Abstract
Adeno-associated virus (AAV) capsid assembly requires expression of the assembly-activating protein (AAP) together with capsid proteins VP1, VP2, and VP3. AAP is encoded by an alternative open reading frame of the cap gene. Sequence analysis and site-directed mutagenesis revealed that AAP contains two hydrophobic domains in the N-terminal part of the molecule that are essential for its assembly-promoting activity. Mutation of these sequences reduced the interaction of AAP with the capsid proteins. Deletions and a point mutation in the capsid protein C terminus also abolished capsid assembly and strongly reduced the interaction with AAP. Interpretation of these observations on a structural basis suggests an interaction of AAP with the VP C terminus, which forms the capsid protein interface at the 2-fold symmetry axis. This interpretation is supported by a decrease in the interaction of monoclonal antibody B1 with VP3 under nondenaturing conditions in the presence of AAP, indicative of steric hindrance of B1 binding to its C-terminal epitope by AAP. In addition, AAP forms high-molecular-weight oligomers and changes the conformation of nonassembled VP molecules as detected by conformation-sensitive monoclonal antibodies A20 and C37. Combined, these observations suggest a possible scaffolding activity of AAP in the AAV capsid assembly reaction.
INTRODUCTION
Adeno-associated virus (AAV) is a nonenveloped single-stranded DNA virus of the Parvoviridae family (15). To date, 13 distinct human or nonhuman primate AAV serotypes have been described and numerous recombinant species have been isolated (10). The AAV assembly pathway proposed by Myers and Carter suggests the rapid formation of empty capsids into which the single-stranded genome is inserted in a slow reaction (16). While the process of genome replication has been elucidated in great detail (15, 22), molecular events underlying capsid formation and genome encapsidation are less well understood (12).
Capsid assembly occurs in the nuclei of infected cells, where capsids are first detectable in the nucleoli but are spread throughout the nucleus at later stages of infection (23). Expression of the cap gene is sufficient for capsid formation. Besides the three capsid proteins, VP1, VP2, and VP3, known to be expressed from open reading frame 1 (ORF1), the cap gene encodes an assembly factor, the assembly-activating protein (AAP), from a second ORF, ORF2 (21). AAP is essential for capsid assembly. It targets newly synthesized capsid proteins to the nucleolus and promotes capsid formation in a still unknown way. AAPs of some, but not all, AAV serotypes can cross-complement each other in the assembly reaction (20). AAP is a rather unstable protein but becomes stabilized upon the coexpression of capsid protein VP3. However, this stabilizing effect depends very much on the serotype of the coexpressed capsid protein, indicating specific AAP-VP protein interactions (20). AAP amino acid sequence alignment of serotypes 1 to 13 shows a high degree of homology. Only AAPs from serotypes 4, 5, 11, and 12 show noticeable amino acid sequence differences (20), suggesting that these serotypes belong to a different assembly group, which is also evident in the evolutionary relationship of the corresponding capsid proteins (20).
In order to unravel the role of AAP in the assembly process, we determined the requirement of conserved AAP amino acid sequence motifs for AAV2 capsid assembly and propose an interaction domain between AAP and the VP proteins crucial for capsid assembly. Surprisingly, AAP shows unprecedented molecular oligomerization behavior and is able to induce a conformational change in low-molecular-weight VP oligomers. Combined, the described characteristics contribute to our understanding of the role of AAP in AAV capsid assembly.
MATERIALS AND METHODS
AAP structure and sequence analysis.
The nucleotide sequences of 13 AAV serotypes were retrieved from GenBank. AAP sequences were defined as beginning at a conserved translation initiation codon, CTG, in ORF2 of the cap gene and ending at the subsequent stop codon. Protein sequences were aligned using the MUSCLE multiple-alignment tool (7). Secondary structural elements were predicted using the JPred tool (5).
Plasmids and cloning.
Plasmids pBS (pBluescript; Stratagene, Amsterdam, Netherlands), pVP2N-gfp (here referred to as pAAP-L1-T177), and pCMV-VP3/2809 (here referred to as pCMV-VP3) have been described previously (21). Plasmid pCMV-VP3 was used for the expression of the VP3 protein of AAV2 under the control of the cytomegalovirus (CMV) promoter. In plasmid pAAP-L1-T177, AAP was expressed under the control of the CMV promoter using the nonconventional translation initiation codon CTG. N-terminal AAP deletions starting with an ATG start codon instead of CTG were generated by PCR amplification and subsequent cloning of the respective amplicons into the pAAP-L1-T177 backbone plasmid via EcoNI/BsiWI restriction. C-terminal deletions of AAP were generated by PCR amplification introducing a stop codon after the indicated amino acid position in the AAP sequence and subsequent cloning into the pAAP-L1-T177 backbone plasmid via EcoNI/BsiWI. All resulting expression constructs are referred to here as pAAP. Point mutant AAPs were generated by site-directed mutagenesis of the previously described backbone plasmid pORF2/ATG-AU1 (21), here referred to as pAAP-AU1, using the QuikChange site-directed mutagenesis kit (Stratagene). PCR amplification and mutagenesis of the AAP sequence was followed by cloning of the respective fragments into pAAP-AU1 via HindIII/XhoI restriction.
C-terminal deletion mutant constructs of VP3 were generated by the introduction of stop codons into pCMV-VP3 by site-directed mutagenesis. XcmI/XhoI fragments containing the respective mutations were subsequently cloned into the backbone plasmid pCMV-VP3, replacing the nonmutated sequence.
Cell culture.
293T cells were maintained in Dulbecco's modified Eagle medium supplemented with 10% heat-inactivated calf serum, 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-glutamine. Cells were constantly kept in a humidified atmosphere at 37°C and 5% CO2.
Transfection and analysis of protein expression.
293T cells were seeded at a density of 5 × 105/6-cm dish or 1 × 106/10-cm dish 1 day prior to transfection by calcium phosphate precipitation (17). At 48 h posttransfection, cells were harvested and washed twice in phosphate-buffered saline (PBS; 18.4 mM Na2HPO4, 10.9 mM KH2PO4, 125 mM NaCl) before identical portions of harvested cells were processed for SDS-PAGE. Expression of VP3 and AU1-tagged AAP was analyzed by Western blot assay using monoclonal antibody (MAb) B1 (24) or anti-AU1 (Covance, Emeryville, CA), respectively. C-terminal deletion mutant constructs of VP3 were detected by the polyclonal AAV2 antiserum VP#51. Polyclonal AAP antiserum GKD-1 (21) was used for detection of AAP expressed without the AU1 tag.
For Western blot analysis of sucrose density gradient fractions, 15 μl (AAP) or 7.5 μl (VP3) of each fraction was processed for SDS-PAGE and analyzed by using the respective anti-AU1 and B1 MAbs for detection.
Quantification of AAV2 capsids.
Titers of assembled AAV2 capsid particles (numbers of capsids per milliliter) were determined as previously described (11) using an A20 antibody-based AAV2 titration enzyme-linked immunosorbent assay (ELISA; Progen). For this purpose, 293T cells were transfected as described above, harvested at 48 h posttransfection in the medium, and lysed by three freeze-thaw cycles (−80°C and 37°C). Cell debris was removed by centrifugation at 10,000 × g for 5 min, before serial dilutions of the supernatant were analyzed according to the manufacturer's protocol.
Sucrose density gradient analysis.
