Abstract
Modified vaccinia virus Ankara (MVA) is a safe, attenuated orthopoxvirus that is being developed as a vaccine vector but has demonstrated limited immunogenicity in several early-phase clinical trials. Our objective was to rationally improve the immunogenicity of MVA-based HIV/AIDS vaccines via the targeted deletion of specific poxvirus immune-modulatory genes. Vaccines expressing codon-optimized HIV subtype C consensus Env and Gag antigens were generated from MVA vector backbones that (i) harbor simultaneous deletions of four viral immune-modulatory genes, encoding an interleukin-18 (IL-18) binding protein, an IL-1β receptor, a dominant negative Toll/IL-1 signaling adapter, and CC-chemokine binding protein (MVAΔ4-HIV); (ii) harbor a deletion of an additional (fifth) viral gene, encoding uracil-DNA glycosylase (MVAΔ5-HIV); or (iii) represent the parental MVA backbone as a control (MVA-HIV). We performed head-to-head comparisons of the cellular and humoral immune responses that were elicited by these vectors during homologous prime-boost immunization regimens utilizing either high-dose (2 × 108 PFU) or low-dose (1 × 107 PFU) intramuscular immunization of rhesus macaques. At all time points, a majority of the HIV-specific T cell responses, elicited by all vectors, were directed against Env, rather than Gag, determinants, as previously observed with other vector systems. Both modified vectors elicited up to 6-fold-higher frequencies of HIV-specific CD8 and CD4 T cell responses and up to 25-fold-higher titers of Env (gp120)-specific binding (nonneutralizing) antibody responses that were relatively transient in nature. While the correlates of protection against HIV infection remain incompletely defined, our results indicate that the rational deletion of specific genes from MVA vectors can positively alter their cellular and humoral immunogenicity profiles in nonhuman primates.
INTRODUCTION
The development of a safe and efficacious human immunodeficiency virus (HIV) vaccine remains a high priority in order to provide a sustainable solution to control the current AIDS pandemic (82). However, correlates of protection against HIV infection and disease progression remain incompletely defined and therefore pose a challenge to vaccine development with regard to defining the precise nature of protective immune responses that should be emulated through immunization (29, 31). While the generation of antibodies that exhibit broad neutralization activity against diverse HIV strains holds the promise of informing efforts to confer sterilizing immunity against HIV infection, relevant HIV envelope (Env)-derived immunogens that are capable of eliciting such broadly neutralizing antibodies in the context of immunization represent a still-unrealized vision (84). Interestingly, the RV144 efficacy trial evaluated a vaccination regimen consisting of priming with a series of recombinant canarypox virus (ALVAC-HIV) immunizations followed by boosting immunizations with recombinant gp120 (AIDSVAX-B/E) and demonstrated an overall vaccine efficacy of 31% for the reduction of HIV acquisition. Because this immunization regimen induced predominantly nonneutralizing antibodies, rather than T cell immunity, these results suggest that nonneutralizing antibodies that target the HIV envelope may contribute more to the prevention of HIV acquisition than previously appreciated (41, 68).
Alternatively, the development of AIDS vaccines whose mechanism of action is predicated upon eliciting T cell responses to control HIV infection, rather than to prevent infection per se, has progressed in preclinical models (8, 37, 43, 50, 51, 85), but these vaccines have not yet demonstrated efficacy in humans (14, 53). While the efficacy of the ALVAC-HIV–AIDSVAX-B/E vaccine evaluated in the RV144 trial was limited, and the mechanism of protection is not known with certainty, results from this trial suggest that an immunization regimen comprised of priming with a poxvirus vector followed by boosting with recombinant gp120 is superior to immunization with recombinant gp120 alone (49, 67). As such, these results provide an additional rationale for our work to improve the immunogenicity of poxvirus-based AIDS vaccines.
In light of currently available clinical trial data, it is widely thought that an effective HIV vaccine will need to elicit both humoral and cellular immunity and at higher levels, while targeting larger numbers of diverse viral targets, than have otherwise been achieved to date (19, 53). Toward this goal, we have conducted the current study with the aim of improving the immunogenicity of HIV vaccines derived from modified vaccinia virus Ankara (MVA), a safe and highly attenuated orthopoxvirus that is actively being developed as an AIDS vaccine (22, 26, 35, 46, 61, 66, 69, 83).
