Abstract
Here we show that the number of activating killer cell immunoglobulin-like receptor (KIR) copies in rhesus monkeys is associated with the extent of release of cytotoxic granules by cytolytic NK cells during primary simian immunodeficiency virus SIVmac251 infection. These findings suggest that NK cells expressing high levels of activating KIRs efficiently kill SIVmac251-infected cells, and this efficient killing contributes to the NK cell-mediated control of replication of this virus during early infection.
TEXT
While it is well established that NK cells play a central role in the early control of a number of viral infections (11, 18, 22, 23), their role in containing HIV-1 replication is only now being clarified. Studies have shown that activating killer cell immunoglobulin-like receptors (KIRs) that are expressed on NK cells are involved in controlling HIV-1 replication and slowing HIV-1 disease progression (1, 2, 15, 17). While there is reason to suppose that NK cells affect HIV-1 replication early following infection, these cells are difficult to study during primary infection because it is difficult to obtain early blood samples from infected individuals. The simian immunodeficiency virus (SIV)-infected rhesus monkey provides an important nonhuman primate animal model for studying NK cell biology during a primary AIDS virus infection. We have recently demonstrated an association between specific NK cell subpopulations and the early containment of SIV replication in rhesus monkeys and a contribution of activating KIRs in stimulating NK cells to control the spread of SIV (8). In that study, we evaluated copy number variation (CNV) of KIR3DH, which represents an activating KIR receptor family in rhesus monkeys (4, 9), and showed that KIR3DH copy numbers were negatively associated with SIV replication at the time of the early peak of viral replication in Mamu-A*01− rhesus monkeys that express restrictive TRIM5 alleles. This observation implicated NK cells that express activating KIRs in controlling viral replication during early SIV infection. However, the mechanism underlying this phenomenon remains unclear.
The present study was initiated to determine how activating KIRs might affect SIV replication during primary infection. One important function of NK cells is to kill virus-infected target cells, and the present study assessed whether the cytolytic activity of NK cells is affected by KIR3DH CNV. In this study, the cytotoxicity of peripheral blood NK cells of Mamu-A*01− rhesus monkeys was evaluated by measuring intracellular granzyme B and perforin levels and assessing CD107a surface expression on NK cells following in vitro stimulation with K562 cells, a cell line that does not express major histocompatibility complex (MHC) class I molecules (3, 13, 14). Peripheral blood mononuclear cells (PBMCs) were isolated from EDTA-anticoagulated whole blood by Ficoll-Paque (GE Healthcare, Piscataway, NJ) gradient separation and either stained immediately or cryopreserved in liquid nitrogen. After thawing, cryopreserved cells were rested at 37°C and 5% CO2 for 6 h. The viability of these thawed cells was >90%. For measuring intracellular granzyme B and perforin levels, PBMCs were first stained with monoclonal antibodies specific for cell surface molecules that delineate NK cells (CD3, CD8α, NKG2A, CD56, and CD16). Cells were then fixed and permeabilized with Cytofix/Cytoperm solution (BD Biosciences, Franklin Lakes, NJ) and stained with monoclonal antibodies specific for granzyme B and perforin. CD107a surface expression on NK cells was measured following exposure of the NK cells to K562 cells. PBMCs were incubated in the presence of RPMI 1640 medium (Cellgro, Manassas, VA) supplemented with 10% fetal calf serum (Thermo Fisher Scientific, Foster City, CA) alone (unstimulated), with K562 at an effector-to-target ratio of 10:1, or with phorbol myristate acetate (PMA) and ionomycin (both Sigma-Aldrich, St. Louis, MO) as a positive control. An anti-CD107a antibody was directly added to each sample. The cells were incubated for 1 h at 37°C and 5% CO2. Then monensin (GolgiStop; BD Biosciences) and brefeldin (GolgiPlug; BD Biosciences) were added to all samples and the cells were incubated for an additional 5 h at 37°C. Cells were next stained with monoclonal antibodies specific for the cell surface molecules CD3, CD8α, NKG2A, CD56, and CD16. The viability of cells was evaluated using the LIVE/DEAD fixable aqua dead cell stain kit (Invitrogen, Carlsbad, CA), which allowed the distinction of live and dead cells in all flow cytometric analyses. Labeled cells were acquired using an LSR II flow cytometer (BD Biosciences) and analyzed using FlowJo software (TreeStar Inc., Ashland, OR).
