Abstract
Extensive glycosylation of the envelope spikes of human and simian immunodeficiency virus (HIV and SIV) is an important factor for the resistance of these viruses to neutralization by antibodies. SIVmac239 gp41 has three closely spaced sites for N-linked carbohydrate attachment. Rhesus macaques experimentally infected with mutant versions of SIVmac239 lacking two or three of these carbohydrate sites developed strong serum reactivity against mutated peptide sequences at the site of these glycosylations, as well as high titers of neutralizing activity to the mutant viruses (E. Yuste et al., J. Virol. 82:12472–12486, 2008). However, whether antibodies that recognize these underlying peptides have neutralizing activity has not been directly demonstrated. Here we describe the isolation and characterization of three gp41-specific monoclonal antibodies (4G8, 6G8, and 7D6) from one of these mutant-infected monkeys. All three antibodies reacted with mutant gp41 from viral particles and also with peptides corresponding to mutated sequences. Slight differences in peptide specificities were observed among the three antibodies. Sequence analysis revealed that the heavy chains of all three antibodies were derived from the same germ line heavy-chain segment (IGHV4-59*01), but they all had very different sequences in complementarity-determining region 3. The light chains of all three antibodies were very closely related to one another. All three antibodies had neutralizing activity to mutant viruses deficient in gp41 carbohydrate attachment, but they did not neutralize the parental SIVmac239. These results demonstrate unambiguously that antibodies with specificity for peptide sequences underlying gp41 carbohydrates can effectively neutralize SIV when these carbohydrates are absent. However, the presence of these gp41 carbohydrates effectively shields the virus from antibodies that would otherwise neutralize viral infectivity.
INTRODUCTION
The ineffectiveness of human immunodeficiency virus (HIV) vaccines based on the elicitation of cellular immune responses (28) and some correlation of antibody responses with the weakly protective effects in the Thai trial (11a) have refocused efforts on the elicitation of antibody responses. Some HIV type 1 (HIV-1)-infected individuals make antibodies with unusually broad, potent neutralizing activity (33). However, our ability to elicit such antibodies with immunogens has so far remained elusive. Certainly, one of the problems is the enormous sequence variability in the HIV-1-encoded envelope glycoprotein. Another is the extent to which the envelope glycoprotein is shielded by carbohydrate. Carbohydrate constitutes about 50% of the mass of the HIV-1 surface subunit gp120. In fact, a survey performed by Myers and Lenroot of approximately 10,000 protein sequences with at least one potential N-linked glycosylation site in the Swiss-Prot library showed that the number of glycosylation sites on HIV-1 gp120 ranks in the top 10 for the extent of this kind of modification (19). These carbohydrates limit both the immunogenicity and antigenicity of the gp120 envelope glycoprotein (4, 32, 34). The evidence suggests that an ever-evolving glycan shield serves to protect conserved regions of the gp120 molecule that might otherwise be targets of antibodies that could neutralize viral infectivity (21). SIVmac239, a commonly used cloned virus of defined sequence that is infectious and pathogenic for rhesus monkeys, has 23 sites for N-linked carbohydrate attachment (Asn-X-Ser/Thr) on gp120 and three closely spaced sites on gp41. The location and number of such sites on gp41 are in general highly conserved between simian immunodeficiency virus (SIV) and HIV-1.
Although carbohydrates on gp120 have been rigorously shown to shield epitopes that would otherwise be the target of neutralizing antibodies (NAbs) (4, 7, 11, 17, 24, 32, 34), corresponding information on the carbohydrates of gp41 is sparse. Sera from SIVmac239-infected monkeys have consistently failed to show reactivity to peptide sequences underlying the gp41 sites of carbohydrate attachment (38). Moreover, according to the Los Alamos Molecular Immunology Database for HIV-1, while there are a large number of antibodies that recognize sequences that flank the area of HIV-1 gp41 where the glycosylations are attached, there is only one antibody (P2D2, of mouse origin) whose epitope maps to the underlying sequences (8, 37). In line with this, Yuste et al. (38) have shown that rhesus macaques experimentally infected with mutants of SIVmac239 lacking two or three of the N-linked sites in gp41, unlike monkeys infected with the parental virus, made antibodies with high titers of neutralizing activity against the gp41 mutant viruses. Sera from these mutant-infected animals showed reactivity with peptides corresponding to the mutated sequences, again unlike monkeys infected with the wild-type virus. However, whether antibodies that recognize these underlying peptide sequences are actually capable of neutralizing gp41 mutant virus has not been rigorously demonstrated. Using newly isolated monoclonal antibodies (MAbs) of rhesus monkey origin, we now show unambiguously that antibodies with specificity for peptide sequences underlying the gp41 carbohydrates can effectively neutralize SIV when these glycans are absent. Since none of the three monoclonal antibodies neutralized the parental virus, the presence of these glycans on gp41 efficiently shields the virus from antibodies that would otherwise neutralize viral infectivity.
MATERIALS AND METHODS
Animal samples.
In order to successfully obtain cell lines secreting monoclonal antibodies against the peptide sequences underlying the carbohydrate sites of SIVmac239 gp41, we selected animal 323-92, a rhesus macaque (Macaca mulatta) infected with SIVmac239-gp41/g23 (i.e., SIVmac239 lacking the second and third glycosylation sites of gp41). In a previous study from our group, this animal showed high neutralizing titers in plasma against this carbohydrate mutant virus (SIVmac239-gp41/g23) as well as virus lacking all three glycosylation sites of gp41 (SIVmac239-gp41/g123) (38). All samples used in this report (from this monkey or others) were obtained from Indian-origin rhesus macaques housed at the New England Primate Research Center in a biosafety level 3 animal containment facility in accordance with the standards of the Association for Assessment and Accreditation of Laboratory Animal Care and the Harvard Medical School Animal Care and Use Committee. Research was conducted according to the principles described in the Guide for the Care and Use of Laboratory Animals and was approved by the Harvard Medical School Animal Care and Use Committee (20).
Generation of monoclonal antibodies against the sequences underlying the carbohydrate sites of SIVmac239 gp41.
Three different MAbs specific for the sequences underlying the carbohydrate sites of SIVmac239 gp41 (4G8, 6G8, and 7D6) were isolated in the present study. As starting material, we used freshly drawn blood from rhesus macaque 323-92. Peripheral blood mononuclear cells (PBMCs) were isolated by standard Ficoll density gradient (Accuspin System-Histopaque-1077; Sigma-Aldrich, Saint Louis, MO), and immunoglobulin G (IgG)-expressing cells were subsequently sorted with anti-IgG magnetic beads (Miltenyi Biotec Inc., Auburn, CA) according to the manufacturer's instructions. The IgG+ cell fraction was spinoculated (1,200 relative centrifugal force [RCF] for 1 h at room temperature) with supernatant containing rhesus Epstein-Barr virus (EBV)-like herpesvirus in the presence of type-B stimulatory oligonucleotides (1 μM TLR9 agonist CpG; Invivogen, San Diego, CA) and then cultured over irradiated feeder cells on 96-well plates in RPMI 1640 medium supplemented with 10 to 15% fetal bovine serum (FBS), 2 mM l-glutamine, 25 mM HEPES, 0.1 mM nonessential amino acids, 1 mM sodium pyruvate (all from Invitrogen, Carlsbad, CA), 100 μg/ml Primocin (InvivoGen, San Diego, CA), and 10% hybridoma-enhancing supplement (Sigma-Aldrich) at 37°C in a humid atmosphere of 5% CO2. As feeder cells, NIH 3T3 cells stably transfected with human CD40-Ligand (tCD40L cells) were used. This cell line was kindly provided by Gordon Freeman (26). By using tCD40L as feeder cells, thus providing efficient costimulation for the B cells (1, 26), together with the CpG synthetic oligonucleotides, which mimic bacterial DNA and stimulate B cells via toll-like receptors, the infection and immortalization of B cells could be optimized (31). To ensure the feeder cells would stop dividing but still provide CD40L-mediated costimulation and secretion of growth factors, tCD40L cells were gamma irradiated the day before spinoculation at 5 × 105 cells/ml for 50 min, for a total dose of 96 Gy, before seeding on 96-well plates at 37°C in a humid atmosphere of 5% CO2. The mixture of feeder and rhesus cells was incubated as described above, replacing one-half of the medium once per week (supplemented RPMI without hybridoma enhancer) for approximately 4.5 weeks until B-cell transformation occurred. Supernatants were then screened by standard enzyme-linked immunosorbent assay (ELISA) for reactivity against a synthetic peptide corresponding to the mutated sequence underlying the glycosylation sites of SIVmac239 gp41. Three B-cell lines with positive ELISA readings were isolated by classical sequential-dilution cloning over fresh irradiated feeder cells until monoclonality of the population of interest was ensured. These monoclonal cell lines were expanded and transferred to serum-free medium (Invitrogen). Afterwards, the antibody-containing medium was harvested, precleared by centrifugation, and filtered through 0.22-μm-pore-size membrane. Then, IgG was affinity purified with protein A+G Sepharose 4 Fast Flow (GE Healthcare, Piscataway, NJ) followed by protein quantification with a Nanodrop UV spectrometer (Thermo Fisher Scientific, Inc., Rockford, IL). Antibody purity was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and subsequent Coomassie blue staining (data not shown). The specificity of all three antibodies was confirmed after purification, and their characterization followed. The monoclonal B-cell lines secreting antibodies 4G8, 6G8, and 7D6 will be made available through the NIH AIDS Research and Reference Reagent Program.