The freeze-thaw supernatant from three 10-cm dishes of transfected 293T cells, lysed in 600 μl TNEM (150 mM NaCl, 50 mM Tris-HCl [pH 8.0], 10 mM EDTA, and 5 mM MgCl2 supplemented with Complete Mini EDTA-free protease inhibitor cocktail [Roche]), was loaded onto a linear 10% to 30% sucrose gradient (sucrose in TNEM and protease inhibitors) in Beckman centrifuge tubes (14 by 89 mm). After centrifugation at 160,000 × g for 4 h at 10°C (SW41 rotor; Beckman), fractions of 500 μl were collected and further analyzed by Western blot assay, dot blot immunoassay, or electron microscopy.
Dot blot immunoassays.
Fractions obtained after sucrose density gradient centrifugation were analyzed under native conditions by transferring 30 μl of each fraction onto a nitrocellulose membrane (Whatman), followed by immunodetection as described before (13), by using MAbs A20, B1, C24, and C37 to detect assembly products.
Immunoprecipitation.
For immunoprecipitation, A20, B1, or anti-AU1 MAb or nonspecific control antibody 4A7 was coupled to protein A Sepharose CL-4B (GE Healthcare) at 4°C overnight. After three washes with NET-N buffer (100 mM NaCl, 1 mM EDTA, 20 mM Tris-HCl [pH 7.5], and 0.5% NP-40 supplemented with Complete Mini EDTA-free protease inhibitor cocktail), sucrose density gradient fractions or freeze-thaw cell lysates were incubated for 6 h at 4°C with the respective Sepharose-coupled antibodies. Bound proteins were recovered after three PBS washes by centrifugation at 6,000 × g for 3 min and processed for SDS-PAGE analysis.
Electron microscopy.
For electron microscopic analysis, 10 μl of each sucrose density gradient fraction was negatively stained with 2% aqueous uranyl acetate on glow-discharged 300-mesh copper grids. Electron micrographs were taken with a Zeiss EM10 electron microscope at 80 kV.
RESULTS
Analysis of conserved AAP domains.
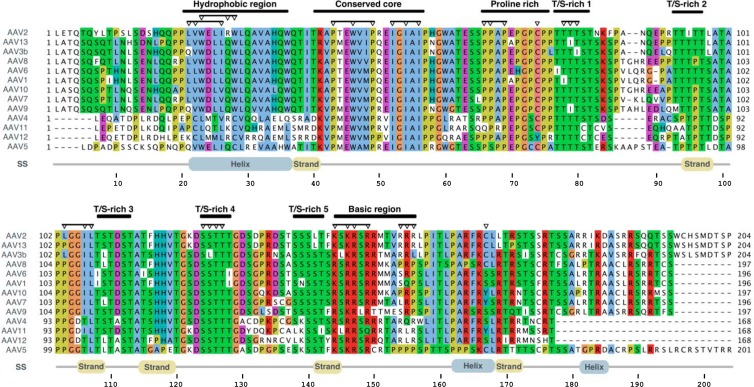
In order to determine the domains of AAP involved in the AAV2 capsid assembly process, we aligned the amino acid sequences of all known AAPs and color coded conserved amino acids according to the physicochemical properties of their side chains (Fig. 1). With this approach, a number of regions appeared to be under strong functional constraint. The N-terminal part of AAP contains two large hydrophobic regions. The first area can be divided into four short hydrophobic motifs interrupted by polar amino acids and spans a region of approximately 15 amino acids (aa) for which an α-helical secondary structure is predicted. This sequence motif is only partially conserved in serotypes 4, 11, 12, and 5. The second hydrophobic area consists of two conserved stretches, each of three hydrophobic amino acids, which are flanked by highly conserved prolines, a glycine, and, at the N-terminal site, a lysine or arginine. We named this region the “conserved core” of AAP.
Fig 1.
Sequence alignment of the AAPs from AAV serotypes 1 to 13. An annotated multiple-sequence alignment of the AAP sequences of 13 AAV serotypes is shown. Sequences are visualized according to the ClustalX coloring scheme. Colored residues are evolutionarily conserved, and the coloring reflects the physicochemical properties of the residues. Regions of interest are annotated above the alignment. Inverted triangles denote the residues experimentally mutated in this study. Predicted secondary structural (SS) elements (strand, helix) for the sequence and amino acid numbering for the AAV serotype 2 AAP are displayed below the alignments.
Furthermore, a proline-rich sequence is noticeable between amino acid positions 66 and 76 of the AAP sequence of AAV2. This motif is followed by five S/T-rich sequence clusters which are 8 to 10 aa apart from each other and conserved among all AAV serotypes. The C terminus of AAP has a more basic character, with a group of lysines and arginines at positions 144 to 150. Secondary-structure predictions indicate several β strands in the center of the molecule, as well as two short helices in the C terminus.
To characterize the function of conserved AAP domains, we introduced N-terminal and C-terminal deletions or site-directed conversions of amino acids (Fig. 1, arrowheads). In a first set of experiments, the capsid assembly activity of mutant AAPs was analyzed by coexpression of the respective AAPs with capsid protein VP3, which requires AAP for capsid formation (21).
Capsid assembly promoted by N-terminal and C-terminal deletion mutant constructs of AAP.
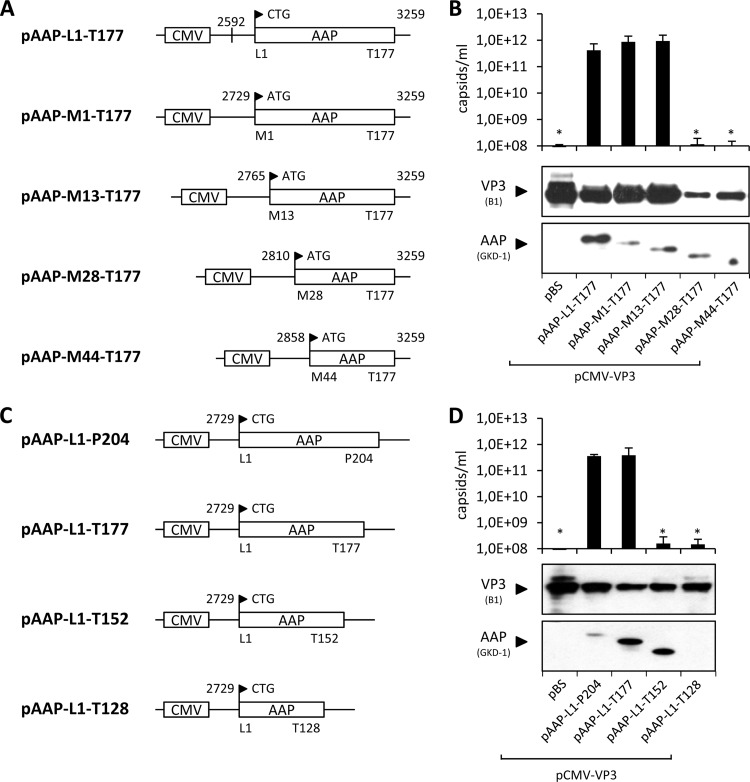
A series of N-terminal-deletion-containing mutant AAPs was generated by PCR amplification and subsequent cloning of the shortened fragments into the pAAP-L1-T177 backbone plasmid (21), resulting in the expression constructs shown in Fig. 2A. The respective amino acids at the N-terminal start of each AAP mutant were converted into methionine. The assembly activity of the mutant AAPs generated was subsequently compared to that of AAP expressed from a construct with the authentic AAP 3′ upstream sequences harboring a CTG translation initiation codon (construct pAAP-L1-T177 [21]), as well as AAP starting with ATG instead of CTG (pAAP-M1-T177). All constructs comprised a C-terminal deletion of 27 amino acids that, however, has no influence on the assembly activity of AAP, as previously shown (21). Western blot analysis revealed remarkable variations in the levels of VP3 expression from the unmodified pCMV-VP3 plasmid, depending on the coexpressed AAP variant (Fig. 2B). Detection of VP3 by antibody B1 was 3- to 5-fold lower when it was coexpressed with AAP M28-T177 and AAP M44-T177 than when it was coexpressed with the other constructs. This was also the case for the steady-state level of mutant AAPs M28-T177 and M44-T177, which had to be loaded in four times larger amounts in order to be detectable. These observations suggest that interactions—or lack of interactions—of VP3 with deletion mutant AAPs make them more susceptible to degradation, most likely by the proteasome, as observed in a previous study (20). VP3 capsid assembly in the presence of AAP with the authentic 5′ untranslated sequences and the CTG start codon was comparable to assembly in the presence of AAP M1-T177 and AAP M13-T177, within the error range of the capsid ELISA (Fig. 2B). Assembly activities of AAP M28-T177 and AAP M44-T177 led to capsid levels in or below the range of detection by the ELISA of 108 capsids/ml, indicating the presence of critical amino acids between positions 13 and 44, which comprises the first hydrophobic cluster of AAP, for the stability and assembly activity of AAP.
