Abstract
It has been proposed that most drug-resistant mutants, resulting from a single-nucleotide change, exist at low frequency in human immunodeficiency virus type 1 (HIV-1) and simian immunodeficiency virus (SIV) populations in vivo prior to the initiation of antiretroviral therapy (ART). To test this hypothesis and to investigate the emergence of resistant mutants with drug selection, we developed a new ultrasensitive allele-specific PCR (UsASP) assay, which can detect drug resistance mutations at a frequency of ≥0.001% of the virus population. We applied this assay to plasma samples obtained from macaques infected with an SIV variant containing HIV-1 reverse transcriptase (RT) (RT-simian-human immunodeficiency [SHIV]mne), before and after they were exposed to a short course of efavirenz (EFV) monotherapy. We detected RT inhibitor (RTI) resistance mutations K65R and M184I but not K103N in 2 of 2 RT-SHIV-infected macaques prior to EFV exposure. After three doses over 4 days of EFV monotherapy, 103N mutations (AAC and AAT) rapidly emerged and increased in the population to levels of ∼20%, indicating that they were present prior to EFV exposure. The rapid increase of 103N mutations from <0.001% to 20% of the viral population indicates that the replicating virus population size in RT-SHIV-infected macaques must be 106 or more infected cells per replication cycle.
INTRODUCTION
Studies have shown that human immunodeficiency virus type 1 (HIV-1) rapidly develops drug resistance in infected patients who undergo monotherapy or incompletely suppressive combination antiretroviral therapy (cART) (13, 15, 18, 28). This rapid emergence of resistance lends support to the hypothesis that HIV-1 variants capable of conferring drug resistance are already present before therapy (8, 10). The high HIV-1 replication rate, high mutation rates (1, 10, 22), and strong host selective pressures that occur during infection and the resulting swarm of nonidentical viral variants (8, 24) also support this hypothesis. This high evolutionary rate of HIV-1 can be used to study the emergence of drug resistance, which in turn can provide insights into the size of the replicating population, information that is important for developing improved treatment strategies.
In the present study, we assessed the levels of preexisting drug-resistant mutants and their emergence, using as a model macaques that were infected with a simian immunodeficiency virus (SIV) containing HIV-1 reverse transcriptase (RT) (RT-simian-human immunodeficiency virus [SHIV]mne) and treated with a short course of efavirenz (EFV) monotherapy followed by a commonly used cART regimen of tenofovir (TNV), emtricitabine (FTC), and EFV (2). As described by Ambrose et al. (2), plasma was collected and allele-specific PCR (ASP) and single-genome sequencing (SGS) were conducted on samples prior to and following EFV monotherapy and after cART. In samples collected after the initiation of cART, M184I and K65R, which encode FTC and TNV resistance, were detected in one of the macaques, which experienced virologic failure. The K103N resistance mutation was readily detectable after the administration of EFV monotherapy; however, neither ASP nor SGS was sufficiently sensitive to detect EFV drug-resistant mutations in the pretherapy samples. To assess the prevalence of resistant variants before drug selection, we therefore developed an ultrasensitive method able to detect mutation frequencies down to 0.001% of the viral population. Using our new ultrasensitive ASP (UsASP) assay on pretherapy plasma samples from 2 RT-SHIVmne-infected macaques, we were able to assess the frequency of 184I and 65R mutations, but the expected mutations at codon 103, which increased rapidly within a few days of the initiation of EFV monotherapy, could not be detected even at this level of sensitivity, revealing a large replicating virus population.
MATERIALS AND METHODS
Infection of macaques and exposure to antiretrovirals.
