Abstract
The nonstructural protein (NS1) of influenza A virus performs multiple functions in the virus life cycle. Proteomic screening for cellular proteins which interact with NS1 identified the cellular protein RAP55, which is one of the components of cellular processing bodies (P-bodies) and stress granules. To verify whether NS1 interacts with cellular P-bodies, interactions between NS1, RAP55, and other P-body-associated proteins (Ago1, Ago2, and DCP1a) were confirmed using coimmunoprecipitation and cellular colocalization assays. Overexpression of RAP55 induced RAP55-associated stress granule formation and suppressed virus replication. Knockdown of RAP55 with small interfering RNA (siRNA) or expression of a dominant-negative mutant RAP55 protein with defective interaction with P-bodies blocked NS1 colocalization to P-bodies in cells. Expression of NS1 inhibited RAP55 expression and formation of RAP55-associated P-bodies/stress granules. The viral nucleoprotein (NP) was found to be targeted to stress granules in the absence of NS1 but localized to P-bodies when NS1 was coexpressed. Restriction of virus replication via P-bodies occurred in the early phases of infection, as the number of RAP55-associated P-bodies in cells diminished over the course of virus infection. NS1 interaction with RAP55-associated P-bodies/stress granules was associated with RNA binding and mediated via a protein kinase R (PKR)-interacting viral element. Mutations introduced into either RNA binding sites (R38 and K41) or PKR interaction sites (I123, M124, K126, and N127) caused NS1 proteins to lose the ability to interact with RAP55 and to inhibit stress granules. These results reveal an interplay between virus and host during virus replication in which NP is targeted to P-bodies/stress granules while NS1 counteracts this host restriction mechanism.
INTRODUCTION
The NS1 protein of influenza A virus plays important roles in antagonizing the host antiviral response and supporting virus replication (18). It is suggested that the NS1 protein has the ability to suppress host antiviral defenses at multiple levels (25). NS1 has been identified to bind to the 30-kDa subunit of cleavage and polyadenylation specificity factor (CPSF30) and the poly(A)-binding protein II (PABPII) to regulate cellular mRNA processing, leading to general inhibition of the host antiviral response (10, 38). However, the NS1 proteins of some influenza virus strains, such as PR8 and the pandemic H1N1 2009 virus, are unable to interact with CPSF30 (19, 25), suggesting that influenza A viruses may have evolved to use different mechanisms to counter host antiviral responses. Besides the interaction with CPSF30, several mechanisms have been described for NS1 inhibition of cellular interferon (IFN) expression. NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the RIG-I-mediated host antiviral response (14, 39). The NS1 protein was identified to block activation of 2′,5′-oligoadenylate synthetase (OAS) and protein kinase R (PKR) (35, 36), which are key regulators of influenza virus transcription/translation processes that are both known to be activated by double-stranded RNA (dsRNA) (35, 36). While the N-terminal RNA binding domain of the NS1 protein is essential for blocking the expression and effects of host interferon (35), the NS1 C-terminal effector domain has been found to interact with numerous host factors (7, 18). NS1 also directly stimulates phosphoinositide 3-kinase (PI3K) signaling through binding to the p85β subunit in the early phase of virus infection (16, 17). There is evidence suggesting that the NS1 protein may activate translation of influenza virus mRNA by recruiting eukaryotic initiation factor 4GI (eIF4GI) during virus replication (3, 9). The biological significance of the NS1 protein is also indicated by its evolutionary signatures in avian and human lineages of influenza A viruses, suggesting an association with host adaptation (19, 31).
The NS1 protein is not essential for virus replication but was found to be an important virulence factor for influenza A viruses. In animal studies using reassortant viruses, NS1 was identified as one of the key viral elements associated with the highly virulent features of some influenza A viruses in mammalian hosts, although mechanistic details remain to be elucidated (5, 22, 43). While previous studies on NS1 have focused mainly on its function in counteracting host antiviral innate immunity, evidence suggests that the NS1 protein may be involved directly in other processes during the virus replication process, with an interaction between NS1 and NP of influenza virus having been reported (32, 41). Influenza A virus replicates its genome in the cell nucleus by using the viral RNP polymerase complex, which is composed of PA, PB1, and PB2 subunits and ribonucleoprotein (NP). Translation of viral mRNA depends on cellular machinery, and the mechanism for regulation of translation of influenza virus proteins is poorly understood. In response to changing environments or stimuli/stress signals, a key mechanism for regulating gene expression in cells is the formation of stress granules and processing bodies (P-bodies), which modulate the rate of mRNA and protein synthesis (4, 8, 47). P-bodies and stress granules are two closely related types of cytoplasmic RNA granules that are responsible for storage, silencing, and degradation of translationally stalled mRNA (8). However, it has been suggested that mRNA accumulated in P-bodies may return to polysomes for translation (4, 13). Cellular stress granules and P-bodies have been documented extensively in recent reviews to play an important role in the dynamics of the life cycle of some viruses (6, 52). Most recently, inhibition of formation of stress granules by the NS1 protein of influenza A virus was reported (24).
Using mass spectrometry (matrix-assisted laser desorption ionization–time of flight mass spectrometry [MALDI-TOF MS]) analysis and cellular and molecular characterization, we identified and confirmed an interaction between the NS1 protein and the cellular RNA-associated protein 55 (RAP55). RAP55 was first identified in newts (Pleurodeles waltl) and belongs to an evolutionarily conserved protein family possessing an LSm14 domain (30, 33). It is found in both P-bodies and stress granules, although its role in these RNA granules is still not clear (8). Molecular analysis shows that RAP55 contains multiple functional domains which are responsible for translational repression, RNA binding, and protein-protein interaction (33). It was reported that the N terminus, which is composed of an LSm-like motif, is necessary for stress granule localization, while the C terminus, containing FDF and RCG motifs, is required for targeting proteins to P-bodies (34, 53). This study examined the interaction between the NS1 protein and RAP55-associated P-bodies and stress granules during influenza A virus replication. We found that in response to influenza A virus infection, and in particular to the expression of NP, P-bodies and stress granules were induced in infected cells. Increasing levels of RAP55 expression significantly increased the amount of NP-harboring stress granules. Our results show that expression of NS1 is essential for antagonizing the host defense mechanism of P-body and stress granule formation in influenza virus-infected cells. The interplay between NS1 and RAP55-associated RNA granules containing NP during influenza A virus infection may represent another, still-uncharacterized aspect of the virus-host interaction and may also influence virus virulence.
MATERIALS AND METHODS
Cells and viruses.
Human embryonic kidney (HEK) 293T cells and A549 cells were maintained in Dulbecco minimal essential medium, and Madin-Darby canine kidney (MDCK) cells were cultured in Eagle minimal essential medium (MEM), with both media supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin sulfate, and the cells were cultured at 37°C with 5% CO2. For transfection studies, subconfluent (70 to 80%) monolayers were transiently transfected or cotransfected with plasmids by use of TransIT-LT1 transfection reagent (Mirus Bio). Influenza virus strains A/Vietnam/1194/2004 (1194; H5N1) and A/Wisconsin/1933 (WSN; H1N1) were propagated in 10- to 12-day-old embryonated specific-pathogen-free chicken eggs at 35°C for 30 and 48 h, respectively.
Generation of recombinant viruses.
MDCK and HEK 293T cell cocultures were seeded in equal proportions at a density of 1 × 106 cells per well in a 6-well plate a day before transfection. TransIT-LT1 transfection reagent was used for transfection in accordance with the manufacturer's instructions. Briefly, 1 μg each of eight pHW2000 plasmids containing individual influenza virus genome segments was mixed with 16 μl of TransIT-LT1, incubated at room temperature for 45 min, and then added to the cell cocultures, for which the overnight culture medium had been replaced with 1 ml of Opti-MEM I medium on the day of transfection. After 6 h of incubation at 37°C, the transfection mixture was removed and replaced with 1 ml of Opti-MEM I medium. Twenty-five hours after transfection, 1 ml of Opti-MEM I containing antibiotics and tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-trypsin (2 μg/ml) was added to the cultures. After 3 days, the culture supernatant was collected and used to infect MDCK cells or embryonated eggs for further propagation and amplification of the recombinant viruses. Following propagation, hemagglutination assays were performed to detect the presence of recombinant viruses, and successfully generated viruses were then sequenced.
Plasmids.