Fig 2.
Assembly-promoting activities of deletion mutant AAPs. N-terminal (A) and C-terminal (C) deletion mutant constructs of AAP were generated by site-directed mutagenesis. pAAP-L1-T177 was used as a control for the expression of AAP comprising the authentic, nonconventional translation initiation codon CTG. All N-terminal deletion constructs started with an ATG codon instead. The respective N- and C-terminal amino acids of each AAP construct are indicated together with their positions, as well as the respective nucleotide positions. (B and D) Capsid assembly was analyzed by cotransfection of 293T cells with pCMV-VP3 and the respective AAP expression plasmids and analyzed by A20 antibody-based capsid ELISA at 48 h posttransfection. Transfection of pCMV-VP3 together with an empty vector (pBS) served as a negative control. Protein expression was analyzed by Western blot assay using MAb B1 (detection of VP3) and the polyclonal anti-AAP serum GKD-1. Bars represent the average capsid titers of at least three independent experiments. Asterisks indicate capsid titers significantly lower than that obtained with pAAP-L1-T177 (P < 0.01).
In a similar approach, we analyzed C-terminal deletion mutant AAPs that were generated by the introduction of stop codons at the indicated positions (Fig. 2C). VP3 protein levels in the presence of AAP were slightly lower than those in the presence of the pBS control; however, strong variations in the steady-state levels were not observed in combination with any of the C-terminal deletion-containing mutant AAPs (Fig. 2D). The C-terminal deletion mutant AAPs were detectable with the GKD-1 antipeptide antiserum, with the exception of the AAP L1-T128 mutant construct, in which the peptide epitope recognized by the antiserum is deleted. The capsid assembly activities of full-length AAP and AAP L1-T177 were indistinguishable (Fig. 2D). However, the assembly activity of the AAP L1-T152 and AAP L1-T128 mutant constructs dropped below the ELISA detection level, indicating the presence of assembly-relevant amino acids between residues T128 and T177.
Capsid assembly by amino acid exchange mutant AAPs.
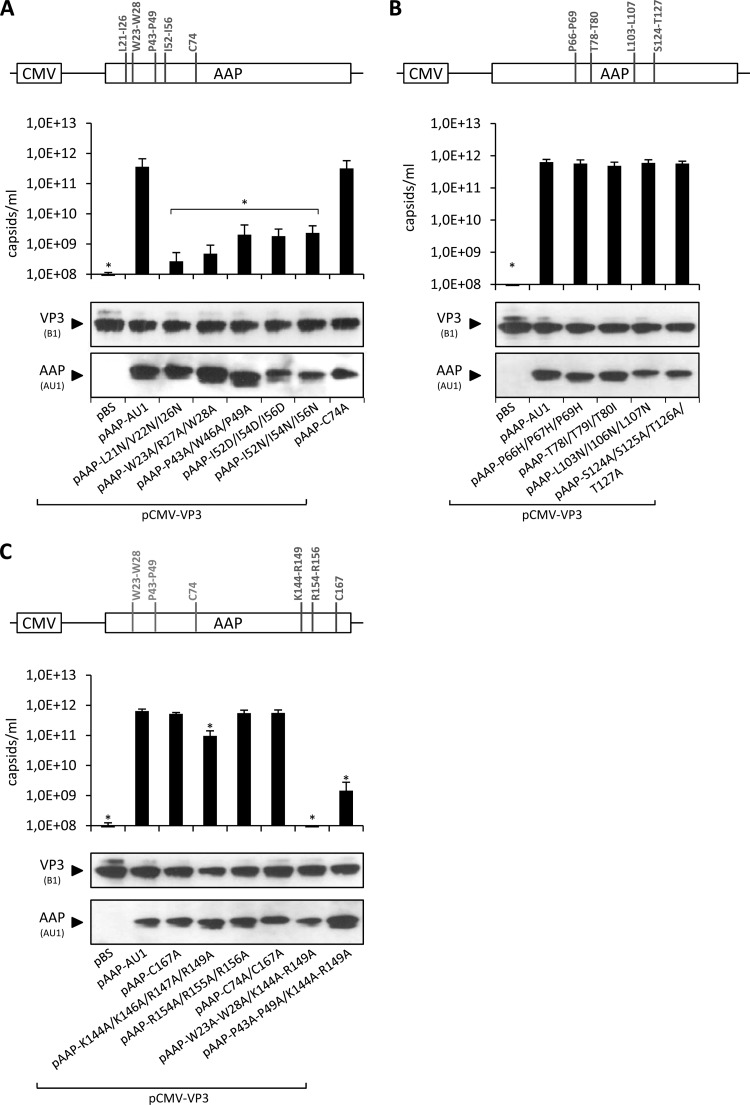
In order to characterize assembly-relevant sequence motifs of AAP in more detail, we introduced amino acid exchanges at selected sequence motifs in the N-terminal, C-terminal, and central parts of AAP and subsequently tested whether these motifs are crucial to the retention of assembly-activating activity (Fig. 3). Regarding the N-terminal part of AAP, mutations of hydrophobic amino acids in the hydrophobic region (L21 to I26, W23 to W28), two conserved prolines together with a tryptophan (P43 to P49), three conserved isoleucines (I52, I54, and I56) of the conserved core, and a cysteine at position 74 were chosen as schematically depicted in Fig. 3A. The cysteine is conserved among all serotypes except AAV12, which, however, has two cysteines at positions 85 and 87 instead. The levels of mutated AAPs and coexpressed VP3 were not altered (Fig. 3A); however, the assembly-promoting activity of the majority of the mutant AAPs analyzed was strongly reduced (Fig. 3A). Mutation of the hydrophobic clusters between L21 and I26 and between W23 and W28, respectively, had the strongest effect, with a more-than-1,000-fold reduction in assembly-promoting activity. Exchange of prolines 43 to 49 and the hydrophobic isoleucines at positions 52 to 56 of the conserved core reduced the assembly activity of AAP by >100-fold. Strikingly, exchange of cysteine 74 had no influence on AAV capsid assembly. Combined, these results provide evidence for a critical role of prominent hydrophobic regions in the N terminus of AAP for its assembly-activating function.