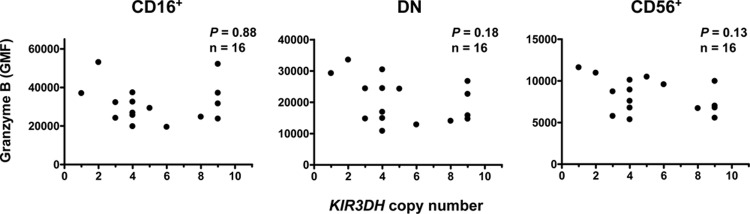
Using these assays, we first evaluated degranulation by circulating NK cells from naïve Mamu-A*01− rhesus monkeys. PBMCs were stained intracellularly for granzyme B, and the geometric mean of the fluorescence intensity (GMF) of granzyme B staining in peripheral blood NK cells was determined. Rhesus monkey NK cells were defined as CD3− CD8α+ NKG2A+ and were further delineated into subsets by their expression of CD56 and CD16 (19, 24). These NK cell subsets differ in their effector functions: CD16+ NK cells have high cytotoxic potential, CD56+ NK cells produce large amounts of cytokines, and double-negative (DN) NK cells mediate both of these functions, producing cytokines and mediating cytotoxicity (19). Due to these functional differences, it is possible that an effect of KIR3DH CNV on NK cell function may be observed only in particular NK cell subsets, and therefore, these NK cell subsets were evaluated individually. Copy numbers of the activating KIR3DH genes were determined using a quantitative real-time PCR assay (qPCR), as previously described (8). First, we determined the GMF of granzyme B in CD16+, CD56+, and DN NK cells of naïve rhesus monkeys (Fig. 1). Consistent with their expected cytotoxic function, CD16+ NK cells expressed the highest levels of granzyme B (19), DN NK cells expressed intermediate granzyme B levels, and CD56+ NK cells expressed low levels of granzyme B. There was, however, no association between KIR3DH copy numbers and granzyme B content in any of the three NK cell subsets sampled from naïve rhesus monkeys using linear regression analysis. Intracellular perforin levels or CD107a expression on stimulated NK cells was also not associated with KIR3DH copy numbers (data not shown). These findings suggest that KIR3DH copy numbers do not influence the intracellular levels of cytolytic proteins in NK cells from naïve rhesus monkeys.
Fig 1.
KIR3DH copy number is not associated with intracellular granzyme B levels in NK cell subsets of naïve rhesus monkeys. Peripheral blood NK cells from naïve rhesus monkeys were evaluated for their intracellular granzyme B content by monoclonal antibody staining and flow cytometric analysis. The association between KIR3DH copy numbers and the geometric mean of the fluorescence intensity (GMF) of granzyme B in each of three rhesus monkey NK cell subsets, CD16+, CD56+, and double-negative (DN) NK cells, was evaluated using linear regression analysis. Each dot represents one animal.
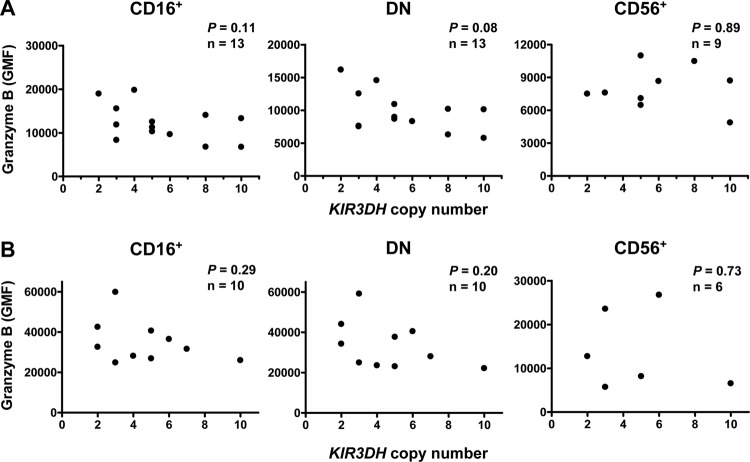
We then assessed the relationship between KIR3DH copy numbers and granzyme B expression by NK cells during the course of an SIV infection. We first sampled blood from Mamu-A*01− rhesus monkeys during the chronic phase of SIVmac251 infection and measured the intracellular granzyme B levels of NK cells. Sampling was done at the set point of the infection (day 70 postinfection) (Fig. 2A) and late during chronic infection (days 231 to 259 postinfection) (Fig. 2B). The GMF of granzyme B staining is shown for the CD16+, CD56+, and DN NK cell subsets. We observed no association between KIR3DH copy numbers and intracellular granzyme B levels in any of the NK cell subsets in rhesus monkeys that were chronically infected with SIVmac251 in a linear regression analysis. Perforin content and CD107a expression on NK cells were also not associated with KIR3DH copy numbers during this late phase of SIVmac251 infection (data not shown). These results indicate that KIR3DH CNV does not affect NK cell content of cytolytic proteins during chronic SIV infection.
