Abstract
Noroviruses (NVs) cause the majority of cases of epidemic nonbacterial gastroenteritis worldwide and contribute to endemic enteric disease. However, the molecular mechanisms responsible for immune control of their replication are not completely understood. Here we report that the transcription factor interferon regulatory factor 1 (IRF-1) is required for control of murine NV (MNV) replication and pathogenesis in vivo. This led us to studies documenting a cell-autonomous role for IRF-1 in gamma interferon (IFN-γ)-mediated inhibition of MNV replication in primary macrophages. This role of IRF-1 in the inhibition of MNV replication by IFN-γ is independent of IFN-αβ signaling. While the signal transducer and activator of transcription STAT-1 was also required for IFN-γ-mediated inhibition of MNV replication in vitro, class II transactivator (CIITA), interferon regulatory factor 3 (IRF-3), and interferon regulatory factor 7 (IRF-7) were not required. We therefore hypothesized that there must be a subset of IFN-stimulated genes (ISGs) regulated by IFN-γ in a manner dependent only on STAT-1 and IRF-1. Analysis of transcriptional profiles of macrophages lacking various transcription factors confirmed this hypothesis. These studies identify a key role for IRF-1 in IFN-γ-dependent control of norovirus infection in mice and macrophages.
INTRODUCTION
Noroviruses (NVs) are a genus of positive-sense RNA viruses that are the major etiologic agent of epidemic nonbacterial gastroenteritis worldwide (11, 12). Symptoms of human norovirus infection are generally self-limiting and typically resolve within 72 h (12), which is consistent with a role for innate antiviral mechanisms such as interferons (IFNs) in control of symptoms and replication. Both type I (IFN-αβ) and II (IFN-γ) IFNs decrease viral RNA and protein expression from a human norovirus replicon (6, 7). Murine noroviruses (MNVs) are culturable viruses that share structural, biochemical, and genetic features with human noroviruses and provide a tractable small-animal model for investigation of NV immunity, pathogenesis, and IFN-mediated control of NV replication (4, 8, 17, 41, 42).
MNVs replicate in myeloid cells in culture and in vivo (40, 41). Both IFN-γ and IFN-αβ play key roles in control of MNV infection in vivo, since mice deleted of both the IFN-γ receptor (IFN-γR−/−) and the IFN-αβ receptor (IFN-αβR−/−) are more susceptible to lethal infection than mice deleted of either IFN receptor individually (15, 17). Pretreatment with either IFN-αβ or IFN-γ limits MNV replication in cultured macrophages and dendritic cells (8, 15). These studies show, together with studies of human norovirus replicons (6, 7), that IFNs exert direct antiviral effects that inhibit NV replication in permissive cells. Furthermore, IFN-αβ exerts autocrine control of MNV replication in primary cell cultures, as MNV replicates to a higher titer in cells lacking the IFN-αβR than in control cells (41).
IFNs exert direct antiviral effects via induction of the transcription of multiple potentially antiviral genes (IFN-stimulated genes [ISGs]). After binding to their respective receptors, IFN-αβ and IFN-γ induce cellular transcription via Janus kinase (JAK)-signal transducer and activator of transcription (STAT) signaling pathways (1, 25, 33). STAT-1 is an essential component of both the IFN-αβ and IFN-γ signaling pathways, and a key role for STAT-1 in control of MNV replication is well established. STAT-1-deficient mice (STAT-1−/−), like mice lacking both the IFN-αβR and the IFN-γR (IFN-αβγR−/−), succumb to MNV infection and have elevated viral titers in multiple tissues (17, 27). Moreover, similar to cells lacking the IFN-αβR, cells lacking STAT-1 replicate MNV to a higher level than wild-type (WT) cells (41). In addition to STAT-1, the autophagy proteins Atg5, Atg16, and Atg7, but not the degradative function of autophagy, are required for IFN-γ-dependent control of MNV replication in macrophages (15). However, factors other than STAT-1 and specific autophagy proteins that are involved in IFN-mediated control of MNV replication are less well defined.
Interferon regulatory factor 1 (IRF-1) is induced by IFNs via STAT-1 signaling (1). IRF-1 is a transcription factor that has a role in the stimulation of cytokines, chemokines, and some ISGs in response to IFN-γ (18–20). Importantly, IRF-1 is capable of activating antiviral programs against a diverse range of RNA viruses (16, 29, 31) and is required for IFN-γ-mediated inhibition of encephalomyocarditis virus (EMCV) and West Nile virus (WNV) replication in cultured cells (2, 18). In addition, IRF-1 has been implicated in IFN-αβ-mediated direct antiviral effects against EMCV (18) and is important for IFN-αβ production in response to poly(I)-poly(C) in vitro but not in response to Newcastle disease virus (24), WNV (2), or poly(I)-poly(C) (30) in vivo. IRF-1 plays additional roles in innate and adaptive immunity. IRF-1-deficient (IRF-1−/−) mice exhibit defects in CD8 T cell and natural killer (NK) cells (24, 28) and exhibit increased TH2 skewing and decreased production of IFN-γ by CD4 T cells as a result of dysregulation of interleukin-12 (22). Thus, the in vivo role of IRF-1 during a given viral infection could reflect a combination of direct effects on viral replication in permissive cells and effects on the cellular innate and adaptive immune responses to infection.
Here we demonstrate that IRF-1 has important in vivo effects on MNV replication and pathogenesis that are independent of effects of IFN-αβ and are consistent with a direct antiviral role for IRF-1 in IFN-γ-mediated inhibition of MNV replication in primary macrophages. Studies in cultured macrophages confirm a key cell-autonomous role for IRF-1 in IFN-γ-mediated inhibition of MNV replication, and this role does not require autocrine secretion of IFN-αβ. Importantly, the role for IRF-1 is highly specific, since IFN-γ does not require other signaling pathways, including those dependent on class II transactivator (CIITA), interferon regulatory factor 3 (IRF-3), or interferon regulatory factor 7 (IRF-7), to elicit an antiviral state for MNV. Confirming this specificity, IFN-γ induces changes in the expression of many genes, but only a subset of these depends on both STAT-1 and IRF-1 signaling.
MATERIALS AND METHODS
Virus stocks and plaque assay.
Plaque-purified MNV-1.CW3 was used for all experiments (38). Concentrated virus stocks were prepared by infecting RAW 264.7 cells (ATCC, Manassas, VA) (4). Viral titers were determined by a previously described plaque assay (41), with the following modifications. Suspension-adapted RAW 264.7 cells were plated at a density of 2 × 106 cells per well in 6-well plates and allowed to adhere overnight at 37°C and 5% CO2. Cells were inoculated for 1 h at room temperature on a rocker, after which the inoculum was removed and cells were overlaid with 2 ml/well of Eagle minimum essential medium (Mediatech, Manassas, VA) containing 10% fetal bovine serum (FBS), 2 mM l-glutamine, 100 μg/ml penicillin, 100 μg/ml streptomycin, 10 mM HEPES, and 1% methylcellulose (viscosity, 1,500 centipoise; Sigma, St. Louis, MO). After 2 to 4 days of incubation at 37°C and 5% CO2, the overlay was removed and plaques were visualized with 1% crystal violet in 20% ethanol.
Mice, inoculations, and tissue preparation.