Expression vectors containing a Flag epitope or a V5 tag were constructed using the pCMV-script (Stratagene) or pCAGGS vector, respectively. All of the plasmids constructed in the present study contained C-terminally tagged epitopes and a Kozak consensus sequence (CCACC) to optimize expression of the fusion proteins. Mutagenesis of the NS1 gene was performed using a QuikChange multisite-directed mutagenesis kit (Stratagene) as described by the manufacturer. The mutagenesis primers were designed using the QuikChange primer design program (Stratagene). GFP-Ago1 (29), GFP-hAgo2 (23), and pT7-eGFP-C1-HsDCP1a (48) were obtained through Addgene (Cambridge, MA). A mutant of RAP55 (GFP_RAP55Δ7) which does not form P-bodies and stress granules was described previously (34).
Protein identification by MALDI-TOF/TOF MS/MS analysis.
Pulldown products obtained using pCMV-VNM1194NS1_Flag as the bait were separated by one-dimensional SDS-polyacrylamide gel electrophoresis (SDS-PAGE). Each protein band was excised, destained, reduced, alkylated, and digested with trypsin. The polypeptides were extracted, purified, and then analyzed using an ABI 4800 Plus MALDI-TOF/TOF analyzer (Applied Biosystems) as described previously (50).
Pulldown and coimmunoprecipitation (co-IP) assay.
HEK 293T cells were transfected and incubated for 48 h. Transfected cells were lysed in a buffer containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% Triton X-100, and protease inhibitor cocktail (Roche). Cell lysates were incubated with 1 μg of specific antibody as described in the figure legends, together with Dynabeads protein G (Invitrogen), in accordance with the manufacturer's manual. The mixtures were incubated at room temperature for 30 min, and the beads were then washed three times with lysis buffer, using a magnet to separate beads from the supernatant at each washing step. Proteins were eluted by boiling in SDS sample buffer and then detected by Western blotting using the relevant secondary antibody. For RNase treatment, 200 μg/ml of RNase A was added to cell lysates and incubated on ice for 30 min prior to addition of antibody.
Luciferase reporter assay for determining polymerase complex activity.
Full-length genomic segments of NP, PA, PB1, and PB2 derived from the H5N1 virus 1194 were cloned into pHW2000 (21). These RNP complex components were cotransfected into HEK 293T cells with a luciferase reporter plasmid, pYH-Luci, which contains a noncoding sequence from the M segment of influenza A virus and the luciferase gene, driven by polymerase I (Pol I) (a kind gift from Robert Webster, St. Jude Children's Research Hospital). The plasmid phRL_TK (Promega), which expresses Renilla luciferase, was also cotransfected in each experiment for data normalization purposes. At 20 to 24 h posttransfection, cell lysates were prepared using a luciferase assay system (Promega), and luciferase activity was measured using a Victor3 multilabel plate reader (PerkinElmer).
Indirect immunofluorescence assay.
HEK 293T and A549 cells were grown on Millicell EZ slides (Millipore) and transiently transfected with expression plasmids for 20 to 24 h. For indirect immunofluorescence assay of virus infection, HEK 293T and A549 cells were used as indicated in the figure legends. Cells were washed with phosphate-buffered saline (PBS) and fixed for 15 min using 4% formaldehyde in PBS, followed by permeabilization with 0.2% Triton X-100 in PBS for 5 min. Cells were then washed with PBS and blocked with 2% bovine serum albumin (BSA) or 10% normal donkey serum in PBS for 30 min at 37°C. After blocking, cells were incubated with mouse or rabbit anti-Flag (Sigma), mouse anti-V5 (Invitrogen), rabbit anti-NS1, mouse anti-NP (50), mouse or rabbit anti-PABP1 (Abcam), mouse anti-G3BP (BD Biosciences), mouse anti-DCP1a (Abnova), or mouse anti-RAP55 (46) at a 1:100 dilution in PBS for 1 h at 37°C (45, 50). After incubation, cells were washed twice with PBS and then incubated with secondary antibodies conjugated with different fluorophores at a 1:200 dilution for 30 min at room temperature. Finally, the cells were washed several times in PBS and the signals visualized using a Carl Zeiss LSM 700 confocal microscope.
Western blotting.
Co-IP products or cell lysates were fractionated by 12% SDS-PAGE and then blotted onto nitrocellulose membranes (Bio-Rad). Membranes were incubated with primary antibodies diluted as follows: mouse anti-Flag, mouse anti-V5, mouse anti-beta-actin, and mouse anti-beta-tubulin (Sigma) were used at a 1:5,000 dilution; mouse anti-NP, rabbit anti-NS1, mouse anti-RAP55, mouse anti-DCP1a, and mouse anti-PABP1 were used at a 1:1,000 dilution; and rabbit anti-PKR (phospho-T446) (Abcam), rabbit anti-PKR (Abcam), rabbit anti-phospho-eIF2 (Ser51) (Cell Signaling Technology), and rabbit anti-eIF2 (Cell Signaling Technology) were used at a 1:500 dilution. Membranes were then incubated with horseradish peroxidase-labeled sheep anti-mouse or anti-rabbit IgG secondary antibody (GE Healthcare) or IRDye 680-labeled donkey anti-mouse or anti-rabbit IgG(H+L) (Li-Cor Biosciences) at a 1:5,000 dilution. Blots were visualized using ECL Plus Western blotting detection reagents (GE Healthcare) or an Odyssey imaging system (Li-Cor Biosciences), respectively.
Fluorescence in situ hybridization (FISH) assay.
For viral mRNA and viral RNA (vRNA) detection, riboprobes (corresponding to nucleotides 572 to 1101 of the NA mRNA) were generated using a FISH Tag RNA multicolor kit (Invitrogen) in accordance with the manufacturer's protocol. After indirect immunofluorescence assay staining (see description above), cells were permeabilized by being incubated on ice with 0.5% Triton X-100 in PBS for 5 min and then fixed in 4% paraformaldehyde (PFA) for 10 min. Cells were then washed twice with PBS, followed by two washes with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Cells were then hybridized in 40 μl of hybridization buffer (50% formamide, 2× SSC, 10% dextran sulfate, 1 mg/ml Escherichia coli tRNA, and 2 mM vanadyl-ribonucleoside complex) containing 100 ng of riboprobe at 37°C overnight. After hybridization, cells were washed with 2× SSC for 15 min, followed by successive washings for 15 min with 1× SSC and 0.2× SSC at room temperature. Cells were then mounted and examined by confocal laser scanning microscopy.
Knockdown of RAP55 expression with siRNA.
Gene-specific small interfering RNAs (siRNAs) (siRAP55_1, SASI_Hs02_00326194, siRAP55_2, and SASI_Hs02_00326195) were purchased from Sigma. The siControl oligonucleotide was obtained from Invitrogen (negative control 2). HEK 293T or A549 cells were transfected with 50 nM siRNA by use of Lipofectamine RNAiMax transfection reagent (Invitrogen) and then incubated for 48 h. The efficiency of RAP55 expression knockdown was confirmed by Western blotting.
RESULTS
The NS1 protein interacts with RAP55 and other P-body components in cells.
To identify host factors interacting with the NS1 protein, we employed MALDI-TOF MS analysis by using a Flag-tagged NS1 protein as bait, transiently expressing it in HEK 293T cells, and then coimmunoprecipitating host factors as previously described (50). The identities of proteins pulled down by the NS1 protein derived from the H5N1 A/Vietnam/1194/2004 strain were verified in at least three repeated experiments (Fig. 1A). Confirmation of direct physical interaction between these cellular proteins and NS1 is ongoing. One of the cellular proteins confirmed to associate with NS1 was RAP55 (RNA-associated protein 55), also referred to as LSm14A in the literature (2) (Fig. 1A). Interaction between NS1 and RAP55 was verified by a coimmunoprecipitation assay. As shown in Fig. 1B, Flag-tagged NS1 coprecipitated with V5-tagged RAP55 from HEK 293T cell lysates, but pretreatment of lysates with RNase abolished the interaction, suggesting that the interaction between NS1 and RAP55 is RNA associated. An RNA binding requirement for interaction was also verified by the observation that NS1 proteins containing R38A and R41A mutations could not precipitate RAP55 (Fig. 1B). The R35A and R46A NS1 mutants, which are unable to form functional dimers (51), did not interact with RAP55. A Y89F mutation in the effector domain, which abolishes interaction between NS1 and p85β (17), a subunit of the PI3K pathway, did not influence the interaction between NS1 and RAP55. PKR is the primary host target of the NS1 protein. We found that interaction between NS1 and RAP55 was not affected when dual mutations were introduced into the 123/124 or 126/127 sites, which are critical for PKR binding or activation, respectively (26). However, interaction between NS1 and RAP55 was completely abolished for a mutant containing mutations at all four of these sites (I123A/M124A/K126A/M127A) (Fig. 1B). It is notable that NS1 proteins containing mutations which only partially affect PKR binding sites retained the ability to interact with RAP55 and inhibit RAP55-associated stress granules (Fig. 2; see below). To verify if the RNA binding domain determines the interaction between NS1 and RAP55, we conducted an RNA binding assay using wild-type (WT), R35A mutant, and quadruple PKR mutant proteins. While the PKR quadruple mutant lost the ability to interact with RAP55 (Fig. 1B, left panel), it still retained the ability to bind RNA (Fig. 1B, right panel). These results indicate that both the RNA binding properties and PKR interacting regions of NS1 are involved in its interaction with RAP55.
