Abstract
Similar to other positive-sense, single-stranded RNA viruses, hepatitis C virus (HCV) replicates its genome in a remodeled intracellular membranous structure known as the membranous web (MW). To date, the process of MW formation remains unclear. It is generally acknowledged that HCV nonstructural protein 4B (NS4B) can induce MW formation through interaction with the cytosolic endoplasmic reticulum (ER) membrane. Many host proteins, such as phosphatidylinositol 4-kinase IIIα (PI4KIIIα), have been identified as critical factors required for this process. We now report a new factor, the cytosolic phospholipase A2 gamma (PLA2G4C), which contributes to MW formation, HCV replication, and assembly. The PLA2G4C gene was identified as a host gene with upregulated expression upon HCV infection. Knockdown of PLA2G4C in HCV-infected cells or HCV replicon-containing cells by small interfering RNA (siRNA) significantly suppressed HCV replication and assembly. In addition, the chemical inhibitor methyl arachidonyl fluorophosphonate (MAFP), which specifically inhibits PLA2, reduced HCV replication and assembly. Electron microscopy demonstrated that MW structure formation was defective after PLA2G4C knockdown in HCV replicon-containing cells. Further analysis by immunostaining and immunoprecipitation assays indicated that PLA2G4C colocalized with the HCV proteins NS4B and NS5A in cells infected with JFH-1 and interacted with NS4B. In addition, PLA2G4C was able to transport the HCV nonstructural proteins from replication sites to lipid droplets, the site for HCV assembly. These data suggest that PLA2G4C plays an important role in the HCV life cycle and might represent a potential target for anti-HCV therapy.
INTRODUCTION
Hepatitis C virus (HCV) was identified in 1989 as the major pathogen causing posttransfusion and sporadic non-A, non-B hepatitis. According to estimates from the World Health Organization, approximately 130 to 170 million people are chronically infected with HCV, and more than 350,000 people die from hepatitis C-related liver diseases each year (18, 34, 65). HCV primarily infects human hepatocytes and frequently causes chronic hepatitis, which may lead to the development of cirrhosis and hepatocellular carcinoma (43, 60).
HCV is a small, enveloped, positive-stranded RNA virus that belongs to the family Flaviviridae and has an RNA genome with a length of approximately 9.6 kb (18). Similar to other positive-stranded RNA viruses, HCV genome replication occurs in association with altered cellular membranes called the membranous web (MW) (33, 64). HCV nonstructural (NS) proteins are associated with the NS4B-induced MW to form a membrane-associated multiprotein complex, which is where HCV RNA replication occurs (20, 24, 32, 61, 66). The MW is thought to be at least partly derived from the endoplasmic reticulum (ER) membrane (20, 48, 49). The membrane structures are lipid rafts recruited from the intracellular membranes, and these structures protect the HCV RNA and NS proteins against RNase and protease digestion (1, 66, 76).
A number of host factors have been identified as components of membrane-associated sites of HCV RNA replication, such as early endosomal proteins (Rab5, EEA1, rabaptin5, and Rab4), late endosomal proteins (RAB7A and Rab9), ER proteins (RAB1B and TBC1D20), the Golgi apparatus-associated protein RAB7L1, and vesicle-associated proteins A and B (VAP-A and -B) (3, 8, 12, 22, 25, 62, 68, 71, 74). Thus, these data suggested that the HCV replication complex may consist of components of early endosomes and ER-to-Golgi apparatus transport vesicles. However, the mechanism of complex formation remains unknown.
Virion assembly is initiated on the surface of lipid droplets (LDs) (11, 30, 46, 59). The HCV core protein, which associates with LDs, interacts with viral RNA to form the virion capsid (10, 16, 44, 46). According to the current model, replication complexes where HCV RNA is synthesized are embedded within the MW (5, 20, 24). Newly synthesized HCV RNA molecules and nonstructural proteins are thought to be transported from the MW to LDs to interact with the HCV core proteins (44, 46). Current data suggest that NS5A is integral to this process (46, 67). Appel et al. showed that domain I (DI) of NS5A could mediate the binding to LDs and that another domain, DIII, could be responsible for the interaction between NS5A and the core proteins on the LD surface (6).
We found that HCV infection upregulated cytosolic phospholipase A2 gamma (PLA2G4C) expression. PLA2G4C, like phosphatidylinositol 4-kinase IIIα (PI4KIIIα), is an enzyme involved in lipid metabolism (70, 73). PI4KIIIα has been found to be essential for HCV replication, and the direct activation of a lipid kinase by HCV NS5A contributes critically to the integrity of the membranous viral replication complex (12, 37, 57). Because of these findings, we examined the roles of PLA2G4C in HCV infection. In this study, we showed that either knockdown of PLA2G4C by small interfering RNA (siRNA) or the use of a specific chemical inhibitor significantly suppressed HCV RNA replication and virion assembly in Huh7.5.1 cells. Electron microscopy revealed that NS4B-induced membranous structures were absent from Lunet-Con1 cells after PLA2G4C knockdown, indicating that PLA2G4C regulates HCV replication by orchestrating MW formation. Consistent with this hypothesis, PLA2G4C colocalized with HCV NS4B in cells infected with HCV replicon JFH-1 or in cells that expressed HCV NS4B alone. Interestingly, PLA2G4C overexpression caused the recruitment of NS4B to LDs in cells containing the Con1 replicon and in cells expressing HCV NS4B alone. Additionally, PLA2G4C induced the translocation of HCV nonstructural proteins, such as NS4B, from replication sites to LDs, which are thought to be the sites for HCV assembly. These findings suggested that PLA2G4C may be an important host factor that interacts with HCV proteins for both the formation of the MW and the process of virion assembly.
MATERIALS AND METHODS
Cells and virus.
Lunet cells (kindly provided by Ralf Bartenschlager) and Huh7.5.1 cells (kindly provided by Frank Chisari) were cultured in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) supplemented with 10% fetal bovine serum (FBS) (Invitrogen), 2 mmol/liter glutamine, 100 U/ml penicillin, and 100 U/ml streptomycin at 37°C in a 5% CO2 atmosphere (13, 14). The subgenomic HCV replicon pFKI389neo/NS3-3′ (Con1, genotype 1b) was kindly provided by Ralf Bartenschlager, and the subgenomic HCV replicon pSGR-JFH-1 (JFH-1, genotype 2a) was constructed as previously described (31). Lunet-Con1 cells (Con1, genotype 1b) and Huh7.5.1-SGR-JFH-1 cells (JFH-1, genotype 2a), harboring the subgenomic HCV replicons, were derived from Lunet cells and Huh7.5.1 cells and maintained in the same medium as Lunet cells supplemented with 0.5 mg/ml G418 (Geneticin; Cytogen) (13, 14, 31, 45). The JFH-1 virus used in this study was based on the pJFH-1 plasmid, kindly provided by Takaji Wakita (75). The HCV J399EM strain was derived from the JFH-1 virus by insertion of enhanced green fluorescent protein (EGFP) into the HCV NS5A region (26). The JFH-1-luc reporter virus was kindly provided by Xunlin Chen (78).
Construction of plasmids.
The coding sequences of HCV NS4B from JFH-1 (amino acids [aa] 1716 to 1976) and PLA2G4C (GenBank accession number NM_003706.2) were amplified with the primer pairs NS4B-FLAGF/NS4B-FLAGR and PLA2G4C-HAF/PLA2G4C-HAR, respectively (see Table S1 in the supplemental material). The FLAG and hemagglutinin (HA) tags were added to the coding sequences of HCV NS4B and PLA2G4C by extension of the respective primers. The PCR fragments were inserted into pcDNA3.1(+) via the restriction sites HindIII/EcoRI and EcoRI/EcoRV to generate pNS4B-FLAG and pPLA2G4C-HA, respectively. A catalytically inactive mutant of PLA2G4C with a mutation at 82 aa (S to A) was introduced by fusion PCR and named pPLA2G4C(S82A) (55). The primers are listed in Table S1.
The primer pair NS4B-EGFPF/NS4B-EGFPR (see Table S1 in the supplemental material) was used to amplify the HCV NS4B fragment. The PCR product was cloned into the BglII and EcoRI restriction sites of pEGFPN1 to generate pEGFPN1-NS4B.
The coding sequences of HCV core, core-p7, E1-p7, NS2, and NS3-NS5B (JFH-1, genotype 2a) were amplified with the primer pairs Core-F/Core-R, Core-F/p7-R, E1-F/p7-R, NS2-F/NS2-R, and NS3-F/NS5B-R, respectively (see Table S1). The PCR fragments were then cloned into the EcoRI and XbaI restriction sites of pcDNA3.1(+) to generate the expression vectors pCore, pCore-p7, pE1-p7, pNS2, and pNS3-NS5B.
The coding sequences of Renilla luciferase, firefly luciferase, and HCV internal ribosome entry site (IRES) were amplified with the primer pairs RenillaF/RenillaR, FireflyF/FireflyR, and IRESF/IRESR, respectively (see Table S1). The coding sequences of Renilla luciferase and firefly luciferase were inserted into pcDNA3.1(+) via the restriction sites EcoRI/EcoRV and NotI/XhoI, respectively. A bicistronic reporter, pHCV-IRES, was generated by linking Renilla luciferase and firefly luciferase with the HCV IRES (nucleotides [nt] 43 to 409) via the restriction sites EcoRV/NotI (35).
Transfection of plasmid DNA and siRNA.