Mice were bred and housed under specific-pathogen-free conditions in accordance with federal and university regulations (3). All mice used were either derived from C57/BL6 embryonic stem (ES) cells or extensively backcrossed to C57/BL6 mice. IRF-1−/− and CIITA-deficient (CIITA−/−) mice were purchased from The Jackson Laboratory, Bar Harbor, ME. IFN-αβR−/− and IFN-γR−/− (17, 26) mice were originally obtained from Michel Aguet (Swiss Institute for Experimental Cancer Research, Lausanne, Switzerland). IRF-3 and IRF-7 doubly deficient (IRF3×7−/−) mice (10) were a generous gift from Michael Diamond (Washington University, St. Louis, MO). Seven- to 9-week-old mice were orally inoculated with 3 × 105 PFU of MNV-1.CW3 (38). Five days after inoculation, tissues were harvested into 2-ml sterile, screw-top tubes containing 250 μl of 1-mm zirconia-silica beads (BioSpec Products, Bartlesville, OK) and stored at −80°C. Tissues were homogenized in 1 ml of complete Dulbecco modified Eagle medium (DMEM) by use of a MagNA Lyser machine (Roche Applied Science, Hague Road, IN), and virus titers were determined by plaque assay. For lethality and in vivo titer experiments involving blocking of IFN-αβ signaling, 2.5 mg of purified endotoxin-free IFNAR-1-blocking antibody (MAR1-5A3) (32) or isotype-matched MOPC-21 antibody (BioXcell, West Lebanon, NH) was administered intraperitoneally 1 day prior to infection. For lethality experiments, a second dose of 0.5 mg was administered intraperitoneally 4 days after infection.
Cells, infections, and IFN treatment.
Primary bone marrow-derived macrophages were prepared as described previously (15). Briefly, bone marrow (43) was incubated in culture medium (DMEM supplemented with 10% FBS, 5% defined equine serum, 10% CMG14-12 supernatant [37], 1% MEM nonessential amino acids, 1 mM sodium pyruvate, 2 mM l-glutamine) for 7 days, after which adherent cells were harvested and replated on fresh culture medium. For in vitro infections, cells were plated at a density of 1 × 105 cells/well in 24-well dishes or 1 × 106 cells/well in 6-well dishes. Infections were performed at day 10 of in vitro culture after 12 to 16 h of treatment with recombinant IFN-γ or IFN-β (R&D Systems, Minneapolis, MN) at the indicated doses (15, 41). After 30 min of absorption at 4°C, cells were washed and IFNs added back into treated cultures. Cells were harvested by freezing at −80°C.
Microarray assay and meta-analysis of differentially expressed genes.
To identify genes that are regulated by IFN-γ in a STAT-1- and IRF-1-dependent manner, RNAs collected from IFN-γ-treated and untreated C57BL/6 (WT), IRF-1−/−, STAT-1−/−, IRF3×7−/−, and CIITA−/− bone marrow-derived macrophages were profiled using Affymetrix Mouse 1.0 Gene ST arrays. Briefly, bone marrow-derived macrophages were prepared as described above. After 7 days of in vitro culture, adherent cells were harvested and replated at a density of 5 × 106 cells/well in 10-cm dishes. RNA was collected at day 10 of in vitro culture after 12 h of treatment with 10 U/ml of recombinant IFN-γ. The microarray assays were performed in three independent experiments. Microarray data were normalized using the robust multiarray average normalization routine in Affymetrix's Expression Console software. Differential expression analysis was carried out in Matlab. Three different comparison schemes were employed. First, the gene expression profile of untreated WT bone marrow-derived macrophages was compared to the profiles of untreated IRF-1−/−, STAT-1−/−, IRF3×7−/−, and CIITA−/− bone marrow-derived macrophages. Second, the gene expression profile of IFN-γ treated WT bone marrow-derived macrophages was directly contrasted with that of untreated WT bone marrow-derived macrophages. Lastly, the gene expression profiles of the various IFN-γ-treated macrophages were normalized to the respective control profiles from untreated macrophages and then evaluated for significant differences in fold change compared to the similarly normalized profile of IFN-γ-treated WT macrophages. The statistical significance of the difference in mean expression levels for each gene was assessed by performing a standard two-tailed, two-sample unpaired t test, assuming unknown and unequal variances between the samples. Genes were also independently ranked based on their significance in separating two labeled groups, which was assessed by computing the area between the empirical receiver operating characteristic (ROC) curve and a random binary classifier slope. Gene expression differences were considered significant if the t test-based nominal P value was <0.05 and the area between the ROC curves was >0.25. Differentially expressed genes were evaluated for pathway enrichment by gene set overlap analysis using mSigDB (34a).
Validation of microarray analyses.
To validate our microarray findings, a subset of genes was selected for analysis by quantitative reverse transcription-PCR (qRT-PCR) (primer sequences are given in Table 1). Total RNAs were isolated from IFN-γ-treated and untreated C57BL/6, IRF-1−/−, STAT-1−/−, IRF3×7−/−, and CIITA−/− bone marrow-derived macrophages, and cDNAs were synthesized (13, 14). qRT-PCR was performed with SYBR green (Invitrogen, Carlsbad, CA). Viral transcript levels were normalized to the level of transcription of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) within each sample, using the ΔΔCT method (where CT is the threshold cycle) (21).
Table 1.
Primers used in this study
| Gene | Orientation | Primer sequence (5′–3′) |
|---|---|---|
| EG634650 | Forward | AGTGGAGCAGTGCCTTGTCTGC |
| Reverse | TGGAGCGTTTCTGTGGGAAGCC | |
| CH25H | Forward | ACGTCCTGATCTGCCTGCTGC |
| Reverse | TATTGGGTCGCCAGCGCGAAG | |
| PXMP2 | Forward | TGCTGGATCGCCTGTTCTTTGCC |
| Reverse | TTTGCAGTGCGGGCCAGAAG | |
| COLEC12 | Forward | ACCACCTGGTCCAACTGGCAAC |
| Reverse | ATCCTTTGCTGCCACGCTCTCC | |
| FABP4 | Forward | TGCTGCAGCCTTTCTCACCTGG |
| Reverse | TTGTGGCAAAGCCCACTCCC | |
| GPR33 | Forward | CATCAGCAACGGCCTCTATCT |
| Reverse | AGACCCAGTGATTGTCCTGAA | |
| GBP2 | Forward | CTGCACTATGTGACGGAGCTA |
| Reverse | GAGTCCACACAAAGGTTGGAAA | |
| CCL3 | Forward | TTCTCTGTACCATGACACTCTGC |
| Reverse | CGTGGAATCTTCCGGCTGTAG | |
| GBP1 | Forward | ACAACTCAGCTAACTTTGTGGG |
| Reverse | TGATACACAGGCGAGGCATATTA | |
| STAP1 | Forward | TCACTGCTCTGCCCCTGTA |
| Reverse | AGTAAGGCACACGAGGTCTATTA | |
| SELL | Forward | TACATTGCCCAAAAGCCCTTAT |
| Reverse | CATCGTTCCATTTCCCAGAGTC |
Statistical analysis.
All data other than microarray data were analyzed using GraphPad Prism software (GraphPad Software, San Diego, CA). For in vitro experiments, statistical analyses were performed by one-way analysis of variance (ANOVA) (with Tukey's posttest), unless specifically stated otherwise. In vivo viral titer data were analyzed with the nonparametric Mann-Whitney test. Survival curves were analyzed using the log rank test. All differences not specifically stated to be significant were insignificant (P > 0.05).
RESULTS
IRF-1 restricts MNV replication in vivo.