Antibody characterization.
Antibodies 4G8, 6G8, and 7D6 were isolated for their ability to react with the peptide m6687-9 (WQEWERKVDFLEEQITALLEEAQ), corresponding to the mutated sequence underlying the glycosylations of wild-type SIVmac239 gp41 (Fig. 1A). To confirm reactivity against gp41, all antibodies were tested by ELISA at 1 μg/ml against recombinant SIVmac239 protein gp120 (which does not have gp41) and gp140 (which includes the extracellular portion of gp41); both were obtained from Immune Technology (New York, NY) and used at 1 μg/ml to coat ELISA plates.
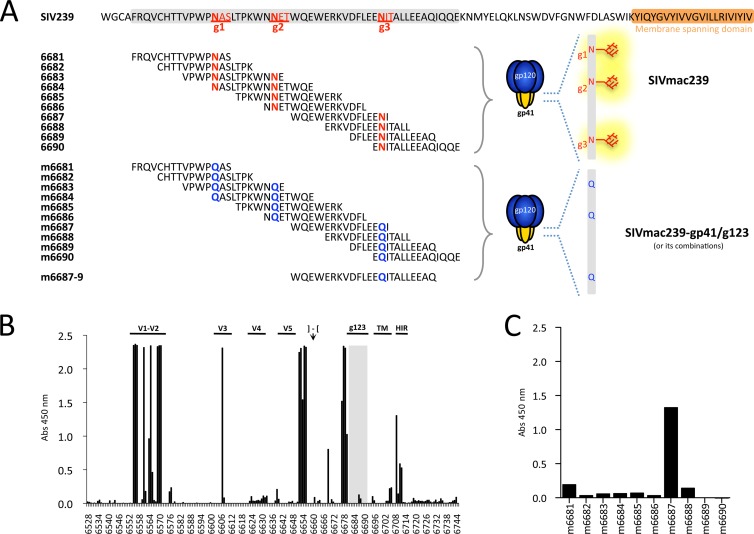
Fig 1.
Analysis of antibody responses against peptide sequences underlying the N-linked carbohydrates sites of SIVmac239 gp41 in rhesus macaque 323-92. (A) Three closely spaced N-linked-glycosylation consensus sites are found in SIVmac239 gp41, here depicted as g1, g2, and g3. The area surrounding glycosylation sites is shadowed in gray. The overlapping peptides 6681 to 6690 match the sequence of the parental virus SIVmac239, while peptides m6681 to m6690 represent the amino acid sequence of mutant virus SIVmac239-gp41/g123, in which the asparagine residue in the first, second, and third canonical N-X-S/T consensus sequence for carbohydrate attachment was changed to the structurally similar glutamine (N-X-S/T to Q-X-S/T). The longer peptide used for subsequent screening of antibodies is designated m6687-9. (B) Representative PepScan (ELISA against overlapping peptides spanning the whole envelope sequence for SIVmac239) with serum from 323-92, a rhesus monkey experimentally infected with SIVmac239-gp41/g23 (glycan-deficient mutant lacking g2 and g3). Sequences underlying the N-linked carbohydrate sites are shadowed in gray. Regions of interest are indicated as follows: variable regions 1-2, 3, 4, and 5 are labeled V1-V2, V3, V4, and V5, respectively; the cleavage site and beginning of gp41 are represented by an arrow and brackets; g123 indicates the location of the glycosylations; TM is the transmembrane domain of gp41; and HIR stands for highly immunogenic region (23). ELISA reactivity was measured as absorbance at 450 nm. (C) Serum from animal 323-92 tested against peptides corresponding to the mutated viral sequences of SIVmac239-gp41/g123 covering the area for the N-linked glycosylations. This monkey was selected as a B-cell source for isolation of monoclonal antibodies with such specificity. Representative results of three independent experiments are shown.
The variable regions of the heavy and light chains of the MAbs were sequenced at the DNA level (Syd Labs, Malden, MA). The nucleotide sequences were then analyzed with IMGT/V-QUEST (International ImMunoGeneTics information system/V-QUEry and STandardization; http://www.imgt.org/) (3, 10), an integrated software that analyzes rearranged immunoglobulin nucleotide sequences. With this tool, the V(D)J [variable(diversity)joining] genes and alleles were identified by alignment with the human immunoglobulin germ line gene and allele sequences of the IMGT reference directory. The complementarity-determining regions (CDRs) and framework regions were identified on the basis of the IMGT unique numbering, and somatic mutations were identified. Two-dimensional graphical representations of the variable regions were created, also using IMGT/V-QUEST: the IMGT Collier de Perles; and three-dimensional structures of the variable domains were obtained with RosettaAntibody, an antibody-modeling server (29).
PepScan.
To map and characterize the epitopes of the three monoclonal antibodies 4G8, 6G8, and 7D6, we tested their reactivity by ELISA against overlapping peptides (PepScan). Fifteen-mer-length peptides representing the entire SIVmac239 envelope (Env), each successive peptide overlapping by 11 amino acids (aa), were obtained from the NIH AIDS Research and Reference Reagent Program. Mutant Env 15-mer peptides were synthesized at the Massachusetts General Hospital peptide core facility (Charlestown, MA). Single wells of 96-well half-area, high-binding polystyrene microplates (Corning Costar, Lowell, MA) were coated with 50 μl of each peptide at 40 μg/ml in phosphate-buffered saline (PBS) overnight at 4°C. The wells were then blocked with 150 μl of 5% nonfat powdered milk in PBS at 37°C for 1 h. Fifty microliters of each corresponding monoclonal antibody diluted at 1 μg/ml with 5% nonfat powdered milk in PBS was added to each well and incubated at 37°C for 1 h. Wells were then washed five times with PBS 0.05%-Tween 20 (PBS-Tween), and 50 μl of horseradish peroxidase (HRP)-conjugated goat anti-human IgG antibody (Santa Cruz Biotechnology, Santa Cruz, CA) diluted 1:1,000 in 5% nonfat powdered milk in PBS was added to each well for further incubation at 37°C for 1 h. The plates were then washed 10 times with PBS-Tween and developed with 50 μl of soluble tetramethylbenzidine reagent (Calbiochem, Gibbstown, NJ). The reaction was stopped by the addition of 50 μl of acidic stop solution (SouthernBiotech, Birmingham, AL), and the optical density at 450 nm was measured using a Wallac Victor plate reader (Perkin-Elmer, Waltham, MA).