Huh7.5.1 or Lunet-Con1 cells were seeded at approximately 30% confluence and transfected with 20 nM FlexiTube siRNAs specific to PLA2G4C (siPLA2G4C-4 [product number SI00685776], siPLA2G4C-5 [product number SI04140647], siPLA2G4C-6 [product number SI04238066], and siPLA2G4C-7 [product number SI04259423]; Qiagen), AllStars negative-control siRNA (siControl [product number 1027281]; Qiagen), or siRNA specific to HCV (siHCV, target sequence GGU CUC GUA GAC CGU GCA C). Transfections of plasmid DNA or siRNA were carried out with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Cells were examined at various time points after transfection as indicated in the figures.
HCVcc infection.
For HCV infection, Huh7.5.1 cells were incubated with HCV for 6 h and then maintained in new medium at 37°C. The absolute titer values of infectious HCV particles in the cell lysates and culture supernatants were determined by limiting dilution analysis (56). To measure intracellular infective HCV particles, cells were harvested by trypsinization, rinsed with phosphate-buffered saline (PBS), centrifuged at 1,200 rpm for 5 min, resuspended in complete medium, and subjected to three rounds of freezing (in liquid nitrogen) and thawing (37°C). Cellular debris was removed by centrifugation at 10,000 rpm for 5 min, and the supernatants were tested for infectivity by limiting dilution analysis.
IP and co-IP.
Cells were lysed in immunoprecipitation (IP) buffer containing 50 mM Tris, pH 7.5, 1 mM EGTA, 1 mM EDTA, 1% Triton X-100, 150 mM NaCl, 100 μM phenylmethylsulfonyl fluoride (PMSF), and a protease inhibitor cocktail (Complete Mini; Roche) for 30 min. Cell lysates were centrifuged at 12,000 × g for 1 min at 4°C. The supernatants were recovered and mixed with 4 μg of primary antibody and then incubated for 6 h at 4°C. The following antibodies were used for IP and co-IP analysis: anti-HA antibody (catalog number H9658; Sigma) and anti-PLA2G4C antibody (catalog number HPA043083; Sigma). The reaction mixtures were then mixed with protein A/G agarose (Santa Cruz Biotechnology) and incubated for an additional 6 h at 4°C. Protein A/G agarose-bound immune complexes were collected by centrifugation at 12,000 × g for 1 min, washed 3 times with IP buffer, and boiled in loading buffer. The samples were centrifuged at 12,000 × g for 1 min, and the proteins in the supernatants were analyzed by SDS-PAGE and Western blotting.
Western blot analysis.
Cells were lysed in IP buffer for 30 min, and cell lysates were centrifuged at 12,000 × g for 1 min at 4°C. The supernatants were recovered and boiled in loading buffer. Protein samples were separated on 12% SDS–PAGE gels and transferred onto Immobilon-P membranes (Millipore). Membranes were probed with primary antibodies as indicated in the figures. The following antibodies were used for Western blotting: anti-NS3 (product code ab65407; Abcam), anti-NS4B (Covance; kindly provided by Kouacou V. Konan), anti-FLAG (catalog number F1804; Sigma), anti-HA (catalog number H9658; Sigma), anti-PLA2G4C (catalog number HPA043083; Sigma), and anti-beta-actin (catalog number A2066; Sigma). The proteins were visualized using suitable horseradish peroxidase (HRP)-conjugated secondary antibodies (Jackson Immuno Research) and SuperSignal-Femto chemiluminescent substrate (Pierce).
Immunofluorescence assay.
Huh7.5.1 cells were grown on glass coverslips and infected with HCVcc and/or transfected with plasmids expressing different HCV and/or host proteins by using Lipofectamine 2000 (Invitrogen). At various time points after infection or transfection, as indicated in the figures, Huh7.5.1 cells were fixed in 3.7% paraformaldehyde for 15 min, permeabilized with 0.5% Triton X-100 for 10 min, washed in PBS, and blocked with 5% bovine serum albumin (BSA). Samples were incubated overnight with the following primary antibodies in various combinations as indicated in the figures in 5% BSA: anti-FLAG antibody (F1804, diluted 1:100 [Sigma]); anti-HA antibody (3724, diluted 1:100 [Cell Signaling Technology]); anti-NS4B antibodies (ab24283, diluted 1:100 [Abcam], and sc52414, diluted 1:50 [Santa Cruz Biotechnology]); anti-NS5A antibody (9E10, diluted 1:100 [kindly provided by Charles M. Rice]); or anti-PLA2G4C antibody (HPA043083 [Sigma]). Alexa Fluor 488- and 561-conjugated secondary antibodies (diluted 1:1,000; Invitrogen) in 5% BSA were added for 1 h. The coverslips were then washed three times with PBS and stained with HCS LipidTOX red and Hoechst 33258 (Invitrogen). Images of the samples were taken using a Perkin Elmer UltraView Vox confocal microscope.
Quantitative real-time RT-PCR.
Total RNA in cells or in the supernatant was prepared by using TRIzol or TRIzol LS reagent (Invitrogen), respectively, according to the manufacturer's protocols. Moloney murine leukemia virus (MMLV) reverse transcriptase (Promega) and random primers were used to generate cDNAs from cellular RNAs and HCV RNAs. The quantification of specific mRNAs and HCV RNAs was performed using SYBR green PCR master mix (Toyobo) and the StepOne real-time PCR System (Applied Biosystems). The levels of mRNAs or HCV RNAs were normalized to the levels of beta-actin with the standard curve method. The quantitative reverse transcription (RT)-PCR primers are listed in Table S1 in the supplemental material. The quantification of HCV RNAs in the supernatant was performed using a one-step quantitative HCV RT-PCR kit (Shenzhen Piji Co. Ltd., China) with the standard-curve TaqMan probe method.
Generation of HCVpp and HCVpp entry assay.
293T cells were seeded into 60-mm dishes at a density of 3 × 106 cells 1 day before transfection. Cells were transfected with the pNL4.3.lucR−E− (27) and pcDNA3.1-E1E2 (a gift of Jin Zhong) plasmids using Lipofectamine 2000 (Invitrogen). Seventy-two hours after transfection, culture supernatants of transfected cells were collected, filtered with a 0.45-μm filter (Millipore), and stored at −80°C before use. Huh7.5.1 cells pretreated with siRNA or methyl arachidonyl fluorophosphonate (MAFP) were seeded into a 96-well plate at a density of 5 × 103 cells per well in 100 μl of DMEM containing 10% FBS. Twenty-four hours later, the medium was replaced with 100 μl of HCV pseudoparticle (HCVpp) stock. After 6 h of incubation at 37°C, HCVpp-containing medium was discarded, and fresh medium was added to the wells. Forty-eight hours later, luciferase assays were performed using the Steady-Glo luciferase assay kit (Promega).
Assays for HCV IRES-dependent translation and HCV RNA replication.
Huh7.5.1 cells were transfected with either siControl or siPLA2G4C for 48 h and then transfected with the pHCV-IRES plasmid. Alternatively, Huh7.5.1 cells were transfected with the pHCV-IRES plasmid and treated with 160 μM MAFP. After 72 h, firefly luciferase (F-Luc) and Renilla luciferase (R-Luc) activities were measured using a dual-luciferase reporter assay system from Promega (catalog number E1910). Lunet-Con1 cells containing a subgenomic HCV replicon were cultured and harvested in TRIzol at the indicated time points after siRNA transfection or treatment with MAFP. HCV RNAs were determined by quantitative RT-PCR. The copy numbers of human beta-actin mRNAs were determined and used for normalization.
The influence of arachidonic acid on HCV replication.
Arachidonic acid was diluted in 100% ethanol in airtight glass vials purged with N2 and stored at −80°C. A fresh vial was used for each experiment to avoid loss of activity (69). Huh7.5.1 cells pretreated with siPLA2G4C were infected with HCV, and then 50 μM arachidonic acid was added (catalog number A3555; Sigma). Alternatively, Huh7.5.1 cells were infected with HCV and were treated 6 h later with 160 μM MAFP (catalog number M2939; Sigma) and 50 μM arachidonic acid. HCV RNAs were quantified by quantitative RT-PCR after 72 h of incubation and the results normalized against the copy numbers of beta-actin mRNAs.
RESULTS
HCV infection upregulates host PLA2G4C protein levels.
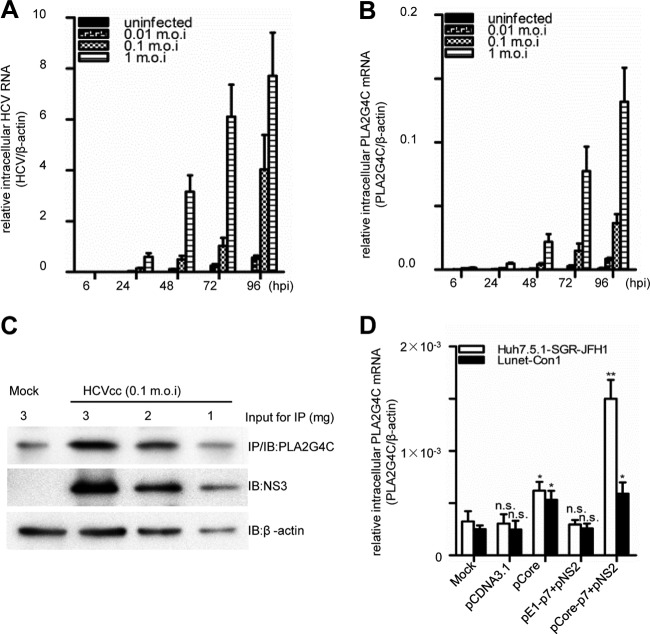
Previous studies have shown that HCV infection modulates the expression of host genes. Some genes, such as the amphiregulin (AREG) gene, are required for HCV replication (54). A comparison of the transcriptomes of HCV-infected Huh7.5.1 cells with that of the uninfected control revealed that PLA2G4C expression was enhanced approximately 10-fold in JFH-1-infected Huh7.5.1 cells (data not shown). To validate the microarray data, quantitative RT-PCR and Western blot assays were performed to determine PLA2G4C mRNA and protein levels in HCV-infected or mock-infected Huh7.5.1 cells. PLA2G4C mRNA levels increased continuously after HCV infection with time, in parallel with the increase of the HCV mRNA levels (Fig. 1A and B). The expression of PLA2G4C in Huh7.5.1 cells was correlated with HCV replication and showed strong induction by a high multiplicity of infection (MOI) of 1. Infection of Huh7.5.1 cells with HCV at an intermediate MOI of 0.1 resulted in an increase of over 20-fold at 96 hours postinfection (h.p.i.) compared to the results for the mock-infected cells (Fig. 1A and B).