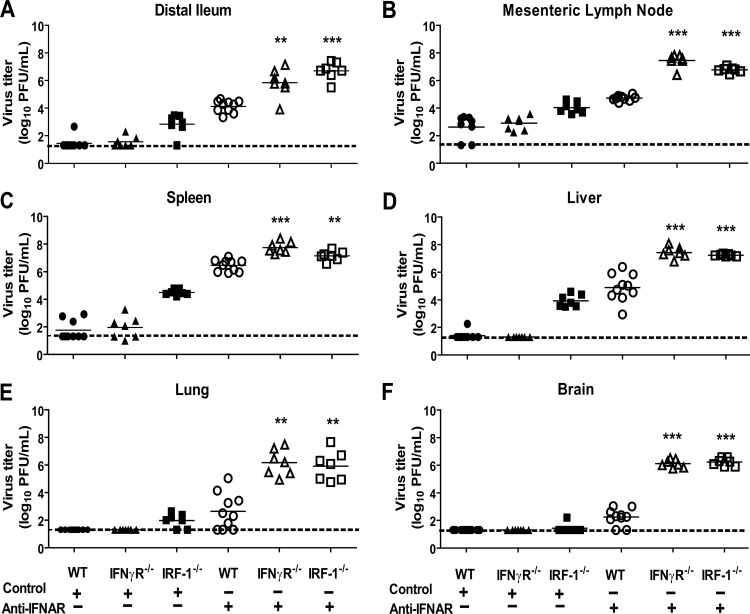
Previous studies demonstrated that MNV is controlled in vivo by a combination of redundant and unique IFN-αβ-dependent and IFN-γ-dependent mechanisms (15, 17). IFN-γ-mediated antiviral effects are revealed and can be studied when IFN-αβ signaling is inhibited (15, 17).To assess the possible contribution of IRF-1 to both IFN-αβ- and IFN-γ-dependent control of MNV in vivo, we compared viral titers in IRF-1−/−, IFN-γR−/−, and C57BL/6 (WT) control mice treated with either a monoclonal antibody (MAb) against the IFNAR-1 subunit of the IFN-αβR (anti-IFNAR) (32) or an isotype control MAb prior to oral (p.o.) inoculation with MNV. It has been shown previously that MNV replication is restricted to the intestine, spleen, and mesenteric lymph nodes (MLN) in WT mice but is disseminated and present at increased levels in mice with various immune deficiencies, including deficiencies in IFN signaling (17). IRF-1−/− mice treated with control MAb had significantly increased viral titers in the intestine (P < 0.05), spleen (P < 0.01), MLN (P < 0.01), liver (P < 0.01), and lung (P < 0.05) compared to WT and IFN-γR−/− mice treated with control MAb (Fig. 1A to F) at day 5 postinoculation. Furthermore, untreated IRF-1−/− mice had significantly increased virus titers (P < 0.001) in the intestine, spleen, MLN, and liver compared to WT and IFN-γR−/− mice at day 3 postinoculation (data not shown). Additionally, WT mice treated with the anti-IFNAR MAb had detectable levels of MNV in the brain, lung, and liver, in addition to increased replication in the distal ileum, spleen, and MLN (Fig. 1A to F). Notably, however, deficiency of either IRF-1 or the IFN-γR resulted in significant enhancement of viral replication in all organs compared to treatment of WT mice with the anti-IFNAR MAb (Fig. 1A to F). Together, these data demonstrate that IRF-1 is important for restriction of MNV replication in vivo. Furthermore, when compensatory IFN-αβ signaling is inhibited by administration of blocking MAb to reveal potential IFN-γ effects, both IRF-1 and the IFN-γR are critical for control of MNV replication.
Fig 1.
IRF-1 and the IFN-γR restrict MNV replication in mice with compromised IFN-αβ signaling. WT, IRF1−/−, and IFN-γR−/− mice were treated with a control or anti-IFNAR MAb before inoculation with 3 × 105 PFU of MNV. Viral titers were determined in the distal ileum (A), mesenteric lymph nodes (B), spleen (C), liver (D), lung (E), and brain (F). Seven to 10 mice were used per group, and data represent at least three independent experiments. Statistically significant differences in the anti-IFNAR-treated IRF1−/− and IFN-γR−/− mice relative to WT mice are indicated by asterisks (**, P < 0.01; ***, P < 0.001). Other significant differences are discussed in the text. The limit of detection is indicated by a dashed line. Bars indicate arithmetic means.
IRF-1 is critical for protection against lethal MNV infection when IFN-αβ signaling is inhibited.
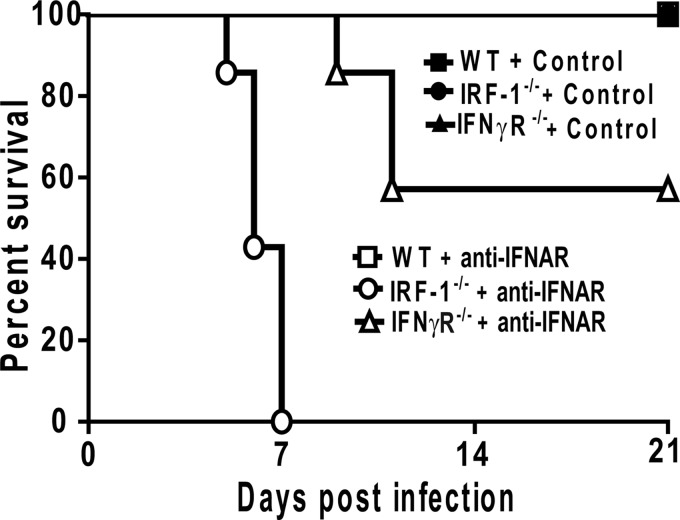
In addition to elevated viral titers, mice deficient in IFN signaling (STAT-1−/− and IFN-αβγR−/− mice) succumb to MNV infection (17, 27). To evaluate whether IRF-1 deficiency also results in increased susceptibility to lethal MNV infection, we monitored the survival of MNV-infected WT, IRF-1−/−, and IFN-γR−/− mice that had been treated with control or anti-IFNAR MAb. None of the mice treated with control MAb succumbed to MNV infection (Fig. 2). Unlike IFN-αβγR−/− mice (17), anti-IFNAR-treated IFN-γR−/− mice did not display significantly increased lethality after MNV infection (Fig. 2). Together with the data in Fig. 1 indicating significant effects of anti-IFNAR antibody treatment on MNV pathogenesis, these data indicate that treatment with this antibody only partially blocks the in vivo effects of type 1 IFNs. Nevertheless, IRF-1−/− mice treated with anti-IFNAR MAb displayed significant lethality after MNV infection compared to control MAb-treated IRF-1−/− mice and control MAb or anti-IFNAR MAb-treated WT or IFN-γR−/− mice (Fig. 2). Therefore, when IFN-αβ signaling is inhibited, there is an essential requirement for IRF-1 for survival after MNV infection.
Fig 2.
IRF-1 is critical for resistance to lethal infection with MNV when IFN-ɑβ signaling is inhibited. WT, IRF1−/−, and IFN-γR−/− mice were treated with a control or anti-IFNAR MAb, inoculated with 3 × 105 PFU of MNV, and monitored for survival for 21 days. Seven mice were used per group, and data represent at least three independent experiments. Survival differences between anti-IFNAR MAb-treated WT and IRF-1−/− mice were significant (P = 0.0003).
While IRF-1 plays a role in multiple aspects of viral immunity (18–20, 22, 24, 28), our observations in vivo that IRF-1 is important for control of MNV both with and without inhibition of IFN-αβ signaling raise the possibility that IRF-1 is important for IFN-αβ- as well as IFN-γ-mediated control of MNV replication in permissive cells.
IRF-1 is required for IFN-γ- but not IFN-αβ-mediated inhibition of MNV replication in primary macrophages.