Western blot analysis.
SIVmac239-gp41/g23 and wild-type SIVmac239 (both prepared by transient transfection of HEK293T cells) were pelleted by centrifugation (10,000 RCF) for 1 h at 4°C. The pellets were washed twice with cold PBS, and the final concentration of p27 capsid was determined by an antigen capture assay (Advanced BioScience Laboratories, Inc., Kensington, MD). An equal volume of 2× Laemmli loading buffer with 10% β-mercaptoethanol was added to each virus, and samples were incubated for 10 min at 99°C before separation by SDS-PAGE. Identical amounts of p27-normalized viruses were loaded per lane (50 ng of p27). Next, the proteins from the gels were transferred to a polyvinylidene difluoride (PVDF) membrane (Bio-Rad Laboratories, Hercules, CA). The membranes were blocked with 5% nonfat powdered milk in PBS (blocking solution) for 1 h at room temperature and incubated with the indicated antibody (4G8, 6G8, or 7D6) at 1 μg/ml in blocking solution overnight at 4°C. Antibody 4B2, a rhesus monoclonal antibody that binds the highly immunogenic region of the cytoplasmic domain of gp41 (the area indicated as HIR in Fig. 1B) was used at the same concentration as a positive control (23). The membranes were then incubated with goat anti-human IgG HRP-conjugated secondary antibody (Santa Cruz Biotechnology) diluted 1:1,000 in blocking solution for 1 h at room temperature. Specific signals were detected by an enhanced-chemiluminescence system using SuperSignal West Femto maximum sensitivity chemiluminescent substrate (Thermo Fisher Scientific). Similar conditions were used when analyzing recombinant proteins (SIVmac239 gp120 and SIVmac239 gp140; both used at 200 ng per lane) with the following changes: the samples were incubated 5 min at 95°C with 5% β-mercaptoethanol, and the blots were developed using SuperSignal West Pico chemiluminescent substrate (Thermo Fisher Scientific). Pooled serum from SIV-infected macaques was used at a 1:10,000 dilution in blocking buffer as a positive control.
Neutralization assays.
The neutralizing activities of MAbs 4G8, 6G8, and 7D6 against various strains of SIVmac were measured using the immortalized human CD4+ T-cell line C8166-45 SIV-SEAP (15). C8166-45 SIV-SEAP cells harbor a Tat-inducible, secreted engineered alkaline phosphatase (SEAP) reporter construct enabling the quantification of SIV infection by measuring SEAP production in the culture supernatant. Virus amounts equivalent to 2 ng of p27 capsid protein for SIVmac239, SIVmac239-gp41/g23, and HIV-1 NL4-3 and 5 ng for SIV239-gp41/g123 and SIVmac316 were chosen as the lowest levels of viral input that would be sufficient to give a clear SEAP signal within the linear range. SEAP activity was measured when levels were sufficiently over background to give reliable measurements (at least 10-fold). Irrelevant purified rhesus IgG was used as a negative control, and serum samples from animal 323-92 were used as a positive control. The neutralization assays were performed in 96-well plates and were set up as follows: outer wells were filled with PBS to maintain hydration. Then, 25 μl of medium (RPMI, 10% FBS) was added to the first two inner columns and 25-μl aliquots of successive 2-fold dilutions in medium of test antibody or serum were added to each of the other columns. All sera had previously been heat inactivated at 56°C for 30 min. Each virus, in a total volume of 75 μl at the indicated concentration, was then added to each well in columns 3 through 11. Virus-free medium was added to column 2 (mock). The plate was incubated for 1 h at 37°C, and subsequently 100 μl of C8166-45 SIV-SEAP was added to each well at 50,000 cells/ml for a final volume of 200 μl/well. The plate was then incubated at 37°C in a humid atmosphere of 5% CO2 for the indicated time (3 to 5 days), and SEAP activity was measured using the chemiluminescent Phosphalight SEAP assay system (Applied Biosystems, Carlsbad, CA) according to the manufacturer's recommendations, with minor modifications as described previously (15). The plate reader was a Victor3 V multilabel counter (Perkin-Elmer), and neutralization was reported as a percentage of SEAP activity.
Cell culture and virus stock preparation.
HEK293T, C8166-45 SIV-SEAP, and tCD40L cells were maintained as previously described (16, 18, 26). Viruses were prepared by transient transfection of HEK293T cells with plasmids bearing the full-length proviral genomes. Cells were seeded at 1.5 × 106 cells per flask the day before transfection, and each flask was transfected with 3 μg of DNA using the calcium phosphate method (Promega, Madison, WI) according to the manufacturer's instructions. The culture medium was replaced with fresh medium on day 2 after transfection, and supernatants were harvested on day 3. To quantify the virus, the concentration of p27 capsid in the supernatant was determined by an antigen capture assay (Advanced BioScience Laboratories).
RESULTS
Isolation of monoclonal antibodies against peptides underlying the carbohydrates of SIVmac239 gp41 from a rhesus monkey infected with glycan-deficient virus.
SIVmac239 has three glycosylation attachment sites in gp41, namely, g1, g2, and g3 (Fig. 1A). In order to test whether antibodies directed against the peptides underlying these glycosylations are those responsible for neutralization of the gp41 glycan-deficient mutants, we aimed at isolating monoclonal antibodies with such specificity. For this purpose, we chose one rhesus macaque (animal 323-92) previously infected with glycan-deficient SIVmac239gp41/g23, a mutant that lacks sites g2 and g3 (38). To confirm the presence of the antibodies of interest, serum from 323-92 was obtained (48 months after infection) and tested by PepScan against a panel of 218 peptides overlapping the entire SIVmac239 Envelope protein. As shown, antibodies in the serum from this animal did not react appreciably with wild-type peptide sequences corresponding to the region covered by the glycosylation sites (Fig. 1B; peptides shadowed in gray and labeled g123). Conversely, strong reactivity was shown to established immunodominant regions of Env, i.e., the variable loops V1-V2 and V3, the C terminus of gp120, some peptides in the ectodomain of gp41, and the highly immunogenic region (HIR) at the beginning of the cytoplasmic tail of gp41 (23, 38). When the same serum was tested against another set of overlapping peptides that had mutated glycosylation consensus sites, strong reactivity to peptide m6687 was observed (Fig. 1C), which represents a region just beneath g2 and g3. From this animal, three monoclonal B-cell lines secreting antibodies specific for this region were generated by EBV immortalization as described in Materials and Methods: 4G8, 6G8, and 7D6. As specificity marker, the longer oligopeptide m6687-9 was used, which corresponds to a region of 23 aa starting immediately C terminal to g2 and including g3 (Fig. 1A).
Rhesus monoclonal antibodies directed against peptides underlying the carbohydrates of SIVmac239 gp41 show different reactivity patterns.