Fig 1.
HCV infection upregulates host PLA2G4C expression. Huh7.5.1 cells infected with HCV JFH-1 (MOI = 0.01, 0.1, and 1) or mock infected were harvested at the indicated time points after infection. (A and B) Relative intercellular HCV RNA levels (A) and PLA2G4C mRNA (B) in HCVcc-infected or uninfected Huh7.5.1 cells were determined by quantitative RT-PCR. (C) The whole-cell lysates of HCVcc-infected (MOI = 0.1) and uninfected Huh7.5.1 cells were collected at 96 h.p.i. The PLA2G4C was concentrated by IP using an anti-PLA2G4C antibody. Three milligrams, 2 mg, and 1 mg of the whole-cell lysate were submitted to IP/Western blot assay for PLA2G4C detection. Thirty micrograms, 20 μg, and 10 μg of the whole-cell lysate were loaded for immunoblot (IB) detection of NS3 and β-actin. (D) Huh7.5.1-SGR-JFH-1 (genotype 2a) and Lunet-Con1 (genotype 1b) cells were cotransfected with different expression plasmids and pCDNA3.1 as the control. Relative intercellular PLA2G4C mRNA levels were measured by quantitative RT-PCR at 72 hours posttransfection. The bars indicate the standard deviations of triplicates. The significant differences of the different groups are shown as * (P < 0.05), ** (P < 0.01), and n.s. (no significant difference).
The expression level of PLA2G4C protein in Huh7.5.1 cells was low and barely detectable by directly loading the whole-cell lysate on the SDS-PAGE gel. Therefore, we first enriched this protein from infected and uninfected cell lysates by immunoprecipitation with an anti-PLA2G4C antibody, and then the enriched PLA2G4C was detected by Western blot assay. Figure 1C shows that PLA2G4C protein levels were increased after HCV infection. Interestingly, PLA2G4C expression was not elevated in Lunet-Con1 and Huh7.5.1-SGR-JFH-1 cells that harbor an HCV 1b or 2a subgenomic replicon (13, 14, 29). The PLA2G4C mRNA levels both in Lunet-Con1 cells and in Huh7.5.1-SGR-JFH-1 cells remained at levels similar to those of the parental cells (Fig. 1D). Thus, HCV structural proteins may be required for the upregulation of PLA2G4C expression. Transfection with the expression vector encoding HCV core proteins could induce about 2-fold upregulation of PLA2G4C expression in both Huh7.5.1-SGR-JFH-1 and Lunet-Con1 cells (Fig. 1D). The coexpression of HCV core-p7 polyprotein and NS2 protein (JFH-1, genotype 2a) could induce more than a 4-fold upregulation of PLA2G4C expression after transfection in Huh7.5.1-SGR-JFH-1 cells and a 2-fold upregulation in Lunet-Con1 cells (Con1, genotype 1b). The coexpression of HCV E1-p7 polyprotein and NS2 protein in these cell lines could not enhance PLA2G4C expression (Fig. 1D). Notably, when each HCV protein was individually expressed in Huh7.5.1 cells, only HCV core protein expression could slightly enhance the PLA2G4C mRNA level (data not shown). Thus, the HCV core protein is essential but not sufficient for the upregulation of PLA2G4C expression in HCV-infected cells.
PLA2G4C is required for HCV replication.
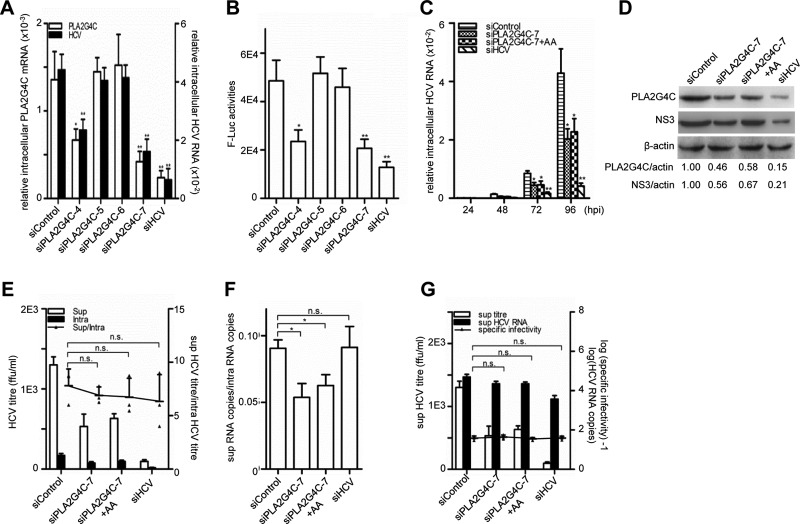
The enhanced PLA2G4C expression during HCV infection indicated a potential role of PLA2G4C in the HCV life cycle. To test this hypothesis, we transfected Huh7.5.1 cells with four siRNAs targeted to the PLA2G4C sequence (siPLA2G4C4-7) and then infected these cells with HCV J399EM, a JFH-1-derived HCVcc with an EGFP reporter (26). siControl and siHCV were included as negative and positive controls, respectively. Quantitative RT-PCR indicated that transfection with siPLA2G4C-7 and -4 decreased the PLA2G4C mRNA levels to 50% and 38% of the level for the control in Huh7.5.1 cells, respectively (Fig. 2A). These two siRNAs also reduced the HCV RNA levels to 62% and 46% compared to the level for the control. Two other siRNAs, siPLA2G4C-5 and -6, did not reduce either the expression of PLA2G4C or the HCV RNA level. Transfection with siHCV led to a 70% decrease of the PLA2G4C mRNA levels and repressed the intracellular HCV RNA levels by approximately 80% (Fig. 2A), consistent with the fact that HCV infection stimulates PLA2G4C expression. The effect on HCV replication of silencing PLA2G4C was further confirmed by using a reporter virus with a luciferase gene (Fig. 2B) (78). However, silencing PLA2G4C had no measurable impact on cell viability (data not shown).
Fig 2.
The knockdown of PLA2G4C decreases HCV replication. Huh7.5.1 cells were pretransfected with 20 nM siRNA for 48 h before being infected with HCV J399EM or HCV JFH1-luc (MOI = 0.1). (A) The relative intracellular PLA2G4C mRNA levels and HCV RNA levels in cells infected with J399EM were examined by quantitative RT-PCR at 96 h.p.i. (B) The luciferase activity in cells infected with HCV JFH1-luc were measured at 96 h.p.i. (C) The cells infected with J399EM after siRNA transfection were then either treated or not with arachidonic acid (AA) (50 μM). The relative intracellular HCV RNA levels were examined by quantitative RT-PCR at the indicated time points. (D) Whole-cell lysates were collected for Western blot analysis for HCV NS3 and β-actin at 96 h.p.i. PLA2G4C was concentrated by IP and then detected by Western blot assay using an anti-PLA2G4C antibody. The protein levels were quantified by densitometry, normalized against beta-actin, and expressed in arbitrary units. (E to G) The intracellular (intra) and supernatant (sup) HCV titers and the intracellular and supernatant HCV RNA levels at 96 h.p.i. were measured. The budding efficiency (sup HCV titer/intra HCV titer) (E), the assembly efficiency (sup HCV copies/intra HCV copies) (F), and the specific infectivity in the supernatant (sup HCV titer/sup HCV copies) (G) were calculated. The bars indicate the standard deviations of triplicates. The statistically significant differences of the different groups (referred to as sup/intra or specific infectivity in panels E to G) are shown as * (P < 0.05), ** (P < 0.01), and n.s. (no significant difference). ffu, focus-forming units.
The effect of PLA2G4C knockdown with siPLA2G4C on HCV replication was further analyzed in detail. Huh7.5.1 cells transfected with siPLA2G4C-7 reduced the relative intracellular PLA2G4C mRNA and HCV RNA levels by approximately 50% at different time points (Fig. 2C). Western blot analysis confirmed the decreased PLA2G4C protein level after siPLA2G4C-7 transfection at 96 h.p.i. Accordingly, the HCV NS3 protein expression was also reduced (Fig. 2D).
The inhibition of HCV replication by silencing PLA2G4C was further confirmed by determining the infective titers of virus in the supernatants and cell lysates. The samples were collected at 96 h.p.i., and the titers of infectious HCV particles were quantified by limiting dilution (Fig. 2E). Although the respective infective titers in the supernatants and cell lysates were apparently different, the ratios of the titers in the supernatants to those in the lysates were comparable (∼6.9-fold) under all conditions. This finding indicated that neither the siPLA2G4C-7 nor the siHCV treatment altered viral budding. However, the ratio of HCV RNA copy numbers in the supernatant to that in the intracellular fractions was significantly lower after siPLA2G4C-7 treatment (0.058, P < 0.05), while the ratios were comparable after siControl and siHCV treatments (0.090 and 0.091, respectively) (Fig. 2F). Obviously, the treatment with siHCV inhibited HCV replication without affecting HCV assembly, while silencing of PLA2G4C had a negative effect on HCV assembly. The specific infectivity in the supernatants, defined as the ratio of the infective titer to the HCV RNA copy number (38), was similar in all three treatments (Fig. 2G).
