Abstract
We have determined the crystal structure of the broadly neutralizing antibody (bnAb) AP33, bound to a peptide corresponding to hepatitis C virus (HCV) E2 envelope glycoprotein antigenic site 412 to 423. Comparison with bnAb HCV1 bound to the same epitope reveals a different angle of approach to the antigen by bnAb AP33 and slight variation in its β-hairpin conformation of the epitope. These structures establish two different modes of binding to E2 that antibodies adopt to neutralize diverse HCV.
TEXT
Structural characterization of conserved neutralizing epitopes provides critical information for the design of vaccines to counteract genetic diversity of pathogens (2, 4, 7). The E2 antigenic site 412 to 423 is a highly conserved neutralizing determi-nant of HCV and is a prime target for vaccine design (1, 9, 11). We recently determined the crystal structure of this conserved site in complex with a human broadly neutralizing antibody (bnAb), HCV1 (6). The antibody-bound epitope forms a β-hairpin displaying a hydrophilic face and a hydrophobic face on opposing sides of the hairpin. The antibody predominantly interacts with the E2 residues Leu413 and Trp420 on the hydrophobic face of the epitope that are nearly 100% conserved (1, 6). Nevertheless, HCV can escape this antibody through mutations at other positions on the binding face, e.g., N415K (in ∼1% of circulating HCV) (1, 6).
To further characterize this important neutralizing determinant, we report a second structure of this antigenic site in complex with the bnAb AP33 (8, 9). The murine monoclonal antibody (MAb) AP33 was discovered by Patel and coworkers (8), and the antibody was found to have broad neutralizing activity to diverse HCV isolates (9). In this study, the antibody was expressed as a chimeric mouse-human antibody to facilitate expression and purification (see Fig. S1 in the supplemental material). The antibody epitope has been mapped and extensively studied by overlapping peptide scanning (8), phage-display mimotope panning (11), selection of in vitro escape mutants (3, 5), and site-directed mutagenesis (3). The E2 mutations N415Y, N415D, N417S, and G418D enable viral escape from neutralization by the MAb AP33 (3, 5).
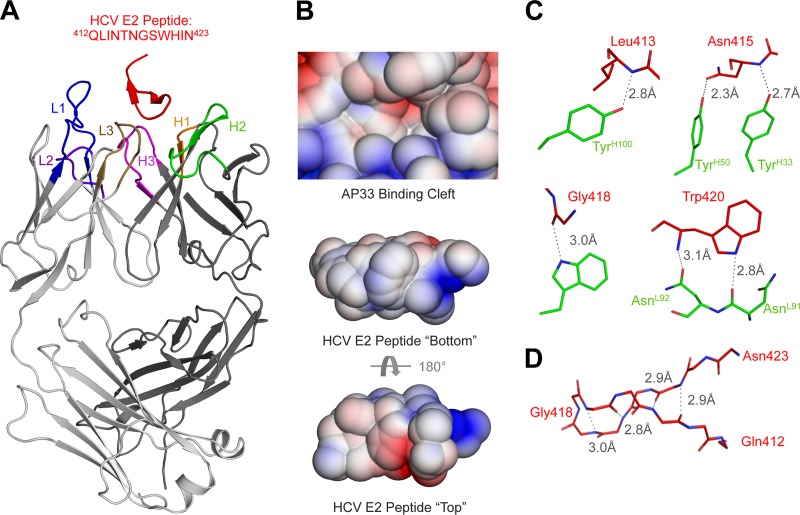
The crystal structure reveals that, similar to the binding site for the bnAb HCV1, the AP33 epitope also forms a β-hairpin sandwiched between the heavy chain (HC) and light chain (LC) of the antibody (Fig. 1A) (detailed methods are provided in the supplemental material). Most of the binding is mediated by hydrophobic interactions along the hydrophobic face of the epitope (Fig. 1B; see also Table S2 in the supplemental material). A number of hydrogen bonds also stabilize the interaction, mostly between side chains on the Fab and main chain of the peptide (Fig. 1C; see also Table S4 in the supplemental material). Overall, there are many similarities between the AP33 and HCV1 epitopes (6). The same type of β-turn (type I′) is found in both structures, and both antibodies bind the hydrophobic face of the β-hairpin (Fig. 1B; see also Table S2 in the supplemental material). However, the anti-parallel β-sheet in the β-hairpin in the AP33 epitope splays apart at the end distal from the β-turn, resulting in only 4 intrapeptide hydrogen bonds stabilizing the hairpin instead of 5 found in the HCV1 epitope (Fig. 1D) (6). Accordingly, AP33 buries less surface area around the termini than HCV1 (Fig. 2D).