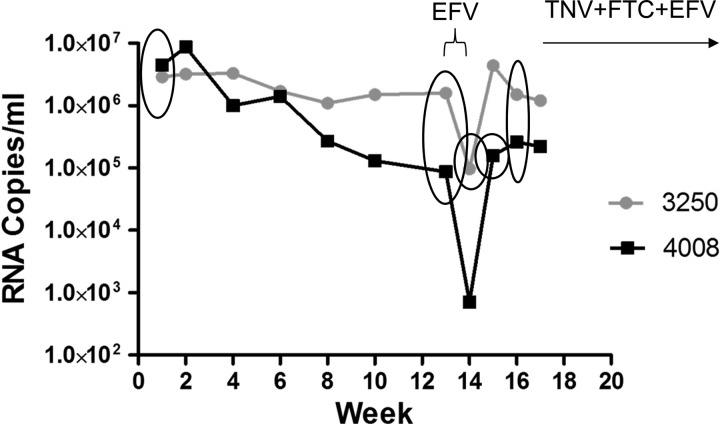
The treatment regimens for six pigtail macaques inoculated with RT-SHIVmne has been described (2). In short, animals were treated with three doses of EFV over 4 days starting 13 weeks postinoculation; two of these animals developed EFV resistance as detected by ASP and SGS (M03250 and M04008) (2). Starting at week 17, all animals were given daily oral doses of TNV plus FTC plus EFV. Eight plasma samples from two macaques, M03250 and M04008, were assessed by UsASP, two from pre-EFV monotherapy (weeks 1 and 13 postinoculation) and two from post-EFV monotherapy (weeks 14 and 16 for M03250 and weeks 15 and 16 for M04008) (Fig. 1).
Fig 1.
Plasma viral RNA shown for two pigtail macaques infected with RT-SHIVmne at week 0 and viremia monitored by RT-PCR. Circled symbols indicate time points measured in this study for each animal. After plasma was collected at week 13, three doses of EFV monotherapy were administered over 4 days as indicated by the bracket. Daily cART containing TNV, FTC, and EFV was initiated at week 17. Adapted from Fig. 2A in reference 2.
Ultrasensitive allele-specific PCR.
UsASP was based on a modification of our previously described allele-specific PCR (ASP) assay (5, 25), which reliably detects specific mutations at a frequency of ≥0.1%. By starting with >100,000 amplifiable copies of cDNA derived from the virus RNA and running numerous assays initiated with 1,000 copies of cDNA in parallel, we greatly increased the sensitivity of our assay. RNA was extracted from each sample as previously described (25, 26), and cDNA was synthesized in 40-μl reaction mixtures per the Life Technologies protocol first-strand synthesis system (catalog number18080051). One hundred microliters of 5 mM Tris HCl (pH 8.0) was added to a total of 140 μl of cDNA/sample. Five microliters of each sample was tested in duplicate by quantitative PCR (qPCR) to quantify the number of amplifiable cDNA molecules in each reaction mixture as described previously (25). Based on this quantification, each sample was then diluted accordingly and amplified by dispersing ∼1,000 copies of cDNA into each of 80 wells in a 96-well plate for the first round of UsASP and also was run under qPCR conditions to ensure that 1,000 cDNA copies were amplified. After this initial round of qPCR, 1/10,000 of the sample from each well (about 107 copies) was used as the template for measuring mutation frequencies by ASP as described previously (5, 25). Using primers that discriminate between the mutant and wild-type (WT) alleles at each codon, we measured the frequency of 103N, 65R, and 184I mutants in each of 80 wells, and we were able to detect a single mutant copy in 1,000 cDNA copies (0.1%) or a single mutant copy in an average of 80,000 cDNA copies per plate (ca. 0.001%).
The following criteria were established to minimize the possibility of obtaining false positives. First, each positive well was required to have a mutation frequency of ≥0.1%. Second, the estimated frequency in a positive well had to be at least 3.29 standard deviations (SD) above the average background in all negative wells run from the same sample, corresponding to a 99.9% probability that the positive test result did not occur by chance (P = 0.001). Third, each positive well was retested to confirm the result.
454 sequencing.
The virus inocula used to infect macaques, the plasma from week 13 (pre-EFV monotherapy) and the plasma from week 24 (post-cART) from animal M03250, were sequenced using 454 Life Sciences GS-FLX pyrosequencing technology as described previously (32, 33). To investigate regions in the HIV-1 RT that included the drug resistance mutations described here, two amplicons were produced for sequencing. The first was a 265-bp amplicon encoding amino acids 41 through 103 of RT, and the second was a 160-bp amplicon encoding amino acids 181 through 219. Resulting sequences were compared to the reference sequence of HxB2 RT, and changes associated with drug resistance mutations were noted.
RESULTS
Validation of ultrasensitive ASP.