Fig 3.
Capsid assembly by amino acid exchange mutant AAPs. Amino acids at selected sequence motifs in the N-terminal (A), central (B) and C-terminal (C) parts of AAP were exchanged as schematically depicted. Capsid assembly was analyzed after cotransfection of 293T cells with the respective mutant AAPs or pAAP-AU1 together with capsid protein VP3 (pCMV-VP3) via A20 antibody-based capsid ELISA. Coexpression of VP3 with the empty pBS vector served as a negative control. Protein expression was analyzed by Western blot assay using MAb B1 (detection of VP3) and the AU1 tag MAb. Bars represent the average capsid titers of at least three independent experiments. Asterisks indicate capsid titers significantly lower than that obtained with pAAP-AU1 (P < 0.01).
In the central part of AAP, a proline-rich region between positions 66 and 69 is conserved among the known AAV serotypes and five S/T-rich clusters are evident (Fig. 1). We converted the respective amino acids of the proline-rich region to histidine and two S/T-rich regions at T78 to T80 and S124 to T127 to isoleucine and alanine, respectively (Fig. 3B), and analyzed the effects on assembly activity. Unexpectedly, none of the mutant constructs was impaired in the ability to support VP3 capsid assembly, although the respective motifs were highly conserved. In addition, a group of hydrophobic amino acids in the center of AAP—although not conserved among all serotypes—were converted to the polar amino acid asparagine (L103 to L107), which had also no effect on capsid assembly. Possibly, the five S/T motifs have redundant functions. They seem not to be a consequence of the superimposed VP protein sequence.
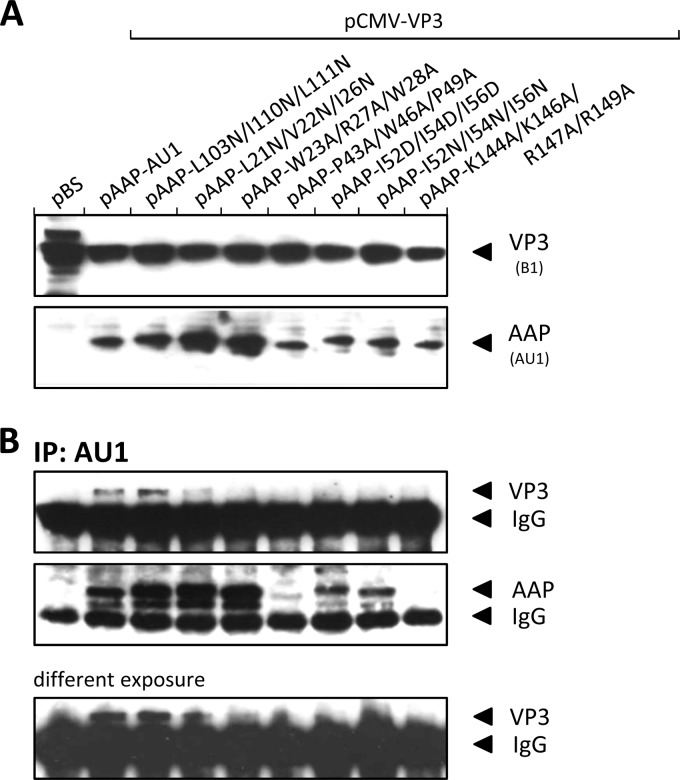
For characterization of the C-terminal domain, we chose two basic sequence motifs that might potentially be involved in the nuclear and nucleolar localization of AAP, as well as the role of another cysteine at position 167 (Fig. 3C). Additionally, several double mutant constructs carrying exchanges in the N-terminal sequence motifs and the C-terminal part of AAP were included in this set of experiments (Fig. 3C; light gray labeled mutations). To our surprise, exchange of K144, K146, R147, and R149 for alanines reduced the assembly activity only moderately, by 5- to 10-fold. Respective exchanges of R154, R155, and R156—which are not conserved—produced no measurable defect in capsid assembly. Also, conversion of cysteine 167 to alanine did not influence the assembly competence of AAP as well as a double mutant construct encompassing both cysteines (C74A and C167A). Combination of W23A, R27A, and W28A and K147A, K146A, and R149A in a double mutant construct had an additive effect, while mutations of P43A, W46A, and P49A together with K144A, K146A, and R149A produced no additive assembly deficiency. Exchange of the basic sequence motifs also did not prevent nucleolar localization of VP3 and AAP (data not shown). Taken together, mutation of several sequence motifs in the C-terminal part of the AAP molecule had only a minor or no effect on the assembly activity of the protein.
VP and AAP oligomerization and AAP-VP interactions.
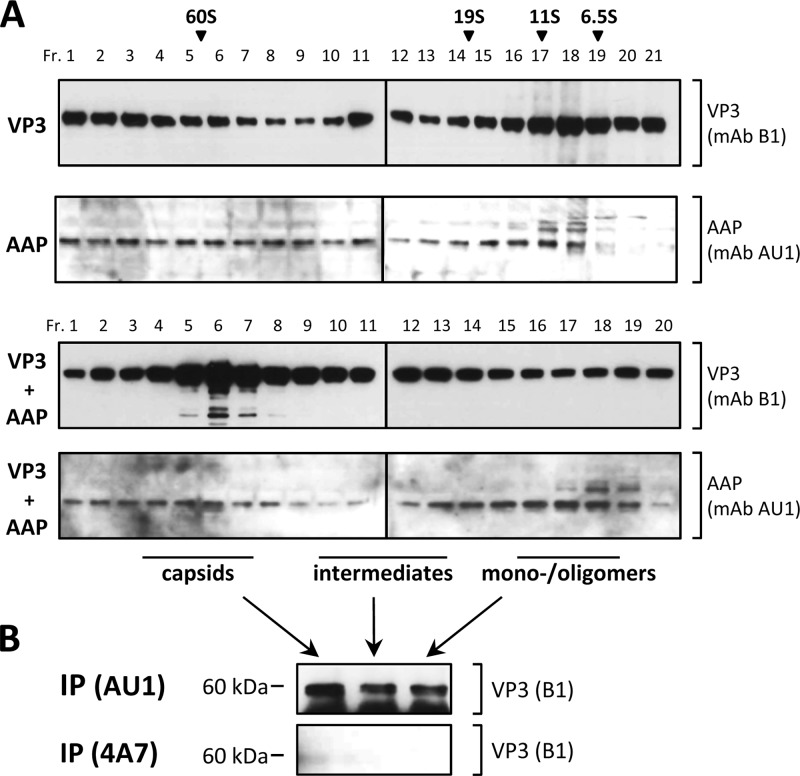
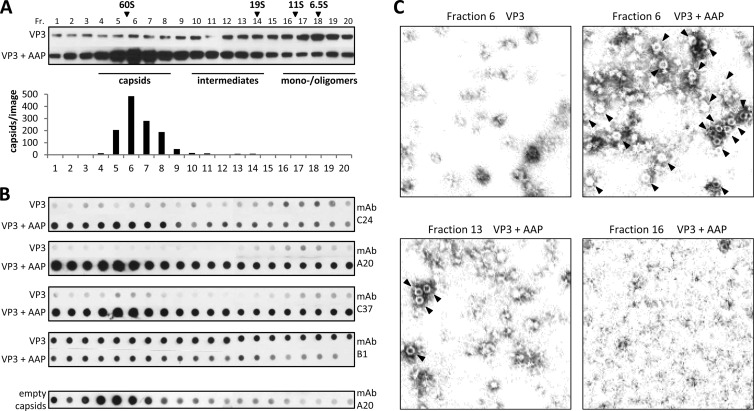
Prior to the analysis of AAP-VP protein interactions, we tested the molecular oligomerization status—monomers, oligomers, or assembly products—of the two proteins expressed alone or in combination. To do this, we subjected lysates of cells transfected with plasmids expressing either VP3 or AAP alone or combinations thereof to fractionation by sucrose density gradients, followed by Western blot analysis (Fig. 4A). Expression of VP3 alone resulted in oligomerization products distributed over the whole gradient, which however, did not show capsid formation, as analyzed by electron microscopy and capsid-specific antibodies (see below). AAP was also found to form high-molecular-weight oligomers of no distinct size, most likely indicating self-oligomerization of the protein. In addition, mutations in the hydrophobic region and conserved core of AAP (L21N/V22N/I26N and I52D/I54D/I56D, respectively) produced oligomerization indistinguishable from that of nonmutated AAP, indicating that these residues are not involved in AAP self-association (data not shown). Sedimentation analysis of cell lysates that had VP3 and AAP coexpressed showed a peak of VP3 at the 60S position, indicating the formation of AAV empty capsids. This was confirmed by electron microscopy and reaction with antibodies detecting capsid formation (see below). The sedimentation behavior of AAP did not change in the presence of VP3.