MVA was originally developed as a smallpox virus vaccine and was derived through the extensive serial passaging of its parental strain of vaccinia virus on primary chicken embryo fibroblast cultures (52). As a result of this classical attenuation, MVA underwent a deletion of 31 kb (∼15%) of its genome, compared to its parental strain, and harbors numerous gene deletions, including deletions of genes that otherwise contribute to virus evasion from host immune responses and that determine host range (6, 56). As a result, MVA does not replicate productively in most mammalian cell types, with infection being aborted at a relatively late stage of virion assembly and maturation following the expression of viral early and late genes (11, 15, 27). The resulting inability of MVA to replicate in human hosts likely contributes substantially to its excellent safety profile as both a smallpox vaccine as well as a recombinant vaccine vector (17, 22, 64, 79). Given its high level of safety and ability to accommodate large foreign gene inserts, MVA has been an attractive candidate as a vector for clinical development into vaccines against a number of infectious diseases in addition to HIV/AIDS, including malaria (9, 28, 42, 59, 60) and tuberculosis (TB) (39, 54, 55, 63), as well as cancers (20, 23, 48, 72; J.-P.L. Bory, J.-L. Brun, V. Dalstein, and M. Baudin, presented at Eurogin 2006, Paris, France). Of particular relevance to clinical development is MVA85A, an MVA-based TB vaccine that expresses antigen 85A of Mycobacterium tuberculosis and has demonstrated safety and immunogenicity in Mycobacterium bovis BCG prime-MVA85A boost immunization strategies (10, 39, 65).
However, MVA-based HIV vaccines have exhibited variable, but generally limited, immunogenicity in clinical evaluations, particularly when utilized as a single modality (22, 34, 35, 46, 47, 61, 66, 83). Several known factors contribute to this suboptimal immunogenicity profile of MVA vectors. These factors include the immunodominance of the T cell responses to the large number of vector-derived gene products over the inserted immunogen of interest as well as the generation of vector-specific neutralizing antibodies that result in a diminished capacity for effective booster immunization (16, 44, 70, 73). While prime-boost immunization regimens that utilize MVA-based vaccines in combination with heterologous vaccine vectors, such as plasmid DNA (34, 69) and recombinant adenovirus vectors (8), can improve vaccine immunogenicity, the identification of modifications to the MVA vector itself that render these vaccines intrinsically more immunogenic than those derived from the parental MVA backbone should prove beneficial.
One approach toward improving the immunogenicity of MVA-based vaccines that we have pursued is based upon the deletion of an essential viral replication gene (udg) to abrogate viral late gene expression and thereby reduce the complexity of the repertoire of expressed vector antigens that may otherwise compete against antigens of interest (expressed from early viral promoters) for eliciting T and B cell responses (30). A complementary approach, which we report here, is to delete MVA immune-modulatory genes that are present in the vector genome and whose gene products may be predicted to interfere with the optimal generation of vaccine-induced cellular and humoral immunity.
Despite the loss of numerous viral immune evasion activities from MVA during its classical attenuation, including genes encoding soluble decoy receptors for type I and II interferons and tumor necrosis factor alpha (TNF-α) (1, 4, 6, 81), MVA still encodes a number of immune-modulatory factors, and all MVA-based vaccines that have advanced into clinical evaluation to date have been derived from MVA vectors that express these immune-modulatory genes. We therefore targeted for deletion those genes encoding a secreted interleukin-18 (IL-18) binding protein (MVA008L) (75, 80), a soluble IL-1β receptor (MVA184R) (3, 76), a CC-chemokine binding protein (MVA153L) (7, 62), and a dominant negative Toll/IL-1 signaling adapter (MVA159R) (13, 77). The biological activities of these factors can be inferred by the fact that deletions of their orthologs from replication-competent strains of vaccinia virus result in altered pathogenic phenotypes in murine models (2, 62, 74, 77, 80). Additionally, the observation that MVA recombinants lacking MVA184R and/or MVA153L may elicit modestly augmented CD8 T cell responses in mice indicates that the deletion of immune-modulatory genes from MVA-based vaccines is a relevant approach toward improving vaccine immunogenicity (18, 21, 32, 78).