Fig 1.
Interaction of NS1 with RNA-associated protein 55 (RAP55). (A) Identification of association of RAP55 and other host factors with influenza virus NS1 by MS analysis. HEK 293T cells were transfected with an expression vector encoding NS1-Flag from A/Vietnam/1194/2004 (H5N1) and then cultured for 48 h. Cell lysates were precipitated with anti-Flag M2 (Sigma) monoclonal antibody, and precipitates were then separated by SDS-PAGE and visualized by silver staining. Each of the separated protein bands was individually excised and analyzed by MS. Only host factors reproducibly identified to associate with NS1 in repeated experiments are shown. (B) (Left) Coimmunoprecipitation of NS1 and RAP55 proteins. V5-tagged RAP55 and Flag-tagged wild-type (WT) and mutant (R35A, R38A/K41A, R46A, Y89F, I123A/M124A, K126A/M127A, and I123A/M124A/K126A/M127A) NS1 expression vectors were cotransfected into HEK 293T cells. RAP55 was used as bait to pull down the NS1 proteins, as described in Materials and Methods. For RNase treatment, 200 μg/ml of RNase A was added to cell lysates, which were then incubated on ice for 30 min prior to addition of antibody. Proteins coprecipitated with mouse anti-V5 antibody (IP) were probed with anti-NS1 rabbit polyclonal and anti-V5 mouse monoclonal antibodies (IB). IP, immunoprecipitation; IB, immunoblotting. (Right) HEK 293T cells were transfected with an expression vector encoding NS1-Flag derived from WSN virus. Cell lysates were collected at 48 h posttransfection and incubated with poly(I-C) immobilized on cyanogen bromide-activated Sepharose beads. Poly(I-C)-bound proteins were isolated and analyzed by Western blotting (IB) using antibodies against NS1. (C) Colocalization of endogenous RAP55 (red) and wild-type or R35A mutant NS1 (green) was assessed by cotransfection of RAP55-V5 and NS1-Flag expression vectors into HEK 293T cells. Expression of RAP55 and NS1 was examined by double immunostaining of cells with anti-RAP55 and anti-Flag mouse monoclonal antibodies (for NS1) at 24 h posttransfection. Signals were visualized using a confocal microscope. (D) Association of NS1 with RAP55 and other P-body-associated proteins. HEK 293T cells were cotransfected with NS1-Flag and Ago1-GFP, Ago2-GFP, or DCP1a-GFP expression vector. Colocalization of NS1 with these P-body components was examined by double immunostaining, using anti-Flag for NS1, anti-RAP55 for endogenous RAP55, and anti-GFP for Ago1, Ago2, and DCP1a. (E) NS1 association with P-bodies in RAP55 knockdown and control HEK 293T cells. Flag-tagged NS1 was expressed in control knockdown (siControl) and RAP55 knockdown (siRAP55) HEK 293T cells, and cells were examined with anti-Flag and anti-RAP55 antibodies. (F) NS1 fails to localize to P-bodies in cells which overexpress a dominant-negative RAP55 mutant (34). Flag-tagged NS1 and wild-type or mutated GFP-fused RAP55 expression vectors were transfected into HEK 293T cells, which were later examined with anti-Flag and anti-GFP antibodies.
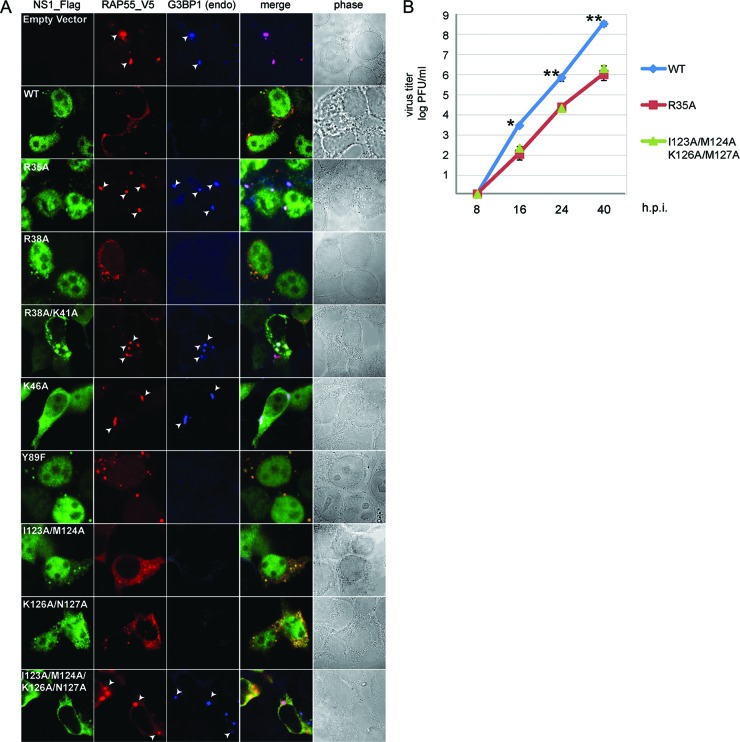
Fig 2.
Colocalization of wild-type or mutant NS1 proteins with overexpressed RAP55 and suppression of RAP55-associated stress granule formation by different versions of NS1. HEK 293T cells were cotransfected with V5-tagged RAP55 and wild-type and mutated Flag-tagged NS1 expression vectors (NS1 mutants are shown in Fig. 1B). Signals were examined by double immunostaining using anti-Flag and anti-V5 antibodies at 16 to 20 h posttransfection. Formation of stress granules was detected using an anti-mouse monoclonal antibody specific for endogenous G3BP1. (B) Growth kinetics of WSN wild-type and mutant (R35A and I123/A/M124A/K125A/M127A) viruses. A549 cells were infected with WSN WT, R35A mutant, or I123/A/M124A/K125A/M127A mutant virus at an MOI of 0.05. Viral titers were determined by plaque assay at the indicated time points. Values represent the means for three independent experiments, and error bars show standard deviations. *, P < 0.05; **, P < 0.005.
Indirect immunofluorescence assay revealed that NS1 and RAP55 colocalized in transfected cells coexpressing NS1 and RAP55, and consistent with the co-IP data, no colocalization was observed with the R35A mutant form of NS1 (Fig. 1C). RAP55 is a known component of P-bodies and stress granules (53). Since NS1 has been reported to inhibit stress granule formation (24), and because RAP55 is associated with stress granules as well, we determined whether the NS1 protein interacts with P-bodies by examining the RAP55-NS1 complexes formed in HEK 293T cells for the presence of other P-body-associated proteins, including Ago1, Ago2, and DCP1a. HEK 293T cells were cotransfected with expression vectors encoding green fluorescent protein (GFP)-tagged Ago1, Ago2, or DCP1a, along with Flag-tagged NS1, and then stained with antibodies specific for GFP, Flag, and RAP55. Colocalization of NS1 with Ago1, Ago2, and DCP1a was observed in cells, and the punctate structures where the colocalizations occurred were found to overlap the areas of endogenous RAP55 expression (Fig. 1D). To further confirm the association of NS1 and RAP55 in P-bodies, we examined NS1 localization in RAP55 knockdown cells. NS1 colocalized nicely with RAP55 in the cytoplasmic granules, and staining with DCP1a confirmed that these granules were P-bodies. However, no NS1-associated P-bodies were observed in RAP55 knockdown cells (Fig. 1E). When a dominant-negative mutant of RAP55 which inhibits formation of P-bodies was coexpressed with NS1, no NS1-associated P-bodies were observed (Fig. 1F). These results indicate that the interaction between influenza A virus NS1 protein and RAP55 does occur in cellular cytoplasmic P-bodies and that RAP55 is required for NS1 to localize to the P-bodies. Given the known role of RNA granules in modulating mRNA protein synthesis, we suggest that this interaction may be involved in host regulation of viral replication.
Characterization of NS1 mutations which affect inhibition of stress granules.