Fig 4.
VP and AAP oligomerization. (A) Analysis of the molecular oligomerization status—monomers, oligomers, or assembly products—of VP3 and AAP expressed alone or in combination. 293T cells transfected with a plasmid expressing VP3 (pCMV-VP3) or AAP harboring a C-terminal AU1 tag (pAAP-AU1) alone or combinations thereof were subjected to sucrose density gradient fractionation and subsequent Western blot analysis. Proteins were detected by MAb B1 (VP3) and anti-AU1 (AAP) and suitable secondary antibodies. Sucrose density gradient fractionations of purified proteins of known sizes (bovine serum albumin, 6.5S; bovine catalase, 11S; thyroglobulin, 19S; empty AAV2 capsids, 60S) served as a reference to determine the size distribution. (B) Pooled gradient fractions of coexpressed VP3 and AAP containing assembled capsids, assembly intermediates, or monomers/oligomers were used for immunoprecipitation by anti-AU1 antibody or the nonrelated 4A7 antibody. Precipitates were analyzed by Western blot assay detecting coprecipitated VP3 via MAb B1.
Immunoprecipitation of AAP with a MAb directed against the fused AU1 tag coimmunoprecipitated VP3 from gradient fractions containing assembled capsids, as well as capsid proteins from the intermediate-size VP3 oligomers and from low-molecular-weight fractions, suggesting an association of AAP with all sizes of VP oligomers, at least to some extent (Fig. 4B). Respective precipitations with a nonrelated antibody, 4A7, did not result in coprecipitation of VP from any of the fractions. The immunoprecipitation assay was also used for analysis of AAP-VP interactions of the assembly-defective mutant AAPs from whole-cell lysates. For comparison, we included the L103N/I106N/L107N mutant AAP, which showed no assembly defect. All mutant AAPs, with the exception of the assembly-positive mutant AAP, showed no or strongly reduced capsid protein coprecipitation (Fig. 5B). Interestingly, VP coprecipitation of the L21N/V22N/I26N and W23A/R27A/W28A mutant AAPs, which showed the strongest assembly defect, revealed only reduced VP3 coprecipitation (Fig. 5B, different exposure). It is noteworthy that despite similar expression levels of all of the mutant AAPs analyzed (Fig. 5A), apparent variations in the amount of precipitated AAP were observed, suggesting that a portion of these molecules was inaccessible for immunoprecipitation, lost, or degraded during the precipitation procedure despite the constant presence of protease inhibitors. Experiments with reverse precipitation of VP3 by the B1 antibody did not yield well-defined coprecipitation of wild-type or mutant AAP in any case. However, this does not have to be in contrast to the described coprecipitation of VP3 with some of the mutant AAPs but might instead by caused by a substoichiometric representation of AAP in VP-AAP complexes that is not sufficient for Western blot assay detection or by masking of the B1 epitope by AAP (see below).
Fig 5.
Analysis of VP-AAP interactions. (A) Western blot analysis of 293T cells transfected with amino acid exchange mutant VP3 and AAP. Protein expression was analyzed by B1 MAb (detects VP3) and anti-AU1 MAb (detects AAP-AU1). Transfections with an empty pBluescript vector (pBS) or AAP expressed from pAAP-AU1 served as controls. (B) Immunoprecipitation (IP) of AAP-VP complexes from lysates of transfected 293T cells using anti-AU1 antibody-coupled protein A Sepharose. AAP and coprecipitated VP3 were detected by Western blot analysis using anti-AU1 and B1 MAbs, respectively.
A capsid protein domain involved in interaction with AAP.
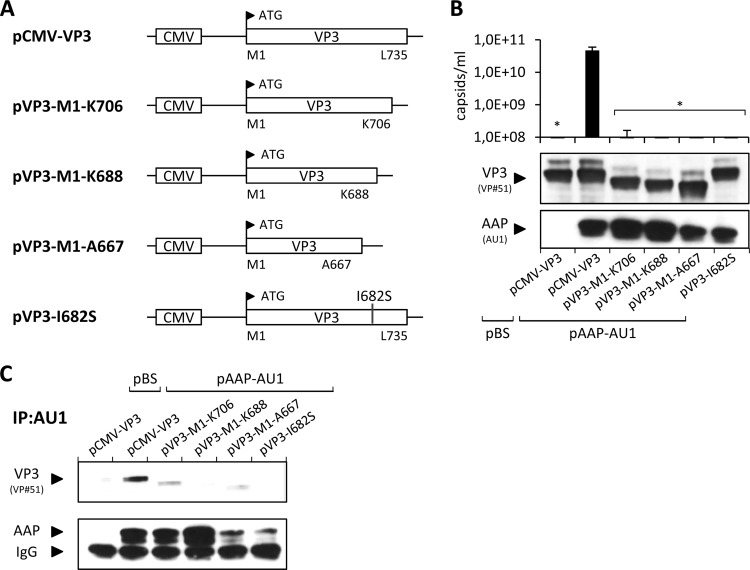
It was previously reported that the C-terminal portion of VP proteins is necessary for capsid assembly (19) and contains several assembly-relevant amino acids (26). We retested this by making a series of C-terminal deletions in VP3 and a point mutation at position I682 (Fig. 6A). VP3 proteins with even small deletions of 29 aa showed no capsid assembly in the presence of AAP (Fig. 6B). Also, the I682S point mutant construct failed to form capsids. Despite similar levels of expression of all of the VP deletion mutant constructs, immunoprecipitations with the AAP-AU1 tag antibody showed no coprecipitation of VP3 for the I682S point mutant construct and the capsid proteins lacking the C-terminal 47 or 68 amino acids (VP3-M1-K688 and VP3-M1-A667, respectively). Furthermore, substantially reduced coprecipitation was observed for the remaining mutant construct, VP3-M1-K706 (Fig. 6C), suggesting that AAP is not able to interact with the C-terminally mutated VP proteins or that it does not remain stably associated with the capsid proteins.