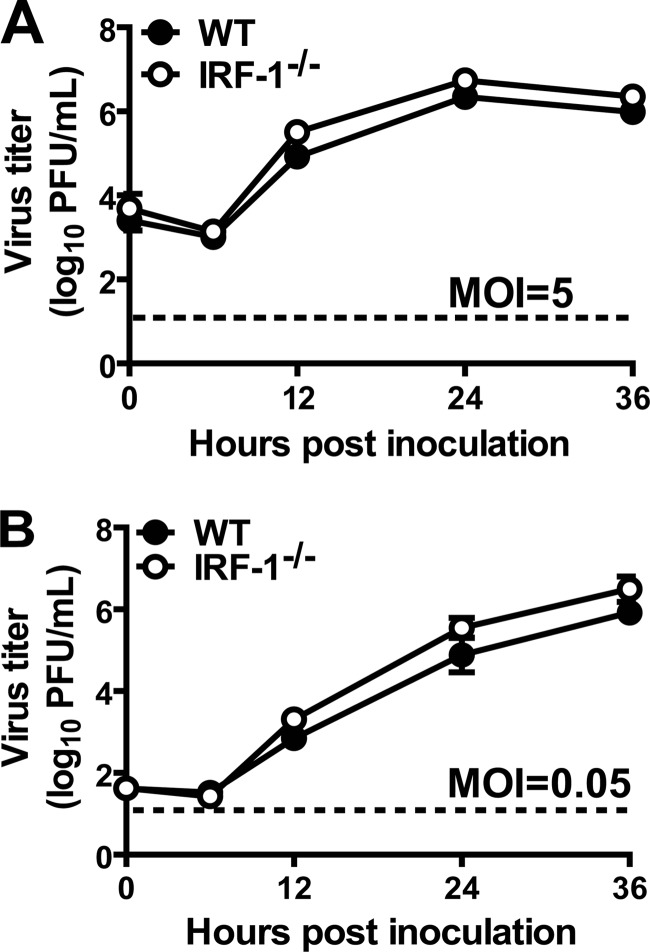
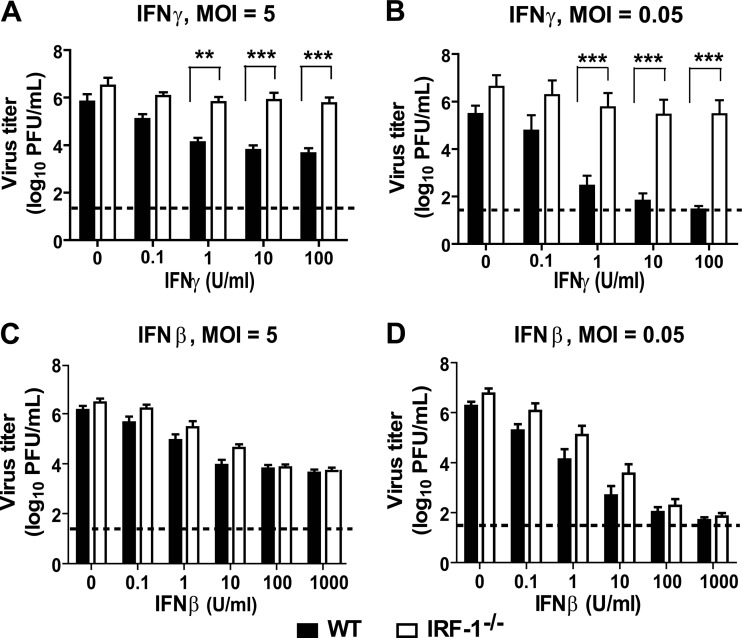
Based on these results and the tropism of MNV for macrophages (41), we hypothesized that IRF-1 would be required for IFN-mediated control of MNV in macrophages. To address this hypothesis, we generated bone marrow-derived macrophages from WT and IRF-1−/− mice and inoculated them with MNV at a high (5) or low (0.05) multiplicity of infection (MOI). There was no statistically significant difference in MNV replication in untreated WT and IRF-1−/− macrophages over 36 h (Fig. 3A and B). However, while treatment of WT macrophages with IFN-γ significantly inhibited MNV replication, in a dose-dependent manner (P < 0.05 for 100 and 10 U/ml IFN-γ at an MOI of 5 and for 100, 10, and 1 U/ml IFN-γ at an MOI of 0.05), there was a significant defect in IFN-γ-mediated inhibition of MNV replication in IRF-1−/− macrophages (Fig. 4A and B). In contrast, treatment with IFN-β inhibited MNV comparably in IRF-1−/− and WT macrophages (Fig. 4C and D). Thus, IRF-1 is required for IFN-γ to control MNV in macrophages but is dispensable for its control by IFN-β.
Fig 3.
Replication of MNV in WT and IRF-1−/− macrophages is similar. Macrophages were inoculated with MNV at an MOI of 5 (A) or 0.05 (B). Viral titers shown represent data from three independent experiments and are means ± standard errors of the means (SEM). No significant differences between the growth curves were detected by the unpaired t test.
Fig 4.
IRF-1 is required for IFN-γ-mediated control of MNV replication in primary macrophages. WT and IRF-1−/− macrophages were treated with increasing doses of IFN-γ (A and B), IFN-β (C and D), or medium alone, inoculated with MNV at an MOI of 5 (A and C) or 0.05 (B and C), and harvested at 12 or 24 h postinfection, respectively. Data were collected from three to eight independent experiments and are presented as mean titers ± SEM. Asterisks indicate significant differences in viral titer between WT and IRF-1−/− cells (**, P < 0.01; ***, P < 0.001). No significant differences were determined between titers in WT and IRF-1−/− cells for any dose of IFN-β used.
Inhibition of MNV replication occurs in IFN-γ-activated macrophages independent of the IFN-αβ receptor.
According to some reports, the antiviral effect of IFN-γ can require the IFN-αβR (36), and IRF-1 is important for production of IFN-αβ in response to certain stimuli (24). Therefore, we examined the potential involvement of autocrine production of IFN-αβ in IFN-γ-induced, IRF-1-dependent inhibition of MNV replication in macrophages. Macrophages were generated from IFN-αβR−/− mice and treated with increasing doses of IFN-γ or medium alone before inoculation with MNV at high and low MOIs. As observed for WT macrophages (Fig. 4), treatment of IFN-αβR−/− macrophages with IFN-γ resulted in significant inhibition of MNV replication (Fig. 5A and B), in a dose-dependent manner (P < 0.05 for 100 and 10 U/ml IFN-γ at an MOI of 5 and for 100, 10 and 1 U/ml IFN-γ at an MOI of 0.05). The effects of IFN-γ were comparable between WT and IFN-αβR−/− macrophages. These data demonstrate that the IFN-αβR is not required for IFN-γ-mediated inhibition of MNV replication in macrophages. This is consistent with our finding in vivo that IFN-γ and IRF-1 have functions independent of the effects of IFN-ɑβ (Fig. 1).
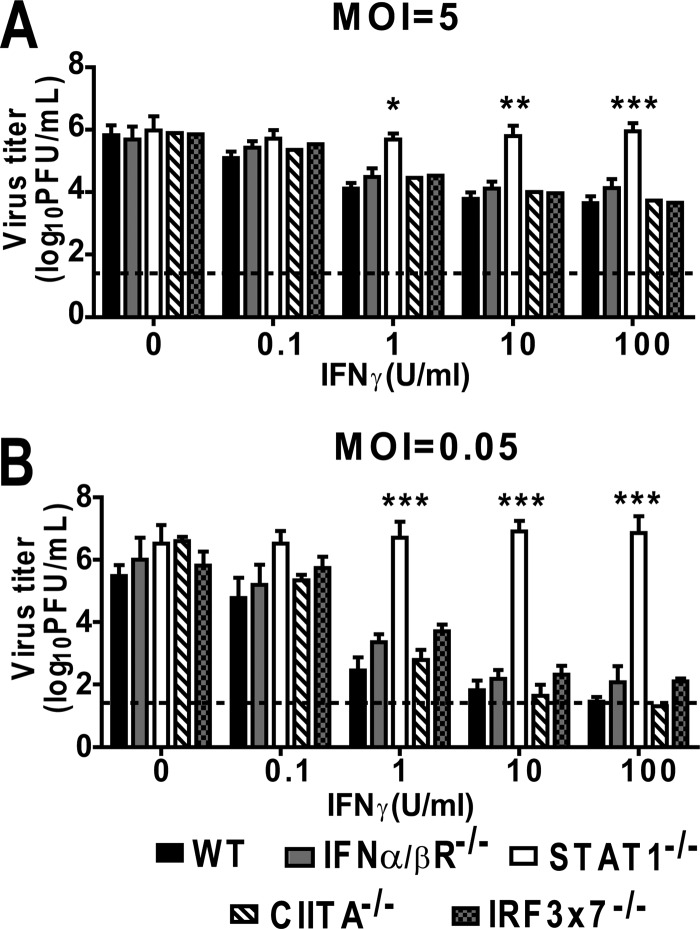
Fig 5.