Fig 1.
Crystal structure of the MAb AP33 in complex with its HCV E2 epitope. (A) The overall structure of the AP33 complex is shown with a cartoon representation. The peptide epitope (red) is bound between the heavy (dark gray) and light (light gray) chains of the Fab. (B) The adaptive Poisson-Boltzmann solver (APBS; http://www.poissonboltzmann.org/apbs/) was used to calculate the surface potential across the solvent-accessible surfaces of both the paratope (top) and the epitope (bottom) (surface potential from −3 kT/e [red] to 3 kT/e [blue] for comparison with the HCV1 antibody interaction [6]). For the peptide, the surface potential is shown looking from above the antibody (top) and for the peptide epitope (middle) viewed from the paratope and a 180° rotation below (bottom). (C) Residues of the epitope that form hydrogen bonds with the epitope are shown with a ball-and-stick representation. Hydrogen bonds are displayed as dashed lines. (D) Backbone hydrogen bonding that stabilizes the β-hairpin of the epitope is shown.
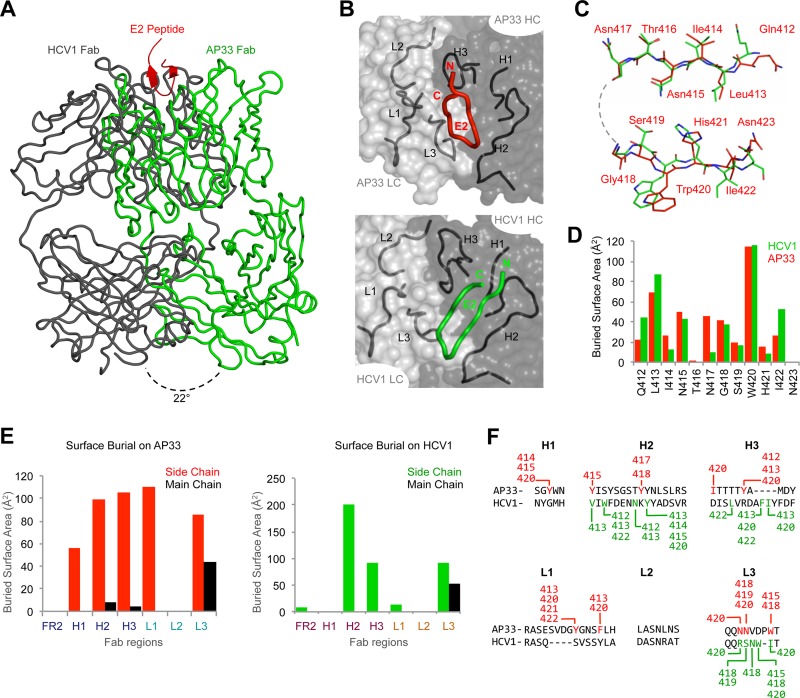
Fig 2.
Comparison of the MAb AP33 and HCV1 binding to the antigenic region. (A) Different angles of approach for AP33 and HCV1 are shown, with the superposed antigenic regions as flat strands and the antibodies as Cα ribbon traces. The angle was calculated between the VL-VH pseudo 2-fold axes in each Fab. (B) The orientations of the bound antigenic region in the AP33 (top) and HCV1 (bottom) structures are shown as ribbons relative to the combining regions. The antibody is presented as a transparent Connelly molecular surface, with the CDR loops shown as ribbons. (C) The backbone atoms of the antigenic regions in the AP33 and HCV1 structures were superimposed and are shown with a ball-and-stick representation. Larger differences are observed on the C-terminal side, which interacts with different portions of the antibodies in the two structures. (D) Burial of residues on the E2 epitope by AP33 and HCV1. (E) Side-by-side comparison of CDR loop usage of AP33 (left) and HCV1 (right), shown by buried surface area. (F) All contacts between the epitope and the CDR loops of AP33 and HCV1 are shown. The sequences of the CDR loops are aligned, and the specific E2 residues interacting with them are highlighted in red (AP33) and green (HCV1).