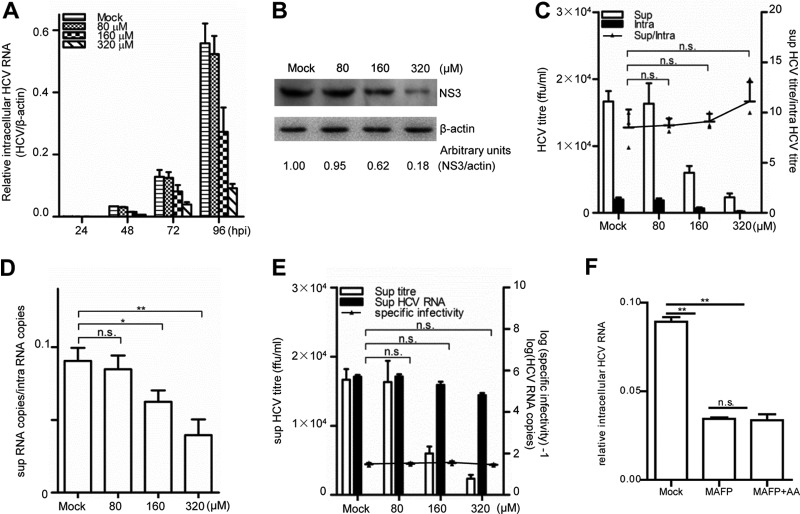
To further validate the requirement of PLA2G4C for HCV replication, a chemical inhibitor of PLA2G4C, methyl arachidonyl fluorophosphonate (MAFP) (7, 21, 70), was used to treat Huh7.5.1 cells. MAFP did not affect the viability of the Huh7.5.1 cells, even at a high concentration of 400 μM (data not shown). The treatment of Huh7.5.1 cells with MAFP at concentrations over 160 μM significantly reduced HCV RNA levels and HCV NS3 protein levels (Fig. 3A and B). In addition, MAFP treatment significantly reduced the virus titers in the supernatants in a dose-dependent manner (Fig. 3C). Furthermore, the efficiency of virion budding (Fig. 3C), the efficiency of viral assembly (Fig. 3D), and specific infectivity (Fig. 3E) were also examined. Similar to PLA2G4C knockdown, MAFP treatment also inhibited HCV assembly in a dose-dependent manner but had no effect on viral budding or the specific infectivity of HCV particles.
Fig 3.
Inhibition of PLA2G4C decreases HCV replication. Huh7.5.1 cells were infected with HCV J399EM (MOI = 0.1) for 6 h before being incubated with MAFP at various concentrations. Intracellular HCV RNA (A) was measured at the indicated time points after HCV infection by quantitative RT-PCR. NS3 protein (B) was detected by Western blot assay at 96 h.p.i. The protein levels were quantified by densitometry, normalized against the beta-actin level, and expressed in arbitrary units. (C to E) The intracellular (intra) and supernatant (sup) HCV titers and the intracellular and supernatant HCV RNA levels at 96 h.p.i. were measured. The budding efficiency (sup HCV titer/intra HCV titer) (C), the assembly efficiency (sup HCV copies/intra HCV copies) (D), and the specific infectivity in the supernatant (sup HCV titer/sup HCV copies) (E) were calculated. The bars indicate the standard deviations of triplicates. (F) Huh7.5.1 cells were infected with HCVcc (MOI = 0.01) for 6 h before incubation with MAFP (160 μM) or MAFP (160 μM) and AA (50 μM). Total RNAs were extracted and submitted for quantitative RT-PCR determination of HCV RNA at 72 h.p.i. The statistically significant differences of the different groups (referred to as sup/intra or specific infectivity in panels C and E, respectively) are shown as * (P < 0.05), ** (P < 0.01), and n.s. (no significant difference).
Taken together, these data indicate that PLA2G4C is required for HCV replication and assembly.
Inhibition of PLA2G4C does not impair HCV entry and translation but does influence HCV RNA replication.
PLA2 is involved in the process of intracellular membrane-bound vesicle fusion, a common step in the transport of macromolecules, via endocytosis or secretory pathways (23, 42). Because the HCV viral genome is transported in this process, PLA2G4C may play a role in the early steps of HCV entry and translation. To test this possibility, an HCV pseudoparticle (HCVpp) transduction system was used after Huh7.5.1 cells were either transfected with PLA2G4C siRNA or treated with the inhibitor MAFP. Huh7.5.1 cells were transfected with 20 nM siControl, siPLA2G4C-4, and siPLA2G4C-7 for 48 h or pretreated with MAFP at a concentration of 160 μM for 2 h, and then the cells were transduced with HCVpp. Intracellular luciferase activities were measured at 48 h after transduction. Neither siPLA2G4C-mediated knockdown nor MAFP treatment led to a significant change in the luciferase activities of treated cells compared to those of mock-treated cells (Fig. 4A). siCD81 transfection as the positive control successfully reduced the HCVpp-mediated luciferase activity. These data suggest that HCV entry was not influenced by PLA2G4C inhibition.
Fig 4.
Inhibition of PLA2G4C does not impair HCV entry and translation but does inhibit HCV RNA replication. (A) Huh7.5.1 cells were transfected with 20 nM siRNA for 48 h or pretreated with MAFP at a concentration of 160 μM and then transduced with HCVpp. The luciferase activities were assessed 48 h later. (B) Huh7.5.1 cells were transfected with 20 nM siRNA and then were transfected with the pHCV-IRES plasmid 48 h later. The ratio of firefly luciferase (F-Luc) activities to Renilla luciferase (R-Luc) activities was determined 48 h later. Alternatively, Huh7.5.1 cells were transfected with pHCV-IRES and treated with MAFP at a concentration of 160 μM. The error bars indicate the standard deviations of triplicates. (C and D) Lunet-Con1 cells were transfected with siRNA for 48 h to establish the knockdown effect before being replated. (C) Total RNAs were extracted and submitted for quantitative RT-PCR determination of PLA2G4C mRNA and HCV RNA levels after 96 h. (D) Whole-cell lysates were collected for Western blot analysis of PLA2G4C, HCV NS3, and β-actin at 96 h. PLA2G4C was concentrated by IP using an anti-PLA2G4C antibody before the Western blot assay. (E and F) Con1 cells were incubated in the presence of MAFP at various concentrations. At the indicated time points after treatment, cells were collected and submitted to a quantitative RT-PCR determination of HCV RNA levels (E) and to Western blot analysis of HCV NS3 and β-actin after 96 h of treatment with MAFP (F). The bars indicate the standard deviations of triplicates. The significant differences of the different groups are shown as * (P < 0.05), ** (P < 0.01), and n.s. (no significant difference).
Next, we investigated whether PLA2G4C influences HCV protein translation. HCV internal ribosome entry site (IRES) activity was monitored with the bicistronic reporter pHCV-IRES as described in Materials and Methods. The Renilla and firefly luciferase activities represented the cap- and IRES-dependent translation, respectively. The HCV IRES-dependent translation level was calculated by the normalization of the firefly luciferase activities against the Renilla luciferase activities. Compared with the results for the control cells, silencing of PLA2G4C by siRNA or inhibition of PLA2G4C activity by MAFP had no significant impact on HCV IRES-dependent translation (Fig. 4B).
To further analyze the influence of PLA2G4C on HCV RNA replication, the HCV replication level after siPLA2G4C transfection and MAFP treatment was determined. PLA2G4C expression is not enhanced in Lunet-Con1 cells that harbor an HCV subgenomic replicon (Fig. 1D). However, basal PLA2G4C expression may still support HCV RNA replication in Lunet-Con1 cells. Quantitative RT-PCR demonstrated that the transfection of Lunet-Con1 cells with siPLA2G4C-4 and siPLA2G4C-7, which could knock down PLA2G4C efficiently, significantly reduced the HCV RNA levels (∼50% decrease) (Fig. 4C). Accordingly, Western blot analysis demonstrated that the protein levels of PLA2G4C and HCV NS3 were decreased in siPLA2G4C-treated cells compared with the levels in the control cells (Fig. 4D). Consistent with PLA2G4C knockdown, MAFP treatment for 96 h also reduced the levels of HCV RNA and NS3 protein (∼50% to 70% decreased) in Lunet-Con1 cells (Fig. 4E and F). These results demonstrated the requirement of PLA2G4C for HCV RNA replication.
Arachidonic acid fails to cure MAFP-induced HCV replication deficiency.
PLA2G4C hydrolyzes phosphatidylcholine and releases many kinds of free fatty acid, such as arachidonic acid (19, 70). Arachidonic acid and/or some of its metabolites have different functions in intracellular signaling and are associated with various cellular processes (19, 69, 70). Therefore, we addressed the question of whether PLA2G4C regulates HCV RNA replication through the production of arachidonic acid by treating cells with this free fatty acid. As shown in Fig. 2, the addition of arachidonic acid did not prevent the reduction in RNA genome replication and virion assembly after PLA2G4C silencing by siRNA. Furthermore, exogenous arachidonic acid could not reverse the inhibitory effect of MAFP on HCV replication in Huh7.5.1 cells (Fig. 3F). Even a high concentration of arachidonic acid of 200 μM did not enhance HCV replication but did induce apoptosis of Huh7.5.1, which is consistent with the previous report (data not shown) (58, 77).
PLA2G4C is involved in the formation of MWs induced by HCV.
It is well established that HCV RNA replication occurs within MWs. The inhibition of PI4KIIIα, which is involved in phospholipid metabolism, affects the formation of MWs and blocks the formation of HCV replication complexes (12, 37, 57). PLA2G4C is also known to metabolize phospholipids and change the membrane curvature (17). Therefore, we hypothesized that PLA2G4C could be required for HCV-induced MW formation in host cells.