Inhibition of MNV replication in IFN-γ-activated macrophages is STAT-1 dependent but IFN-αβR, CIITA, IRF-3, and IRF-7 independent. IFN-αβR−/−, CIITA−/−, IRF3×7−/−, and STAT-1−/− macrophages were treated with increasing doses of IFN-γ or medium alone before inoculation at an MOI of 5 (A) or 0.05 (B). These experiments were conducted concurrently with the experiments for Fig. 4; therefore, the data for WT macrophages are repeated in the figure for comparison. Data were collected from at least three independent experiments. Asterisks indicate significant differences in viral titers relative to those in WT cells (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
IFN-γ-mediated control of MNV replication requires STAT-1 but not IRF-3, IRF-7, or CIITA.
The transcription factors STAT-1, IRF-1, and CIITA are important for induction of specific ISGs (1) in response to IFN-γ. To investigate the involvement of other IFN-γ-related signaling molecules in IFN-γ-mediated control of MNV replication, we generated macrophages from STAT-1−/− and CIITA−/− mice and evaluated their ability to control MNV replication after treatment with IFN-γ. As controls, we also analyzed IFN-γ-mediated control of MNV in macrophages generated from mice deficient in the IRF-1-related transcription factors IRF-3 and IRF-7 (IRF3×7−/− mice). Treatment of CIITA−/− and IRF3×7−/− macrophages with IFN-γ significantly inhibited MNV replication, in a dose-dependent manner, at both high and low MOIs (Fig. 5A and B). However, STAT1−/− macrophages were unable to control MNV replication even after stimulation with 100 U/ml of IFN-γ. These data demonstrate that the signaling molecules IRF-1 and STAT-1, but not CIITA, IRF-3, or IRF-7, are required for IFN-γ-mediated inhibition of MNV replication in macrophages.
Identification of STAT-1- and IRF-1-dependent subset of IFN-γ-regulated genes.
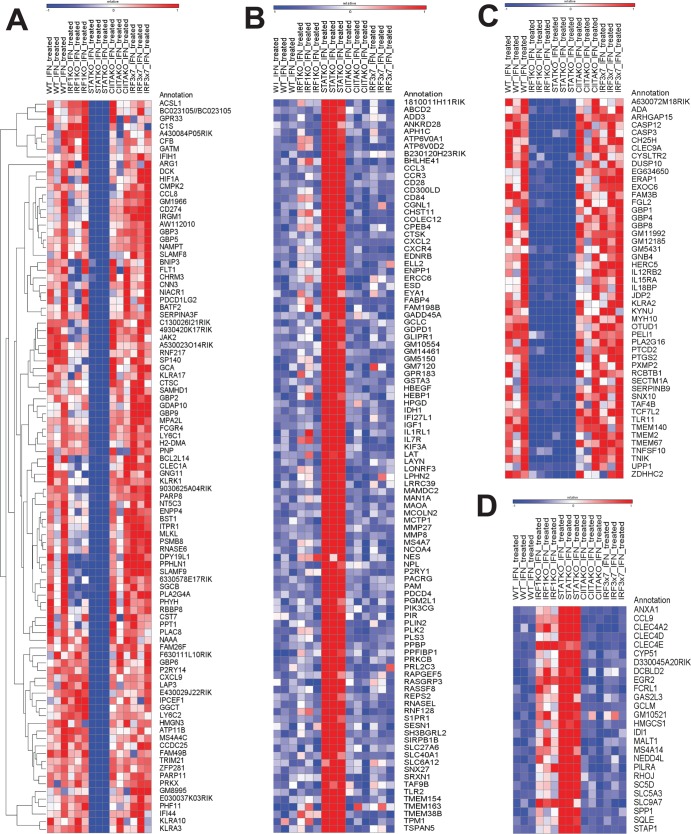
Based on the above observations, we hypothesized that there would be a subset of IFN-γ-regulated genes that depend on STAT-1 and IRF-1 but not on IRF-3, IRF-7, or CIITA for their expression. To address this hypothesis, we performed microarray analyses comparing steady-state transcript levels of 28,853 mouse gene transcripts in WT, IRF-1−/−, STAT-1−/−, CIITA−/−, and IRF3×7−/− macrophages after 12 h of treatment with IFN-γ, using three independent biological replicates. The 12-h time point was selected because this is sufficient to induce an antiviral state that limits MNV replication (Fig. 4 and 5). We identified 359 genes whose expression was changed >2-fold in WT macrophages by IFN-γ treatment (216 were upregulated and 143 were downregulated). We then determined which of these genes depend on both STAT-1 and IRF-1 but not on CIITA or IRF-3/7 for regulation by IFN-γ. We found that 38 genes were differentially regulated by IFN-γ when either CIITA or IRF-3/IRF-7 was missing. We focused on the remaining CIITA- and IRF-3/IRF-7-independent genes (Table 2; Fig. 6). Ninety-seven genes required STAT-1 but not IRF-1 for upregulation by IFN-γ (group A) (Table 2; Fig. 6). One hundred genes were downregulated in a manner dependent on STAT-1 but not IRF-1 (group B) (Table 2; Fig. 6). Consistent with our hypothesis, a set of genes did depend on both STAT-1 and IRF-1 (groups C and D) (Table 2; Fig. 6) for their regulation by IFN-γ. Ten genes required just IRF-1 for downregulation by IFN-γ. Eighteen genes did not meet statistical criteria for assignment to any of the designated groups in Table 2.
Table 2.
Transcription factor requirements of genes that were responsive to IFN-γ in macrophagesa
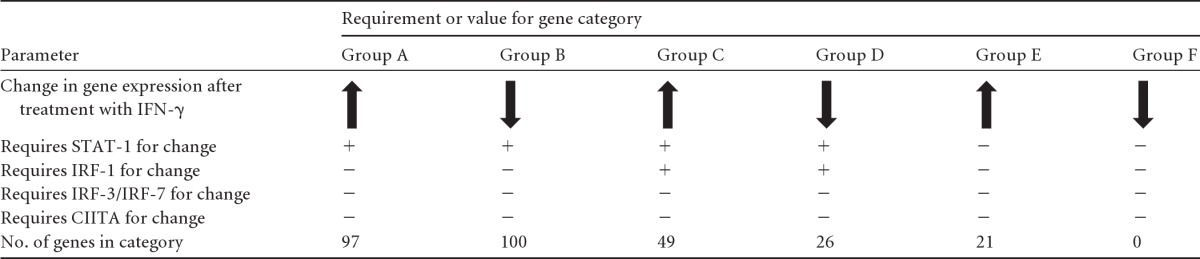
Genes whose expression profiles were changed by IFN-γ were identified and grouped based on the requirement of STAT-1, IRF-1, CIITA, IRF-3, or IRF-7 for this change.
Fig 6.