NS1's function in counteracting multiple cellular antiviral signaling pathways associated with regulation of interferon expression during virus infection is well characterized. To examine whether the function associated with RAP55 interaction is associated with other properties of the NS1 protein, we used a panel of NS1 mutants to study the NS1-RAP55 interaction in cells. The R38A mutant has lost the ability to bind RNA, is unable to suppress cellular expression of interferon, and no longer interacts with TRIM25 in RIG-I-induced antiviral activity (14). The Y89F mutation is reported to be associated with a loss of interaction with p85β, a subunit of the PI3K pathway (17). NS1 is normally able to interact with the PKR pathway and interfere with the host antiviral response, but mutations at positions 123/124 and 126/127 were found to cause this function to be lost (36). When the R38A, Y89F, I123A/M124A, and K126A/N127A NS1 mutants were expressed in HEK 293T cells overexpressing RAP55, we found that they retained the ability to colocalize with RAP55, as demonstrated by indirect immunofluorescence assay (Fig. 2), suggesting that the function of the NS1 protein associated with RAP55/P-bodies is independent of its previously characterized properties. However, the R35A, R46A, R38A/K41A, and I123A/M124A/K126A/N127A NS1 mutants, which cannot interact with RAP55 (Fig. 1), were observed to not colocalize with RAP55. It was notable that in cells where no NS1 was expressed or in those expressing NS1 mutants which had lost the ability to interact with RAP55, overexpression of RAP55 induced formation of stress granules, as indicated by colocalization of RAP55 and G3BP1 (Fig. 2). To verify whether the ability to inhibit stress granules may be associated at least partly with virus growth properties, we examined the growth kinetics of WSN WT, R35A mutant, and PKR quadruple mutant (I123A/M124A/K126A/N127A) viruses in A549 cells. As shown in Fig. 2B, both R35A and PKR quadruple mutants exhibited significantly attenuated replication. While the R35A mutation and the mutations in the PKR interacting site may attenuate the ability of NS1 to antagonize other host antiviral responses, it is possible that the attenuation also affects viral inhibition of host stress granules. These results reveal that NS1 possesses functions associated with RAP55 that are required for inhibition of formation of the stress granules normally induced by RAP55 overexpression.
RAP55 inhibits influenza virus replication.
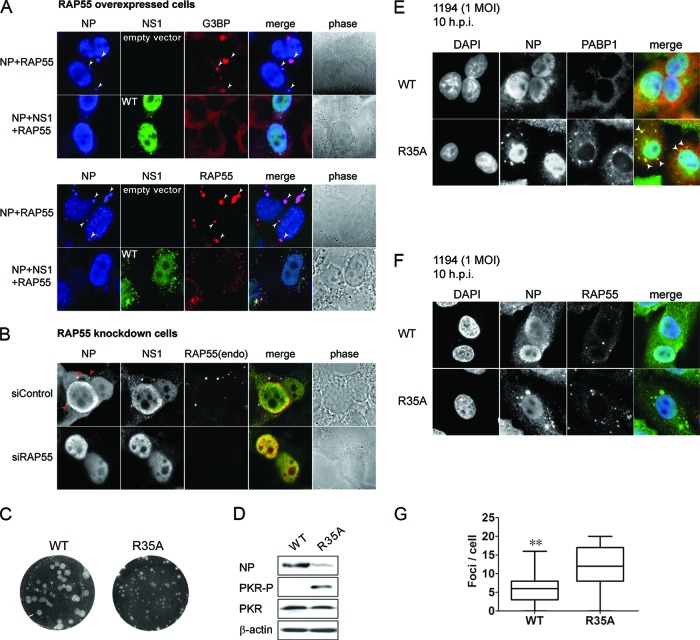
RAP55 is associated with both stress granules and P-bodies, structures which have important regulatory roles in the virus life cycle (6). To understand the biological relevance of the interaction between the NS1 protein and RAP55, we examined the effects of RAP55, Ago1, and Ago2 on viral RNP polymerase activity by using a viral minigenome system described in an earlier study (50). Overexpression of RAP55 and, to a lesser extent, Ago2, but not that of Ago1, significantly inhibited RNP polymerase activity, while coexpression of NS1 restored the RNP polymerase activity (Fig. 3A). Consistent with the observation that overexpression of RAP55 downregulates RNP polymerase activity, knockdown of RAP55 increased the activity about 2-fold compared to that with control cells (Fig. 3A, RAP55 knockdown cells). We showed above that overexpression of RAP55 induces stress granules and that coexpression of WT NS1 or an NS1 mutant which retains the ability to interact with RAP55 inhibits formation of stress granules (Fig. 2). NS1 has been described to inhibit formation of stress granules through interference of PKR activation (24). We found that overexpression of RAP55, but not Ago1 or Ago2, activates PKR and that coexpression of NS1 inhibits RAP55-induced PKR phosphorylation (Fig. 3A). To further examine the effect of RAP55 on virus replication, HEK 293T cells were transfected with an expression vector encoding RAP55 and then infected with the influenza virus strain WSN at a multiplicity of infection (MOI) of 0.005 at 24 h posttransfection. Expression of NP was examined in cells with or without RAP55 overexpression (Fig. 3B). Consistent with the effect on RNP polymerase activity (Fig. 3A), transient expression of RAP55 inhibited virus replication, as demonstrated by a significant reduction in the number of NP-expressing cells; the only NP-positive cells were those in which RAP55 overexpression was undetectable (Fig. 3B, lower panels). Cytoplasmic P-bodies are associated with a cellular machinery that facilitates repression of translation, accumulation, storage, and degradation of mRNA and represents one of the key host antiviral responses to infection. We examined the dynamics of RAP55 inhibition of virus replication in cells by tracking WSN virus infection in A549 cells and found that RAP55-associated P-bodies were observed mainly in the early hours of infection and were markedly reduced in the later phase of infection (Fig. 3C and D). Analysis of RAP55 and other P-body- or stress granule-associated proteins revealed that expression of RAP55 was downregulated along the course of virus replication, with correspondingly increased levels of NP and NS1 in virus-infected cells present later in infection, while expression of the P-body-associated protein DCP1a and the stress granule-associated protein PABP1 was not affected (Fig. 3E, left panel). As a control, cells were infected with the R35A mutant virus; these cells did not exhibit a decrease in RAP55 expression over time (Fig. 3E, right panel). Examination of virus titers in cells with or without overexpression of RAP55 also supported the notion that RAP55 exhibits an inhibitory effect on influenza virus replication, as demonstrated by decreased virus titers at 8 h postinfection and lower levels of viral protein (NS1) in RAP55-overexpressing cells (Fig. 3F and G).
Fig 3.
RAP55 negatively regulates influenza A virus replication. (A) Effects of RAP55, Ago1, and Ago2 on influenza A virus polymerase complex activity. Plasmids expressing RNP complex components (PB1, PB2, PA, and NP), with or without an expression vector for NS1 derived from A/Vietnam/1194/2004, were cotransfected into HEK 293T cells along with a vector expressing one of the P-body components (RAP55, Ago1, or Ago2), plus a luciferase reporter, as described in Materials and Methods. Knockdown of RAP55 cells was also used to measure the effect of RAP55 on RNP polymerase activity in this experiment. A plasmid expressing Renilla luciferase was cotransfected as an internal control for data normalization. (Top) Luciferase activities were estimated at 20 h posttransfection. Results shown are averages for three independent experiments, and error bars indicate standard deviations. *, P < 0.05; ***, P < 0.001. (Bottom) Whole-cell lysates were analyzed by immunoblotting with antibodies specific for phospho-PKR, total PKR, and NS1, as indicated. (B) RAP55 inhibits influenza virus replication. Immunostaining of NP expression is shown for vector-transfected (upper panels) or RAP55-overexpressing (middle and lower panels) HEK 293T cells infected with the A/Wisconsin/1933 (WSN) strain at an MOI of 0.005. The average percentage of infected cells was calculated from five randomly selected fields (magnification, ×200). NP expression was not detected in cells where RAP55 granules were observed, as indicated by white arrows in the high-power images (lower panels). h.p.i, hours postinfection. (C) Degradation of RAP55 granules during the course of virus infection. Visualization of RAP55 granule dynamics is shown for A549 cells infected with WSN virus at an MOI of 1. Cells were fixed at 0, 3, and 7 h postinfection, as indicated, and then stained with anti-NP and anti-RAP55 mouse monoclonal antibodies. To enhance the image contrast, single staining of NP (green), RAP55 (red), and DAPI (blue) was adjusted to a monochrome presentation in this panel of images. (D) The number of RAP55-associated granules per cell was quantitated for 100 randomly selected WSN-infected A549 cells at each indicated time point. Data observed are shown with a box-and-whisker plot. *, P < 0.05; ***, P < 0.001. (E) Effects of WT and R35A mutant virus infection on expression of RAP55 and other P-body and stress granule components. Cell lysates collected at different time points were subjected to Western blot analysis with various antibodies. Beta-tubulin was used a loading control. (F) Mock-transfected or RAP55-transfected HEK 293T cells were infected with WSN virus at an MOI of 1, and the replication efficiency was analyzed by plaque assay. The average number of PFU was quantified for three individual sets of experiments. **, P < 0.01. (G) Expression of viral NS1 protein in WSN-infected and mock- or RAP55-transfected HEK 293T cells was analyzed by Western blot analysis at the indicated hours postinfection (h.p.i.). Numbers below the α-NS1 panel indicate the relative expression levels of NS1 at 24 h.p.i. compared to the levels in mock-transfected cells. β-Actin was used as a loading control. Quantification of band intensities was performed using Image J software (developed at the National Institutes of Health).