Fig 6.
Capsid assembly by VP deletion mutant constructs. (A) C-terminal deletions of VP3 and an I682S point mutation were generated by site-directed mutagenesis. pCMV-VP3 was used as a control for the expression of full-length VP3. The respective N- and C-terminal amino acids of each VP3 construct are indicated together with their positions. (B) Capsid assembly was analyzed by A20 antibody-based capsid ELISA of freeze-thaw lysates of 293T cells transfected with pAAP-AU1 and the respective mutant VP3 proteins. Coexpression of the empty pBluescript vector (pBS) served as a negative control. Protein expression was confirmed by Western blot analysis using VP#51 polyclonal serum (detection of VP) and anti-AU1 MAb (detection of AAP-AU1 tag). Bars represent the average capsid titers of at least three independent experiments. Asterisks indicate capsid titers significantly lower than that obtained with pCMV-VP3 (P < 0.01). (C) Immunoprecipitation (IP) of AAP-VP complexes from lysates of transfected 293T cells using anti-AU1 antibody-coupled protein A Sepharose. AAP and coprecipitated VP3 were detected by Western blot analysis using the anti-AU1 MAb and VP#51 polyclonal serum, respectively.
Analysis of VP3 conformations in the presence or absence of AAP.
Analysis of density gradient fractions by Western blotting showed that VP3 expressed alone formed high-molecular-weight protein oligomers but not capsids (Fig. 4A and 7A), while in the presence of AAP, capsids were well detectable by electron microscopy (Fig. 7C). We therefore asked whether AAP induces a conformation of VP3 that allows capsid formation. To analyze this question, we fractionated lysates of cells expressing VP3 either alone or together with AAP on sucrose density gradients and applied them under nondenaturing conditions to nitrocellulose membranes. The reactions of different MAbs against AAV2 capsid proteins (24, 25) with proteins in the respective gradient fractions were used to draw conclusions about possible AAP-induced conformational changes in VP3. Antibody C24 reacted weakly and to the same extent with VP3 in the monomer/oligomer fractions in the presence and absence of AAP. However, the reaction with VP3 was increased in the intermediate-size fractions and strongly enhanced in the 60S fraction, indicating capsid assembly upon the coexpression of VP3 and AAP. The application of MAbs A20 and C37 showed similar reactions in the intermediate-size fraction and the 60S fractions but revealed, in addition, a significant reaction with VP3 in the monomer/oligomer fractions at around 6.5S when AAP was coexpressed. Purified empty capsids showed no comparable reaction with A20 in the low-S-value range. Western blot assays with B1 confirmed that similar amounts of VP3 were present throughout the 6.5S gradient fractions in the presence and absence of AAP (Fig. 7A). This suggests that AAP induces a conformational alteration of VP3 monomers and/or oligomers. This interpretation was supported by the observation that MAb B1 showed a decreased reaction with VP3 monomer/oligomer polypeptides in the presence of AAP. Interestingly, the B1 epitope is located at the C terminus, close to the potential AAP binding site. Thus, binding by AAP could competitively inhibit the access of B1 to its binding epitope at the VP3 C terminus.
Fig 7.
Analysis of VP3 conformations in the presence or absence of AAP. (A) Western blot analysis of sucrose density gradient fractions (Fr. 1 to 20) of lysates from 293T cells expressing VP3 alone or in combination with AAP. VP3 was detected using MAb B1. The same amount of each fraction was analyzed under the same conditions. A reference gradient with purified proteins with known S values was used for calibration. The distribution of assembled AAV2 capsids (number of capsids per image) was analyzed by quantification of at least three representative electron microscopy images from each gradient fraction. (B) Native dot blot analysis of sucrose density gradient fractions from cell lysates containing VP3 or VP3 and AAP, respectively. The same amount of each fraction was dotted onto nitrocellulose membranes under the same nondenaturing conditions and detected by the capsid-specific antibodies C24, A20, and C37 or the B1 antibody, which is specific for a linear epitope at the VP3 C terminus, in order to draw conclusions about AAP-induced conformational changes. The distribution of purified empty AAV2 capsids served as a control. (C) Electron microscopy images of sucrose density gradient fractions containing VP3 alone (fraction 6) or in combination with AAP (fractions 6, 13, and 16). Capsids are indicated by arrowheads.
DISCUSSION
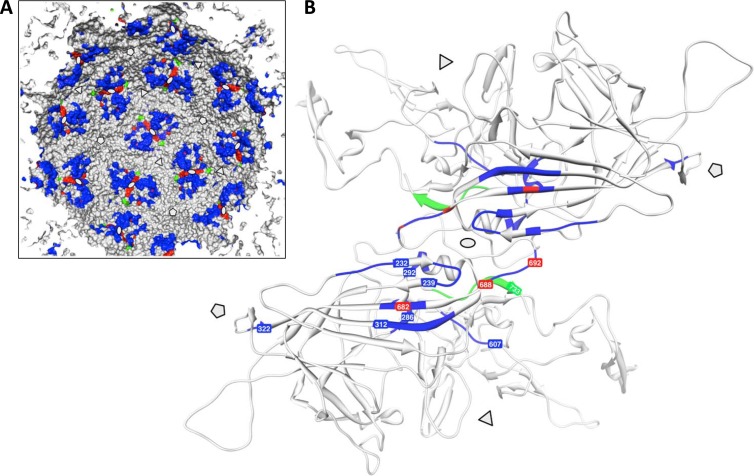
AAV capsid assembly requires the coordinated formation of three types of capsid protein subunit interactions (1, 27). At the 2-fold symmetry axis, the overlapping C-terminal loops extending from the βI sheet to the C terminus (residues 687 to 735) form a relatively weak interaction. It is further stabilized by two conserved helices (residues 712 to 717) localized on opposite sides of a VP dimer flanking the loop interaction (Fig. 8B). VP subunits surrounding the 3-fold axis of the capsid form subunit interface interactions with large invasions of the GH loops into the neighboring subunits. This leads to a complicated but very stable interdigitating trimeric structure. The subunits around the 5-fold symmetry axis also form interactions along the subunit surface which are supported by sling-like connections of the GI loops around the pores at the 5-fold symmetry axes. While the interactions at the 2-fold axes are relatively simple, interactions at the 5-fold axes and even more at the 3-fold axes seem to be sophisticated and likely require defined conformational constraints during assembly. Most likely, these restrictions prevent the self-assembly reaction of VP3, and AAP may provide some guidance to overcome them. The characterization of a number of AAP properties provided hints for understanding the role of AAP in the capsid assembly reaction. Determination of hydrophobic AAP-VP interaction sites, AAP oligomerization, and the influence of AAP on capsid protein conformations suggests a role for AAP as a scaffolding protein in the AAV assembly reaction.
Fig 8.
Capsid protein domains contributing to AAV2 capsid assembly. (A) Schematic representation of the inner capsid surface of AAV2. Positions of capsid assembly defect mutant constructs described previously (3, 26) are shown in blue. Mutant constructs described in this study and by Popa-Wagner et al. (18) are shown in red, while the B1 antibody epitope is shown in green. Note the clustering of assembly mutant constructs at the 2-fold symmetry axes. (B) Ribbon drawing of a VP dimer highlighting the positions of assembly defect mutant constructs described previously (blue) and in this study (red). The B1 epitope is shown in green. The 3-fold and 5-fold axes of symmetry are indicated by triangles and pentamers, respectively.