Initially, we validated the UsASP assay for proof of concept and accuracy. One thousand copies (as confirmed by endpoint dilution PCR) of a WT DNA standard, described previously (25), were plated into each of 40 wells. Replicates of 10 wells were each spiked with 100 copies, 10 copies, or 1 copy (verified by endpoint dilution) of the K103N DNA standard, also described previously (5, 25). The remaining 10 wells, containing 100% WT DNA, were used to determine the assay background. The first-round qPCR was conducted, and then the products were diluted and used as the template for ASP to determine the frequency of 103N in each well. The expected frequencies of the 103N variants in the spiked wells were 9%, 1%, and 0.1%. The average assay background/well was 0.005% mutant. The UsASP frequency for the 9% expected-frequency group was 10/10 positive wells for 103N, with a mean frequency of 15% (standard deviation [SD], 4). The UsASP frequency for the 1% expected-frequency group was 10/10 positive wells for 103N, with a mean frequency of 1.8% (SD, 1.7). In the 0.1% expected-frequency group, there were 5/10 positive wells for 103N, with a mean frequency of 0.11% (SD, 0.13) (Table 1), consistent with the values of 6/10 and 0.33% expected from a Poisson distribution. This result validates the accuracy and sensitivity of UsASP to detect and quantitate 1 mutant copy against a background of 1,000 wild-type copies per reaction. By running multiple reactions in parallel, we can therefore detect as few as 1 mutation per copy number equal to 1,000 times the number of wells assayed.
Table 1.
Percentages of mutant DNA measured in 10 replicates of 40 wells spiked with different percentages of mutant 103N AAC HIV DNA in a WT 103K backgrounda
| Well | % mutant DNA in wells spiked with indicated % of 103N DNA |
|||
|---|---|---|---|---|
| 9% | 1% | 0.1% | 0% | |
| 1 | 9.5 | 1.3 | 0.38b | 0.016 |
| 2 | 10.5 | 1.1 | 0.01 | 0.013 |
| 3 | 13.9 | 0.98 | 0.28b | 0.009 |
| 4 | 15.9 | 1.1 | 0.13b | 0.001 |
| 5 | 15.9 | 1.2 | 0.02 | 0.007 |
| 6 | 15.4 | 4.4 | 0.006 | 0.006 |
| 7 | 17.4 | 0.4 | 0.18b | 0.001 |
| 8 | 21.9 | 0.4 | 0.10b | 0.001 |
| 9 | 19.5 | 5.5 | 0.01 | 0.004 |
| 10 | 11.2 | 1.3 | 0.007 | 0.007 |
| Mean (SD) | 15 (4) | 1.8 (1.7) | 0.11 (0.13) | 0.006 (0.005) |
Each replicate consisted of 4 groups of 10 wells. The actual distribution was 5 positive wells out of 10; the expected Poisson frequency was 6 positive wells out of 10.
The well was positive; the percentage of mutant DNA measured is above the background value.
Detection of resistance mutations in macaques.
We applied the UsASP technique to better understand the evolution of resistance in two RT-SHIV-infected macaques, both of which developed resistance (K103N) following a 4-day course of EFV monotherapy and one of which (M03250) developed resistance (K65R and M184I) during a subsequent period of cART (2). The effect of the monotherapy is shown in Fig. 1. UsASP detected K65R (AAA to AGA) mutants in all samples from both macaques pre- and post-EFV monotherapy. The week 1 sample from M03250 was determined to have an average 65R frequency over 80 wells of 0.014%. Multiplying this frequency of occurrence (1.44 × 10−4) by the viral load (2.9 × 106 copies/ml), we calculated the average number of 65R mutants to be 418 copies/ml after 1 week of infection. Similarly, M04008 was calculated to have 551 copies of the K65R mutant per ml. Throughout the time course of this study, the 65R mutant was maintained at similar fractions of the total virus population in both macaques (Tables 2 and 3), although there were greater fluctuations in the total copies/ml due to fluctuations in the viral load (Fig. 1).
Table 2.
Ultrasensitive ASP results for macaque M03250
| Mutation | Average no. of mutant copies/ml (average % mutant frequency across 80 wells) |
|||
|---|---|---|---|---|
| Week 1 (2.9 × 106)a | Week 13 (1.6 × 106) | Week 14 (9.7 × 104) | Week 16 (1.5 × 106) | |
| K65Rb | 418 (0.01) | 78 (0.005) | 4 (0.004) | 304 (0.02) |
| K103N (AAC)b | <29 (<0.001) | <17 (<0.001) | 11,154 (11.5) | 139,500 (9.3) |
| K103N (AAT)b | <29 (<0.001) | <17 (<0.001) | 6,111 (6.3) | 39,000 (2.6) |
| M184Ib | <29 (<0.001) | 520 (0.004) | <1 (<0.001) | 356 (0.015) |
Viral RNA copy number/ml in plasma.