Taken together, these results indicate that RAP55 negatively influences influenza A virus replication and that inhibition occurs in the early phase of infection, presumably through recruitment of RAP55-associated P-bodies/stress granules in virus-infected cells.
RAP55 colocalizes with NP in P-bodies and stress granules.
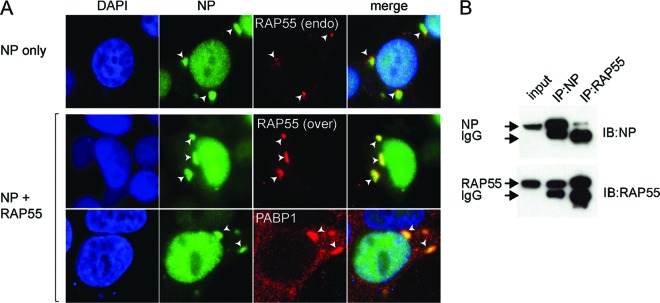
Stress granules and P-bodies are closely related cytoplasmic granules associated with the cellular regulation of mRNA translation in response to stimuli/stress signals and virus infection (6, 8, 52). It was observed here that RAP55-associated P-bodies gradually diminished along the course of virus infection in cells (Fig. 3C and D). We examined whether any particular viral proteins were targeted to RAP55-associated P-bodies and/or stress granules during virus infection. While formation of stress granules and P-bodies was commonly observed during cell growth, we found that viral nucleoprotein (NP) was present in RAP55-associated granules in cells (Fig. 4A, upper panels) but that this effect was not observed with the other RNP subunits, i.e., PB1, PB2, and PA (data not shown). Because RAP55 is present in both P-bodies and stress granules, we characterized NP- and RAP55-associated granules by using an antibody recognizing PABP1, a protein specific to stress granules (8). In cells that expressed only NP, the RAP55-associated granules were comparatively small, with endogenous RAP55 colocalized at the edges of the NP-associated granules (Fig. 4A, upper panels); no PABP1-positive stress granules were observed (data not shown). However, when RAP55 was overexpressed via plasmid transfection in NP-expressing cells, stress granules formed, as indicated by the presence of PABP1, and both RAP55 and PABP1 colocalized with the NP-associated granules (Fig. 4A, middle and lower panels). Interaction between RAP55 and NP was confirmed by coimmunoprecipitation (Fig. 4B). These results indicate that viral NP colocalizes with RAP55 in cells and that increased levels of RAP55 can promote recruitment of other proteins to form NP-associated stress granules.
Fig 4.
Interaction of NP with RAP55. (A) Colocalization patterns of NP and endogenous and plasmid-expressed RAP55 were examined in HEK 293T cells transfected with expression vectors for V5-tagged RAP55 and Flag-tagged NP. NP and RAP55 complexes were detected by double staining with anti-Flag antibody for NP and anti-RAP55 antibody for endogenous RAP55 (upper panels) or anti-V5 for overexpressed RAP55 protein (middle panels). Formation of NP-associated stress granules was detected by double staining with anti-Flag antibody for NP and anti-PABP1 antibody (lower panels). (B) Coimmunoprecipitation of RAP55 and NP proteins. V5-tagged RAP55 and Flag-tagged NP proteins were coexpressed in HEK 293T cells by plasmid transfection. NP was used as bait to pull down plasmid-expressed RAP55, and vice versa, as described in Materials and Methods.
NS1 inhibits formation of RAP55-associated stress granules and P-bodies during virus replication.
Building on our observation of colocalization of RAP55 and NP in stress granules, we were able to examine the composite effects of coexpression of NS1, RAP55, and NP on the formation of stress granules and P-bodies in cells. In HEK 293T cells expressing NP and RAP55, staining of the G3BP1 protein, which is specifically associated with stress granules, clearly showed that stress granules were formed (Fig. 5A, upper panels). However, stress granules were not apparent in cells when the NS1 protein was coexpressed (Fig. 5A, lower panels). Because RAP55 may be present in either stress granules or P-bodies, we further examined the involvement of RAP55 in different subcellular cytoplasmic granules in the presence and absence of NS1 expression. Double staining with antibodies to NP and RAP55 showed that in the absence of NS1, NP and RAP55 colocalized in cytoplasmic granules in cells (Fig. 5B, upper panels). However, only sporadic and smaller RAP55-associated P-bodies, where NS1 and RAP55 colocalized, were observed in cells when NS1 was coexpressed (Fig. 5B, lower panels). To investigate the relevance of RAP55 association with NS1 and NP in P-bodies, we coexpressed NP and NS1 in RAP55 knockdown and control cells and examined colocalization of NP and RAP55. NP was observed to colocalize with NS1 and RAP55 in P-bodies only in control cells. No NP-associated P-bodies were apparent in RAP55 knockdown cells (Fig. 5B). To further investigate the dynamics of RAP55 in stress granules and P-bodies during virus replication, we conducted an infection experiment using a wild-type–mutant pair of H5N1 viruses where the mutant virus contained the R35A form of NS1, which is unable to interact with RAP55 (Fig. 1). Replication of the R35A virus was attenuated, as shown by formation of smaller plaques than those of WT virus, and it was unable to inhibit PKR phosphorylation in virus-infected cells (Fig. 5C and D). A549 cells were infected with either WT or R35A mutant virus at an MOI of 1 and then stained with antibodies specific for PABP1 or RAP55 at 10 h postinfection. Consistent with the results obtained in the transfection experiment described above, stress granules were clearly observed, as indicated by PABP1 staining, and colocalized with NP in A549 cells infected with the R35A mutant virus (Fig. 5E, lower panels), but no PABP1-expressing stress granules were observed in cells infected with wild-type virus (Fig. 5E, upper panels). Further examination of RAP55 in R35A mutant virus-infected cells revealed that RAP55 formed distinct speckles and colocalized with NP granules (Fig. 5F, lower panels), leading us to believe that these RAP55-associated granules were stress granules. Because no stress granules were observed in cells infected with wild-type virus, we believe that the RAP55-associated granules observed in wild-type virus-infected cells are P-bodies, based on their comparatively small size. The numbers of RAP55-associated granules were also observed to be reduced significantly in cells infected with wild-type virus (Fig. 5E and F). These results suggest that NS1 may dissociate or inhibit formation of NP-induced stress granules and RAP55-associated P-bodies. As RAP55-associated granules were shown to be reduced markedly along the course of virus replication (Fig. 3C), it may be more appropriate to suggest a mechanism through which expression of NS1 dissociates the RAP55 granules.
Fig 5.
NS1 inhibits formation of RAP55-associated stress granules. (A and B) NP and RAP55 expression vectors were cotransfected with an NS1 expression plasmid or control vector into HEK 293T cells. Formation of stress granules was examined using antibodies specific for G3BP and RAP55. (A) (Top) Formation of stress granules was revealed by immunostaining for endogenous G3BP protein, as shown by white arrows. (Bottom) Formation of RAP55-associated granules was examined by immunostaining of RAP55, as indicated by white arrows. (B) Localization of NP to P-bodies in the presence of NS1 and RAP55. NP and NS1 expression vectors were cotransfected into RAP55 knockdown and control siRNA-treated HEK 293T cells. Red arrows indicate the localization of NP to cytoplasmic P-bodies. (C) Plaque phenotypes of wild-type NS1 and R35A mutant NS1 viruses. (D) Western blot analysis of A549 cells infected with WT or R35A mutant NS1 virus, using antibodies specific for NP, phospho-PKR, total PKR, and β-actin, as indicated. (E and F) A549 cells were infected with wild-type NS1 or the R35A NS1 mutant of the A/Vietnam/1194/2004 strain at an MOI of 1. At 10 h postinfection, NP and RAP55 colocalization patterns were examined by double immunostaining analysis. Formation of stress granules and P-bodies was detected using anti-PABP1 (E) and anti-RAP55 (F), respectively. Stress granules are indicated by white arrows in the bottom row of panel E. (G) Numbers of RAP55-associated granules were estimated by examining 100 randomly selected cells infected with wild-type or R35A mutant A/Vietnam/1194/2004 virus at 10 h postinfection. Data observed are shown with a box-and-whisker plot. **, P < 0.005.