Lunet-Con1 cells, harboring an HCV subgenomic replicon, were transfected with siControl or siPLA2G4C-7 and then cultured for 48 h. The cells were reseeded, fixed, and processed for electron microscopy. Lunet-Con1 cells with active HCV replication harbored MWs, as characterized by the clustering of heterogeneous vesicles in the cytoplasm (Fig. 5A). MWs were found in approximately 67% (20 of 30) of Lunet-Con1 cells treated with siControl (Fig. 5A). After transfection with siPLA2G4C-7, only 13% (4 of 30) of Lunet-Con1 cells contained MWs (Fig. 5B).
Fig 5.
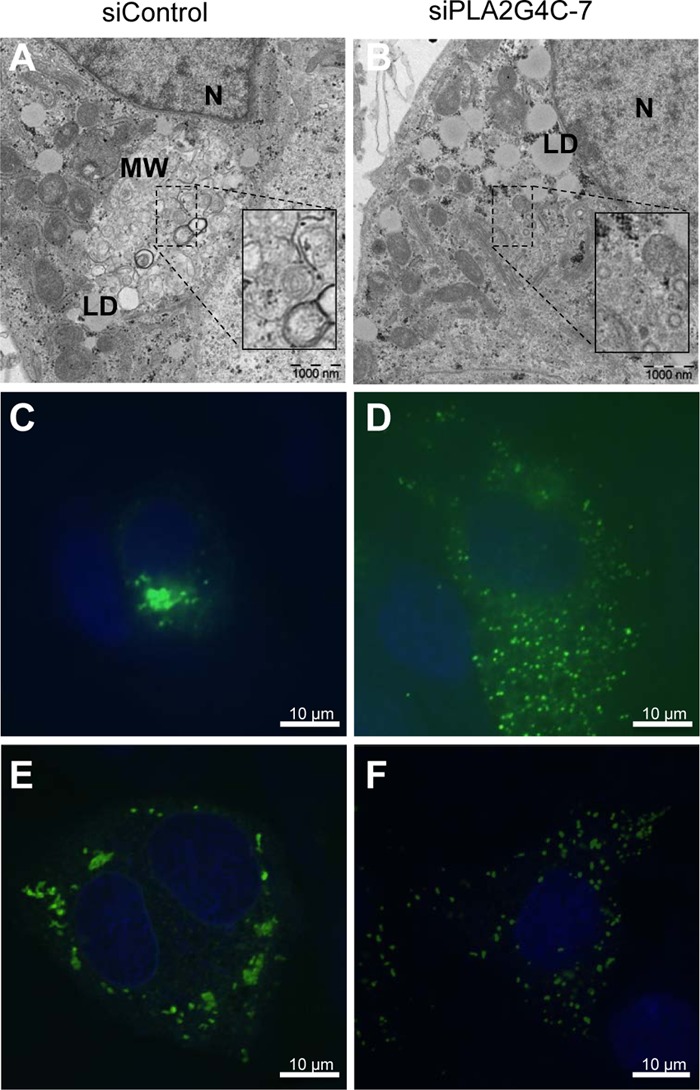
PLA2G4C is needed in the rearrangement of the cellular membrane by HCV. (A and B) Lunet-Con1 cells were transfected with siControl (A) or siPLA2G4C-7 (B) for 48 h. At 72 hours posttransfection, cells were fixed and processed for electron microscopy. N, nucleus; MW, membranous web; LD, lipid droplet. (C and D) Huh7.5.1 cells cotransfected with pEGFPN1-NS4B and siControl (C) or siPLA2G4C-7 (D) were fixed and stained with Hoechst 33258 and processed for confocal microscopy. (E and F) Huh7.5.1 cells cotransfected with pNS3-5B and siControl (E) or siPLA2G4C-7 (F) and were fixed and immunostained with anti-NS4B antibody (green). Cellular DNA was labeled with Hoechst 33258 (blue). The experiments were repeated two times, and a total of 30 cells for each treatment were randomly analyzed. Representative images are shown.
It has been reported that NS4B alone can induce the distinct ultrastructural membrane alteration called the membranous web (20, 32), which appears as dots or foci in fluorescence microscopy (2, 24, 39). To exclude the possibility that siPLA2G4C blocked the formation of MWs by inhibiting HCV replication, Huh7.5.1 cells were cotransfected with pEGFPN1-NS4B and either siControl or siPLA2G4C-7. In about 63% of GFP-positive cells, NS4B-EGFP protein showed a focal and perinuclear distribution as previously reported (Fig. 5C) (2, 36, 71). However, although NS4B-EGFP protein was expressed in a similar percentage of cells, in about 70% of GFP-positive cells, NS4B-EGFP protein formed sporadic smaller foci and widespread distribution in the cytoplasm of cells transfected with siPLA2G4C-7 (Fig. 5D). In addition, Huh7.5.1 cells were cotransfected with the expression plasmid pNS3-5B, encoding HCV NS3-NS5B polyprotein, in the presence of siControl or siPLA2G4C-7. Similarly, immunofluorescence staining with anti-NS4B antibody showed that in about 60% of cells NS4B protein formed typical foci in control cells (Fig. 5E). Transfection with siPLA2G4C-7 led to the localization of NS4B protein in sporadic smaller foci in 73% of cells (Fig. 5F). These results demonstrated that knockdown of PLA2G4C changed NS4B cellular distribution and that PLA2G4C may be involved in the formation of MWs.
PLA2G4C colocalizes with NS4B and HCV replication complexes.
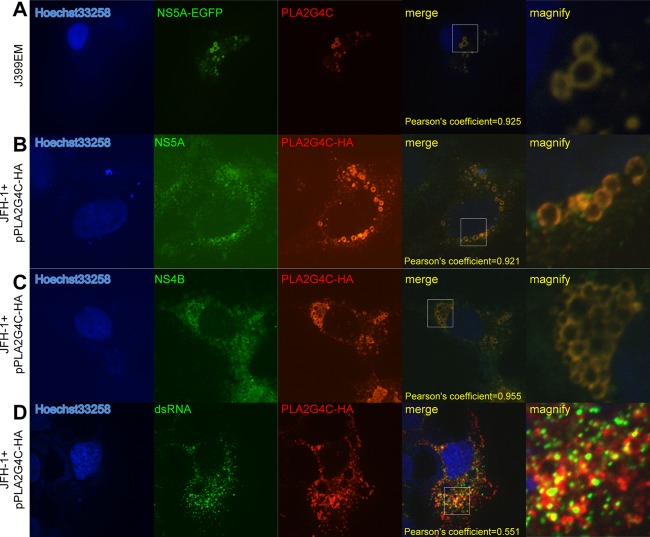
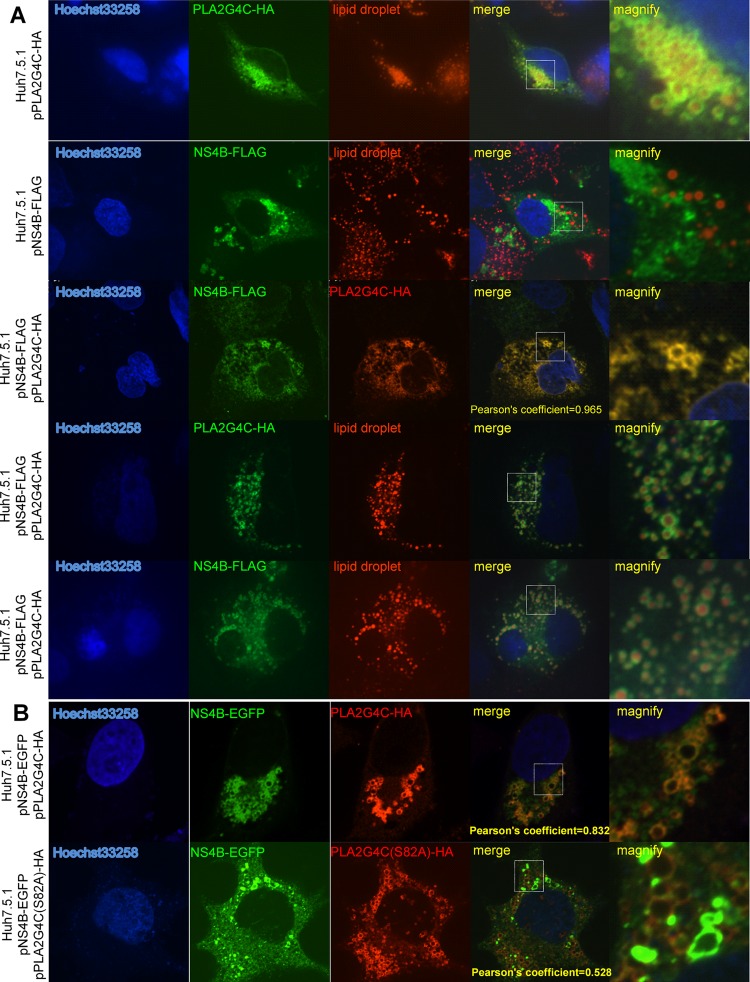
We further analyzed the subcellular localization of PLA2G4C to address whether it is part of the HCV replication complexes. PLA2G4C was not detectable in uninfected Huh7.5.1 cells (data not shown). After infection with J399EM, a JFH-1-derived construct expressing NS5A-EGFP, PLA2G4C was detected in infected Huh7.5.1 cells by immunostaining with anti-PLA2G4C antibody and was found to be colocalized with NS5A (Fig. 6A). PLA2G4C-HA, like HCV NS5A, displayed a circular pattern. The colocalization of PLA2G4C and HCV NS5A was confirmed by transfection of JFH-1-infected Huh7.5.1 cells with the expression plasmid pPLA2G4C-HA (Fig. 6B). Huh7.5.1 cells were infected with JFH-1 for 48 h and then transfected with pPLA2G4C-HA.The increased amount of PLA2G4C facilitated the observation of its colocalization with HCV NS5A. Interestingly, PLA2G4C-HA and HCV NS4B were also found to be at least partially colocalized in JFH-1-infected Huh7.5.1 cells (Fig. 6C). Staining of double-stranded RNA (dsRNA) with the antibody J2 confirmed the partial colocalization of PLA2G4C with HCV dsRNAs (Fig. 6D).