Fig 2.
KIR3DH copy number is not associated with intracellular granzyme B levels in NK cell subsets of rhesus monkeys during chronic SIV infection. GMF of granzyme B staining was assessed in peripheral blood NK cells on day 70 (A) and days 231 to 259 (B) following SIVmac251 infection in three NK cell subsets in rhesus monkeys, CD16+, CD56+, and DN NK cells. Associations were analyzed using linear regression models. Each dot represents one animal.
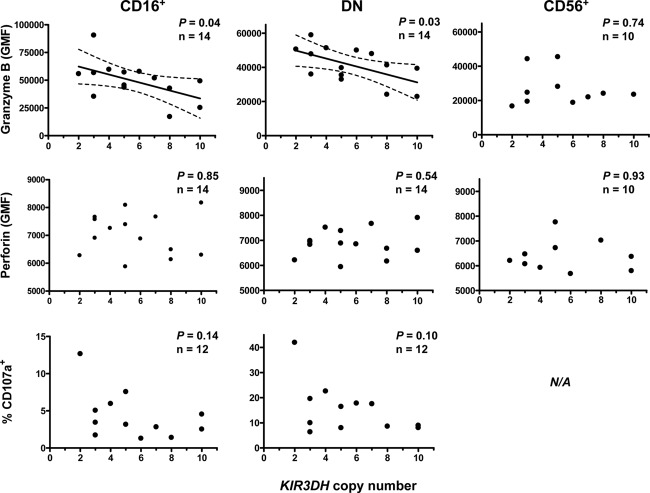
We finally assessed the cytotoxicity of NK cells during primary SIV infection. We sampled PBMCs from Mamu-A*01− rhesus monkeys on day 28 following SIVmac251 infection and stained these cells for intracellular granzyme B and perforin as well as CD107a surface expression following K562 stimulation. The GMF of granzyme B and perforin staining and the percentage of CD107a+ cells were determined in the CD16+, CD56+, and DN subsets of NK cells. Strikingly, we observed a significant negative association between intracellular granzyme B levels and KIR3DH copy numbers using linear regression analysis in both CD16+ (P = 0.04) and DN (P = 0.03) NK cells (Fig. 3), the two rhesus monkey NK cell subsets that mediate strong cytotoxicity. No association between KIR3DH copy numbers and expression levels of granzyme B was observed in the CD56+ NK cells of these monkeys (P = 0.74). Interestingly, while the intracellular granzyme B levels were associated with KIR3DH copy numbers in CD16+ and DN NK cells, the percentages of granzyme B-positive CD16+ NK cells and granzyme B-positive DN NK cells did not associate with differences in KIR3DH copy numbers (data not shown), a finding which may suggest that lower intracellular levels of granzyme B were the consequence of an increased release of granzyme B during primary SIV infection rather than the expansion of NK cells lacking granzyme B expression. In contrast to the expression of granzyme B, only low levels of intracellular perforin were detected in these three NK cell subsets, a finding which is consistent with observations reported in the literature (19). KIR3DH copy numbers were not associated with perforin content in CD16+ and DN NK cells. KIR3DH copy numbers were not associated with the percentage of CD107a+ cells in the CD16+ or DN NK cell subset following in vitro stimulation with K562 cells. We also evaluated the in vitro inhibition of SIV replication in CD4+ T cells after coculture with autologous, bulk NK cells from rhesus monkeys with different KIR3DH copy numbers but did not observe a statistically significant association between KIR3DH copy numbers and inhibition of SIV replication in CD4+ T cells (data not shown). These findings suggest that higher copy numbers of KIR3DH are associated with decreased intracellular levels of granzyme B in cytotoxic NK cells during primary SIV infection.