Heat maps of the genes assigned to group A (A), group B (B), group C (C), and group D (D) by microarray analyses (see Table 2). Group A represents genes that are upregulated by IFN-γ in a STAT-1-dependent manner, group B represents genes that are downregulated by IFN-γ in a STAT-1-dependent manner, group C represents genes that are upregulated by IFN-γ in an IRF-1- and STAT-1-dependent manner, and group D represents genes that are downregulated by IFN-γ in an IRF-1- and STAT-1-dependent manner. The heat maps represent row-normalized Z-score values per gene. Expression values of >1 standard deviation above the expression level in untreated WT macrophages are indicated in red, and values of >1 standard deviation below the WT expression level are indicated in blue.
Validation of microarray analysis by qRT-PCR.
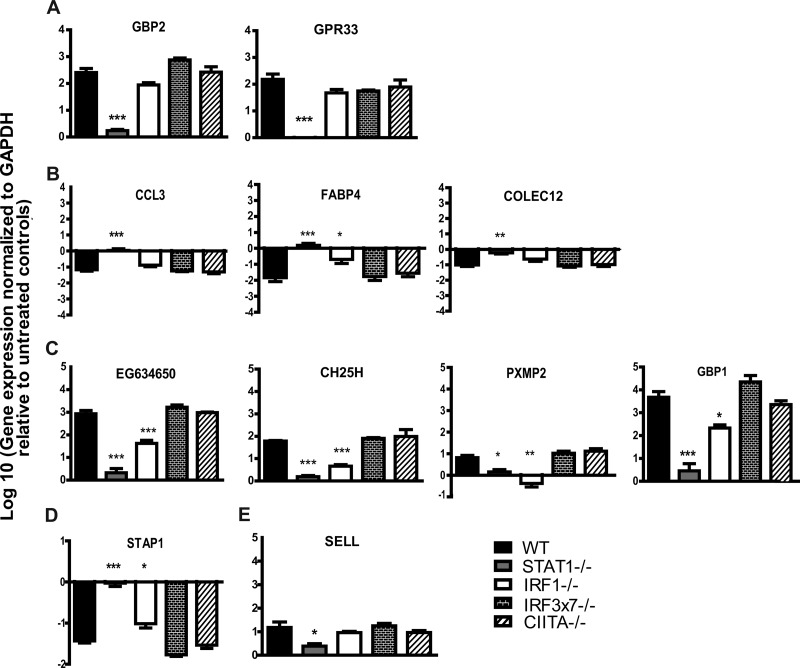
To assess whether the gene expression changes we detected by microarray analysis could be verified by an independent methodology, we selected 11 genes with various patterns of transcription factor dependence and measured their transcript levels by qRT-PCR. To this end, we isolated RNAs from untreated and IFN-γ-treated WT, IRF-1−/−, IRF3×7−/−, STAT-1−/−, and CIITA−/− macrophages for analysis (Fig. 7). When we compared the fold changes in expression induced by IFN-γ in the various gene-deficient macrophages to those observed in WT macrophages, we found that for 82% (9 of 11 genes) of the genes, our microarray analyses accurately predicted the gene's dependence on the various transcription factors for responsiveness to IFN-γ (Fig. 7). These data support the bioinformatic analysis of microarray data indicating patterns of transcription factor dependence for IFN-γ-regulated genes.
Fig 7.
Validation of microarray analyses by qRT-PCR. WT, IRF-1−/−, CIITA−/−, IRF3×7−/−, and STAT-1−/− macrophages were treated with 10 U of IFN-γ or with medium alone. Transcription levels of select genes were examined after 12 h of treatment. Data were collected from three independent experiments. Genes assigned to group A (A), group B (B), group C (C), group D (D), and group E (E) by microarray analyses (see Table 2) were examined. Asterisks indicate significant differences in transcript levels relative to those in WT cells (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Pathway analysis of gene sets identified by microarray analysis.
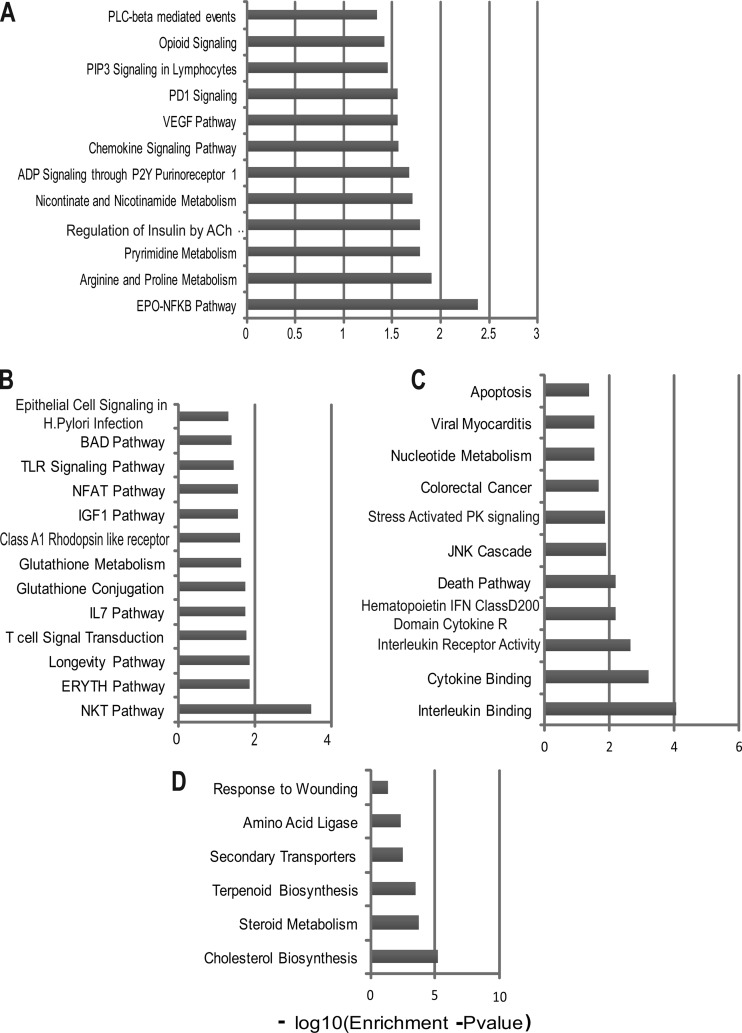
To identify the gene pathways that these STAT-1- and IRF-1-dependent genes contribute to in IFN-γ-activated macrophages, we analyzed groups A to D (Table 2) separately by gene set overlap analysis using mSigDB. We found enrichment for several metabolic pathways, such as pyrimidine metabolism, arginine and proline metabolism, and nicotinate and nicotinamide metabolism, among genes that were regulated by IFN-γ in a manner dependent on STAT-1 but not on IRF-1, IRF-3, IRF-7, or CIITA (Fig. 8). Conversely, we found enrichment of several immune response pathways in genes that were inhibited by STAT-1 (Fig. 8). These included genes involved in the Toll-like receptor (TLR) signaling pathway, the IL-7 pathway, the NFAT pathway, and glutathione metabolism. The genes that required both STAT-1 and IRF-1 for upregulation in response to IFN-γ were enriched for stress-activated protein kinase signaling, JNK cascade, and interleukin/cytokine binding pathways, while the genes that were inhibited by IFN-γ in a manner dependent on both STAT-1 and IRF-1 were enriched for lipid metabolism, steroid metabolism, cholesterol biosynthesis, and terpenoid biosynthesis.
Fig 8.
Gene overlap analyses of the genes assigned to group A (A), group B (B), group C (C), and group D (D) by microarray analyses (see Table 2). The x axis represents the P value, indicating the significance of enrichment for any given gene set. The values are plotted on a negative log10 scale. All gene sets with values of >1.3 on the x axis were significantly enriched.