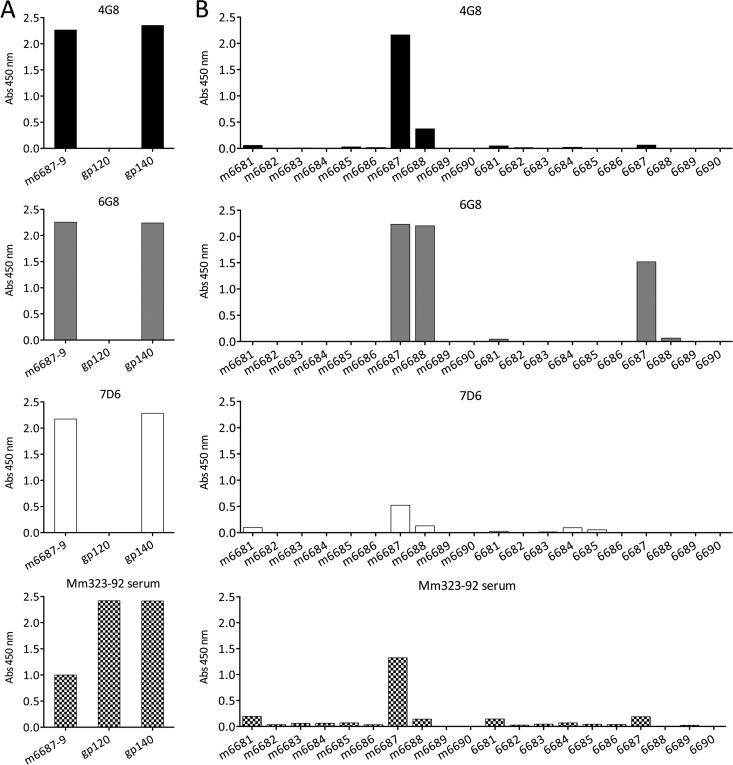
Isotyping of 4G8, 6G8, and 7D6 revealed that they are all IgG1 with kappa light chain. The specificity of the antibodies for the glycosylation region of gp41 was verified by ELISA. All three antibodies bound efficiently to the longer peptide used for the screening peptides m6687 to m6689 (which covers the sequence between g2 and g3), while they did not bind recombinant protein gp120 (which lacks gp41), thus further demonstrating its specificity (Fig. 2A). All three MAbs bound recombinant gp140, which has the extracellular portion of gp41. Serum from monkey 323-92 was used as a positive control. To fine map the epitopes of 4G8, 6G8, and 7D6, they were tested by ELISA against 15-aa overlapping peptides, thus spanning the glycosylation region of SIVmac239 gp41 in either the mutant or the wild-type version (sequences are detailed in Fig. 1A). Antibody 4G8 showed strong binding to peptide m6687 and moderate binding to peptide m6688 (Fig. 2B; black bars), but it did not significantly react with any of the wild-type peptides. Antibody 6G8 bound strongly to both mutant peptides m6687 and m6688 and robustly cross-reacted with wild-type 6687 (Fig. 2B; gray bars). Antibody 7D6 bound poorly to m6687 and slightly over the background to m6688, but like 4G8 it did not detectably bind to wild-type peptides (Fig. 2B; white bars). Serum from 323-92 was used in parallel as comparison (Fig. 2B; cross-hatched bars). The reactivity of all three monoclonal antibodies to wild-type SIVmac239 gp140 may seem surprising given the relative inability of two of these to react appreciably with peptides corresponding to the wild-type gp41 sequence. However, it should be noted that 50 ng of gp140 was used for each ELISA well and the data in Fig. 2A do not measure the relative efficiency of binding to the gp140. We show in the next section (Fig. 3) by Western blotting that the efficiency of binding is much weaker to wild-type sequences than to the mutated gp41 sequences.
Fig 2.
Reactivity and epitope mapping of rhesus monoclonal antibodies 4G8, 6G8, and 7D6. Three MAbs were isolated by B-cell immortalization as described in Materials and Methods from rhesus macaque 323-92 infected with SIVmac239-gp41/g23 (lacking g2 and g3). Screening was based on ELISA reactivity to peptide m6687-9, which corresponds to mutated glycan-deficient sequences underlying N-linked carbohydrate sites 2 and 3 of SIVmac239 gp41 (Fig. 1A). ELISA reactivity was measured as absorbance at 450 nm. (A) All three antibodies were tested by ELISA for reactivity with peptide m6687-9 and recombinant gp120 and gp140 protein. Serum from animal 323-92, from which the antibody-producing B-cell lines originated, was used as a positive control (bottom). (B) In order to further characterize the binding of 4G8, 6G8, and 7D6, their reactivity against overlapping peptides spanning either the mutant (left half, peptides m6681 to m6690) or wild-type (right half, peptides 6681 to 6690) glycosylation region of gp41 was assayed. Representative results of three independent experiments are shown.
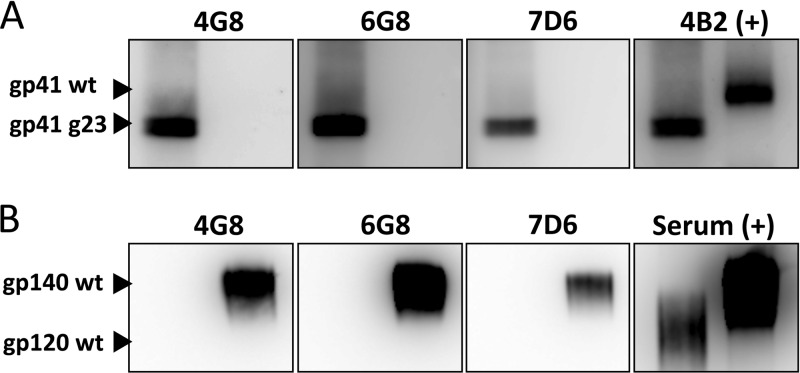
Fig 3.
Antibodies 4G8, 6G8, and 7D6 efficiently react with carbohydrate-deficient mutant but not wild-type gp41 as shown on a Western blot. The reactivity of 4G8, 6G8, and 7D6 against pelleted whole-virus and recombinant proteins was tested by Western blotting. (A) SIVmac239-gp41/g23 (indicated as g23) and SIVmac239 (wt) virions were obtained by transient transfection of HEK293T cells and then pelleted and normalized for p27 content. Fifty nanograms of p27 was loaded per lane under reducing conditions. Antibody 4B2, a monoclonal antibody that binds to the highly immunogenic region of the cytoplasmic domain of gp41 (23), was used as a positive control. Notice the lower molecular weight in the mutant due to the absence of two glycosylations. (B) Binding of 4G8, 6G8, and 7D6 to commercially available recombinant SIVmac239 gp140 (6×His-tagged protein purified from HEK293 cell culture; 200 ng loaded per lane; aa 20 to 692) and gp120 (6×His-tagged protein purified from HEK293 cell culture; 200 ng loaded per lane; aa 34 to 518) was also tested by Western blotting. A serum pool from rhesus macaques infected with wild-type SIV was used as positive control. Representative results of two independent experiments are shown.
Western blotting with 4G8, 6G8, and 7D6 shows efficient binding of glycosylation-deficient gp41 and inefficient binding of wild-type gp41 obtained from viral particles.
To test whether our monoclonal antibodies are able to react by Western blotting against gp41, infectious viral particles of wild-type SIVmac239 and glycan-deficient SIVmac239-gp41/g23 were pelleted, normalized by amount of p27, and used for Western blot analysis. The g23 double mutant was picked for this assay because it lacks the glycosylation sites located in the vicinity of the sequences recognized by 4G8, 6G8, and 7D6. All three antibodies efficiently bound the glycan-deficient version of gp41, whereas reactivity against the wild-type version of the protein was undetectable (Fig. 3A). Consistent with the ELISA results, 6G8 gave the stronger signal, closely followed by 4G8, while 7D6 showed the weakest signal. An anti-gp41 that we had previously isolated, rhesus monoclonal antibody 4B2, whose epitope is located in the cytoplasmic domain of the protein and therefore not influenced by extracellular glycosylations (23), was used as a positive control to demonstrate equivalent levels of gp41 protein per lane. Indeed, image analysis demonstrated equivalent levels of wild-type and g23 mutant gp41 per ng virion p27 [Fig. 3A, image labeled 4B2 (+)]. The Western blots were run in parallel, and equal exposure times were chosen to allow direct comparison. Although the low amounts of gp41 protein present in the virion preparations did not result in detection of wild-type gp41 by the monoclonal antibodies, reactivity by Western blot was detected when much larger amounts of recombinant, commercially-available, wild-type gp140 protein were used (Fig. 3B), consistent with the ELISA data in Fig. 2A. Again, 6G8 showed the strongest reactivity, followed by 4G8 and then 7D6. A pool of sera from SIV-infected animals was used as a positive control.