Point mutations of AAP sequence motifs allowed us to identify amino acids critical for capsid assembly, unlike the cruder deletion analysis, and revealed a dominant influence of two domains—which we named the hydrophobic region and the conserved core—on the assembly-promoting activity of AAP. Coimmunoprecipitation studies suggest that these domains are involved in the VP-AAP interaction. Point mutations in this region confirmed the observation made by the N-terminal deletion analysis that critical amino acids are located between positions 13 and 28. In addition, the mutational analysis revealed the requirement of two sequence motifs in the conserved core downstream of the first assembly-negative deletion mutant construct from amino acid position 28, which are completely conserved among the serotypes analyzed. The characters of these motifs are different, with two prolines and a tryptophan (P43, W46, and P49) at one site and three hydrophobic isoleucines (I52, I54, and I56) at the other site. Moreover, the fact that the W23A-W28A/K144A-R149A double mutant construct showed an additive assembly defect while the P43A-P49A/K144A-R14A mutant construct showed no additive decrease in capsid assembly indicates that conserved amino acids in the hydrophobic region and the conserved core act in different ways in the assembly reaction. The C-terminal deletions suggest functionally relevant sequences between M152 and T177. Conserved amino acids in this region are R/K residues at position 166 and T and P at positions 160 and 162, respectively. Mutation of conserved basic amino acids located farther upstream (residues 144, 146, and 147) had only a moderate influence on AAP activity, suggesting an accessory but not essential function. The basic residues do not act as nucleolar localization motifs, as we initially speculated, and localization of AAP to the nucleolus was not altered by the mutation of these amino acids (data not shown). Rather unexpected was the finding that mutation of a completely conserved S/T motif (residues 124 to 127) and the highly conserved S/T motif at positions 78, 79, and 80 had no effect on capsid assembly. The five prominent S/T-rich clusters appear at rather regular distances from each other, which is suggestive of a functional meaning. Maybe their contributions to the assembly process are redundant and mutations combining several, if not all, of the S/T motifs at the same time are necessary to reveal an effect on capsid assembly.
On the basis of previous observations, we focused on the role of the VP C terminus in capsid assembly (19, 26) by introducing deletions or a point mutation into VP3. Analysis of capsid assembly and coimmunoprecipitation suggest an essential role for this capsid protein sequence in the assembly process and in the AAP-VP interaction. The I682S point mutation (Fig. 8B) abolished not only capsid assembly but also AAP-VP coprecipitation, indicating a critical role for this hydrophobic amino acid in AAP-VP interactions. I682 is surrounded by a group of hydrophobic amino acids (V678, V680, W684, and I686) that may provide a hydrophobic patch for interaction with one of the hydrophobic N-terminal domains of AAP. Interestingly, I682 is located on the βI sheet, which directly continues into the HI loop, which plays an essential role in capsid assembly (6). However, a C-terminal deletion of VP, leaving I682 and the surrounding hydrophobic amino acids intact, also abolished AAP-VP interactions. This result points to a broader interaction between AAP and VP involving other parts of the VP C terminus. A large number of capsid assembly-defective mutant constructs cluster structurally at the 2-fold symmetry axis (Fig. 8A and B), although they are far away from the C terminus in terms of the linear protein sequence (3, 26). This implies that the structural surrounding of the VP C terminus is important for the capsid assembly process either by contributing to AAP-VP interactions or by providing conformational information for the assembly reaction. It has to be determined whether these mutant VPs are also defective in AAP binding. A very interesting VP mutation in this respect is K688A/K692A in the protein's C terminus, which produces a strong assembly defect (18). Although AAP is able to bind to the mutated VP (data not shown) and accumulates in the nucleus, it does not accumulate in the nucleoli and is deposited in nuclear speckles. This mutant protein clearly indicates the need for a specific conformation—independent of AAP binding—for successful intranuclear transport, localization, and finally capsid assembly. It is also important to note that one mutation leading to capsid assembly defects is structurally located close to the 5-fold symmetry axis (Fig. 8).
Analysis of AAP-VP interactions by coimmunoprecipitation requires a note of caution. There are a number of unexpected observations associated with the results of immunoprecipitation and coimmunoprecipitation of AAP and VP. Although coprecipitation of VP with antibodies against the AAP-fused AU1 tag was reproducible, the recovery of AAP in the immunoprecipitate was low for some mutant proteins although the expression of these AAPs detected by Western blotting was similar to that of other mutant proteins that showed stronger immunoprecipitation levels. It is not clear whether the low AAP recovery level is due to the degradation of unstable AAP during the precipitation procedure or the inaccessibility of the antibody epitope in a fraction of the respective mutant protein. A lack of coprecipitated VP in such cases cannot unequivocally be related to a lack of AAP-VP interaction. Independently, the amount of coprecipitated VP was variable. Nevertheless, in most cases, it was reproducibly low, as shown, for example, in Fig. 4 for VP from different sucrose density gradient fractions. In some experiments, however, we noted large amounts of coprecipitated VP, suggesting the coprecipitation of capsids or oligomers (data not shown). Because we could not find conditions to reproducibly show that, even for the 60S fraction containing assembled capsids, we cannot firmly conclude that AAP remains associated with capsids. The 60S fractions may contain a low background level of nonassembled VPs associated with AAP which are recovered by coimmunoprecipitations. Also, precipitation of AAP with VP-directed antibodies B1 and A20 did not give rigorous proof of the association of AAP with capsids (data not shown). One problem was the cross-reaction of an immunoprecipitation product with the AAP antibody in the size range of AAP. Despite all of these caveats, an interaction of AAP with VP can be assumed on the basis of the observation of relocalization of VP to the nucleolus in the presence of AAP (21) and demonstrated by coimmunoprecipitation of VPs with antibodies against the AAP-AU1 tag.
The analysis of AAP and VP oligomerization by sucrose density gradient fractionation revealed the formation of high-molecular-weight oligomers of VP3 alone, indicating VP-VP interactions that are, however, nonproductive in terms of capsid formation. This has been concluded from the lack of reaction with capsid-specific antibodies and was confirmed by visualization of the proteins in the 60S fractions by electron microscopy (Fig. 7C). The unchanged oligomerization of C-terminally truncated VP3 (data not shown) indicates that oligomer formation involves not interactions that occur at the 2-fold symmetry axis but rather surface interactions as they occur at the 3-fold and 5-fold symmetry axes. Most likely, the sophisticated loop interdigitations cannot be formed in the absence of AAP. Surprising was the strong oligomerization of AAP over the whole range of the gradient, which did not visibly change in the presence or absence of VP coexpression. This oligomerization behavior is suggestive of an assembly scaffolding function of AAP. In this case, AAP has to be continuously removed in the course of the assembly reaction because no intermediates with increased amounts of AAP could be detected. The oligomerization of AAP opens the opportunity to study a possible trans-dominant negative effect of different mutant AAPs on this process.