Fig 3.
Higher KIR3DH copy numbers are associated with lower intracellular granzyme B levels in CD16+ and DN NK cells of rhesus monkeys during acute SIV infection. Peripheral blood NK cells from rhesus monkeys evaluated for their granzyme B and perforin content. PBMCs from these monkeys were also stimulated with K562 cells at an effector-to-target ratio of 10:1 and stained for CD107a. GMF of the granzyme B and perforin staining and the percentages of CD107a+ cells were measured on day 28 post-SIVmac251 infection for each of three rhesus monkey NK cell subsets, CD16+, CD56+, and DN NK cells. Scatter plots show an association between GMF of granzyme B and KIR3DH copy numbers in CD16+ (P = 0.04, R2 = 0.29) and DN (P = 0.03, R2 = 0.33) NK cells during acute SIV infection using linear regression. For the CD107a staining, insufficient CD56+ NK cells could be acquired to allow this analysis. Each dot represents one animal. The 95% confidence interval is indicated by dashed lines.
In the present study, we demonstrated that granzyme B content was significantly lower during primary SIVmac251 infection in cytotoxic NK cells from rhesus monkeys with high numbers of KIR3DH copies than in the same NK cell subsets from rhesus monkeys with low numbers of KIR3DH copies. These findings suggest that higher numbers of KIR3DH copies are associated with a greater release of granzyme B by cytotoxic NK cells during primary SIV infection, resulting in lower levels of intracellular granzyme B content during this early phase of SIV infection. Hence, NK cells from animals with high KIR3DH copy numbers have increased cytolytic activity, and copy numbers of the activating KIR3DH directly affect the functionality of cytolytic NK cells.
In contrast to the negative association in rhesus monkeys between KIR3DH copy numbers and intracellular granzyme B levels in the two cytotoxic NK cell subsets CD16+ and DN, there was no association between KIR3DH copy numbers and NK cell function in CD56+ NK cells, the subset of NK cells that produces large amounts of cytokines. This observation is consistent with studies of humans showing that KIRs are expressed predominantly on the cytotoxic CD16+ NK cells (6). Our results are also consistent with a study by Huntington et al. that suggested that cytokine production by NK cells might be independent of interactions between KIRs and their ligands while NK cell cytotoxicity is regulated by KIRs that are expressed on NK cells (10).
Since there is no KIR3DH-specific monoclonal antibody available, KIR3DH CNV was used as an indirect measure of KIR3DH surface expression. We used a qPCR assay that recognized a conserved region of the KIR3DH genes that encodes the transmembrane domain of the KIR3DH proteins to determine KIR3DH copy numbers. In addition, we used this assay to quantify the expression of KIR3DH alleles that contain a transmembrane domain, molecules that are likely to be expressed on the surface of the NK cell, and we have previously shown that higher KIR3DH copy numbers were positively associated with higher KIR3DH transcript levels (8). Higher KIR3DH copy numbers therefore likely result in increased surface expression of KIR3DH on subpopulations of NK cells, as reported for the activating KIR3DS1 in humans (12, 16). These latter studies showed that individuals with higher gene counts of KIR3DS1 had higher percentages of KIR3DS1-expressing NK cells. The association between KIR3DH copy numbers and the intracellular granzyme B levels in the cytotoxic NK cell subsets therefore suggests that NK cells with increased surface expression of activating KIR receptors degranulated more potently than other NK cells in response to SIV-infected cells. This finding is consistent with a report that the surface density of the activating NK cell receptor NKp46 directly correlated with efficient cytolytic activity against NK cell-sensitive target cells (21).
