristics of viruses that are best able to engage the macaque CD4 receptor sensitivity to soluble CD4 and the ability to infect cells expressing low levels of human CD4, two biological properties that often go hand in hand (12, 57, 58). The use of these phenotypic markers may permit a more strategic approach to developing SHIV/macaque models based on circulating, recently transmitted HIV-1 variants from diverse subtypes.
The basis for the species-specific differences in CD4 receptor function was mapped to a single amino acid change from N to I at position 39 (of the mature CD4 protein) in macaque CD4. The introduction of asparagine at position 39 in ptCD4 increased infection >30-fold by viruses carrying acute/early HIV-1 Envs from all subtypes tested. Structural analyses suggest that amino acid N39 is in close proximity to the F43 or R59 residues in huCD4 that are thought to form hydrogen bonds with gp120 to stabilize binding (described in reference 29 and depicted in Fig. 8). Residue 39 of CD4 has been found to be under positive selection (61), suggesting the intriguing possibility that the function of CD4 as a viral receptor may have contributed to its evolution. It is possible that other residues that are under positive selection that were not directly examined in the present study, such as S52 or C65, may have coevolved with I39 to allow proper folding and/or expression of macaque CD4. In support of this model, we found that huCD4 encoding the reciprocal N39I change did not express well on the cell surface, suggesting that other amino acid differences in macaque versus human CD4 are needed to compensate for the isoleucine at position 39. This model is further supported by the fact that chimeric ptCD4 encoding the C-terminal portion of huD1, which includes the S52 and C65 residues, conferred a partial increase in entry by an early-stage HIV-1 variant.
Fig 8.
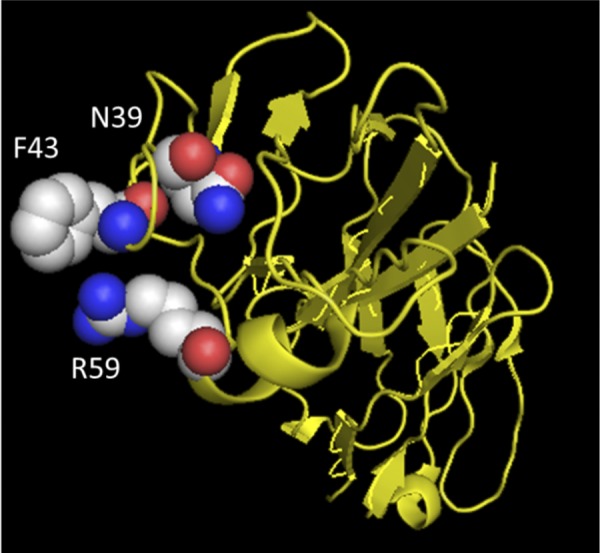
Location of N39 in relation to other amino acid residues in CD4 important for HIV-1 envelope-CD4 interaction. Residues F43 and R59 (29) and N39 are shown in space-filling mode, while the rest of CD4 is shown as a yellow ribbon diagram. The crystal structure coordinates were obtained from Huang et al. (25).
The critical contact positions between human CD4 and Env were identified in structural studies using a lab-adapted, CXCR4 envelope protein, HXBc2 (29). A SHIV containing the HXBc2 envelope replicates to high levels in macaque peripheral blood mononuclear cells (50), suggesting that macaque CD4 functions as a receptor for this lab-adapted subtype B envelope. This is consistent with our findings demonstrating Envs encoded by macaque-infectious SHIVs, such as YU2, BaL, SF162, and 89.6, can engage the macaque CD4 receptor for entry. These results indicate that such envelopes—all of which were cloned from virus amplified in culture—do not require interaction with N39 in the same manner as the circulating envelope variants obtained directly from infected individuals soon after their infection. This difference may explain why the N39 amino acid was not identified in prior structural studies using the lab-adapted HxBc2 envelope that defined the points of interaction between HIV-1 envelope and human CD4. Our findings suggest that the N39 position may be an important residue for CD4 interaction of circulating, transmitted HIV-1 variants, a hypothesis that could be tested by examining the structure of envelope from an early-stage virus in complex with human CD4.