Sequencing of 4G8, 6G8, and 7D6 shows similar variable regions but different CDR-H3s.
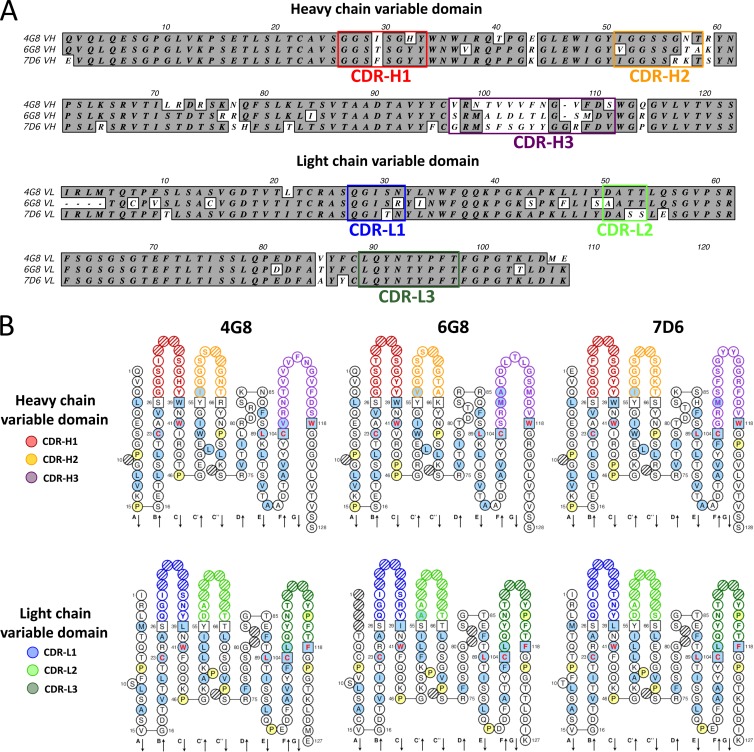
To further characterize 4G8, 6G8, and 7D6, nucleotide sequences were obtained for each monoclonal antibody by reverse transcription (RT)-PCR and cDNA amplification. The sequencing confirmed the monoclonality of the cell lines, as only one sequence was found for each of the antibodies (Fig. 4A). As the information on rhesus macaques is incomplete, we analyzed the variable-domain sequences of the heavy and light chains based on the human sequences and nomenclature (13), which are highly homologous to those of the rhesus macaque (9, 27, 30). Analysis with IMGT/V-QUEST (3, 10) confirmed that all three were derived from productively rearranged sequences with no stop codons and proper in-frame junction. The software was also used to identify the CDRs for heavy chain (CDR-H1, CDR-H2, and CDR-H3) and kappa light chain (CDR-L1, CDR-L2, and CDR-L3) together with the respective framework regions. Alignment of the three amino acid sequences for heavy and light chains revealed quite different CDR-H3s among the three monoclonal antibodies (Fig. 4A). Conversely, the CDR-H1/H2 and CDR-L1/L2 regions were very similar to one another and the CDR-L3 regions were identical. Sparse amino acid differences were characteristically revealed along the framework regions for each antibody (Fig. 4A). We also generated a two-dimensional structure representation of the variable domains of 4G8, 6G8, and 7D6 using the IMGT Collier de Perles (Fig. 4B) (10, 25). This representation clearly identifies the CDRs with a color code (Fig. 4B). The five characteristically conserved positions in the variable domain were confirmed: Cys 23 (first Cys), Trp 41, hydrophobic 89 (in this case Leu for both heavy and light chain), Cys 104 (second Cys), and the J-Trp 118 or J-Phe 118 (for heavy and light chain, respectively) at the end of the junction. For further comparison, the CDR sequences were analyzed separately (Table 1). The lengths (in amino acids) for the heavy chain CDRs (H1, H2, and H3) were very similar in all of them (4G8, 8, 8, and 14 aa; 6G8, 8, 7, and 14 aa; and 7D6, 8, 8, and 15 aa) and equal in the light chain CDRs (L1, L2, and L3 had common lengths of 6, 3, and 9 aa, respectively). Additional analysis of the variable domains with IMGT/V-QUEST revealed that all three CDR-H2s had a three-nucleotide insertion (1 aa) corresponding to the first Gly in both 4G8 and 6G8 and to a Lys in 7D6 (underlined in Table 1). By comparison with published human sequences, we used the IMGT/V-QUEST to identify the equivalent closest germ line gene from which the antibodies likely originated. As shown in Table 1, the variable segment (V) selected for the heavy chain during the VDJ recombination corresponds to the same gene and allele in all three monoclonal antibodies: IGHV4-59*01. Furthermore, 4G8 and 6G8 also share the same diversity (D) and joining (J) segments, IGHD2-08*02 (in reading frame 3) and IGHJ4*02, respectively, while 7D6 derives from IGHD3-22*01 (reading frame 2) and IGHJ5*02, correspondingly. The closest germ line genes for the V and J segments of the kappa light chains were the same in all three monoclonal antibodies: IGKV1-17*01 and IGKJ3*01, respectively (note that light chains do not have D segments).
Fig 4.
Sequence analysis of 4G8, 6G8, and 7D6 reveals starkly different CDR-H3 regions and similar light chains. The variable domains of all three MAbs were sequenced. (A) Alignment of amino acid sequences. Identical residues are boxed in gray, and complementarity-determining regions (CDRs) are indicated with colored frames. (B) Two-dimensional representation of the variable domains or IMGT Collier de Perles (10, 25). Numbering according to IMGT unique numbering for V-domain residues. CDRs for heavy chain (H1, H2, and H3) and light chain (L1, L2, and L3) are colored as indicated on the left; hatched-circle positions indicate gaps according to the IMGT numbering. Light blue indicates positions that are either hydrophobic amino acids (hydropathy index with positive value) or a tryptophan (W) as in 50% or more of analyzed variable domains, according to Pommié et al. (22). Anchor positions are shown as squares and prolines in yellow. Amino acids with red and bold letters indicate the five conserved positions of a variable domain. Arrows indicate the β-strands and their direction.
Table 1.
Analysis of complementarity-determining regionsa
| Antibody | CDRs |
Closest germ line gene and allele |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CDR1 | n | CDR2 | n | CDR3 | n | Vb | D (Rf)b | Jb | ||
| HVb | 4G8 | GGSISGHY | 8 | IGGSSGNT | 8 | VRNTVVVFNG-VFDS | 14 | IGHV4-59*01 | IGHD2-08*02 (3) | IGHJ4*02 |
| 6G8 | GGSTSGYY | 8 | -VGSSGTA | 7 | SRMALDLTLG-SMDV | 14 | IGHV4-59*01 | IGHD2-08*02 (3) | IGHJ4*02 | |
| 7D6 | GGSFSGYY | 8 | IGGSSRKT | 8 | GRMSFSGYYGGRFDV | 15 | IGHV4-59*01 | IGHD3-22*01 (2) | IGHJ5*02 | |
| KVb | 4G8 | QGISNY | 6 | DAT | 3 | LQYNTYPFT | 9 | IGKV1-17*01 | NAb | IGKJ3*01 |
| 6G8 | QGISRY | 6 | AAT | 3 | LQYNTYPFT | 9 | IGKV1-17*01 | NAb | IGKJ3*01 | |
| 7D6 | QGITNY | 6 | DAS | 3 | LQYNTYPFT | 9 | IGKV1-17*01 | NAb | IGKJ3*01 | |
Complementarity-determining regions (CDRs) were analyzed with IMGT/V-QUEST (International ImMunoGeneTics information system/V-QUEry and STandardization, a software for analysis of rearranged immunoglobulin nucleotide sequences (3, 10). Insertions are underlined, and hyphens have been added to CDRs for proper alignment in the table.