Fig 6.
Colocalization of PLA2G4C and HCV replication complexes. (A) Huh7.5.1 cells were infected with J399EM. The cells were immunostained with anti-PLA2G4C antibody (red) at 96 h.p.i. The nuclei were stained with Hoechst 33258. (B, C and D) Huh7.5.1 cells were infected with JFH-1 for 48 h and then transfected with pPLA2G4C-HA (JFH-1+pPLA2G4C-HA). Forty-eight hours later, the cells were fixed and subjected to indirect immunofluorescence staining for PLA2G4C-HA and HCV NS5A (B), NS4B (C), and dsRNA (D) with the anti-HA, anti-NS5A, anti-NS4B, and J2 primary antibodies, respectively. The nuclei were stained with Hoechst 33258. LDs were stained with HCS LipidTOX red (LD). Higher-magnification images of the selected area are also shown.
A previous report described the association of HCV replication complexes and NS5A around lipid droplets (LDs) (46). Therefore, we addressed the question of whether PLA2G4C is indeed associated with LDs, as indicated by the immunostaining results and the fact that it colocalized with HCV NS5A. PLA2G4C was expressed in Huh7.5.1 cells by transfection with the plasmid pPLA2G4C-HA and detected with anti-HA antibody. As shown in Fig. 7, PLA2G4C-HA displayed a circular pattern around the LDs. This result suggested that PLA2G4C could interact with LDs without the HCV proteins and explained its colocalization with HCV NS5A. Furthermore, we examined why PLA2G4C partially colocalized with HCV NS4B. NS4B-FLAG was expressed in Huh7.5.1 cells by transfection with the respective expression vector and presented a punctuated pattern which was not associated with the LDs, consistent with previous publications (Fig. 7) (2, 36, 71). Interestingly, in 50% of cotransfected cells (10 of 20), NS4B-FLAG changed its distribution pattern, showing colocalization with PLA2G4C-HA around the LDs. This indicated that PLA2G4C might have the ability to recruit NS4B to LDs when coexpressed (Fig. 7A). However, the ability of PLA2G4C(S82A), a catalytic inactive form of PLA2G4C (56), to recruit NS4B to lipid droplets was significantly reduced (Fig. 7B), indicating that the enzyme activity is required.
Fig 7.
Colocalization of PLA2G4C and NS4B. (A) Huh7.5.1 cells were transfected with the plasmids pPLA2G4C-HA and/or pNS4B-FLAG and stained by indirect immunofluorescence with anti-HA and anti-FLAG primary antibodies. (B) Huh7.5.1 cells were transfected with the plasmids pNS4B-EGFP and pPLA2G4C-HA or pPLA2G4C(S82A) and stained by indirect immunofluorescence with anti-HA antibody. The nuclei were stained with Hoechst 33258. LDs were stained with HCS LipidTOX red (LD). Higher-magnification images of the selected area are also shown.
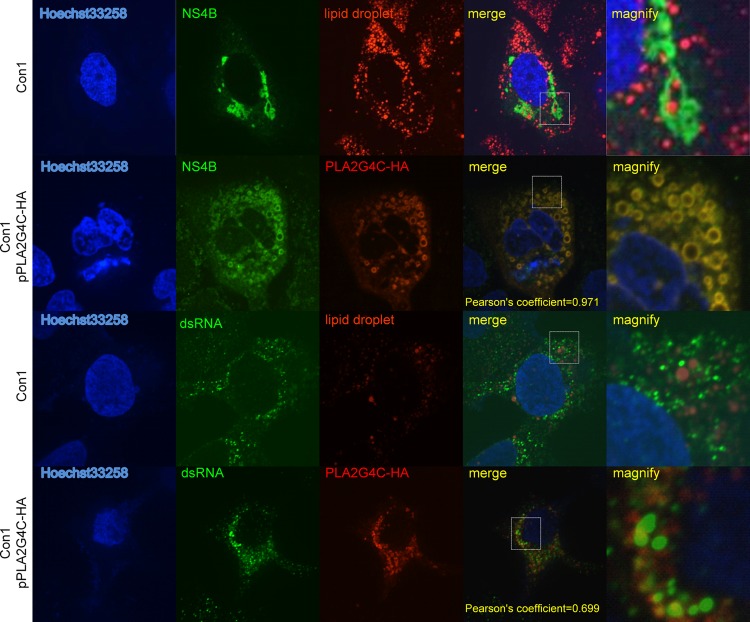
In Lunet-Con1 cells, NS4B was located in the MWs but was not associated with LDs (Fig. 8) (46). In 80% of transfected cells (16 of 20), overexpression of PLA2G4C-HA in Lunet-Con1 cells resulted in a redistribution of NS4B and colocalization of PLA2G4C-HA and NS4B around the LDs (Fig. 8). Staining of dsRNA with the specific antibody J2 indicated that PLA2G4C-HA partially colocalized with HCV dsRNAs (Fig. 8). These results strongly suggested a partial colocalization of PLA2G4C with HCV replication complexes in host cells. In addition, upregulation or overexpression of PLA2G4C resulted in the partial translocation of NS4B to LDs.
Fig 8.
Colocalization of PLA2G4C and replication complexes in Con1 cells. Lunet-Con1 cells were transfected with pPLA2G4C-HA (Con1 pPLA2G4C-HA) or empty vector (Con1) as the control. Forty-eight hours later, the cells were fixed and probed with antibodies against the HA tag, NS4B, and dsRNA for indirect immunofluorescence analysis. The nuclei were stained with Hoechst 33258. LDs were stained with HCS LipidTOX red (LD). Higher-magnification images of the selected area are also shown.
PLA2G4C interacts with the HCV NS4B protein.
The colocalization of PLA2G4C with NS4B implies an interaction between the two molecules. To verify this hypothesis, we performed a coimmunoprecipitation assay. The expression vectors pNS4B-FLAG and pPLA2G4C-HA were transfected alone or together into human HEK 293T cells. The cell lysates were subjected to IP with an anti-HA antibody, followed by immunoblotting with anti-FLAG and anti-HA antibodies (Fig. 9A). The results showed that HCV NS4B could be coimmunoprecipitated with PLA2G4C, indicating an interaction of the NS4B and PLA2G4C proteins. To determine the interaction of NS4B and endogenously expressed PLA2G4C, IP assays were performed in HCVcc-infected cells (Fig. 9B) by using anti-PLA2G4C antibody. The results indicated an interaction between PLA2G4C and NS4B under physiological conditions, likely through the close association of LDs and HCV replication complexes.
Fig 9.
PLA2G4C physically interacts with the HCV NS4B protein. (A) 293T cells were cotransfected with pPLA2G4C-HA and pNS4B-FLAG. The cells were lysed and submitted to IP assay 72 h later. (B) Huh7.5.1 cells infected with J399EM at 1 MOI for 96 h or mock infected were lysed for immunoprecipitation assay. WCL, whole-cell lysate.
DISCUSSION
HCV replicates its RNA genomes in NS4B-induced MWs (33, 64). Several host factors have been identified as important in this process. Here, we found a new host factor, PLA2G4C, that is required for HCV RNA replication and assembly. Our results suggested that PLA2G4C is involved in the formation of MWs. More importantly, we found that PLA2G4C overexpression relocates the NS4B protein to LDs, where virion assembly occurs. Thus, PLA2G4C may bridge the steps of RNA replication and HCV assembly by translocation of replication complexes to LDs.
PLA2G4C is predominantly expressed in the brain, heart, and skeletal muscle and is only expressed at low levels in the liver and the other organs (55, 70, 73). Consistent with these findings, PLA2G4C was barely detectable in the hepatoma cell lines Huh7.5.1 and Lunet. The expression of PLA2G4C in hepatoma cells was enhanced after HCV infection (Fig. 1). The HCV core protein is able to interfere with a whole array of host cell functions, including signal transduction and the transcriptional regulation of cellular genes (50). Here, we found that the HCV core is essential but not sufficient to upregulate PLA2G4C expression in Huh7.5.1 cells (Fig. 1). The regulation of PLA2G4C by multiple HCV proteins is logical because HCV structural proteins, NS proteins, and HCV RNA need to accumulate coordinately above some threshold to switch the HCV life cycle from replication to assembly. PLA2G4C may be one host factor involved in this switch process. An early overexpression of PLA2G4C would prematurely recruit HCV NS proteins to LDs. The mitogen-activated protein kinase–extracellular signal-regulated kinase (MAPK-ERK) pathway has been found to be involved in cPLA2 protein expression in endothelial cells (4). Amphiregulin (AREG) and epidermal growth factor (EGF) are the activators of the MAPK-ERK pathway, but these proteins were not able to enhance PLA2G4C expression (data not shown). Thus, PLA2G4C expression in HCV-infected hepatocyte cells is not mediated by the MAPK-ERK pathway. Further experiments are needed to delineate how HCV stimulates PLA2G4C expression.