The assay background for K65R, K103N AAC and AAT, and 184I was 0.0001% across 80 wells.
Table 3.
Ultrasensitive ASP results for macaque M04008
| Mutation | Average no. of mutant copies/ml (average % mutant frequency across 80 wells) |
|||
|---|---|---|---|---|
| Week 1 (4.5 × 106)a | Week (13 8.7 × 104) | Week 15 (1.6 × 105) | Week 16 (2.6 × 105) | |
| K65Rb | 551 (0.01) | 1 (0.001) | 32 (0.02) | 21 (0.008) |
| K103N (AAC)b | <45 (<0.001) | <1 (<0.001) | 12,800 (8.0) | 29,120 (11.2) |
| K103N (AAT)b | <45 (<0.001) | <1 (<0.001) | 20,800 (13.0) | 16,900 (6.5) |
| M184Ib | 180 (0.004) | 9 (0.01) | <2 (<0.001) | 36 (0.014) |
Viral RNA copy number/ml in plasma.
The assay background for K65R, K103N AAC and AAT, and 184I was 0.0001% across 80 wells.
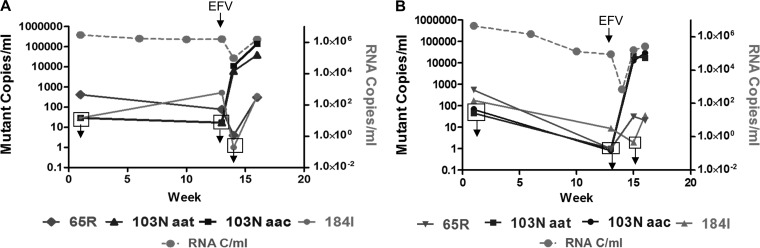
In contrast, the frequencies of M184I mutations (ATG to ATA) in both macaques varied around the limit of detection over the time course investigated. The 184I mutation was not detected in M03250 at week 1 (<0.001%, <29 copies/ml), yet appeared at week 13 before EFV monotherapy was administered (0.033%, 520 copies/ml). One week later (week 14), after a decrease in plasma HIV-1 RNA of 1.5 log10 copies/ml had occurred from treatment with EFV monotherapy, the 184I mutation was again not detectable (<0.001% mutant frequency) but reappeared at week 16 (2 weeks later) at 0.024% (356 copies/ml) (Table 2). Similar fluctuations were seen in M04008 (Fig. 2B and Table 2).
Fig 2.
Mutant copy number versus RNA copy number/ml of plasma virus in the treated RT-SHIV-infected macaques, M03250 (A) and M04008 (B). The frequencies of the indicated mutations were assessed by UsASP. Open squares with downward arrows indicate time points where the mutant copy number was less than the sensitivity of the assay.
In contrast to the pattern observed for the 65R and 184I alleles, neither of the 103N alleles (AAC and AAT) was detectable in either macaque at either time point (week 1 or week 13) prior to EFV monotherapy, indicating a frequency of <0.001% of each allele or <29 and < 45 copies/ml of 103N AAC and AAT at week 1 and <17 and <1 copies/ml of each at week 13. Remarkably, 7 days following the first dose of EFV (week 14), both the 103N AAC and AAT alleles were detected in substantial fractions of the virus population in M03250. The 103N AAC allele was present at an average of 11.5% (11,154 copies/ml), corresponding to a greater than 656-fold increase in 7 days, and the AAT allele at codon 103 was measured at an average of 6.3%, a greater than 360-fold increase. Three weeks after the first dose of EFV (week 16), the frequency of the 103N AAC allele was similar (9%), corresponding to an average of 139,500 copies of mutant genomes/ml due to a 30-fold increase in plasma HIV-1 total RNA (Table 2), but the AAT allele had declined to 2.6% of the virus population, corresponding to 39,000 mutant copies/ml (Table 2). Seventeen weeks postinoculation (week 17), the animals were started on cART (TNV plus FTC plus EFV), and as published previously (2, 20), but not tested by UsASP in this study, at week 26, the 103N AAC mutation and the 184I mutation were fixed at 100% in macaque M03250.
Samples from the week 14 time point (7 days after the first dose of EFV) in M04008 could not be tested because viral RNA levels were too low (Fig. 1). However, at week 15, the AAC allele was present at 8% frequency (12,800 mutant copies/ml), signifying a greater than 12,800-fold increase of mutant viral genomes in 14 days. The AAT allele was present at 13% (20,800 mutant copies/ml), a greater than 20,800-fold increase in 14 days (Table 3). Samples taken from this animal 1 week later, at week 16, revealed similar frequencies of the AAC allele at 11% (29,000 mutant copies/ml) and the AAT allele at 6.5% (16,900 mutant copies/ml).