A direct comparison between AP33 and HCV1 structures reveals that the antibodies approach this antigenic site from different directions. When the epitopes are structurally superposed, the antibodies bind with a 22° difference in the angle of approach (Fig. 2A). Although both peptide epitopes bind in the cleft between VH and VL of the antibodies (Fig. 2B, top), in the HCV1 structure, the tip of the β-hairpin points toward VL (158-Å2 buried surface), while the majority of the β-hairpin interacts with VH (300 Å2) (Fig. 2B, bottom). In contrast, VL and VH of AP33 interact almost equally with the N- and C-terminal β-strands of the antigen (242 Å2 and 273 Å2, respectively). This difference in VL usage is highlighted when the backbone atoms of the epitopes are superposed: Gln412-Asn417, which interact with VH of both AP33 and HCV1, are highly similar between the two structures, while differences are more apparent in Ser419-Asn423, which interact mainly with VL of AP33 (Fig. 2C and F).
Despite these differences, it is clear that two independent antibody selection and maturation pathways arrived at a similar solution to engage the hydrophobic β-hairpin epitope: both events rely on hydrophobic interactions along the cleft between VL and VH. Both antibodies use primarily their side chains for interactions (Fig. 2D); coincidentally, the relatively few backbone interactions are mediated by their L3 loops (Fig. 2E). In both structures, a hydrogen bond is formed with the Gly418 tip of the peptide (Fig. 1C) and the major hydrophobic interactions are with Leu413 and Trp420 (see Tables S2 and S3 in the supplemental material). These interactions define the essential features of this antigenic site for broad neutralization of HCV. Interestingly, these similar solutions were realized using very different antibody complementarity-determining region (CDR) loops (Fig. 2E and F). For the MAb AP33, all CDR loops except L2 are involved in the interactions (Fig. 2E). For the MAb HCV1, CDR H2, H3, and L3 loops account for most interactions, although framework region (FR) 2 and L1 also contribute to some interactions (Fig. 2E). Also, MAb HCV1 binding relies on an insertion in CDR H3 that is not present in MAb AP33 (Fig. 2F). The lack of CDR L2 involvement in antigen binding for both MAbs is not unusual for small-molecule or linear peptide interactions (10, 12). The MAb AP33, on the other hand, has a longer L1, with inserted residues immediately upstream of a contacting residue in the CDR L1 loop of the paratope (Fig. 2F).