PLA2G4C is a membrane-bound, calcium-independent, cytosolic phospholipase (70, 73) that hydrolyzes fatty acid from the sn-2 and sn-1 positions of phosphatidylcholine. The hydrolyzation of phosphatidylcholine by PLA2G4C leads to the generation of lipid signaling molecules, such as arachidonic acid. Arachidonic acid is an inflammatory factor that is capable of activating inflammatory signaling (4, 9, 70). The addition of arachidonic acid did not reverse the inhibitory effects of siPLA2G4C or MAFP on HCV replication in Huh7.5.1 (Fig. 2 and 3F), indicating that PLA2G4C regulation of HCV replication is not through the enzymatic hydrolysate.
All positive-stranded RNA viruses replicate their genomes on intracellular membranes, often in association with rearrangements of the target membrane, such as single- or double-membrane vesicles (33, 63). The expression of the full-length HCV polyprotein or NS4B induces the formation of MWs (20, 48, 49). It has been reported that PLA2G4C is mainly located in the ER (7, 55, 70, 73, 79). In addition to releasing arachidonic acid and other fatty acids, PLA2G4C activity promotes the reconformation and fusion of membranes to participate in membrane trafficking (17, 42, 51). Thus, it is possible that PLA2G4C is involved in the formation of HCV-induced MWs by altering the curve and the shape of the membranes within the cell. One possibility could be that HCV NS4B activates PLA2G4C by direct interaction between the proteins, similar to the case of PI4KIIIα, which is activated by HCV NS5A (57).
It has been proposed that HCV virion assembly is initiated on the surface of LDs (11, 41, 46, 59). The HCV core protein, which associates with LDs, plays an essential role in recruiting NS proteins and newly synthesized HCV RNA molecules from MWs to the LD-associated membranes to produce virions (46). NS5A has been reported to be integral in this process (46, 67). We found that the overexpression of PLA2G4C resulted in the localization of PLA2G4C around LDs. PLA2G4C overexpression also caused the recruitment of NS4B to LDs in the absence of HCV core proteins (Fig. 7 and 8). However, NS4B is a four-transmembrane segment protein and is different from lipid droplet-associated proteins, such as HCV core and the cellular adipose differentiation-related protein (ADRP), which tend to have peripheral membrane associations, or Rab18, which localizes through protein lidipdation. HCV NS4B probably does not interact with lipid droplets directly but is localized in lipid droplet-associated ER membranes by overexpression of PLA2G4C (15, 28, 39, 40, 52, 53, 67). In Lunet-Con1 cells, the overexpression of PLA2G4C also recruited NS proteins and HCV replication complexes to LDs (Fig. 8). Our working hypothesis is that the HCV core protein does not directly recruit HCV replication complexes to LDs (46, 67). Instead, the HCV core protein-mediated upregulation of PLA2G4C expression indirectly causes this recruitment. The replication complexes, surrounded by lipid bilayer membranes, are not directly associated with the phospholipid monolayer membranes of LDs (20, 47, 72). However, this association may occur through the interaction of PLA2G4C and NS4B (46, 67).
Taken together, the results of this study indicate that PLA2G4C is involved in the formation of MWs during the early stages of HCV infection and that it is a part of the HCV replication complex. After the accumulation of HCV proteins during HCV infection, particularly the HCV core protein, a continuous stimulation of PLA2G4C expression occurs. This stimulation leads to the association of PLA2G4C with LDs, initiating the translocation of the HCV replication complex to the place of virion assembly. However, this working model needs further refinement.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Zhiping Zhang for helping us with the electron microscopic assay and to Anna Du for technical support in confocal microscopy.
This work was supported by the National Basic Research Program of China (grant 2009CB118903) and National Nature Science Foundation of China (grant 31200135).
Footnotes
Published ahead of print 26 September 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Aizaki H, Lee KJ, Sung VM, Ishiko H, Lai MM. 2004. Characterization of the hepatitis C virus RNA replication complex associated with lipid rafts. Virology 324:450–461 [DOI] [PubMed] [Google Scholar]
- 2. Aligo J, Jia S, Manna D, Konan KV. 2009. Formation and function of hepatitis C virus replication complexes require residues in the carboxy-terminal domain of NS4B protein. Virology 393:68–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Amako Y, Sarkeshik A, Hotta H, Yates J, III, Siddiqui A. 2009. Role of oxysterol binding protein in hepatitis C virus infection. J. Virol. 83:9237–9246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Anfuso CD, et al. 2007. Endothelial cell-pericyte cocultures induce PLA2 protein expression through activation of PKCalpha and the MAPK/ERK cascade. J. Lipid Res. 48:782–793 [DOI] [PubMed] [Google Scholar]
- 5. Appel N, Schaller T, Penin F, Bartenschlager R. 2006. From structure to function: new insights into hepatitis C virus RNA replication. J. Biol. Chem. 281:9833–9836 [DOI] [PubMed] [Google Scholar]
- 6. Appel N, et al. 2008. Essential role of domain III of nonstructural protein 5A for hepatitis C virus infectious particle assembly. PLoS Pathog. 4:e1000035 doi:10.1371/journal.ppat.1000035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Asai K, et al. 2003. Human group IVC phospholipase A2 (cPLA2gamma). Roles in the membrane remodeling and activation induced by oxidative stress. J. Biol. Chem. 278:8809–8814 [DOI] [PubMed] [Google Scholar]
- 8. Backes P, et al. 2010. Role of annexin A2 in the production of infectious hepatitis C virus particles. J. Virol. 84:5775–5789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Balsinde J, Dennis EA. 1997. Function and inhibition of intracellular calcium-independent phospholipase A2. J. Biol. Chem. 272:16069–16072 [DOI] [PubMed] [Google Scholar]
- 10. Barba G, et al. 1997. Hepatitis C virus core protein shows a cytoplasmic localization and associates to cellular lipid storage droplets. Proc. Natl. Acad. Sci. U. S. A. 94:1200–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bartenschlager R, Penin F, Lohmann V, Andre P. 2011. Assembly of infectious hepatitis C virus particles. Trends Microbiol. 19:95–103 [DOI] [PubMed] [Google Scholar]
- 12. Berger KL, et al. 2009. Roles for endocytic trafficking and phosphatidylinositol 4-kinase III alpha in hepatitis C virus replication. Proc. Natl. Acad. Sci. U. S. A. 106:7577–7582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Blight KJ, McKeating JA, Marcotrigiano J, Rice CM. 2003. Efficient replication of hepatitis C virus genotype 1a RNAs in cell culture. J. Virol. 77:3181–3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Blight KJ, McKeating JA, Rice CM. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 76:13001–13014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Boulant S, et al. 2006. Structural determinants that target the hepatitis C virus core protein to lipid droplets. J. Biol. Chem. 281:22236–22247 [DOI] [PubMed] [Google Scholar]
- 16. Boulant S, Targett-Adams P, McLauchlan J. 2007. Disrupting the association of hepatitis C virus core protein with lipid droplets correlates with a loss in production of infectious virus. J. Gen. Virol. 88:2204–2213 [DOI] [PubMed] [Google Scholar]
- 17. Brown WJ, Chambers K, Doody A. 2003. Phospholipase A2 (PLA2) enzymes in membrane trafficking: mediators of membrane shape and function. Traffic 4:214–221 [DOI] [PubMed] [Google Scholar]
- 18. Choo QL, et al. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359–362 [DOI] [PubMed] [Google Scholar]
- 19. Dong M, et al. 2005. Cytoplasmic phospholipase A2 levels correlate with apoptosis in human colon tumorigenesis. Clin. Cancer Res. 11:2265–2271 [DOI] [PubMed] [Google Scholar]
- 20. Egger D, et al. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 76:5974–5984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Forlenza OV, Mendes CT, Marie SK, Gattaz WF. 2007. Inhibition of phospholipase A2 reduces neurite outgrowth and neuronal viability. Prostaglandins Leukot. Essent. Fatty Acids 76:47–55 [DOI] [PubMed] [Google Scholar]
- 22. Gao L, Aizaki H, He JW, Lai MM. 2004. Interactions between viral nonstructural proteins and host protein hVAP-33 mediate the formation of hepatitis C virus RNA replication complex on lipid raft. J. Virol. 78:3480–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Girod A, et al. 2002. The VP1 capsid protein of adeno-associated virus type 2 is carrying a phospholipase A2 domain required for virus infectivity. J. Gen. Virol. 83:973–978 [DOI] [PubMed] [Google Scholar]
- 24. Gosert R, et al. 2003. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J. Virol. 77:5487–5492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hamamoto I, et al. 2005. Human VAP-B is involved in hepatitis C virus replication through interaction with NS5A and NS5B. J. Virol. 79:13473–13482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Han Q, et al. 2009. Compensatory mutations in NS3 and NS5A proteins enhance the virus production capability of hepatitis C reporter virus. Virus Res. 145:63–73 [DOI] [PubMed] [Google Scholar]
- 27. Hsu M, et al. 2003. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc. Natl. Acad. Sci. U. S. A. 100:7271–7276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hugle T, et al. 2001. The hepatitis C virus nonstructural protein 4B is an integral endoplasmic reticulum membrane protein. Virology 284:70–81 [DOI] [PubMed] [Google Scholar]