Expression of NS1 inhibits RAP55-induced stress granules and increases viral mRNA in the cytoplasm during virus infection.
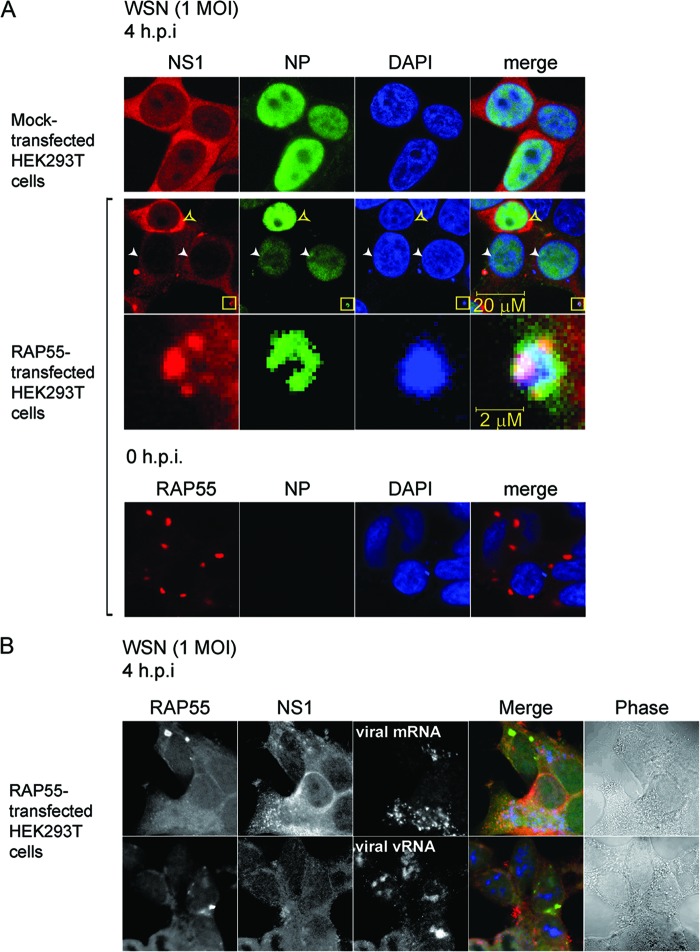
To further characterize how the interplay between NP, RAP55-associated P-bodies/stress granules, and NS1 contributes to the regulation of virus replication during virus infection, we examined the interaction of RAP55 with NP and NP-bound RNA in HEK 293T cells infected with WSN virus at an MOI of 1. Consistent with the above observation (Fig. 5E), no stress granules were observed in HEK 293T cells when NS1 was expressed (Fig. 6A, upper panels). In cells with RAP55 overexpression, we were able to identify cells with different levels of expression of NP and to examine NS1 and NP complexes. In cells where NS1 was normally expressed, no granules were observed (Fig. 6A, second row, yellow arrowheads). We have shown that in cells where RAP55 is overexpressed, no NP expression is observed (Fig. 3B). For this experiment, it is notable that in cells where cytoplasmic granules were observed, NS1 and NP expression levels were decreased (Fig. 6A, second row, white arrowheads). In cells with low-level expression of NS1, examination of interactions between NS1, NP, and NP-bound RNA became possible. Using an antibody to detect NP and DAPI (4′,6-diamidino-2-phenylindole) staining to detect nucleic acids, it was revealed that these granules were composed of NP and nucleic acid (Fig. 6A [the granules in the yellow boxed area are shown in an enlarged view in the third row from the top]). A similar result was observed at 10 h postinfection (data not shown). To further verify if expression of NS1 may stabilize viral mRNA in the cytoplasm for translation, we used FISH to detect viral mRNA and vRNA in RAP55-overexpressing cells infected with WSN virus. It is notable that in cells with low levels of NS1 and apparent RAP55-induced stress granules, little viral mRNA was observed, while in cells with normal NS1 expression, no stress granules were observed and significantly higher levels of viral mRNA were present in the cytoplasm (Fig. 6B, upper panels). At 4 h postinfection, vRNA was detected mainly in the nucleus and did not appear to be influenced by the presence of stress granules (Fig. 6B, bottom panels). These results show that there is a dynamic interplay between NS1 and RAP55 which may control the stability of viral mRNA in the cytoplasm. Early in viral infection, RAP55 could trap NP and NP-bound viral RNP in cytoplasmic stress granules, thereby stalling viral protein translation. However, the increasing level of expression of NS1 during the course of virus replication would then disassociate RAP55 stress granule complexes and thereby stabilize viral mRNA, allowing further viral protein translation.
Fig 6.
Detection of cytoplasmic granules composed of NP, NP-bound nucleic acids, and NS1 in RAP55-overexpressing cells. (A) HEK 293T cells were first transfected with a RAP55 expression vector and then infected with WSN virus at an MOI of 1 at 24 h posttransfection. NS1 and NP expression was examined by immunostaining WSN-infected HEK 293T cells in the presence or absence of RAP55 overexpression. White arrowheads indicate cells with low levels of NP expression in RAP55-overexpressing cell cultures. Yellow boxes indicate a complex where colocalization of NS1, NP, and NP-bound nucleic acids occurs; this area is shown in an enlarged view in the third row. Overexpression of RAP55 in HEK 293T cells was examined prior to virus infection (0 h) to ensure that most of the cells had been transfected successfully (bottom row). (B) HEK 293T cells were transfected with RAP55 expression vector and then infected with WSN virus at an MOI of 1. At 4 h postinfection, the NS1 and RAP55 proteins were detected by costaining with rabbit anti-NS1 polyclonal antibodies (red) and mouse anti-RAP55 monoclonal antibodies (green). Viral mRNA and vRNA were detected by FISH as described in Materials and Methods (blue). To enhance the image contrast, single-color panels were adjusted to a monochrome presentation, while the merge panels are shown in color.
DISCUSSION
Influenza A viruses exhibit different degrees of virulence in humans. While seasonal influenza viruses normally cause mild self-limited upper respiratory tract infections, the highly pathogenic 1918 pandemic H1N1 and avian H5N1 and H7N7 subtypes are known to cause severe, sometimes fatal pneumonia in infected patients (1, 12, 54). It has been suggested that the highly virulent properties of these viruses may be attributed to their high replication efficiency, with moderate- to low-pathogenicity strains exhibiting correspondingly lower replication efficiencies (20, 42, 49). While influenza virus uses viral RNP complexes, which are composed of NP and the three polymerase subunits (PA, PB1, and PB2), for replication of the viral genome, synthesis of viral proteins relies completely on the host machinery. Exactly how interactions between viral elements and the host machinery for mRNA translation regulate influenza virus protein synthesis is less clear. The nonstructural (NS1) protein of influenza A viruses is well recognized as a virulence element and for its function in antagonizing the host immune response (11, 15, 18, 25). It has been suggested that there may be multiple levels of interaction between NS1 and components of the host antiviral response (25, 44). It was also recently reported that NS1 interacts with NP, although the details of the biological significance of this interaction remain unknown (41). NS1 traffics between the nucleus and the cytoplasm during virus replication and localizes predominantly to the cytoplasm soon after virus infection of cells (Fig. 6). A previous report on the NS1 nuclear export process also suggested that there may be an interaction between NS1 and another virus-specific protein in virus-infected cells (28). The NS1 protein is known to inhibit PKR activation during influenza virus infection, but the molecular basis of the antagonism of PKR signaling in virus replication is not clear. These clues provided a strong indication that NS1 may also be involved in other virus replication processes in the cellular cytoplasm. Revealing the mechanism for the regulation of virus protein synthesis in infected cells will therefore provide new insights into the molecular basis of influenza A virus replication.