Viruses expressing acute/early Envs displayed a modest increase in infection in using rhCD4 versus ptCD4 receptors, despite the fact that rhCD4 and ptCD4 are identical in critical regions for gp120 binding, including having the isoleucine at position 39. There have been previous studies identifying amino acid changes outside of the D1 and D2 domains of CD4 that can modulate fusion after binding (5, 39); thus, it is possible that the sole amino acid difference between rhCD4 and ptCD4 at position 324 in the D4 domain may modulate fusion by HIV-1 Env.
To better understand the basis for differences in macaque CD4-mediated entry among HIV-1 variants, we also explored determinants in the HIV-1 envelope that contribute to increased infectivity using the macaque CD4 receptor. In prior studies, we identified mutations at A204E and G312V that increase macaque CD4 tropism in the context of the subtype A envelope (26). We show here that these same changes can also increase macaque CD4-mediated entry of subtypes C and D, suggesting that the amino acid positions impact CD4 interactions in diverse envelope contexts. The effect of the A204E mutation was more pronounced than that seen with G312V, with increased infectivity of 17- to 80-fold of A204E variants versus 2- to 30-fold for G312V variants compared to the corresponding parental virus in cells expressing the macaque CD4 receptor. In some subtype C and D envelope clones, introduction of G312V or A204E amino acid changes resulted in decreased envelope function, as evidenced by substantially decreased infectivity of these viruses in human cells compared to the wild-type virus. Nonetheless, we identified both subtype C and subtype D variants that achieved comparable infection levels with the macaque and human CD4. The infectivity of these viruses was similar to viruses carrying the HIV-1 envelopes from SHIVs that are infectious and pathogenic in macaques, such as Bal, and SF162P3 (36, 45). Thus, these Envs may be good candidates for the development of subtype C and D SHIVs.
Naturally occurring polymorphisms flanking the CD4 binding site at amino acids 279, 283, and 362 have previously been found to be associated with the ability to use low levels of CD4 (14, 57, 58), a property we found to be highly correlated with macaque CD4 usage. Only amino acid 283 showed any correlation with the efficiency of macaque CD4-mediated entry and only with rhCD4. Neither amino acid position 279 nor amino acid position 362 impacted the usage of macaque CD4. In contrast to previous studies (14, 57, 58), we did not observe a statistically significant association between the presence of these polymorphisms and infection of CD4LOW cells (not shown). One reason for this difference may be that these polymorphisms were originally identified in late-stage subtype B variants (14, 57, 58), and it is possible that there are other Env determinants that are required to further modulate the influence of these polymorphisms that are not present in acute/early Envs from other subtypes (12).
SHIV/macaque models currently play central roles in studies of HIV transmission and pathogenesis, and they have been used extensively to evaluate vaccine approaches and other prevention methods. However, the current SHIV models do not reflect HIV-1 variants circulating globally and thus do not fully recapitulate the viral factors that contribute to the infection dynamics being studied. We show here that the major global subtypes do not efficiently mediate entry using the macaque CD4 receptor, and this is the result of just one amino acid difference between the macaque and human receptors. We also show that some natural variants and engineered viruses do replicate to high levels in cells expressing the macaque receptor, paving the way for use of such envelopes to develop more relevant SHIV models. Importantly, we show that the property of sensitivity to sCD4 provides a means of identifying HIV-1 Envs with increased capacity to use macaque CD4, which may allow for the further identification of Envs for use in the construction of SHIVs.
ACKNOWLEDGMENTS
This study was supported by NIH grant R01 AI38518 to J.O.
We thank members of the lab and Michael Emerman and Ron Diskin for helpful discussions. We thank the NIH AIDS Reagents program and the scientists who donated the envelope clones used in this study (J. Mascola, C. Derdeyn, E. Hunter, L. Morris, K. Mlisana, D. Montefiori, F. Gao, M. Li, S. Abdool Karim, G. Ramjee, C. Williamson, B. Hahn, J. Salazar-Gonzalez, D. Kothe, X. Wei, G. Shaw, Y. Li, and R. Collman). We also thank C. Cheng-Mayer for the SF162P3, D. Miller for pLXSH, M. Emerman for the pjK3, E. Platt, and D. Kabat for the JC24 and RC49 cells, and T. Mirzabekov and J. Sodroski (via the NIH AIDS Reagents Program) for the CF2Th/syn CCR5 cells. We thank Betsy Ferguson and the OHSU macaque DNA bank for providing samples for CD4 sequence analyses.