While we observed a negative association between the intracellular granzyme B levels and KIR3DH copy numbers, there was no significant association between KIR3DH copy numbers and intracellular perforin levels or K562-stimulated CD107a surface expression by NK cells. While our inability to demonstrate associations between perforin levels or CD107a expression and KIR3DH copy numbers may reflect true NK cell biology during primary SIVmac251 infection, it may also be a consequence of the assays used to evaluate these cell populations. Intracellular levels of perforin are much lower than intracellular levels of granzyme B in NK cells of naïve and SIV-infected rhesus monkeys (19). A loss of intracellular perforin following NK cell activation would therefore likely not be as readily demonstrable as a loss of granzyme B. The low baseline intracellular stores of perforin might make it difficult to show the effects of activating KIRs on perforin release by NK cells during a primary SIVmac251 infection. Our inability to demonstrate an association between activating KIR copy numbers and NK cell surface expression of CD107a may be a consequence of the method used to evaluate CD107a cell surface expression. While KIRs recognize MHC class I molecules as their ligands, K562 is an NK cell-sensitive cell line that lacks MHC class I surface expression (3, 13, 14). Therefore, the cocultivation of NK cells with K562 cells may not allow the activation of NK cells through their activating KIRs, but rather through non-MHC class I-binding receptors (5, 7, 20). The absence of an association between activating KIR CNV and the percentage of CD107a+ NK cells in the evaluated monkeys might therefore be a consequence of an absence of interactions of the activating KIRs with their ligands on the target cells used in these assays.
Lower granzyme B content in the cytolytic NK cell subsets from monkeys expressing higher numbers of KIR3DH copies was observed during primary SIV infection. These findings are consistent with our previous observations that KIR3DH copy numbers were associated with plasma viral load during the acute phase of SIVmac251 infection (8). A study of SIV-infected monkeys showed that intracellular granzyme B levels increased during SIV infection in the three primary NK cell subsets (19). Whereas that study assessed subpopulations of NK cells during chronic infection, the present study evaluated NK cells during primary SIV infection and is therefore potentially a more accurate representation of the physiological role of NK cells in SIV infection. Moreover, we compared granzyme B contents of NK cells expressing high and low copy numbers of activating KIRs within each NK cell subpopulation, without respect to a potential overall increase of granzyme B levels in the NK cell subsets. In contrast, we show that cytolytic NK cell subsets from monkeys with high copy numbers of KIR3DH genes contained low levels of granzyme B, which may indicate an increased cytotoxicity of these NK cell subsets and may further suggest that the mechanism underlying an effect of KIR3DH copy numbers on plasma SIV RNA levels during acute SIV infection that we previously described (8) may be a more efficient killing of SIV-infected target cells by cytotoxic NK cell subpopulations, with a consequent decrease in SIV replication.
Taken together, the novelty of the presented studies is that copy numbers of KIR3DH directly affect the function of cytolytic NK cell subsets during primary SIV infection, with higher KIR3DH copy numbers being associated with decreased granzyme B content. These findings further emphasize the importance of KIR expression in modulating NK cell responses during the earliest phase of SIV infection.
ACKNOWLEDGMENTS
This work was supported by the NIAID Center for HIV/AIDS Vaccine Immunology grant AI067854.
We thank the NIAID Vaccine Research Center for providing us with the rhesus macaque PBMCs.