Abbreviations: V, variable region; D (Rf), diversity region (reading frame); J, joining region; HV, heavy chain variable region; KV, kappa light chain variable region; NA, not applicable.
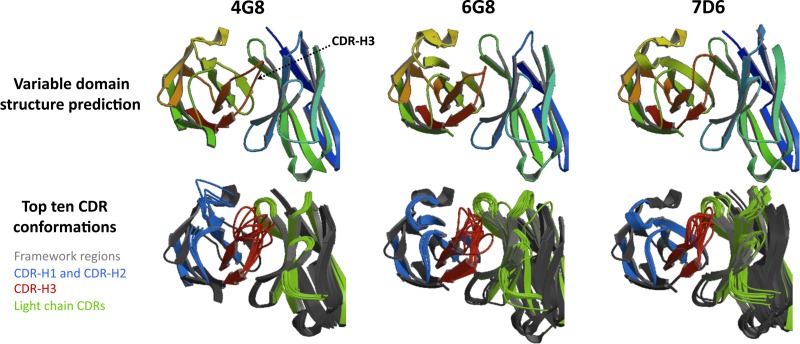
Structure prediction of the variable domains in 4G8, 6G8, and 7D6 reveals a common configuration with disparity in the CDR-H3 disposition.
In order to obtain a prediction for the three-dimensional structure of the variable domains of 4G8, 6G8, and 7D6, the amino acid sequences were submitted to the antibody-modeling server RosettaAntibody (29). This tool is built on the Rosetta structure prediction suite comprising knowledge-based techniques for template selection, grafting for non-H3 CDR loops, de novo loop modeling for creating CDR-H3, and docking to optimize the relative orientation of the light and heavy chains. A full-protocol approach was chosen to achieve refinement of variable heavy and light chain structures together with efficient CDR3 modeling. The predicted model for the variable domain is shown for each antibody (Fig. 5, top). The predicted structures were comparable for all three antibodies, consistent with the similarities found in the primary sequences and the fact that they virtually share a light chain. However, a closer look at the CDR-H3s revealed three fairly different loop structures. CDR-H3 of 4G8 begins angled toward the light chain and then turns to the opposite direction. CDR-H3 of 6G8, despite having a very similar structure, features a loop that does not get so close to the light chain, while the angle of torsion appears more pronounced. CDR-H3 of 7D6 shows a more divergent structure than the other two, with not one but two more pronounced turns that confer a thin, inverted “U” shape. To further evaluate CDR-H3 differences, the 10 most likely CDR conformations were also rendered (Fig. 5, bottom). In these models, CDR-H3s of 4G8 and 6G8 had a wider range of low-energy positions (and therefore more likely conformations) than CDR-H3 of 7D6, which in general maintained the same shape in every combination, just in slightly different angles.
Fig 5.
High-resolution structure prediction model shows similar conformation of the variable domains with variations in CDR-H3. For prediction of the three-dimensional structure of the variable domains, 4G8, 6G8, and 7D6 sequences were analyzed with RosettaAntibody, an antibody-modeling server (29). Full protocol with VL-VH refinement was performed, including detection of templates, grafting of CDRs, modeling of CDR-H3 with simultaneous minimization of CDR backbone conformations, and relative orientation of the heavy chain (left part of molecules) and light chain (right part). The predicted model is shown for each antibody (top), as well as the top 10 most likely CDR conformations (bottom). CDR-H3 is labeled in the first model as a reference. Color codes are indicated in the bottom left.
Capacity of 4G8, 6G8, and 7D6 to neutralize glycan-deficient mutants of SIVmac239 gp41.
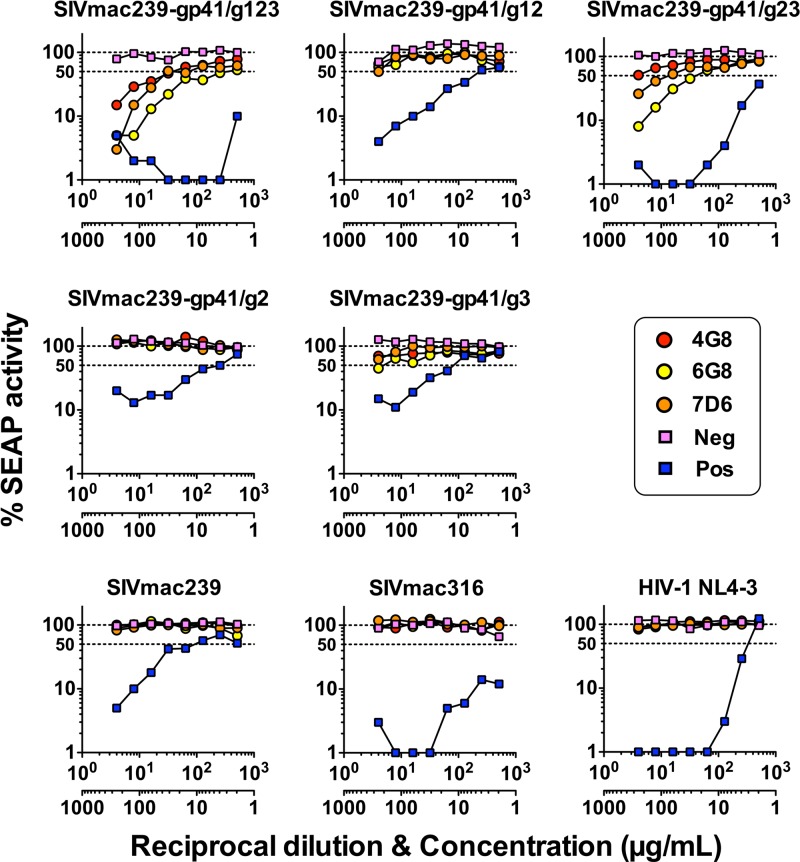
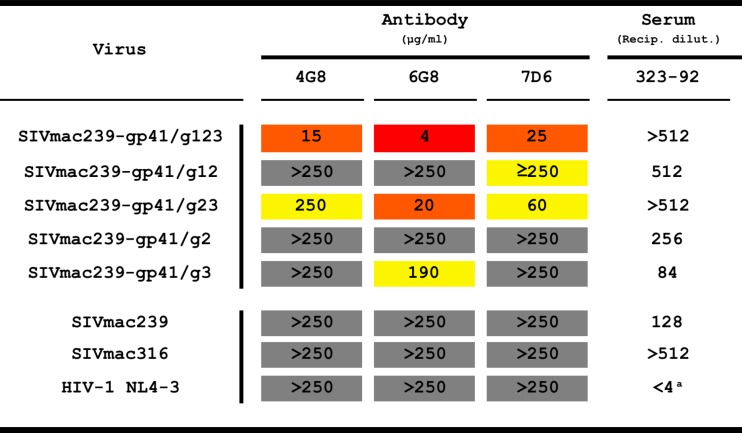
We next tested the ability of 4G8, 6G8, and 7D6 to neutralize wild-type SIVmac239 and its corresponding carbohydrate-deficient mutants. All three monoclonal antibodies were capable of neutralizing SIVmac239-gp41/123 and SIVmac239-gp41/g23 to various degrees (Fig. 6). These mutants lack all three or the g2 and g3 N-linked glycosylation sites in gp41. The triple-glycan-deficient virus was the most sensitive to monoclonal antibody-mediated neutralization. The three monoclonal antibodies exhibited weak or no neutralizing activity against the other mutants. No neutralization was detected with wild-type SIVmac239 or with the closely related but highly neutralization-sensitive strain SIVmac316, which differs in its Env sequence only at eight amino acid positions, none in the vicinity of the glycosylation region. As expected, HIV-1 strain NL4-3, with a very different sequence in the region of the targeted gp41 epitope, was also not neutralized by 4G8, 6G8, or 7D6 (Fig. 6). For every virus, a negative control of irrelevant rhesus IgG was run in parallel at the same concentration as the antibodies. Serum from 323-92 was used as a positive control for all SIVs, while neutralizing antibody anti-gp120 b12 was used for HIV-1. The degree of neutralization varied considerably from one antibody to another, 6G8 being the most potent antibody followed by 7D6 and then 4G8. Doses required for 50% neutralization (IC50) were calculated and are shown in Table 2.