Selective drug pressure on the 103 alleles in both macaques.
In both macaques, both 103N alleles (AAC and AAT) were under strong selective pressure during the period of EFV monotherapy and increased markedly in the population 7 and 14 days after the start of treatment. Assuming a replication cycle of ∼1 day (8) and using the simple population genetics equation of Rn = x, where R is the reproductive ratio (6), n is the number of replication cycles, and x is the fold increase over time, we can calculate the average increase in the number of newly infected cells that produced new infectious progeny with each cycle of replication, i.e., R. For example, in animal M03250, a >656-fold increase of the 103N AAC allele occurred in 7 days (R7 = >656, R = >2.5, on average). Thus, we can estimate that each infected cell harboring a 103N AAC allele gave rise to an average of >2.5 newly infected cells every day between weeks 13 and 14, and that each newly infected cell then produced viruses that in turn infected another >2.5 cells. Using the same simple equation, we determined that each infected cell harboring the 103N AAT allele must have given rise to >2.3 newly infected cells per replication cycle on average during the same 7 days, leading to the >359-fold increase in mutant variants observed. For macaque M04008, similar reproductive ratios occurred for both 103N AAC and AAT, with each infected cell giving rise to averages of >1.9 and >2.0 newly infected cells per day, respectively.
Detection of resistance mutations in M03250 by 454 sequencing.
We used 454 sequencing to further analyze the frequencies of resistance mutations in one of the animals (Table 4). This analysis, which had an assay background significantly higher than that of UsASP, detected no preexisting resistance mutations above background in the inoculum or in the week 13 plasma at codon 65 or at codon 103. However, the M184I mutation was detected at just above background at week 13 by 454 analysis. The week 14 or 16 plasma samples collected from macaque M03250 and described in this report were not available for testing by 454 analyses. However, the week 24 plasma sample from macaque M03250 was tested by 454 analyses (Table 4), and revealed very comparable frequencies of K65R, K103N, and M184I, which have been reported for previous allele-specific PCR and SGS results (2). In addition, other drug resistance mutations were detected in some of the samples by 454 analyses (Table 4). In the week 13 sample, low levels of M184V and Y188C were detected. The inoculum also showed these mutations above the background of the 454 assay along with the 181C mutation; however, the sensitivity of detection by 454 at these positions needs further study. By week 24, macaque M03250 had failed therapy and we also detected mutations 100I, 181C, 188C, and 190A.
Table 4.
454 sequencing of plasma virus in macaque M03250
| Mutation | Percent mutant |
|||
|---|---|---|---|---|
| Assay background | Inoculum | Week 13 | Week 24 | |
| K65Ra | 0.17 | 0.17 | 0.28 | 4.42 |
| L100Ia | 0.00 | 0.00 | 0.02 | 1.36 |
| K101Ea | 0.12 | 0.05 | 0.27 | 0.04 |
| K103N (AAC)a | 0.00 | 0.00 | 0.00 | 78.4 |
| K103N (AAT)a | 0.02 | 0.00 | 0.00 | 0.2 |
| Y181Cb | 0.00 | 0.09 | 0.05 | 0.56 |
| M184Ib | 0.06 | 0.11 | 0.34 | 60.6 |
| M184Vb | 0.00 | 0.15 | 0.66 | 0.6 |
| Y188Cb | 0.00 | 0.14 | 0.58 | 0.45 |
| G190Ab | 0.00 | 0.00 | 0.00 | 0.4 |
Amplicon 1 comprised 265 bp encoding amino acids 41 through 103. The total numbers of sequences obtained for this amplicon were 58,431 for the assay background, 10,836 for the inoculum, 12,130 for the week 13 plasma sample, and 36,544 for the week 24 plasma sample.
Amplicon 2 comprised 160 bp encoding amino acids 181 through 219. The total numbers of sequences obtained for this amplicon were 45,385 for the assay background, 25,831 for the inoculum, 24,636 for the week 13 plasma sample, and 49,236 for the week 24 plasma sample.