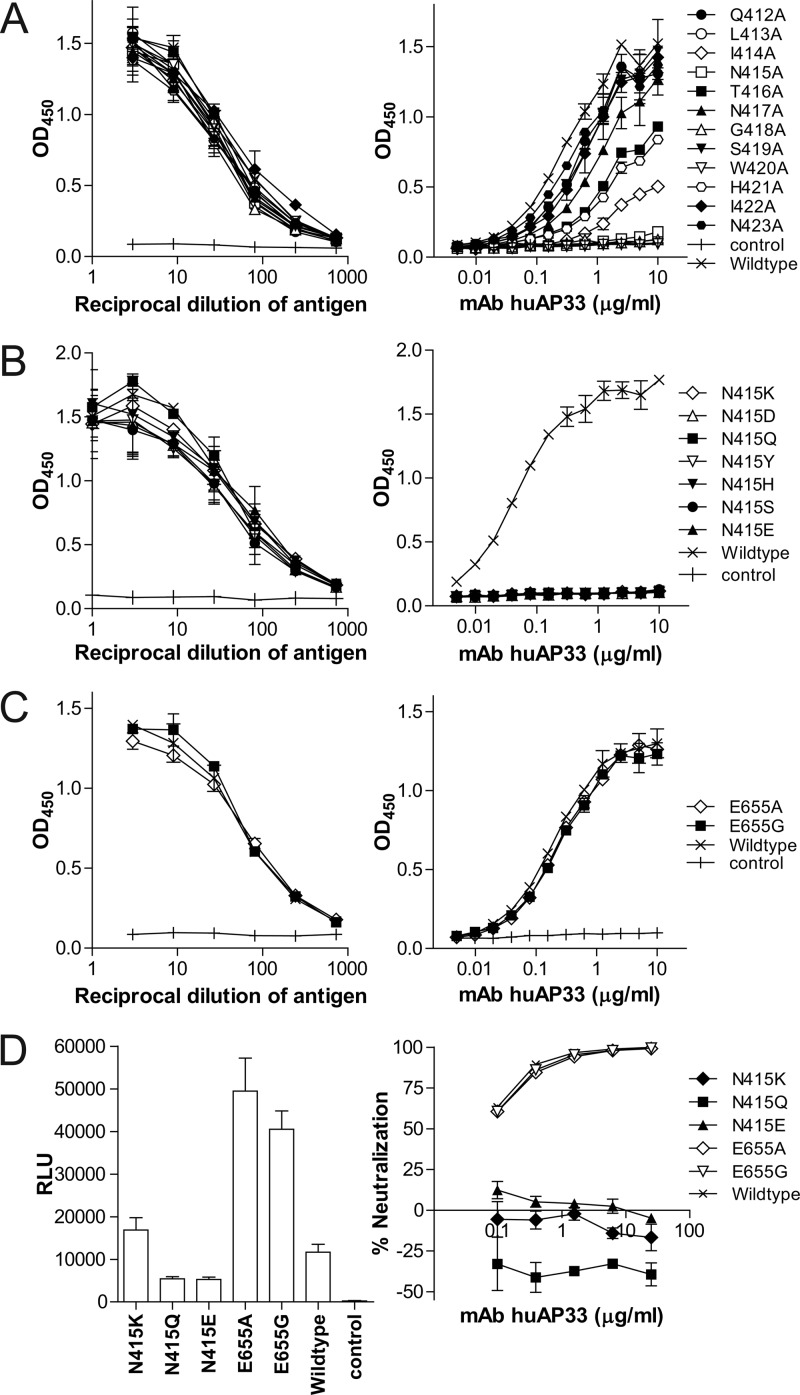
The structure also helps explain how different mutations escape the MAb AP33. In addition to hydrophobic interactions with E2 Leu413 and Trp420, binding of the MAb AP33 also requires hydrogen bonding to Asn415 and Gly418 (Fig. 1C and 3A), thus explaining some of the known escape mutants of the antibody (N415Y and N415D) (3, 5). AP33 also interacts with Gly418 and may not accommodate substitutions with bulkier side chains, such as those for the known escape mutant G418D. Interestingly, the antibody forms two hydrogen bonds with both the main chain and the side chain of Asn415 (Fig. 1C; see also Table S4 in the supplemental material), whereas only one hydrogen bond to the main chain of that residue is found in the HCV1 structure (6). Consequently, the MAb AP33 does not bind mutants with Asn415 replaced by any of the residues found in natural variants (Fig. 3B) and does not neutralize infectious pseudotype virus harboring these mutations, in contrast to HCV1 (Fig. 3D) (6). In addition, an E655G mutation has been reported to help the virus to escape the MAb AP33 (5). E2 Glu655 is found in ∼74% of known HCV isolates, and the E655G mutation does not appear to be directly involved in forming the AP33 epitope (Fig. 3C). Instead, the mutation enhances viral infectivity (Fig. 3D), which may help compensate for reduced viral infectivity due to mutations in the conserved AP33 epitope (5).
Fig 3.
Escape mutants of MAb AP33. Binding of MAb AP33 to alanine scanning mutants of E2 region 412 to 423 (A) or mutants with a substitution at position 415 (B) or 655 (C). (D) Resistance to MAb AP33 by mutations leading to loss of binding. The antibody used in this study (huAP33) is a chimeric form of the MAb, with the murine variable domains fused to human kappa LC and IgG1 HC constant regions. Functional verification of this chimeric MAb is provided in Fig. S1 in the supplemental material.