Fig 6.
Antibodies 4G8, 6G8, and 7D6 neutralize glycan-deficient mutants of SIVmac239. Ability of 4G8 (red), 6G8 (yellow), and 7D6 (orange) to neutralize five different SIVmac239 glycan-deficient gp41 mutants lacking one, two, or three glycosylation sites and parental SIVmac239, SIVmac316, and HIV-1 NL4-3 as a control. Virus stocks virions were obtained by transient transfection of HEK293T cells, normalized for the amount of p27, and used to infect C8166-45 SIV-SEAP cells with series of 2-fold dilutions of the antibodies. SEAP activity was measured at 3 days after infection. Neutralization curves are shown as the percentage of SEAP activity versus activity from cells infected with virus in the absence of antibodies. To fit the logarithmic scale, percentages of <1 where normalized to 1. Irrelevant rhesus IgG at the same concentration was used as negative control (pink). Serum from glycan-deficient SIV-infected animal 323-92 was used as a positive control for SIV infection and antibody b12 for HIV-1 infections (blue). Antibody concentration or serum dilution folds are indicated in two separate abscissas. The dotted lines indicate 100% (upper line) and 50% (lower line) of maximal viral replication. Neg, negative control; Pos, positive control. Representative results of three independent experiments are shown.
Table 2.
Half-maximal neutralizing concentrations (IC50)
aMonoclonal antibody b12 was used as a positive control for HIV-1 NL4-3 neutralization (IC50 = 3 μg/ml). Color codes for relative neutralization potency: red, high; orange, medium; yellow, low; gray, nondetectable neutralization at maximum dose.
DISCUSSION
All lentiviruses possess envelope proteins that are densely glycosylated. N-linked glycosylation of the surface subunit gp120 of HIV-1 and SIV has been demonstrated to restrict the ability of antibodies to access gp120 and to restrict the ability to neutralize infectivity (4, 11, 17, 24, 32, 34). Much less is known about the role of N-linked glycosylation of the transmembrane subunit gp41. SIVmac239 has three sites for N-linked glycosylation in the extracellular domain of its gp41. Based on the earlier work of Yuste et al. (38), we have now isolated three monoclonal antibodies of rhesus macaque origin, namely, 4G8, 6G8, and 7D6, which recognize sequences corresponding to the carbohydrate attachment sites of SIVmac239 gp41. They all bind the amino acid sequence WQEWERKVDFLEEQITALLEEAQ. 4G8, 6G8, and 7D6 all effectively neutralize gp41 glycan-deficient mutants, thus establishing the ability of antibodies directed to this stretch to neutralize infectivity when carbohydrates are not attached. Not only do the gp41 carbohydrates severely limit the ability of the host to mount antibody responses to the underlying peptide sequences, such antibodies when present are able to neutralize the infectivity of virus lacking carbohydrates at this region. While sites g2 and g3 seem to be the key players in the protective effects with these three MAbs, all three sites apparently contribute since the triple mutant SIVmac239-gp41/g123 was consistently the most sensitive to neutralization by all three antibodies.
Interestingly, all three of these monoclonal antibodies exhibited some level of reactivity to gp140 derived from the wild-type sequence both by ELISA (Fig. 2A) and by Western blotting (Fig. 3B). It needs to be noted, however, that relatively large amounts of purified gp140 were used for these assays in Fig. 2A and 3B. Furthermore, the reactivity of 6G8 should not be too surprising given its reactivity to peptide 6687 corresponding to the wild-type sequence (Fig. 2B). These monoclonal antibodies certainly do react considerably more strongly to mutant gp41 sequences than to wild-type gp41 sequences both by Western blotting (Fig. 3A) and by peptide ELISA (Fig. 2B).
The analyses of the variable-domain sequences of the heavy and light chains were based on the human sequences and nomenclature (13), which are highly homologous to those of the rhesus macaques (9, 27, 30). All three antibodies share highly similar light chains, and their heavy-chain variable segment (V) appears to derive from the same germ line allele. The diversity (D) and joining (J) segments of the heavy chains of at least 4G8 and 6G8 seem to originate from the same germ line allele, and all three antibodies show characteristic amino acid changes along their variable domains. While it is possible that the particular V, D, and J segments selected during maturation of these B cells may be especially effective combinations for targeting the glycosylation region of SIVmac239 gp41, the similitude suggests that all three antibodies are likely to have originated from a common B cell.
Despite these similarities at the genetic level, 4G8, 6G8, and 7D6 differ in several characteristics, in particular in their peptide binding profiles. Importantly, they diverge heavily in the sequence of their CDR-H3, indicating considerable hypermutation. While any dissimilarity in the amino acid sequence might account for the specific properties of each antibody, the differences in CDR-H3 are most likely the key determinant of epitope specificity and affinity (35, 36). Consistent with this notion, it has been demonstrated that both CDR-H1 and CDR-H2 and all CDRs of the light chains can assume only a few canonical conformations, while solely CDR-H3 has a wide range of variations in both length and shape (6). This is in agreement with the structural models of the variable domains shown here, in which CDR-H3 had the most variable conformation. In this regard, the same Gly found in the CDR-H3s of 4G8 and 6G8 could be responsible for the similar turn found in both CDR-H3s, while the four Gly found in 7D6's CDR-H3 could account for the thin, inverted “U” shape (14). All three antibodies had glycine at position 2 of the CDR-H2. This glycine arose by insertion in 4G8 and 6G8 and by somatic mutation in 7D6. Thus, this glycine may also well be important for recognition of the nonglycosylated gp41 sequence.
The results described in the Yuste et al. publication (38) demonstrate the appearance of antibodies to underlying peptides in monkeys infected with gp41-mutant virus but not in monkeys infected with the parental wild-type virus. In the same publication (38), the appearance of antibodies with high-titer neutralizing activity against mutant virus only in animals infected with mutant virus was also unambiguously demonstrated. However, whether the antibodies directed to the underlying peptides were responsible for neutralizing activity was not directly demonstrated. Here we show that antibodies directed to the underlying peptides can and do have neutralizing activity specifically to the gp41 mutant viruses. Our data establish conclusively that the presence of glycans on gp41 effectively shields the virus from antibodies that would otherwise neutralize viral infectivity.
ACKNOWLEDGMENTS
We thank Gordon J. Freeman, Scott K. Dessain, Sharad P. Adekar, and Keith A. Reimann for providing reagents and/or useful advice and Thomas S. Postler for critical comments on the manuscript. We also thank Jacqueline Bixby, Jackie Gillis, Michelle Connole, and Fay Eng Wong for technical assistance. We thank as well the veterinarian staff at the NEPRC.
This work was supported by NIH grant AI25328 to R.C.D. and by base grant RR00168 to the NEPRC from NCRR and ORIP of NIH.