DISCUSSION
Understanding the frequencies of preexisting HIV-1 drug resistance mutations and their selection under drug pressure is important for determining their effect on treatment outcome, and for providing insights into the population size of HIV-1. For these reasons, we developed a new ultrasensitive allele-specific assay (UsASP) which can detect very low frequency mutants (≥0.001% of the virus population).
The assay is based on a simple principle; our standard allele-specific PCR assay is sensitive down to 0.1% with a background of about 0.01%. When the ASP reaction mixtures are divided over multiple wells, each with 1,000 starting cDNA molecules, wells with one or more mutant molecules will give a PCR signal significantly (10-fold) over the background signal (0.01%). Using this approach, we can detect, with confidence, mutations present at frequencies as low as 1 in 1,000 times the number of wells analyzed. There are two key assumptions underlying our method. The first is that the initial (round one) PCR step was initiated with at least as many cDNA molecules as the inverse of the estimated mutation frequency, e.g., 105 to measure frequencies down to ca. 0.001%. For this purpose, it is not sufficient to rely (as do some researchers [12, 16, 30]) on the measured level of viremia in copies of RNA in the starting sample, as the efficiency of the reverse transcription step cannot be assumed to be 100%. The second assumption is that the initial PCR is uniform; i.e., that all DNA molecules present are amplified equally. We deal with both these issues by running the first-round amplification step under qPCR conditions, allowing us to directly assess the number of starting copies of cDNA without making any assumptions concerning the efficiency of the previous steps.
Evolutionary theory predicts that drug resistance mutations accumulate in replicating virus populations before the initiation of therapy. It is, therefore, important to understand the frequency of such mutations, as well as their variability and kinetics of appearance in drug-naïve infected individuals. Although we (14, 19, 25) and others (17, 27) have been able to sporadically detect important mutations such as K103N, K65R, and M184V in such individuals, prior methodology is clearly insufficiently sensitive in giving reproducible and reliable values to be useful in this regard. Our results also show that ultradeep methods, such as 454, are useful to screen sequences for moderately low frequency mutations, but less sensitive than standard ASP for detection and quantification of mutations at specific sites.
We have used UsASP to investigate the evolution of drug resistance before and after EFV monotherapy in 2 RT-SHIVmne infected macaques. We were able to reproducibly detect the K65R and M184I drug resistance mutations before and after drug exposure and observe how they were maintained in the same proportion relative to the total virus population, consistent with the predicted balance between mutation and selection at these sites. In contrast, neither of the EFV resistance-associated 103N alleles (AAC or AAT) was detected in either animal prior to drug exposure, indicating that these variants comprised <0.001% of the virus population prior to the initiation of therapy. As confirmation, neither 103N allele was detected by 454 sequencing, although, as noted, this method is much less sensitive than the UsASP assay described here (23, 34). Detection of the K65R and M184I mutations and not the K103N mutations was surprising, considering that the 65R and 184I viral variants have been reported to be less fit, with reduced replication rates compared to the 103N variants (9, 11, 21). However, this result may be explained by the following two possibilities. First, the fitness of the 103N mutation in RT-SHIV may not be the same as its fitness in HIV-1. Second, transitions required for K65R and M184I are 2 to 6 times as likely to occur as the transversions required for both K103N variants (1, 31). In fact, using a cell culture single-cycle HIV-1 replication system, Abram (1) et al. described the in vivo mutation rate of HIV-1 RT to be site specific and observed that A-to-C or A-to-T single-nucleotide substitutions are quite rare, implying that the mutation rates at 103N, and therefore its steady-state levels, are likely to be much lower than the reported average for RT mutations (10, 22).
Both K103N mutant alleles (AAC and AAT) were rapidly selected by EFV monotherapy, with an estimated reproductive number of greater than 2. These two alleles emerged in the viral RNA population for a combined 20% mutation frequency ≤7 days after exposure to EFV monotherapy in M03250 and ≤14 days in M04008. Even if incomplete WT suppression is assumed, and WT replication was allowed to proceed, but at a much reduced rate following the initiation of monotherapy, the chance of both alleles in both macaques occurring spontaneously during this short window of reduced replication is statistically low compared to their accumulation during the ∼91 cycles (13 weeks) prior to monotherapy. Therefore, these results are consistent with the concept that both the 103N AAC and AAT alleles were present in the replicating population at the time of EFV exposure, at <1 in 80,000 viral genomes (undetectable by UsASP). Consequently, and according to the following argument, the number of productive cells contributing alleles to the next generation (the replicating population size) is most likely greater than 106 infectious cells per replication cycle. If a smaller replicating population is assumed, as has occurred in many studies (3, 7), the probability that both mutations would emerge reproducibly in both macaques is extremely low. For example, if the replicating population were 104 infected cells, according to a Poisson distribution, the probability that one of the 103N alleles (present at <1 in 80,000 viral genomes) would propagate the infection in one animal is about 1−e−10∧4/8 × 10∧4 or 11.7%. The expected proportion that both mutations would contribute to propagating the infection in a single animal is even less, at 1.3% (0.1172), and the occurrence of both mutations in both animals is even less probable, at 0.02%. Even with a replicating population size of 105, and again using Poisson calculations, the chance of both 103N alleles emerging in both animals is still only 26%. However, with a replicating population of >106 infectious cells per replication cycle, the probability that both mutations would emerge in both animals is much greater than 99.99%.