The comparative study of two independent structures of the conserved E2 antigenic site 412 to 423 further defines it as a prime vaccine target and demonstrates how antibodies of different genetic origins adopt similar solutions to recognizing this type-I′ β-hairpin on E2 for neutralization. The structures provide useful information for the design of immunogens that will optimally present this site of vulnerability. An immunogen directing neutralizing antibody responses to this conserved antigenic site may form the basis of a broadly effective HCV vaccine.
Protein structure accession number.
The atomic coordinates have been deposited in the Protein Data Bank under PBD identification (ID) code 4G6A.
Supplementary Material
ACKNOWLEDGMENTS
We thank Kristin Cogburn and Arthur Kim for technical support in cell culture and antibody production, Henry Tien and Thomas Clayton for help in setting up initial crystal screens using the Crystalmation robot, and Jane Verenini for manuscript formatting. Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource (SSRL), a Directorate of the SLAC National Accelerator Laboratory and an Office of Science User Facility operated for the U.S. Department of Energy (DOE) Office of Science by Stanford University.
The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, NIH's National Center for Research Resources Biomedical Technology Program (P41RR001209), and the National Institute of General Medical Sciences (NIGMS). This work is supported by NIH grants AI79031 (to M.L.), AI84817, and GM U54 GM094586 (to I.A.W.) and the Skaggs Institute (I.A.W.).
This is TSRI manuscript number 21874.
The authors declare no conflict of interest.
Footnotes
Published ahead of print 12 September 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Broering TJ, et al. 2009. Identification and characterization of broadly neutralizing human monoclonal antibodies directed against the E2 envelope glycoprotein of hepatitis C virus. J. Virol. 83:12473–12482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Burton DR. 2002. Antibodies, viruses and vaccines. Nat. Rev. Immunol. 2:706–713 [DOI] [PubMed] [Google Scholar]
- 3. Dhillon S, et al. 2010. Mutations within a conserved region of the hepatitis C virus E2 glycoprotein that influence virus-receptor interactions and sensitivity to neutralizing antibodies. J. Virol. 84:5494–5507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dormitzer PR, Ulmer JB, Rappuoli R. 2008. Structure-based antigen design: a strategy for next generation vaccines. Trends Biotechnol. 26:659–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gal-Tanamy M, et al. 2008. In vitro selection of a neutralization-resistant hepatitis C virus escape mutant. Proc. Natl. Acad. Sci. U. S. A. 105:19450–19455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kong L, et al. 2012. Structural basis of hepatitis C virus neutralization by broadly neutralizing antibody HCV1. Proc. Natl. Acad. Sci. U. S. A. 109:9499–9504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kwong PD, Wilson IA. 2009. HIV-1 and influenza antibodies: seeing antigens in new ways. Nat. Immunol. 10:573–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Owsianka A, Clayton RF, Loomis-Price LD, McKeating JA, Patel AH. 2001. Functional analysis of hepatitis C virus E2 glycoproteins and virus-like particles reveals structural dissimilarities between different forms of E2. J. Gen. Virol. 82:1877–1883 [DOI] [PubMed] [Google Scholar]
- 9. Owsianka A, et al. 2005. Monoclonal antibody AP33 defines a broadly neutralizing epitope on the hepatitis C virus E2 envelope glycoprotein. J. Virol. 79:11095–11104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stanfield RL, Wilson IA. 1993. X-ray crystallographic studies of antibody-peptide complexes. Immunomethods 3:211–221 [Google Scholar]
- 11. Tarr AW, et al. 2006. Characterization of the hepatitis C virus E2 epitope defined by the broadly neutralizing monoclonal antibody AP33. Hepatology 43:592–601 [DOI] [PubMed] [Google Scholar]
- 12. Wilson IA, Stanfield RL. 1993. Antibody-antigen interactions. Curr. Opin. Struct. Biol. 3:113–118 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.