Footnotes
Published ahead of print 19 September 2012
REFERENCES
- 1. Adekar SP, et al. 2008. Hybridoma populations enriched for affinity-matured human IgGs yield high-affinity antibodies specific for botulinum neurotoxins. J. Immunol. Methods 333:156–166 [DOI] [PubMed] [Google Scholar]
- 2. Bixby JG, Laur O, Johnson WE, Desrosiers RC. 2010. Diversity of envelope genes from an uncloned stock of SIVmac251. AIDS Res. Hum. Retroviruses 26:1115–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brochet X, Lefranc MP, Giudicelli V. 2008. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 36:W503–W308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chaillon A, et al. 2011. The V1V2 domain and an N-linked glycosylation site in the V3 loop of the HIV-1 envelope glycoprotein modulate neutralization sensitivity to the human broadly neutralizing antibody 2G12. J. Virol. 85:3642–3648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Reference deleted.
- 6. Chothia C, et al. 1989. Conformations of immunoglobulin hypervariable regions. Nature 342:877–883 [DOI] [PubMed] [Google Scholar]
- 7. Cole KS, Steckbeck JD, Rowles JL, Desrosiers RC, Montelaro RC. 2004. Removal of N-linked glycosylation sites in the V1 region of simian immunodeficiency virus gp120 results in redirection of B-cell responses to V3. J. Virol. 78:1525–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Derby NR, et al. 2007. Isolation and characterization of monoclonal antibodies elicited by trimeric HIV-1 Env gp140 protein immunogens. Virology 366:433–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gibbs RA, et al. 2007. Evolutionary and biomedical insights from the rhesus macaque genome. Science 316:222–234 [DOI] [PubMed] [Google Scholar]
- 10. Giudicelli V, Brochet X, Lefranc MP. 2011. IMGT/V-QUEST: IMGT standardized analysis of the immunoglobulin (IG) and T cell receptor (TR) nucleotide sequences. Cold Spring Harb. Protoc. 2011:695–715 [DOI] [PubMed] [Google Scholar]
- 11. Hatada M, et al. 2010. Human immunodeficiency virus type 1 evasion of a neutralizing anti-V3 antibody involves acquisition of a potential glycosylation site in V2. J. Gen. Virol. 91:1335–1345 [DOI] [PubMed] [Google Scholar]
- 11a. Haynes BF, et al. 2012. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N. Engl. J. Med. 366:1275–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Johnson WE, et al. 2003. Assorted mutations in the envelope gene of simian immunodeficiency virus lead to loss of neutralization resistance against antibodies representing a broad spectrum of specificities. J. Virol. 77:9993–10003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lefranc MP. 2001. Nomenclature of the human immunoglobulin genes. Curr. Protoc. Immunol. Appendix 1:Appendix 1P. doi:10.1002/0471142735.ima01ps40 [DOI] [PubMed] [Google Scholar]
- 14. McCormack WT, Tjoelker LW, Stella G, Postema CE, Thompson CB. 1991. Chicken T-cell receptor beta-chain diversity: an evolutionarily conserved D beta-encoded glycine turn within the hypervariable CDR3 domain. Proc. Natl. Acad. Sci. U. S. A. 88:7699–7703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Means RE, Greenough T, Desrosiers RC. 1997. Neutralization sensitivity of cell culture-passaged simian immunodeficiency virus. J. Virol. 71:7895–7902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mori K, Ringler DJ, Kodama T, Desrosiers RC. 1992. Complex determinants of macrophage tropism in env of simian immunodeficiency virus. J. Virol. 66:2067–2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mori K, et al. 2001. Quintuple deglycosylation mutant of simian immunodeficiency virus SIVmac239 in rhesus macaques: robust primary replication, tightly contained chronic infection, and elicitation of potent immunity against the parental wild-type strain. J. Virol. 75:4023–4028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Morrison HG, Kirchhoff F, Desrosiers RC. 1993. Evidence for the cooperation of gp120 amino acids 322 and 448 in SIVmac entry. Virology 195:167–174 [DOI] [PubMed] [Google Scholar]
- 19. Myers G, Lenroot R. 1992. HIV glycosylation: what does it portend? AIDS Res. Hum. Retroviruses 8:1459–1460 [DOI] [PubMed] [Google Scholar]
- 20. National Research Council, Commission on Life Sciences 1996. Guide for the Care and Use of Laboratory Animals, 7th ed National Academies Press, Washington, DC [Google Scholar]
- 21. Pikora CA. 2004. Glycosylation of the ENV spike of primate immunodeficiency viruses and antibody neutralization. Curr. HIV Res. 2:243–254 [DOI] [PubMed] [Google Scholar]
- 22. Pommié C, Levadoux S, Sabatier R, Lefranc G, Lefranc MP. 2004. IMGT standardized criteria for statistical analysis of immunoglobulin V-REGION amino acid properties. J. Mol. Recognit. 17:17–32 [DOI] [PubMed] [Google Scholar]
- 23. Postler TS, Martinez-Navio JM, Yuste E, Desrosiers RC. 2012. Evidence against extracellular exposure of a highly immunogenic region in the C-terminal domain of the simian immunodeficiency virus gp41 transmembrane protein. J. Virol. 86:1145–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Reitter JN, Means RE, Desrosiers RC. 1998. A role for carbohydrates in immune evasion in AIDS. Nat. Med. 4:679–684 [DOI] [PubMed] [Google Scholar]
- 25. Ruiz M, Lefranc MP. 2002. IMGT gene identification and Colliers de Perles of human immunoglobulins with known 3D structures. Immunogenetics 53:857–883 [DOI] [PubMed] [Google Scholar]
- 26. Schultze JL, et al. 1995. Follicular lymphomas can be induced to present alloantigen efficiently: a conceptual model to improve their tumor immunogenicity. Proc. Natl. Acad. Sci. U. S. A. 92:8200–8204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Scinicariello F, Engleman CN, Jayashankar L, McClure HM, Attanasio R. 2004. Rhesus macaque antibody molecules: sequences and heterogeneity of alpha and gamma constant regions. Immunology 111:66–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sekaly RP. 2008. The failed HIV Merck vaccine study: a step back or a launching point for future vaccine development? J. Exp. Med. 205:7–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sircar A, Kim ET, Gray JJ. 2009. RosettaAntibody: antibody variable region homology modeling server. Nucleic Acids Res. 37:W474–W479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Thullier P, Chahboun S, Pelat T. 2010. A comparison of human and macaque (Macaca mulatta) immunoglobulin germline V regions and its implications for antibody engineering. MAbs 2:528–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Traggiai E, et al. 2004. An efficient method to make human monoclonal antibodies from memory B cells: potent neutralization of SARS coronavirus. Nat. Med. 10:871–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. van Gils MJ, et al. 2011. Longer V1V2 region with increased number of potential N-linked glycosylation sites in the HIV-1 envelope glycoprotein protects against HIV-specific neutralizing antibodies. J. Virol. 85:6986–6995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Walker LM, et al. 2010. A limited number of antibody specificities mediate broad and potent serum neutralization in selected HIV-1 infected individuals. PLoS Pathog. 6:e1001028 doi:10.1371/journal.ppat.1001028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wei X, et al. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307–312 [DOI] [PubMed] [Google Scholar]
- 35. Xu JL, Davis MM. 2000. Diversity in the CDR3 region of V(H) is sufficient for most antibody specificities. Immunity 13:37–45 [DOI] [PubMed] [Google Scholar]
- 36. Yamagami J, et al. 2010. Homologous regions of autoantibody heavy chain complementarity-determining region 3 (H-CDR3) in patients with pemphigus cause pathogenicity. J. Clin. Invest. 120:4111–4117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yusim K, et al. (ed). 2011. HIV molecular immunology. LA-UR-12-10074. Los Alamos National Laboratory, Theoretical Biology and Biophysics, Los Alamos, NM [Google Scholar]
- 38. Yuste E, et al. 2008. Glycosylation of gp41 of simian immunodeficiency virus shields epitopes that can be targets for neutralizing antibodies. J. Virol. 82:12472–12486 [DOI] [PMC free article] [PubMed] [Google Scholar]