Footnotes
Published ahead of print 26 September 2012
REFERENCES
- 1. Alter G, et al. 2007. Differential natural killer cell-mediated inhibition of HIV-1 replication based on distinct KIR/HLA subtypes. J. Exp. Med. 204:3027–3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alter G, et al. 2009. HLA class I subtype-dependent expansion of KIR3DS1+ and KIR3DL1+ NK cells during acute human immunodeficiency virus type 1 infection. J. Virol. 83:6798–6805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Andersson LC, Nilsson K, Gahmberg CG. 1979. K562—a human erythroleukemic cell line. Int. J. Cancer 23:143–147 [DOI] [PubMed] [Google Scholar]
- 4. Blokhuis JH, Doxiadis GG, Bontrop RE, 2008. A splice site mutation converts an inhibitory killer cell Ig-like receptor into an activating one. Mol. Immunol. 46:640–648 [DOI] [PubMed] [Google Scholar]
- 5. Brandt CS, et al. 2009. The B7 family member B7-H6 is a tumor cell ligand for the activating natural killer cell receptor NKp30 in humans. J. Exp. Med. 206:1495–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cooper MA, Fehniger TA, Caligiuri MA. 2001. The biology of human natural killer-cell subsets. Trends Immunol. 22:633–640 [DOI] [PubMed] [Google Scholar]
- 7. Duchler M, et al. 1995. NKG2-C is a receptor on human natural killer cells that recognizes structures on K562 target cells. Eur. J. Immunol. 25:2923–2931 [DOI] [PubMed] [Google Scholar]
- 8. Hellmann I, Lim SY, Gelman RS, Letvin NL. 2011. Association of activating KIR copy number variation of NK cells with containment of SIV replication in rhesus monkeys. PLoS Pathog. 7:e1002436 doi:10.1371/journal.ppat.1002436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hershberger KL, Shyam R, Miura A, Letvin NL. 2001. Diversity of the killer cell Ig-like receptors of rhesus monkeys. J. Immunol. 166:4380–4390 [DOI] [PubMed] [Google Scholar]
- 10. Huntington ND, et al. 2009. IL-15 trans-presentation promotes human NK cell development and differentiation in vivo. J. Exp. Med. 206:25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lee SH, et al. 2001. Susceptibility to mouse cytomegalovirus is associated with deletion of an activating natural killer cell receptor of the C-type lectin superfamily. Nat. Genet. 28:42–45 [DOI] [PubMed] [Google Scholar]
- 12. Li H, Pascal V, Martin MP, Carrington M, Anderson SK. 2008. Genetic control of variegated KIR gene expression: polymorphisms of the bi-directional KIR3DL1 promoter are associated with distinct frequencies of gene expression. PLoS Genet. 4:e1000254 doi:10.1371/journal.pgen.1000254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lozzio BB, Lozzio CB, Bamberger EG, Feliu AS. 1981. A multipotential leukemia cell line (K-562) of human origin. Proc. Soc. Exp. Biol. Med. 166:546–550 [DOI] [PubMed] [Google Scholar]
- 14. Lozzio CB, Lozzio BB. 1975. Human chronic myelogenous leukemia cell-line with positive Philadelphia chromosome. Blood 45:321–334 [PubMed] [Google Scholar]
- 15. Martin MP, et al. 2002. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat. Genet. 31:429–434 [DOI] [PubMed] [Google Scholar]
- 16. Pascal V, et al. 2007. Detection of KIR3DS1 on the cell surface of peripheral blood NK cells facilitates identification of a novel null allele and assessment of KIR3DS1 expression during HIV-1 infection. J. Immunol. 179:1625–1633 [DOI] [PubMed] [Google Scholar]
- 17. Pelak K, et al. 2011. Copy number variation of KIR genes influences HIV-1 control. PLoS Biol. 9:e1001208 doi:10.1371/journal.pbio.1001208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Reading PC, Whitney PG, Barr DP, Smyth MJ, Brooks AG. 2006. NK cells contribute to the early clearance of HSV-1 from the lung but cannot control replication in the central nervous system following intranasal infection. Eur. J. Immunol. 36:897–905 [DOI] [PubMed] [Google Scholar]
- 19. Reeves RK, et al. 2010. CD16-natural killer cells: enrichment in mucosal and secondary lymphoid tissues and altered function during chronic SIV infection. Blood 115:4439–4446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sivori S, et al. 2002. Early expression of triggering receptors and regulatory role of 2B4 in human natural killer cell precursors undergoing in vitro differentiation. Proc. Natl. Acad. Sci. U. S. A. 99:4526–4531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sivori S, et al. 1999. NKp46 is the major triggering receptor involved in the natural cytotoxicity of fresh or cultured human NK cells. Correlation between surface density of NKp46 and natural cytotoxicity against autologous, allogeneic or xenogeneic target cells. Eur. J. Immunol. 29:1656–1666 [DOI] [PubMed] [Google Scholar]
- 22. Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S. 2008. Functions of natural killer cells. Nat. Immunol. 9:503–510 [DOI] [PubMed] [Google Scholar]
- 23. Wabuke-Bunoti MA, Bennink JR, Plotkin SA. 1986. Influenza virus-induced encephalopathy in mice: interferon production and natural killer cell activity during acute infection. J. Virol. 60:1062–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Webster RL, Johnson RP. 2005. Delineation of multiple subpopulations of natural killer cells in rhesus macaques. Immunology 115:206–214 [DOI] [PMC free article] [PubMed] [Google Scholar]