- 29. Ikeda M, Yi M, Li K, Lemon SM. 2002. Selectable subgenomic and genome-length dicistronic RNAs derived from an infectious molecular clone of the HCV-N strain of hepatitis C virus replicate efficiently in cultured Huh7 cells. J. Virol. 76:2997–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jones DM, McLauchlan J. 2010. Hepatitis C virus: assembly and release of virus particles. J. Biol. Chem. 285:22732–22738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kato T, et al. 2003. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology 125:1808–1817 [DOI] [PubMed] [Google Scholar]
- 32. Konan KV, et al. 2003. Nonstructural protein precursor NS4A/B from hepatitis C virus alters function and ultrastructure of host secretory apparatus. J. Virol. 77:7843–7855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kopek BG, Perkins G, Miller DJ, Ellisman MH, Ahlquist P. 2007. Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PLoS Biol. 5:e220 doi:10.1371/journal.pbio.0050220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kuo G, et al. 1989. An assay for circulating antibodies to a major etiologic virus of human non-A, non-B hepatitis. Science 244:362–364 [DOI] [PubMed] [Google Scholar]
- 35. Laporte J, et al. 2000. Comparative analysis of translation efficiencies of hepatitis C virus 5′ untranslated regions among intraindividual quasispecies present in chronic infection: opposite behaviors depending on cell type. J. Virol. 74:10827–10833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liefhebber JM, Brandt BW, Broer R, Spaan WJ, van Leeuwen HC. 2009. Hepatitis C virus NS4B carboxy terminal domain is a membrane binding domain. Virol. J. 6:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lim YS, Hwang SB. 2011. Hepatitis C virus NS5A protein interacts with phosphatidylinositol 4-kinase type IIIalpha and regulates viral propagation. J. Biol. Chem. 286:11290–11298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lindenbach BD, et al. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623–626 [DOI] [PubMed] [Google Scholar]
- 39. Lundin M, Monne M, Widell A, Von Heijne G, Persson MA. 2003. Topology of the membrane-associated hepatitis C virus protein NS4B. J. Virol. 77:5428–5438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Martin S, Driessen K, Nixon SJ, Zerial M, Parton RG. 2005. Regulated localization of Rab18 to lipid droplets: effects of lipolytic stimulation and inhibition of lipid droplet catabolism. J. Biol. Chem. 280:42325–42335 [DOI] [PubMed] [Google Scholar]
- 41. Martin S, Parton RG. 2006. Lipid droplets: a unified view of a dynamic organelle. Nat. Rev. Mol. Cell Biol. 7:373–378 [DOI] [PubMed] [Google Scholar]
- 42. Mayorga LS, et al. 1993. Inhibition of endosome fusion by phospholipase A2 (PLA2) inhibitors points to a role for PLA2 in endocytosis. Proc. Natl. Acad. Sci. U. S. A. 90:10255–10259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. McGivern DR, Lemon SM. 2011. Virus-specific mechanisms of carcinogenesis in hepatitis C virus associated liver cancer. Oncogene 30:1969–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. McLauchlan J. 2009. Lipid droplets and hepatitis C virus infection. Biochim. Biophys. Acta 1791:552–559 [DOI] [PubMed] [Google Scholar]
- 45. Miyamoto M, Kato T, Date T, Mizokami M, Wakita T. 2006. Comparison between subgenomic replicons of hepatitis C virus genotypes 2a (JFH-1) and 1b (Con1 NK5.1). Intervirology 49:37–43 [DOI] [PubMed] [Google Scholar]
- 46. Miyanari Y, et al. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 9:1089–1097 [DOI] [PubMed] [Google Scholar]
- 47. Miyanari Y, et al. 2003. Hepatitis C virus non-structural proteins in the probable membranous compartment function in viral genome replication. J. Biol. Chem. 278:50301–50308 [DOI] [PubMed] [Google Scholar]
- 48. Moradpour D, et al. 2004. Insertion of green fluorescent protein into nonstructural protein 5A allows direct visualization of functional hepatitis C virus replication complexes. J. Virol. 78:7400–7409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Moradpour D, et al. 2003. Membrane association of hepatitis C virus nonstructural proteins and identification of the membrane alteration that harbors the viral replication complex. Antiviral Res. 60:103–109 [DOI] [PubMed] [Google Scholar]
- 50. Moriya K, et al. 1998. The core protein of hepatitis C virus induces hepatocellular carcinoma in transgenic mice. Nat. Med. 4:1065–1067 [DOI] [PubMed] [Google Scholar]
- 51. Moriya K, et al. 2001. Oxidative stress in the absence of inflammation in a mouse model for hepatitis C virus-associated hepatocarcinogenesis. Cancer Res. 61:4365–4370 [PubMed] [Google Scholar]
- 52. Nakamura N, Fujimoto T. 2003. Adipose differentiation-related protein has two independent domains for targeting to lipid droplets. Biochem. Biophys. Res. Commun. 306:333–338 [DOI] [PubMed] [Google Scholar]
- 53. Ozeki S, et al. 2005. Rab18 localizes to lipid droplets and induces their close apposition to the endoplasmic reticulum-derived membrane. J. Cell Sci. 118:2601–2611 [DOI] [PubMed] [Google Scholar]
- 54. Pei R, et al. 2011. Hepatitis C virus infection induces the expression of amphiregulin, a factor related to the activation of cellular survival pathways and required for efficient viral assembly. J. Gen. Virol. 92:2237–2248 [DOI] [PubMed] [Google Scholar]
- 55. Pickard RT, Strifler BA, Kramer RM, Sharp JD. 1999. Molecular cloning of two new human paralogs of 85-kDa cytosolic phospholipase A2. J. Biol. Chem. 274:8823–8831 [DOI] [PubMed] [Google Scholar]
- 56. Randall G, et al. 2006. Silencing of USP18 potentiates the antiviral activity of interferon against hepatitis C virus infection. Gastroenterology 131:1584–1591 [DOI] [PubMed] [Google Scholar]
- 57. Reiss S, et al. 2011. Recruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartment. Cell Host Microbe 9:32–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rizzo MT, et al. 1999. Induction of apoptosis by arachidonic acid in chronic myeloid leukemia cells. Cancer Res. 59:5047–5053 [PubMed] [Google Scholar]
- 59. Roingeard P, Hourioux C, Blanchard E, Prensier G. 2008. Hepatitis C virus budding at lipid droplet-associated ER membrane visualized by 3D electron microscopy. Histochem. Cell Biol. 130:561–566 [DOI] [PubMed] [Google Scholar]
- 60. Saito I, et al. 1990. Hepatitis C virus infection is associated with the development of hepatocellular carcinoma. Proc. Natl. Acad. Sci. U. S. A. 87:6547–6549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Salonen A, Ahola T, Kaariainen L. 2005. Viral RNA replication in association with cellular membranes. Curr. Top. Microbiol. Immunol. 285:139–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Saxena V, Lai CK, Chao TC, Jeng KS, Lai MM. 2012. Annexin A2 is involved in the formation of hepatitis C virus replication complex on the lipid raft. J. Virol. 86:4139–4150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Schwartz M, et al. 2002. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol. Cell 9:505–514 [DOI] [PubMed] [Google Scholar]
- 64. Schwartz M, Chen J, Lee WM, Janda M, Ahlquist P. 2004. Alternate, virus-induced membrane rearrangements support positive-strand RNA virus genome replication. Proc. Natl. Acad. Sci. U. S. A. 101:11263–11268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Shepard CW, Finelli L, Alter MJ. 2005. Global epidemiology of hepatitis C virus infection. Lancet Infect. Dis. 5:558–567 [DOI] [PubMed] [Google Scholar]
- 66. Shi ST, Lee KJ, Aizaki H, Hwang SB, Lai MM. 2003. Hepatitis C virus RNA replication occurs on a detergent-resistant membrane that cofractionates with caveolin-2. J. Virol. 77:4160–4168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Shi ST, et al. 2002. Hepatitis C virus NS5A colocalizes with the core protein on lipid droplets and interacts with apolipoproteins. Virology 292:198–210 [DOI] [PubMed] [Google Scholar]
- 68. Sklan EH, et al. 2007. TBC1D20 is a Rab1 GTPase-activating protein that mediates hepatitis C virus replication. J. Biol. Chem. 282:36354–36361 [DOI] [PubMed] [Google Scholar]
- 69. Soldati L, et al. 2002. Arachidonic acid increases intracellular calcium in erythrocytes. Biochem. Biophys. Res. Commun. 293:974–978 [DOI] [PubMed] [Google Scholar]
- 70. Stewart A, Ghosh M, Spencer DM, Leslie CC. 2002. Enzymatic properties of human cytosolic phospholipase A(2)gamma. J. Biol. Chem. 277:29526–29536 [DOI] [PubMed] [Google Scholar]
- 71. Stone M, Jia S, Do Heo W, Meyer T, Konan KV. 2007. Participation of Rab5, an early endosome protein, in hepatitis C virus RNA replication machinery. J. Virol. 81:4551–4563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Tauchi-Sato K, Ozeki S, Houjou T, Taguchi R, Fujimoto T. 2002. The surface of lipid droplets is a phospholipid monolayer with a unique fatty acid composition. J. Biol. Chem. 277:44507–44512 [DOI] [PubMed] [Google Scholar]
- 73. Underwood KW, et al. 1998. A novel calcium-independent phospholipase A2, cPLA2-gamma, that is prenylated and contains homology to cPLA2. J. Biol. Chem. 273:21926–21932 [DOI] [PubMed] [Google Scholar]
- 74. Vaillancourt FH, et al. 2009. Identification of a lipid kinase as a host factor involved in hepatitis C virus RNA replication. Virology 387:5–10 [DOI] [PubMed] [Google Scholar]
- 75. Wakita T, et al. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Welsch S, et al. 2009. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5:365–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Wolf LA, Laster SM. 1999. Characterization of arachidonic acid-induced apoptosis. Cell Biochem. Biophys. 30:353–368 [DOI] [PubMed] [Google Scholar]
- 78. Wu Y, Liao Q, Yang R, Chen X, Chen X. 2011. A novel luciferase and GFP dual reporter virus for rapid and convenient evaluation of hepatitis C virus replication. Virus Res. 155:406–414 [DOI] [PubMed] [Google Scholar]
- 79. Yamashita A, et al. 2009. Subcellular localization and lysophospholipase/transacylation activities of human group IVC phospholipase A2 (cPLA2gamma). Biochim. Biophys. Acta 1791:1011–1022 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.