Footnotes
Published ahead of print 12 September 2012
REFERENCES
- 1. Arthos J, et al. 1989. Identification of the residues in human CD4 critical for the binding of HIV. Cell 57:469–481 [DOI] [PubMed] [Google Scholar]
- 2. Bartz SR, Vodicka MA. 1997. Production of high-titer human immunodeficiency virus type 1 pseudotyped with vesicular stomatitis virus glycoprotein. Methods 12:337–342 [DOI] [PubMed] [Google Scholar]
- 3. Blish CA, et al. 2009. Cross-subtype neutralization sensitivity despite monoclonal antibody resistance among early subtype A, C, and D envelope variants of human immunodeficiency virus type 1. J. Virol. 83:7783–7788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Burton DR, et al. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266:1024–1027 [DOI] [PubMed] [Google Scholar]
- 5. Camerini D, Seed B. 1990. A CD4 domain important for HIV-mediated syncytium formation lies outside the virus binding site. Cell 60:747–754 [DOI] [PubMed] [Google Scholar]
- 6. Capon DJ, Ward RH. 1991. The CD4-gp120 interaction and AIDS pathogenesis. Annu. Rev. Immunol. 9:649–678 [DOI] [PubMed] [Google Scholar]
- 7. Chen Z, et al. 2000. Enhanced infectivity of an R5-tropic simian/human immunodeficiency virus carrying human immunodeficiency virus type 1 subtype C envelope after serial passages in pig-tailed macaques (Macaca nemestrina). J. Virol. 74:6501–6510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen Z, Zhou P, Ho DD, Landau NT, Marx PA. 1997. Genetically divergent strains of simian immunodeficiency virus use CCR5 as a coreceptor for entry. J. Virol. 71:2705–2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cheng-Mayer C, Weiss C, Seto D, Levy JA. 1989. Isolates of human immunodeficiency virus type 1 from the brain may constitute a special group of the AIDS virus. Proc. Natl. Acad. Sci. U. S. A. 86:8575–8579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Collman R, et al. 1992. An infectious molecular clone of an unusual macrophage-tropic and highly cytopathic strain of human immunodeficiency virus type 1. J. Virol. 66:7517–7521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Deminie CA, Emerman M. 1993. Incorporation of human immunodeficiency virus type 1 Gag proteins into murine leukemia virus virions. J. Virol. 67:6499–6506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Duenas-Decamp MJ, Peters PJ, Burton D, Clapham PR. 2009. Determinants flanking the CD4 binding loop modulate macrophage tropism of human immunodeficiency virus type 1 R5 envelopes. J. Virol. 83:2575–2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dunfee RL, Thomas ER, Gabuzda D. 2009. Enhanced macrophage tropism of HIV in brain and lymphoid tissues is associated with sensitivity to the broadly neutralizing CD4 binding site antibody b12. Retrovirology. 6:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dunfee RL, et al. 2006. The HIV Env variant N283 enhances macrophage tropism and is associated with brain infection and dementia. Proc. Natl. Acad. Sci. U. S. A. 103:15160–15165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dunfee RL, et al. 2007. Loss of the N-linked glycosylation site at position 386 in the HIV envelope V4 region enhances macrophage tropism and is associated with dementia. Virology 367:222–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fomsgaard A, Hirsch VM, Johnson PR. 1992. Cloning and sequences of primate CD4 molecules: diversity of the. Eur. J. Immunol. 22:2973–2981 [DOI] [PubMed] [Google Scholar]
- 17. Fomsgaard A, et al. 1995. Receptor function of CD4 structures from African green monkey and pig-tail macaque for simian immunodeficiency virus, SIVsm, SIVagm, and human immunodeficiency virus type-1. Viral Immunol. 8:121–133 [DOI] [PubMed] [Google Scholar]
- 18. Gorry PR, et al. 2002. Increased CCR5 affinity and reduced CCR5/CD4 dependence of a neurovirulent primary human immunodeficiency virus type 1 isolate. J. Virol. 76:6277–6292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Harouse JM, et al. 2001. Mucosal transmission and induction of simian AIDS by CCR5-specific simian/human immunodeficiency virus SHIV(SF162P3). J. Virol. 75:1990–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Harouse JM, Gettie A, Tan RC, Blanchard J, Cheng-Mayer C. 1999. Distinct pathogenic sequela in rhesus macaques infected with CCR5 or CXCR4 utilizing SHIVs. Science 284:816–819 [DOI] [PubMed] [Google Scholar]