Scaffolding proteins seem to have two essential functions in viral morphogenesis, (i) facilitating the nucleation of assembly and (ii) mediating the reaction to completion (9). Nucleation involves the increase in statistical probability of getting a sufficient number of molecules together (8) and the promotion of a change from an unassociable to an associable conformation (4). An increase in the local concentration of capsid proteins may be achieved by the nucleolar accumulation of VPs in association with AAP, and the conformational change may also be introduced by AAP, as shown by the analysis of capsid protein conformations with MAbs. This change is detected by antibodies A20 and C37 (24, 25) showing a stronger reaction with VPs sedimenting in the range of 6.5S to 11S when AAP was coexpressed than when VP3 was expressed alone. It may also be the case that the A20 reaction reflects only subunit oligomerization necessary to build up the A20 epitope (14). The decrease in the reaction with antibody B1 in fractions with low S values under nondenaturing conditions may indicate a competition between AAP and the binding of MAb B1 to VP3 (Fig. 8B, epitope shown in green). Binding of AAP to the C terminus may sterically hinder the binding of B1 to its epitope at the very C terminus (25). This study does not show whether AAP, after nucleation of assembly, provides form-determining information for the assembly process. An additional contribution of nucleolar proteins to the assembly process cannot be excluded. However, knockdown of nucleophosmin, which associates with VPs and Rep68 (2), had only a moderate effect on capsid assembly (5-fold reduction of assembly; data not shown). An influence of nucleolin has not been tested so far.
Combined, our data suggest that AAP acts as a scaffolding protein during AAV capsid assembly. Concentration of VPs in the nucleolus may promote nucleation of assembly, and induction of a conformational change by binding to the VP interaction site at the 2-fold symmetry axes may allow more complicated subunit interactions at the 3-fold and/or 5-fold symmetry axes.
ACKNOWLEDGMENTS
We thank Birgit Hub (German Cancer Research Center) for excellent electron microscopy analysis.
F.S., M.N., and R.P.-W. were supported by grants from the Deutsche Krebshilfe (109653) and the Deutsche Forschungsgemeinschaft (KL516/9-1) to J.A.K.
We have no conflicts of interest to declare.
Footnotes
Published ahead of print 26 September 2012
REFERENCES
- 1. Agbandje-McKenna M, Kleinschmidt J. 2011. AAV capsid structure and cell interactions. Methods Mol. Biol. 807:47–92 [DOI] [PubMed] [Google Scholar]
- 2. Bevington JM, et al. 2007. Adeno-associated virus interactions with B23/nucleophosmin: identification of sub-nucleolar virion regions. Virology 357:102–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bleker S, Sonntag F, Kleinschmidt JA. 2005. Mutational analysis of narrow pores at the fivefold symmetry axes of adeno-associated virus type 2 capsids reveals a dual role in genome packaging and activation of phospholipase A2 activity. J. Virol. 79:2528–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Caspar DL. 1980. Movement and self-control in protein assemblies. Quasi-equivalence revisited. Biophys. J. 32:103–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cole C, Barber JD, Barton GJ. 2008. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 36:W197–W201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. DiPrimio N, Asokan A, Govindasamy L, Agbandje-McKenna M, Samulski RJ. 2008. Surface loop dynamics in adeno-associated virus capsid assembly. J. Virol. 82:5178–5189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Erickson HP, Pantaloni D. 1981. The role of subunit entropy in cooperative assembly. Nucleation of microtubules and other two-dimensional polymers. Biophys. J. 34:293–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fane BA, Prevelige PE., Jr 2003. Mechanism of scaffolding-assisted viral assembly. Adv. Protein Chem. 64:259–299 [DOI] [PubMed] [Google Scholar]
- 10. Gao G, Vandenberghe LH, Wilson JM. 2005. New recombinant serotypes of AAV vectors. Curr. Gene Ther. 5:285–297 [DOI] [PubMed] [Google Scholar]
- 11. Grimm D, et al. 1999. Titration of AAV-2 particles via a novel capsid ELISA: packaging of genomes can limit production of recombinant AAV-2. Gene Ther. 6:1322–1330 [DOI] [PubMed] [Google Scholar]
- 12. Kleinschmidt JA, King JA. 2006. Molecular interactions involved in assembling the viral particle and packaging the genome, p 305–319 In Kerr J, Cotmore SF, Bloom ME, Linden RM, Parrish CR. (ed), Parvoviruses. Hodder Arnold, New York, NY [Google Scholar]
- 13. Kuck D, Kern A, Kleinschmidt JA. 2007. Development of AAV serotype-specific ELISAs using novel monoclonal antibodies. J. Virol. Methods 140:17–24 [DOI] [PubMed] [Google Scholar]
- 14. McCraw DM, O'Donnell JK, Taylor KA, Stagg SM, Chapman MS. 2012. Structure of adeno-associated virus-2 in complex with neutralizing monoclonal antibody A20. Virology 431:40–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Muzyczka N, Berns KI. 2001. Parvoviridae: the viruses and their replication, p 2327–2360 In Knipe DM, et al. (ed) Fields virology, 4th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 16. Myers MW, Carter BJ. 1980. Assembly of adeno-associated virus. Virology 102:71–82 [DOI] [PubMed] [Google Scholar]
- 17. Naldini L, et al. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263–267 [DOI] [PubMed] [Google Scholar]
- 18. Popa-Wagner R, Sonntag F, Schmidt K, King J, Kleinschmidt JA. 2012. Nuclear translocation of AAV2 capsid proteins for virion assembly. J. Gen. Virol. 93:1887–1898 [DOI] [PubMed] [Google Scholar]
- 19. Ruffing M, Heid H, Kleinschmidt JA. 1994. Mutations in the carboxy terminus of adeno-associated virus 2 capsid proteins affect viral infectivity: lack of an RGD integrin-binding motif. J. Gen. Virol. 75:3385–3392 [DOI] [PubMed] [Google Scholar]
- 20. Sonntag F, et al. 2011. The assembly-activating protein promotes capsid assembly of different adeno-associated virus serotypes. J. Virol. 85:12686–12697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sonntag F, Schmidt K, Kleinschmidt JA. 2010. A viral assembly factor promotes AAV2 capsid formation in the nucleolus. Proc. Natl. Acad. Sci. U. S. A. 107:10220–10225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ward P. 2006. Replication of adeno-associated virus DNA, p 189–212 In Kerr J, Cotmore SF, Bloom ME, Linden RM, Parrish CR. (ed), Parvoviruses. Hodder Arnold, New York, NY [Google Scholar]
- 23. Wistuba A, Kern A, Weger S, Grimm D, Kleinschmidt JA. 1997. Subcellular compartmentalization of adeno-associated virus type 2 assembly. J. Virol. 71:1341–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wistuba A, Weger S, Kern A, Kleinschmidt JA. 1995. Intermediates of adeno-associated virus type 2 assembly: identification of soluble complexes containing Rep and Cap proteins. J. Virol. 69:5311–5319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wobus CE, et al. 2000. Monoclonal antibodies against the adeno-associated virus type 2 (AAV-2) capsid: epitope mapping and identification of capsid domains involved in AAV-2-cell interaction and neutralization of AAV-2 infection. J. Virol. 74:9281–9293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wu P, et al. 2000. Mutational analysis of the adeno-associated virus type 2 (AAV2) capsid gene and construction of AAV2 vectors with altered tropism. J. Virol. 74:8635–8647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xie Q, et al. 2002. The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy. Proc. Natl. Acad. Sci. U. S. A. 99:10405–10410 [DOI] [PMC free article] [PubMed] [Google Scholar]