In contrast, a much smaller population size is required to yield the observed frequencies of M184I or K65R, reflecting the high RT error rate at these sites compared to that at position 103. The rare A-to-C and A-to-T errors at the third position in codon 103 in HIV-1 RT allowed us to calculate more accurate measurements of the minimum replicating population of RT-SHIV in this model. Further studies are clearly needed to more accurately determine population sizes of HIV-1 in infected individuals. The complex topic of effective population size and HIV-1 evolution has been greatly debated, with many elegant studies and complex mathematical models proposed (3, 4, 7, 8, 29). Here we propose an elementary model for the study of drug resistance that uses fundamental probability to investigate the replicating population size of HIV-1 infections in macaques. Our findings demonstrate how even highly infrequent mutations can affect antiretroviral therapy and suggest that the HIV-1 population size is much larger than other studies have predicted.
ACKNOWLEDGMENTS
We thank Ron Swanstrom, Ann Wiegand, and Jon Spindler for helpful discussions and Sue Toms, Connie Kinna, and Susan Jones for administrative support.
Funding for this research was provided by the National Cancer Institute's Intramural Center for Cancer Research, which supports the HIV Drug Resistance Program, and NIH R01 grant AI080290 (to Z.A.). J.M.C. was a research professor of the American Cancer Society with support from the FM Kirby Foundation.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Published ahead of print 29 August 2012
REFERENCES
- 1. Abram ME, Ferris AL, Shao W, Alvord WG, Hughes SH. 2010. Nature, position, and frequency of mutations made in a single cycle of HIV-1 replication. J. Virol. 84:9864–9878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ambrose Z, et al. 2007. Suppression of viremia and evolution of human immunodeficiency virus type 1 drug resistance in a macaque model for antiretroviral therapy. J. Virol. 81:12145–12155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Balagam R, Singh V, Sagi AR, Dixit NM. 2011. Taking multiple infections of cells and recombination into account leads to small within-host effective-population-size estimates of HIV-1. PLoS One 6:e14531 doi:10.1371/journal.pone.0014531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Batorsky R, et al. 2011. Estimate of effective recombination rate and average selection coefficient for HIV in chronic infection. Proc. Natl. Acad. Sci. U. S. A. 108:5661–5666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boltz VF, et al. 2009. Optimization of allele-specific PCR using patient-specific HIV consensus sequences for primer design. J. Virol. Methods 164:122–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bonhoeffer S, Coffin JM, Nowak MA. 1997. Human immunodeficiency virus drug therapy and virus load. J. Virol. 71:3275–3278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brown AJ. 1997. Analysis of HIV-1 env gene sequences reveals evidence for a low effective number in the viral population. Proc. Natl. Acad. Sci. U. S. A. 94:1862–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Coffin JM. 1995. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science 267:483–489 [DOI] [PubMed] [Google Scholar]
- 9. Collins JA, et al. 2004. Competitive fitness of nevirapine-resistant human immunodeficiency virus type 1 mutants. J. Virol. 78:603–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Domingo E, Menendez-Arias L, Holland JJ. 1997. RNA virus fitness. Rev. Med. Virol. 7:87–96 [DOI] [PubMed] [Google Scholar]
- 11. Dykes C, Demeter LM. 2007. Clinical significance of human immunodeficiency virus type 1 replication fitness. Clin. Microbiol. Rev. 20:550–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Flys T, et al. 2005. Sensitive drug-resistance assays reveal long-term persistence of HIV-1 variants with the K103N nevirapine (NVP) resistance mutation in some women and infants after the administration of single-dose NVP: HIVNET 012. J. Infect. Dis. 192:24–29 [DOI] [PubMed] [Google Scholar]