- 21. Hemelaar J, et al. 2011. Global trends in molecular epidemiology of HIV-1 during 2000–2007. AIDS 25:679–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Himathongkham S, et al. 2002. Species tropism of chimeric SHIV clones containing HIV-1 subtype-A and subtype-E envelope genes. Virology 298:189–199 [DOI] [PubMed] [Google Scholar]
- 23. Himathongkham S, et al. 2000. Simian-human immunodeficiency virus containing a human immunodeficiency virus type 1 subtype-E envelope gene: persistent infection, CD4+ T-cell depletion, and mucosal membrane transmission in macaques. J. Virol. 74:7851–7860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hsu M, et al. 2003. Increased mucosal transmission but not enhanced pathogenicity of the CCR5-tropic, simian AIDS-inducing simian/human immunodeficiency virus SHIV(SF162P3) maps to envelope gp120. J. Virol. 77:989–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang CC, et al. 2005. Structure of a V3-containing HIV-1 gp120 core. Science 310:1025–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Humes D, Overbaugh J. 2011. Adaptation of subtype A human immunodeficiency virus type 1 envelope to pig-tailed macaque cells. J. Virol. 85:4409–4420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Joag SV, et al. 1996. Chimeric simian/human immunodeficiency virus that causes progressive loss of CD4+ T cells and AIDS in pig-tailed macaques. J. Virol. 70:3189–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Korber BT, Letvin NL, Haynes BF. 2009. T-cell vaccine strategies for human immunodeficiency virus, the virus with a thousand faces. J. Virol. 83:8300–8314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kwong PD, et al. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li J, Lord CI, Haseltine W, Letvin NL, Sodroski J. 1992. Infection of cynomolgus monkeys with a chimeric HIV-1/SIVmac virus. J. Acquir. Immune Defic. Syndr. 5:639–646 [PubMed] [Google Scholar]
- 31. Li M, et al. 2005. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 79:10108–10125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li M, et al. 2006. Genetic and neutralization properties of subtype C human immunodeficiency virus type 1 molecular env clones from acute and early heterosexually acquired infections in Southern Africa. J. Virol. 80:11776–11790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li Y, et al. 1992. Complete nucleotide sequence, genome organization, and biological properties of human immunodeficiency virus type 1 in vivo: evidence for limited defectiveness and complementation. J. Virol. 66:6587–6600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li Y, et al. 2006. Characterization of antibody responses elicited by human immunodeficiency virus type 1 primary isolate trimeric and monomeric envelope glycoproteins in selected adjuvants. J. Virol. 80:1414–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Long EM, Rainwater SM, Lavreys L, Mandaliya K, Overbaugh J. 2002. HIV type 1 variants transmitted to women in Kenya require the CCR5 coreceptor for entry, regardless of the genetic complexity of the infecting virus. AIDS Res. Hum. Retrovir. 18:567–576 [DOI] [PubMed] [Google Scholar]
- 36. Luciw PA, Pratt-Lowe E, Shaw KES, Levy JA, Cheng-Mayer C. 1995. Persistent infection of rhesus macaque with T-cell-line-tropic and macrophage-tropic clones of simian/human immunodeficiency viruses (SHIV). Proc. Natl. Acad. Sci. U. S. A. 92:7490–7494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Miller AD, Rosman GJ. 1989. Improved retroviral vectors for gene transfer and expression. Biotechniques 7:980–982 [PMC free article] [PubMed] [Google Scholar]
- 38. Mirzabekov T, et al. 1999. Enhanced expression, native purification, and characterization of CCR5, a principal HIV-1 coreceptor. J. Biol. Chem. 274:28745–28750 [DOI] [PubMed] [Google Scholar]
- 39. Moir S, Perreault J, Poulin L. 1996. Postbinding events mediated by human immunodeficiency virus type 1 are sensitive to modifications in the D4-transmembrane linker region of CD4. J. Virol. 70:8019–8028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Morgenstern JP, Land H. 1990. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 18:3587–3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Naldini L, Blomer U, Gage FH, Trono D, Verma IM. 1996. Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proc. Natl. Acad. Sci. U. S. A. 93:11382–11388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ndung'u T, et al. 2001. Infectious simian/human immunodeficiency virus with human immunodeficiency virus type 1 subtype C from an African isolate: rhesus macaque model. J. Virol. 