In the present study, we targeted four immune-modulatory genes (MVA008L, MVA153L, MVA159R, and MVA184R) for simultaneous deletion (Δ4) alone and in combination with the deletion of udg (Δ5) as experimental vector modifications. We derived vaccines from the Δ4, Δ5, and parental (control) MVA backbones that express identical HIV consensus subtype C Env and Gag antigens and comparatively evaluated the cellular and humoral immune responses that were elicited by these recombinant vaccines in rhesus macaques. As a proof of concept, we demonstrate that these modified vectors elicit frequencies of HIV-specific CD8 and CD4 T cells as well as titers of HIV Env-specific antibodies that are transiently of higher magnitudes than those elicited by a parental control vaccine following prime-boost immunization of rhesus macaques. Our results demonstrate, in a relevant nonhuman primate model, that the cellular and humoral immunogenicity of MVA-based AIDS vaccines can be rationally enhanced and suggest that such genetically modified vectors may be considered alternative platforms to parental MVA vectors for further development toward clinical trial evaluations.
MATERIALS AND METHODS
Animals and immunizations.
Female rhesus macaques, aged 3 to 9 years (average age, 5.4 years), were randomized into six immunization cohorts (n = 10 per group). Immunization regimens were comprised of two serial doses of an identical vaccine vector (MVAΔ4-HIV, MVAΔ5-HIV, or MVA-HIV) at either 2 × 108 PFU (high dose) or 1 × 107 PFU (low dose) administered at weeks 0 and 9 of the study. Immunizations were performed via intramuscular injection (1-ml volume) into the thigh. All macaques were maintained at the Yerkes National Primate Research Center (Field Station) in accordance with institutionally and federally approved animal welfare protocols and standards.
Vectors and inserts.
All recombinant MVA vectors used in this study were derived from an original stock of MVA-1974 (MVA-1974 clone 1 LVDP6 7/22/02), which was kindly provided by B. Moss (NIH). All recombinant MVA vectors express synthetic, codon-optimized, HIV subtype C consensus gag and env genes (GenBank accession numbers JX236707 and JX236708) from independent early (modified H5) viral promoters recombined into the MVA deletion III locus. Salient features of the HIV Env antigen expressed by all vaccine vectors include the deletion of V1/V2 and a truncation at Y712 to enhance genetic stability within recombinant MVA vectors. Additionally, the MVAΔ4-HIV vector harbors deletions of four viral immune evasion genes (MVA008L, MVA153L, MVA159R, and MVA184R), whereas the MVAΔ5-HIV vector harbors deletions of these four immune evasion genes in combination with the deletion of viral uracil-DNA glycosylase (MVA101R) (see Table S1 in the supplemental material). All vectors were generated and propagated and titers were determined by using a udg-complementing DF-1 fibroblast-derived cell line, as previously described (30). Sucrose gradient-purified stocks of recombinant viruses were used for immunizations.
Intracellular cytokine assay.
Cryopreserved peripheral blood mononuclear cells (PBMCs) were thawed and rested overnight at 37°C in RPMI cell culture medium containing 10% heat-inactivated fetal bovine serum (FBS) and DNase. The assay was performed with 96-well V-bottom plates. For stimulation, replicate cultures of 1 × 106 PBMCs were incubated with Gag, Env-1, or Env-2 peptide pools (15-mer, overlapping by 11 that were identically matched to the Gag and Env antigens expressed by recombinant MVA vectors; [NIH AIDS Research and Reference Reagent Program and Bio-Synthesis, Inc., Lewisville, TX]) at a final concentration of 2 μg/peptide/ml or were stimulated with Staphylococcus enterotoxin B (SEB) as a positive control or not stimulated (negative control), all in the presence of brefeldin A (10 μg/ml), for 6 h at 37°C. For staining, cells were washed and then stained with Alexa 430 (Invitrogen) to enable live/dead discrimination, permeabilized (BD fluorescence-activated cell sorter [FACS] permeabilizing solution 2), and stained with a cocktail of fluorescently conjugated antibodies, fixed in 1% formaldehyde, and acquired on a BD LSR-II flow cytometer. Antibody conjugates include CD14/CD20-allophycocyanin (APC)-Cy7, CD3-Pacific blue, CD4-fluorescein isothiocyanate (FITC), CD8-phycoerythrin-Texas Red (ECD), CD69-peridinin chlorophyll protein (PerCP), and gamma interferon (IFN-γ)–APC. Data were analyzed by using FlowJo software (TreeStar, Inc.).
Antibody assays.