DISCUSSION
IFNs are likely vital to the host defense against human noroviruses and are important for inhibition of MNV (8, 15, 17, 41). However, the antiviral mechanisms that IFNs induce to combat NVs and the transcription factors involved in these responses are not well defined. In this report, we demonstrate a critical role for the transcription factor IRF-1 in control of MNV replication in vivo. Furthermore, we show that in the absence of IFN-αβ signaling, IRF-1 is essential for survival after MNV infection. We also demonstrate that IRF-1 is required for the control of MNV by IFN-γ in primary macrophages. Despite previous studies that suggest a role for IRF-1 in IFN-αβ-induced antiviral responses, we found that IRF-1 is dispensable for control of MNV by IFN-β in cultured macrophages. Finally, we demonstrated an additional requirement for STAT-1, but not autocrine IFN-αβ, CIITA, IRF-3, or IRF-7, in IFN-γ-induced control of MNV in macrophages. Consistent with the importance of STAT-1 and IRF-1 in the antiviral effects of IFN-γ, we used microarrays and bioinformatics to show that there is a defined subset of genes whose regulation by IFN-γ requires these two transcription factors.
IRF-1 in IFN-γ-mediated responses in vivo.
Previous studies with IRF-1−/− mice have confirmed the importance of IRF-1 for defense against both EMCV and WNV in vivo (2, 18). However, due to the complex nature of IRF-1 actions in host defense, the in vivo contribution of IRF-1 as a direct effector of IFN-γ signaling in infected cells is unclear. For example, studies of immune responses to WNV found that IRF-1 is required for IFN-γ-mediated control of viral replication in macrophages and for shaping the antigen-specific CD8 T cell response (2). Furthermore, since IRF-1 contributes to both IFN-αβ and IFN-γ responses to EMCV infection in vitro, it is difficult to ascertain the specific contribution of IRF-1 to IFN-γ-dependent responses in vivo in this model (18). Our finding that mice deficient in either IRF-1 or IFN-γR have elevated viral titers when IFN-αβ signaling is blocked is consistent with the idea of shared functions for IRF-1 and IFN-γ in vivo and strongly suggests that the antiviral effects of IFN-γ are mediated at least in part by IRF-1 in vivo.
IFN-γ-independent functions of IRF-1.
Our studies with blocking antibody (Fig. 1) revealed a role for IRF-1 that is consistent with a role in mediating IFN-γ responses in vivo. Interestingly, our studies of viral replication in WT, IRF-1−/−, and IFN-γR−/− mice and our survival studies have also revealed functions for IRF-1 in restricting MNV replication that cannot be explained by its role in IFN-γ signaling. IRF-1−/− mice are highly susceptible to lethal MNV infection under conditions where IFN-γR−/− mice are not. Moreover, IRF-1−/− mice treated with control MAb displayed significantly enhanced replication and viral dissemination of MNV (but not increased lethality) compared to IFN-γR−/− and WT mice treated with control MAb (Fig. 1) for every organ tested except the brain. This enhanced replication, while not of the same magnitude, paralleled the increased replication and viral dissemination induced in WT mice treated with anti-IFNAR and suggests additional functions of IRF-1 in mediating at least some of the effects of IFN-αβ. However, in primary macrophages, we observed that IRF-1 is dispensable for inhibition of MNV by IFN-β (Fig. 4). Furthermore, unlike what is seen for WNV infection, we did not observe a significant increase in replication in untreated IRF-1−/− macrophages compared to WT macrophages (Fig. 3) that could explain elevated viral replication in vivo. Further studies may reveal cell type- and tissue-specific requirements for IRF-1 in IFN-γ-independent but IFN-αβ-dependent control of MNV replication.
STAT-1- and IRF-1-dependent gene pathways.
Although STAT-1 and IRF-1 have well-established activities against a diverse range of RNA viruses, few STAT-1- and IRF-1-dependent individual antiviral effectors have been identified to date. One such effector is viperin (9), whose activity has been demonstrated against several families of viruses, including orthomyxoviruses (39), rhabdoviruses (34), and flaviviruses (35). However, viperin is not required to control MNV replication in primary macrophages or in vivo (L. B. Thackray and H. W. Virgin, unpublished data). Clearly, there is a need to identify STAT-1- and IRF-1-regulated processes that control MNV replication control. Interestingly, we showed that the Atg5-Atg12/Atg16L1 autophagosome elongation protein complex, but not the degradative activity of the autophagy pathway, is required for IFN-γ-mediated inhibition of MNV replication in macrophages (15). This property of the Atg5-Atg12/Atg16L1 complex is shared with IRF-1, as shown here, in that both are required for the effects of IFN-γ but not IFN-αβ on MNV replication in macrophages. Therefore, it is intriguing to speculate that IRF-1 regulates a key early step in an IFN-γ/IRF-1/STAT-1/Atg5-Atg12/Atg16L1-dependent antiviral mechanism. Our recent work showed that IFN-γ blocks the formation of the membranous norovirus replication complex in an Atg5-Atg12/Atg16L1-dependent manner (15). Interestingly, the microarray analyses in this study revealed that IFN-γ downregulates components of lipid and cholesterol biogenesis pathways via an IRF-1- and STAT-1-dependent mechanism. Upregulation of components of cholesterol biogenesis pathways occurs in cells stably expressing a human norovirus replicon or infected with WNV (5, 23). We speculate that changes in cellular lipids mediated by IFN-γ alter membrane rearrangements and trafficking, in an autophagy protein-dependent manner, so as to inhibit formation of the replication complex. Future studies of the IRF-1- and/or STAT-1-dependent IFN-γ-mediated transcriptional regulation identified here may uncover novel antiviral mechanisms that can be harnessed as antimicrobial treatments.
ACKNOWLEDGMENTS
The MAR1-5A3 antibody was a generous gift from R. Schreiber and K. Sheehan. The mM-CSF-producing cell line CMG14-12 was a generous gift from A. Kudo (Tokyo Institute of Technology). We thank the Genome Technology Access Center in the Department of Genetics at Washington University School of Medicine for help with genomic analysis. We thank members of the Virgin lab for their comments on the manuscript and D. Kreamalmeyer for managing mouse colonies.
This work was supported by National Institutes of Health grants ROI AI054483 and U54 AI057160, project 3, to H.W.V. N.S.M. was supported by institutional training grant T32-AI007172. Washington University and H.W.V. receive income based on licenses for MNV technology. The Genome Technology Access Center is partially supported by NCI Cancer Center support grant P30 CA91842 to the Siteman Cancer Center and by ICTS/CTSA grant UL1RR024992 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH), and the NIH Roadmap for Medical Research.
This publication is solely the responsibility of the authors and does not necessarily represent the official view of the NCRR or the NIH.