- 13. Gunthard HF, et al. 1998. Human immunodeficiency virus replication and genotypic resistance in blood and lymph nodes after a year of potent antiretroviral therapy. J. Virol. 72:2422–2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Halvas EK, et al. 2010. Low frequency nonnucleoside reverse-transcriptase inhibitor-resistant variants contribute to failure of efavirenz-containing regimens in treatment-experienced patients. J. Infect. Dis. 201:672–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Havlir DV, Eastman S, Gamst A, Richman DD. 1996. Nevirapine-resistant human immunodeficiency virus: kinetics of replication and estimated prevalence in untreated patients. J. Virol. 70:7894–7899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Johnson JA, et al. 2005. Emergence of drug-resistant HIV-1 after intrapartum administration of single-dose nevirapine is substantially underestimated. J. Infect. Dis. 192:16–23 [DOI] [PubMed] [Google Scholar]
- 17. Johnson JA, et al. 2008. Minority HIV-1 drug resistance mutations are present in antiretroviral treatment-naive populations and associate with reduced treatment efficacy. PLoS Med. 5:e158 doi:10.1371/journal.pmed.0050158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kantor R, et al. 2004. Evolution of resistance to drugs in HIV-1-infected patients failing antiretroviral therapy. AIDS 18:1503–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kearney M, et al. 2008. Frequent polymorphism at drug resistance sites in HIV-1 protease and reverse transcriptase. AIDS 22:497–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kearney M, et al. 2011. Genetic diversity of simian immunodeficiency virus encoding HIV-1 reverse transcriptase persists in macaques despite antiretroviral therapy. J. Virol. 85:1067–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Koval CE, Dykes C, Wang J, Demeter LM. 2006. Relative replication fitness of efavirenz-resistant mutants of HIV-1: correlation with frequency during clinical therapy and evidence of compensation for the reduced fitness of K103N + L100I by the nucleoside resistance mutation L74V. Virology 353:184–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mansky LM, Temin HM. 1995. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J. Virol. 69:5087–5094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mitsuya Y, et al. 2008. Minority human immunodeficiency virus type 1 variants in antiretroviral-naive persons with reverse transcriptase codon 215 revertant mutations. J. Virol. 82:10747–10755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Moir S, Chun TW, Fauci AS. 2011. Pathogenic mechanisms of HIV disease. Annu. Rev. Pathol. 6:223–248 [DOI] [PubMed] [Google Scholar]
- 25. Palmer S, et al. 2006. Selection and persistence of non-nucleoside reverse transcriptase inhibitor-resistant HIV-1 in patients starting and stopping non-nucleoside therapy. AIDS 20:701–710 [DOI] [PubMed] [Google Scholar]
- 26. Palmer S, et al. 2005. Multiple, linked human immunodeficiency virus type 1 drug resistance mutations in treatment-experienced patients are missed by standard genotype analysis. J. Clin. Microbiol. 43:406–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Paredes R, et al. 2010. Pre-existing minority drug-resistant HIV-1 variants, adherence, and risk of antiretroviral treatment failure. J. Infect. Dis. 201:662–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Richman DD, et al. 1994. Nevirapine resistance mutations of human immunodeficiency virus type 1 selected during therapy. J. Virol. 68:1660–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rouzine IM, Coffin JM. 1999. Linkage disequilibrium test implies a large effective population number for HIV in vivo. Proc. Natl. Acad. Sci. U. S. A. 96:10758–10763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rowley CF, Boutwell CL, Lockman S, Essex M. 2008. Improvement in allele-specific PCR assay with the use of polymorphism-specific primers for the analysis of minor variant drug resistance in HIV-1 subtype C. J. Virol. Methods 149:69–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Salemi M, Vandamme A-M. 2003. The phylogenetic handbook: a practical approach to DNA and protein phylogeny. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 32. Shao W, et al. 2009. Presented at the 16th International HIV Dynamics and Evolution Workshop, Oxford, United Kingdom [Google Scholar]
- 33. Shao W, et al. 2009. Presented at the 18th International HIV Drug Resistance Workshop, Fort Myers, FL [Google Scholar]
- 34. Simen BB, et al. 2009. Low-abundance drug-resistant viral variants in chronically HIV-infected, antiretroviral treatment-naive patients significantly impact treatment outcomes. J. Infect. Dis. 199:693–701 [DOI] [PubMed] [Google Scholar]