This study identified an interaction between NS1 and the cellular factor RAP55 and further characterized the relationship between NS1 and the formation of RAP55-associated stress granules and P-bodies during influenza virus replication. RAP55 is an RNA binding protein known to be associated with stress granules and also suggested to have a function in shuttling mRNAs between P-bodies and stress granules (8, 33, 53). We confirmed that NS1 is associated with P-bodies, as evidenced by colocalization with RAP55 and the P-body-specific components Ago1, Ago2, and DCP1a (Fig. 1). As summarized in a recent review, there are different mechanisms for viruses to inhibit the formation of stress granules (52). The functional role of RAP55 in the formation of P-bodies and stress granules remains unclear. One previous study using siRNA to knock down RAP55 found that reducing the cellular levels of RAP55 could alter the structure of P-bodies and decrease the number of RAP55-containing P-bodies (53), suggesting that RAP55 may play an important role in the formation of P-bodies. Here we found that RAP55 negatively regulates RNP polymerase activity and virus replication (Fig. 3A and B), presumably through recruitment of RAP55-associated P-bodies/stress granules, which act to silence viral protein synthesis. However, RAP55-associated negative regulation of virus replication in the early phase of virus infection is later released, as the number of RAP55-associated P-bodies decreases along the course of virus infection (Fig. 3C, D, And E), possibly due to counteraction by the NS1 protein. This study also provides evidence that through interaction with RAP55, NS1 may be able to inhibit formation of NP-associated stress granules, thereby retaining NP in P-bodies, which may serve as storage for stalled mRNA and may release the mRNA when subsequently dissociated by NS1, making the mRNA available for active translation. It remains to be proven whether this effect is mediated solely by interaction between NS1 and RAP55 or if it also involves other stress granule/P-body components. While a recent study suggested that NS1 inhibits stress granule formation through interaction with the cellular PKR pathway (24), this study shows that the RNA binding property of NS1 is important to but does not exclusively determine the interaction between NS1 and RAP55. Viral elements for both RNA binding and interaction with PKR are required for interaction with RAP55, and NS1 mutants which cannot interact with RAP55 also fail to inhibit the formation of stress granules in cells. It has been suggested that NS1 may inhibit PKR activation by relocating bound PKR to the nucleus to prevent phosphorylation of the eIF2α substrate (36). Interestingly, three mutant forms of the NS1 protein, the R38A/K41A, R46A, and I123A/M124A/K126A/N127A mutants, all of which have lost the ability to inhibit formation of RAP55-induced stress granules, localized predominantly within the cytoplasm in a transfection assay (Fig. 2). It remains to be investigated whether nuclear localization of the NS1 protein is required for NS1-mediated inhibition of stress granule formation.
Cellular stress granules and P-bodies are formed in response to stimuli such as virus infection. This study revealed that viral NP, but not the other subunits of the viral polymerase complex, is able to induce stress granule formation in the absence of NS1 expression (Fig. 4A). Characterization of the interaction between NP and RAP55 suggested that RAP55 may play a role in targeting NP, and probably also NP-bound viral RNA, to stress granules, causing viral mRNA translation to be stalled. Results from cells infected with either wild-type virus or a mutated virus containing a nonfunctional R35A form of NS1 demonstrated that NS1 is essential for dissociation of (or inhibiting the formation of) NP-induced P-bodies and stress granules (Fig. 5F). This study reveals a previously undescribed interplay between NS1 and a host restriction strategy. It seems that in response to virus infection, cells have adapted a mechanism to sequester NP and NP-bound RNA in P-bodies/stress granules to either stall translation or cause the decay of virus mRNA, restricting virus replication. However, NS1 antagonizes this host restriction mechanism and restores virus protein synthesis by stabilizing virus mRNA in the cytoplasm to enable replication (Fig. 6B). In this working model (Fig. 7), influenza virus-infected cells are initially stimulated by NP protein through an as yet undefined mechanism to target NP to the RNA granules (stress granules and P-bodies) and stall the translation of viral proteins, as NS1 levels are low early in infection. However, expression of functional NS1, which increases along the course of infection, eventually overcomes host restriction by inhibiting formation of stress granules via inhibition of PKR activation, as described here and in another recent report (24). The results presented here further reveal that NS1 inhibits formation of stress granules and P-bodies through interaction with RAP55, which is a core component of both types of RNA granules.
Fig 7.
Working model for the interplay between NS1 and NP-induced stress granules (SG) and P-bodies (PB) during influenza A virus infection. Influenza virus mRNA is synthesized by a messenger ribonucleoprotein (mRNP) complex in the nucleus and exported to the cytoplasm for protein translation. Expression of NP during virus infection induces assembly of P-bodies and stress granules, which may trap NP and NP-bound viral RNA and thereby stall viral mRNA translation. Expression of NS1 dissociates (or inhibits formation of) NP-associated P-bodies/stress granules and releases NP and NP-bound mRNA, allowing translation of viral proteins.
P-bodies have also been reported to serve as storage vessels for cellular 5′-mRNA caps for the viral cap-snatching mechanism of hantaviruses (37). Influenza virus transcription of viral mRNA also requires cellular 5′-mRNA primers obtained through a cap-snatching mechanism (40). It remains to be investigated if the interaction between NS1 and P-bodies is also involved in the storage of cellular 5′-mRNA caps for influenza virus transcription. While the manuscript for this study was under revision, a study was published which showed that RAP55 (LSm14A) is able to mediate IRF3 activation and induce IFN-β induction in response to virus infection, suggesting that RAP55 may serve as a sensor for detecting virus in P-bodies (27). In animal studies using reassortant viruses, the nonstructural protein of influenza virus, NS1, has been identified as one of the key viral elements associated with the highly virulent features of some influenza A viruses, including 1918 pandemic H1N1 and avian H5N1 viruses, in mammalian hosts (5, 22, 43). How NS1 may contribute to the virulence properties of different influenza viruses is not clear. It will be important to investigate if there is any variation in the activity of NS1 proteins from these highly pathogenic strains in terms of their efficiency in dissociating NP-associated stress granules and P-bodies through interaction with RAP55, leading to enhanced replication efficiency in cells.
ACKNOWLEDGMENTS
We thank Yee-Joo Tan (Department of Microbiology, Yong Loo Lin School of Medicine, National University of Singapore, and Institute of Molecular and Cell Biology, A*STAR, Singapore) for the NS1 polyclonal antibody.
This study was supported partially by the Research Grants Council of the Hong Kong SAR (grants 7500/06M and 7620/10M), the Areas of Excellence Scheme of the University Grants Committee (grant AoE/M-12/06), the Research Fund for the Control of Infectious Diseases of the Health, Welfare and Food Bureau of the Hong Kong SAR, the National Institutes of Health (NIAID contract HHSN2662007 00005C), and the Providence Foundation Limited, in memory of the late Lui Hac Minh.