75:11417–11425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nishimura Y, et al. 2010. Generation of the pathogenic R5-tropic simian/human immunodeficiency virus SHIVAD8 by serial passaging in rhesus macaques. J. Virol. 84:4769–4781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Overbaugh J, Rudensey LM, Papenhausen MD, Benveniste RE, Morton WR. 1991. Variation in simian immunodeficiency virus env is confined to V1 and V4 during progression to simian AIDS. J. Virol. 65:7025–7031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pal R, et al. 2003. Characterization of a simian human immunodeficiency virus encoding the envelope gene from the CCR5-tropic HIV-1 Ba-L. J. Acquir. Immune Defic. Syndr. 33:300–307 [DOI] [PubMed] [Google Scholar]
- 46. Pineda MJ, Orton BR, Overbaugh J. 2007. A TRIM5alpha-independent post-entry restriction to HIV-1 infection of macaque cells that is dependent on the path of entry. Virology 363:310–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. 1998. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophage-tropic isolates of human immunodeficiency virus type 1. J. Virol. 72:2855–2864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Puffer BA, et al. 2002. CD4 independence of simian immunodeficiency virus Envs is associated with macrophage tropism, neutralization sensitivity, and attenuated pathogenicity. J. Virol. 76:2595–2605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Reimann KA, et al. 1996. A chimeric simian/human immunodeficiency virus expressing a primary patient human immunodeficiency virus type 1 isolate Env causes an AIDS-like disease after in vivo passage in rhesus monkeys. J. Virol. 70:6922–6928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Reimann KA, et al. 1996. An env gene derived from a primary human immunodeficiency virus type 1 isolate confers high in vivo replicative capacity to a chimeric simian/human immunodeficiency virus in rhesus monkeys. J. Virol. 70:3198–3206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Roben P, et al. 1994. Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 that show differing abilities to neutralize human immunodeficiency virus type 1. J. Virol. 68:4821–4828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rossi F, et al. 2008. The V1-V3 region of a brain-derived HIV-1 envelope glycoprotein determines macrophage tropism, low CD4 dependence, increased fusogenicity and altered sensitivity to entry inhibitors. Retrovirology 5:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sakuragi S, et al. 1992. Infection of macaque monkeys with a chimeric human and simian immunodeficiency virus. J. Gen. Virol. 73:2983–2987 [DOI] [PubMed] [Google Scholar]
- 54. Shibata R, et al. 1991. Generation of a chimeric human and simian immunodeficiency virus infectious to monkey peripheral blood mononuclear cells. J. Virol. 65:3514–3520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Siddappa NB, et al. 2009. Neutralization-sensitive R5-tropic simian-human immunodeficiency virus SHIV-2873Nip, which carries env isolated from an infant with a recent HIV clade C infection. J. Virol. 83:1422–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Song RJ, et al. 2006. Molecularly cloned SHIV-1157ipd3N4: a highly replication-competent, mucosally transmissible R5 simian-human immunodeficiency virus encoding HIV clade C Env. J. Virol. 80:8729–8738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sterjovski J, et al. 2007. Asn 362 in gp120 contributes to enhanced fusogenicity by CCR5-restricted HIV-1 envelope glycoprotein variants from patients with AIDS. Retrovirology 4:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sterjovski J, et al. 2011. CD4-binding site alterations in CCR5-using HIV-1 envelopes influencing gp120-CD4 interactions and fusogenicity. Virology 410:418–428 [DOI] [PubMed] [Google Scholar]
- 59. Thippeshappa R, et al. 2011. Vif substitution enables persistent infection of pig-tailed macaques by human immunodeficiency virus type 1. J. Virol. 85:3767–3779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wu X, et al. 2010. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329:856–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhang ZD, Weinstock G, Gerstein M. 2008. Rapid evolution by positive Darwinian selection in T-cell antigen CD4 in primates. J. Mol. Evol. 66:446–456 [DOI] [PubMed] [Google Scholar]