Footnotes
Published ahead of print 12 September 2012
REFERENCES
- 1. Boehm U, Klamp T, Groot M, Howard JC. 1997. Cellular responses to interferon-gamma. Annu. Rev. Immunol. 15:749–795 [DOI] [PubMed] [Google Scholar]
- 2. Brien JD, et al. 2011. Interferon regulatory factor-1 (IRF-1) shapes both innate and CD8 T cell immune responses against West Nile virus infection. PLoS Pathog. 7:e1002230 doi:10.1371/journal.ppat.1002230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cadwell K, et al. 2010. Virus-plus-susceptibility gene interaction determines Crohn's disease gene Atg16L1 phenotypes in intestine. Cell 141:1135–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chachu KA, LoBue AD, Strong DW, Baric RS, Virgin HW. 2008. Immune mechanisms responsible for vaccination against and clearance of mucosal and lymphatic norovirus infection. PLoS Pathog. 4:e1000236 doi:10.1371/journal.ppat.1000236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chang KO. 2009. Role of cholesterol pathways in norovirus replication. J. Virol. 83:8587–8595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chang KO, George DW. 2007. Interferons and ribavirin effectively inhibit Norwalk virus replication in replicon-bearing cells. J. Virol. 81:12111–12118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chang KO, Sosnovtsev SV, Belliot G, King AD, Green KY. 2006. Stable expression of a Norwalk virus RNA replicon in a human hepatoma cell line. Virology 353:463–473 [DOI] [PubMed] [Google Scholar]
- 8. Changotra H, et al. 2009. Type I and type II interferons inhibit the translation of murine norovirus proteins. J. Virol. 83:5683–5692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chin KC, Cresswell P. 2001. Viperin (cig5), an IFN-inducible antiviral protein directly induced by human cytomegalovirus. Proc. Natl. Acad. Sci. U. S. A. 98:15125–15130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Daffis S, Suthar MS, Szretter KJ, Gale M, Jr, Diamond MS. 2009. Induction of IFN-beta and the innate antiviral response in myeloid cells occurs through an IPS-1-dependent signal that does not require IRF-3 and IRF-7. PLoS Pathog. 5:e1000607 doi:10.1371/journal.ppat.1000607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Estes MK, Prasad BV, Atmar RL. 2006. Noroviruses everywhere: has something changed? Curr. Opin. Infect. Dis. 19:467–474 [DOI] [PubMed] [Google Scholar]
- 12. Glass RI, Parashar UD, Estes MK. 2009. Norovirus gastroenteritis. N. Engl. J. Med. 361:1776–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Goodwin MM, Canny S, Steed A, Virgin HW. 2010. Murine gammaherpesvirus 68 has evolved gamma interferon and stat1-repressible promoters for the lytic switch gene 50. J. Virol. 84:3711–3717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Goodwin MM, et al. 2010. Histone deacetylases and the corepressor NCoR regulate the lytic-latent switch gene 50 in murine gammaherpesvirus 68-infected macrophages. J. Virol. 84:12039–12047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hwang S, et al. 2012. Nondegradative role of Atg5-Atg12/Atg16L1 autophagy protein complex in antiviral activity of interferon gamma. Cell Host Microbe 11:397–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kanazawa N, et al. 2004. Regulation of hepatitis C virus replication by interferon regulatory factor 1. J. Virol. 78:9713–9720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Karst SM, Wobus CE, Lay M, Davidson J, Virgin HW. 2003. STAT1-dependent innate immunity to a Norwalk-like virus. Science 299:1575–1578 [DOI] [PubMed] [Google Scholar]
- 18. Kimura T, et al. 1994. Involvement of the IRF-1 transcription factor in antiviral responses to interferons. Science 264:1921–1924 [DOI] [PubMed] [Google Scholar]
- 19. Liu J, Cao S, Herman LM, Ma X. 2003. Differential regulation of interleukin (IL)-12 p35 and p40 gene expression and interferon (IFN)-gamma-primed IL-12 production by IFN regulatory factor 1. J. Exp. Med. 198:1265–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu J, Guan X, Ma X. 2005. Interferon regulatory factor 1 is an essential and direct transcriptional activator for interferon gamma-induced RANTES/CCL5 expression in macrophages. J. Biol. Chem. 280:24347–24355 [DOI] [PubMed] [Google Scholar]
- 21. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 22. Lohoff M, et al. 1997. Interferon regulatory factor-1 is required for a T helper 1 immune response in vivo. Immunity 6:681–689 [DOI] [PubMed] [Google Scholar]
- 23. Mackenzie JM, Khromykh AA, Parton RG. 2007. Cholesterol manipulation by West Nile virus perturbs the cellular immune response. Cell Host Microbe 2:229–239 [DOI] [PubMed] [Google Scholar]
- 24. Matsuyama T, et al. 1993. Targeted disruption of IRF-1 or IRF-2 results in abnormal type I IFN gene induction and aberrant lymphocyte development. Cell 75:83–97 [PubMed] [Google Scholar]
- 25. Meraz MA, et al. 1996. Targeted disruption of the Stat 1 gene in mice reveals unexpected physiologic specificity of the JAK-STAT signalling pathway. Cell 84:431–442 [DOI] [PubMed] [Google Scholar]
- 26. Muller U, et al. 1994. Functional role of type I and type II interferons in antiviral defense. Science 264:1918–1921 [DOI] [PubMed] [Google Scholar]
- 27. Mumphrey SM, et al. 2007. Murine norovirus 1 infection is associated with histopathological changes in immunocompetent hosts, but clinical disease is prevented by STAT1-dependent interferon responses. J. Virol. 81:3251–3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ogasawara K, et al. 1998. Requirement for IRF-1 in the microenvironment supporting development of natural killer cells. Nature 391:700–703 [DOI] [PubMed] [Google Scholar]
- 29. Pine R. 1992. Constitutive expression of an ISGF2/IRF1 transgene leads to interferon-independent activation of interferon-inducible genes and resistance to virus infection. J. Virol. 66:4470–4478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Reis LFL, Ruffner H, Stark G, Aguet M, Weissman C. 1994. Mice devoid of interferon regulatory factor 1 (IRF-1) show normal expression of type I interferon genes. EMBO J. 13:4798–4806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schoggins JW, et al. 2011. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472:481–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sheehan KC, et al. 2006. Blocking monoclonal antibodies specific for mouse IFN-alpha/beta receptor subunit 1 (IFNAR-1) from mice immunized by in vivo hydrodynamic transfection 1. J. Interferon Cytokine Res. 26:804–819 [DOI] [PubMed] [Google Scholar]
- 33. Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227–264 [DOI] [PubMed] [Google Scholar]
- 34. Stirnweiss A, et al. 2010. IFN regulatory factor-1 bypasses IFN-mediated antiviral effects through viperin gene induction. J. Immunol. 184:5179–5185 [DOI] [PubMed] [Google Scholar]
- 34a. Subramanian A, et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102:15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Szretter KJ, et al. 2011. The interferon-inducible gene viperin restricts West Nile virus pathogenesis. J. Virol. 85:11557–11566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Takaoka A, et al. 2000. Cross talk between interferon-gamma and -alpha/beta signaling components in caveolar membrane domains. Science 288:2357–2360 [DOI] [PubMed] [Google Scholar]
- 37. Takeshita S, Kaji K, Kudo A. 2000. Identification and characterization of the new osteoclast progenitor with macrophage phenotypes being able to differentiate into mature osteoclasts. J. Bone Miner. Res. 15:1477–1488 [DOI] [PubMed] [Google Scholar]
- 38. Thackray LB, et al. 2007. Murine noroviruses comprising a single genogroup exhibit biological diversity despite limited sequence divergence. J. Virol. 81:10460–10473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang XY, Hinson ER, Cresswell P. 2007. The interferon-inducible protein viperin inhibits influenza virus release by perturbing lipid rafts. Cell Host Microbe 2:96–105 [DOI] [PubMed] [Google Scholar]
- 40. Ward JM, et al. 2006. Pathology of immunodeficient mice with naturally occurring murine norovirus infection. Toxicol. Pathol. 34:708–715 [DOI] [PubMed] [Google Scholar]
- 41. Wobus CE, et al. 2004. Replication of norovirus in cell culture reveals a tropism for dendritic cells and macrophages. PLoS Biol. 2:e432 doi:10.1371/journal.pbio.0020432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wobus CE, Thackray LB, Virgin HW. 2006. Murine norovirus: a model system to study norovirus biology and pathogenesis. J. Virol. 80:5104–5112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhao Z, et al. 2007. Coronavirus replication does not require the autophagy gene ATG5. Autophagy 3:581–585 [DOI] [PubMed] [Google Scholar]