Footnotes
Published ahead of print 12 September 2012
REFERENCES
- 1. Abdel-Ghafar AN, et al. 2008. Update on avian influenza A (H5N1) virus infection in humans. N. Engl. J. Med. 358:261–273 [DOI] [PubMed] [Google Scholar]
- 2. Albrecht M, Lengauer T. 2004. Novel Sm-like proteins with long C-terminal tails and associated methyltransferases. FEBS Lett. 569:18–26 [DOI] [PubMed] [Google Scholar]
- 3. Aragon T, et al. 2000. Eukaryotic translation initiation factor 4GI is a cellular target for NS1 protein, a translational activator of influenza virus. Mol. Cell. Biol. 20:6259–6268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Arribere JA, Doudna JA, Gilbert WV. 2011. Reconsidering movement of eukaryotic mRNAs between polysomes and P bodies. Mol. Cell 44:745–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Basler CF, et al. 2001. Sequence of the 1918 pandemic influenza virus nonstructural gene (NS) segment and characterization of recombinant viruses bearing the 1918 NS genes. Proc. Natl. Acad. Sci. U. S. A. 98:2746–2751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Beckham CJ, Parker R. 2008. P bodies, stress granules, and viral life cycles. Cell Host Microbe 3:206–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bornholdt ZA, Prasad BV. 2006. X-ray structure of influenza virus NS1 effector domain. Nat. Struct. Mol. Biol. 13:559–560 [DOI] [PubMed] [Google Scholar]
- 8. Buchan JR, Parker R. 2009. Eukaryotic stress granules: the ins and outs of translation. Mol. Cell 36:932–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Burgui I, Aragon T, Ortin J, Nieto A. 2003. PABP1 and eIF4GI associate with influenza virus NS1 protein in viral mRNA translation initiation complexes. J. Gen. Virol. 84:3263–3274 [DOI] [PubMed] [Google Scholar]
- 10. Chen Z, Li Y, Krug RM. 1999. Influenza A virus NS1 protein targets poly(A)-binding protein II of the cellular 3′-end processing machinery. EMBO J. 18:2273–2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Egorov A, et al. 1998. Transfectant influenza A viruses with long deletions in the NS1 protein grow efficiently in Vero cells. J. Virol. 72:6437–6441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fouchier RA, et al. 2004. Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proc. Natl. Acad. Sci. U. S. A. 101:1356–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Franks TM, Lykke-Andersen J. 2008. The control of mRNA decapping and P-body formation. Mol. Cell 32:605–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gack MU, et al. 2009. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe 5:439–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Garcia-Sastre A, et al. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252:324–330 [DOI] [PubMed] [Google Scholar]
- 16. Hale BG, Jackson D, Chen YH, Lamb RA, Randall RE. 2006. Influenza A virus NS1 protein binds p85beta and activates phosphatidylinositol-3-kinase signaling. Proc. Natl. Acad. Sci. U. S. A. 103:14194–14199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hale BG, et al. 2010. Structural insights into phosphoinositide 3-kinase activation by the influenza A virus NS1 protein. Proc. Natl. Acad. Sci. U. S. A. 107:1954–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hale BG, Randall RE, Ortin J, Jackson D. 2008. The multifunctional NS1 protein of influenza A viruses. J. Gen. Virol. 89:2359–2376 [DOI] [PubMed] [Google Scholar]
- 19. Hale BG, et al. 2010. Inefficient control of host gene expression by the 2009 pandemic H1N1 influenza A virus NS1 protein. J. Virol. 84:6909–6922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hatta M, Gao P, Halfmann P, Kawaoka Y. 2001. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science 293:1840–1842 [DOI] [PubMed] [Google Scholar]
- 21. Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG. 2000. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc. Natl. Acad. Sci. U. S. A. 97:6108–6113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Imai H, et al. 2010. The HA and NS genes of human H5N1 influenza A virus contribute to high virulence in ferrets. PLoS Pathog. 6:e1001106 doi:10.1371/journal.ppat.1001106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jakymiw A, et al. 2005. Disruption of GW bodies impairs mammalian RNA interference. Nat. Cell Biol. 7:1267–1274 [DOI] [PubMed] [Google Scholar]
- 24. Khaperskyy DA, Hatchette TF, McCormick C. 2012. Influenza A virus inhibits cytoplasmic stress granule formation. FASEB J. 26:1629–1639 [DOI] [PubMed] [Google Scholar]
- 25. Kochs G, Garcia-Sastre A, Martinez-Sobrido L. 2007. Multiple anti-interferon actions of the influenza A virus NS1 protein. J. Virol. 81:7011–7021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li S, Min JY, Krug RM, Sen GC. 2006. Binding of the influenza A virus NS1 protein to PKR mediates the inhibition of its activation by either PACT or double-stranded RNA. Virology 349:13–21 [DOI] [PubMed] [Google Scholar]
- 27. Li Y, et al. 2012. LSm14A is a processing body-associated sensor of viral nucleic acids that initiates cellular antiviral response in the early phase of viral infection. Proc. Natl. Acad. Sci. U. S. A. 109:11770–11775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li Y, Yamakita Y, Krug RM. 1998. Regulation of a nuclear export signal by an adjacent inhibitory sequence: the effector domain of the influenza virus NS1 protein. Proc. Natl. Acad. Sci. U. S. A. 95:4864–4869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lian SL, et al. 2009. The C-terminal half of human Ago2 binds to multiple GW-rich regions of GW182 and requires GW182 to mediate silencing. RNA 15:804–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lieb B, Carl M, Hock R, Gebauer D, Scheer U. 1998. Identification of a novel mRNA-associated protein in oocytes of Pleurodeles waltl and Xenopus laevis. Exp. Cell Res. 245:272–281 [DOI] [PubMed] [Google Scholar]
- 31. Ludwig S, Schultz U, Mandler J, Fitch WM, Scholtissek C. 1991. Phylogenetic relationship of the nonstructural (NS) genes of influenza A viruses. Virology 183:566–577 [DOI] [PubMed] [Google Scholar]
- 32. Marion RM, Zurcher T, de la Luna S, Ortin J. 1997. Influenza virus NS1 protein interacts with viral transcription-replication complexes in vivo. J. Gen. Virol. 78:2447–2451 [DOI] [PubMed] [Google Scholar]
- 33. Marnef A, Sommerville J, Ladomery MR. 2009. RAP55: insights into an evolutionarily conserved protein family. Int. J. Biochem. Cell Biol. 41:977–981 [DOI] [PubMed] [Google Scholar]
- 34. Matsumoto K, et al. 2012. PRMT1 is required for RAP55 to localize to processing bodies. RNA Biol. 9:610–623 [DOI] [PubMed] [Google Scholar]
- 35. Min JY, Krug RM. 2006. The primary function of RNA binding by the influenza A virus NS1 protein in infected cells: inhibiting the 2′-5′ oligo(A) synthetase/RNase L pathway. Proc. Natl. Acad. Sci. U. S. A. 103:7100–7105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Min JY, Li S, Sen GC, Krug RM. 2007. A site on the influenza A virus NS1 protein mediates both inhibition of PKR activation and temporal regulation of viral RNA synthesis. Virology 363:236–243 [DOI] [PubMed] [Google Scholar]
- 37. Mir MA, Duran WA, Hjelle BL, Ye C, Panganiban AT. 2008. Storage of cellular 5′ mRNA caps in P bodies for viral cap-snatching. Proc. Natl. Acad. Sci. U. S. A. 105:19294–19299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nemeroff ME, Barabino SM, Li Y, Keller W, Krug RM. 1998. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′end formation of cellular pre-mRNAs. Mol. Cell 1:991–1000 [DOI] [PubMed] [Google Scholar]
- 39. Pichlmair A, et al. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314:997–1001 [DOI] [PubMed] [Google Scholar]
- 40. Plotch SJ, Bouloy M, Krug RM. 1979. Transfer of 5′-terminal cap of globin mRNA to influenza viral complementary RNA during transcription in vitro. Proc. Natl. Acad. Sci. U. S. A. 76:1618–1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Robb NC, et al. 2011. The influenza A virus NS1 protein interacts with the nucleoprotein of viral ribonucleoprotein complexes. J. Virol. 85:5228–5231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Salomon R, et al. 2006. The polymerase complex genes contribute to the high virulence of the human H5N1 influenza virus isolate A/Vietnam/1203/04. J. Exp. Med. 203:689–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Seo SH, Hoffmann E, Webster RG. 2002. Lethal H5N1 influenza viruses escape host anti-viral cytokine responses. Nat. Med. 8:950–954 [DOI] [PubMed] [Google Scholar]
- 44. Steidle S, et al. 2010. Glycine 184 in nonstructural protein NS1 determines the virulence of influenza A virus strain PR8 without affecting the host interferon response. J. Virol. 84:12761–12770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tan Z, et al. 2010. A new panel of NS1 antibodies for easy detection and titration of influenza A virus. J. Med. Virol. 82:467–475 [DOI] [PubMed] [Google Scholar]
- 46. Tanaka KJ, et al. 2006. RAP55, a cytoplasmic mRNP component, represses translation in Xenopus oocytes. J. Biol. Chem. 281:40096–40106 [DOI] [PubMed] [Google Scholar]
- 47. Thomas MG, Loschi M, Desbats MA, Boccaccio GL. 2011. RNA granules: the good, the bad and the ugly. Cell Signal. 23:324–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tritschler F, et al. 2009. DCP1 forms asymmetric trimers to assemble into active mRNA decapping complexes in metazoa. Proc. Natl. Acad. Sci. U. S. A. 106:21591–21596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tumpey TM, et al. 2005. Characterization of the reconstructed 1918 Spanish influenza pandemic virus. Science 310:77–80 [DOI] [PubMed] [Google Scholar]
- 50. Wang P, et al. 2009. Nuclear factor 90 negatively regulates influenza virus replication by interacting with viral nucleoprotein. J. Virol. 83:7850–7861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang W, et al. 1999. RNA binding by the novel helical domain of the influenza virus NS1 protein requires its dimer structure and a small number of specific basic amino acids. RNA 5:195–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. White JP, Lloyd RE. 2012. Regulation of stress granules in virus systems. Trends Microbiol. 20:175–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yang WH, Yu JH, Gulick T, Bloch KD, Bloch DB. 2006. RNA-associated protein 55 (RAP55) localizes to mRNA processing bodies and stress granules. RNA 12:547–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yuen KY, et al. 1998. Clinical features and rapid viral diagnosis of human disease associated with avian influenza A H5N1 virus. Lancet 351:467–471 [DOI] [PubMed] [Google Scholar]