Recombinant gp120 (corresponding to amino acids [aa] 1 to 446 of the consensus HIV subtype C Env protein expressed from MVAΔ4-HIV) expressing a C-terminal hexahistidine tag was expressed in 293 cells, purified via metal affinity chromatography, and used, in carbonate buffer, to coat Maxisorb plates (Nunc, Rochester, NY) for enzyme-linked immunosorbent assays (ELISAs). Duplicate serial dilutions (2-fold) of macaque plasma samples, beginning at a 1:20 dilution, were incubated in wells of gp120-coated ELISA plates for 1 h at 37°C. The plates were washed four times and incubated with a 1:10,000 dilution of 2° polyclonal anti-monkey IgG-horseradish peroxidase (HRP) antibody (Accurate Chemical and Scientific Corp., Westbury, NY) for 1 h at 37°C. Plates were washed four times with wash buffer and incubated with a TMB (3,3,5,5′-tetramethylbenzidine) Microwell peroxidase substrate system (2-C) (KPL, Inc.) for 20 min at room temperature. The reaction was stopped by the addition of 2 N sulfuric acid. The absorbance (450 nm) values were obtained via a plate reader and were corrected for the blank-well absorbance value. Corrected absorbance data were analyzed as a function of the plasma dilution by nonlinear regression analysis using Prism software (GraphPad Software, Inc.). For each individual sample, the HIV Env-specific ELISA titer is reported as the plasma dilution corresponding to an A450 value of 0.3, as interpolated from the fitted curve. Plasma samples with no detectable Env-specific ELISA titers (i.e., A450 of <0.3) were coded at the threshold of detection. B5R-specific ELISAs utilized a recombinant B5R protein (BEI Resources) as the coating antigen and were performed analogously to those for HIV Env. Titers of MVA-specific binding and neutralizing antibodies were determined as described previously (30).
RESULTS
Modified MVA vectors elicit enhanced T cell responses in immunized macaques.
We sought to determine whether MVA vectors with deletions of four different viral immune-modulatory genes, alone or in combination with a deletion of udg (30), elicit enhanced T cell responses against expressed HIV antigens (Gag and Env) compared to those elicited by a control vector that does not harbor these deletions and represents a parental (i.e., wild-type) MVA vector backbone. Toward this end, we constructed vaccine vectors that express identical synthetic, human codon-optimized, HIV subtype C consensus gag and env genes from early viral promoters at the MVA deletion III locus but that differ with regard to deletions of specific combinations of MVA immune-modulatory and DNA replication genes (MVAΔ4-HIV, MVAΔ5-HIV, and MVA-HIV [control]) (Table 1; also see Table S1 in the supplemental material).
Table 1.
Genotypes of recombinant MVA-based HIV vaccines a
| Vaccine | Genotype |
||||||
|---|---|---|---|---|---|---|---|
| MVA genes |
HIV genes |
||||||
| 008L | 159R | 153L | 184R | 101R | C-env | C-gag | |
| MVAΔ4-HIV | Δ | Δ | Δ | Δ | + | + | + |
| MVAΔ5-HIV | Δ | Δ | Δ | Δ | Δ | + | + |
| MVA-HIV | + | + | + | + | + | + | + |
The recombinant vaccines MVAΔ4-HIV, MVAΔ5-HIV, and MVA-HIV commonly express the HIV subtype C env and gag genes from MVA vector backbones that differ from one another with regard to the presence (+) or absence via deletion (Δ) of the indicated MVA immune-regulatory genes, encoding a secreted IL-18 binding protein (MVA008L), a Toll/IL-1 receptor homolog (MVA159R), a CC-chemokine binding protein (MVA153L), and a secreted IL-1β receptor (MVA184R), and a DNA replication gene (MVA101R) encoding uracil-DNA glycosylase. HIV C-env (GenBank accession number JX236708) and C-gag (GenBank accession number JX236707) are synthetic genes, codon optimized for maximal rev-independent expression in human cells, that are devoid of early vaccinia virus transcription termination signals (5′-TTTTTNT-3′) and are expressed from independent early (modified H5) viral promoters from the MVA deletion III locus. The HIV genes express antigenic subtype C consensus Gag (492 amino acids) and Env (647 amino acids) antigens. HIV C-Env harbors a V1/V2 deletion (aa 138 to 195 with respect to HXB2) and a deletion of the C-terminal 151 amino acids of gp41 (corresponding to a truncation at Y712 with respect to HXB2). All recombinant MVA vectors were derived from an original stock of plaque-purified MVA-1974 (MVA-1974 clone 1 P572 LVDP6 7/22/02), which was kindly provided by B. Moss (NIH). Additional details regarding the genomic boundaries of MVA gene deletions and flanking regions are provided in Table S1 in the supplemental material.
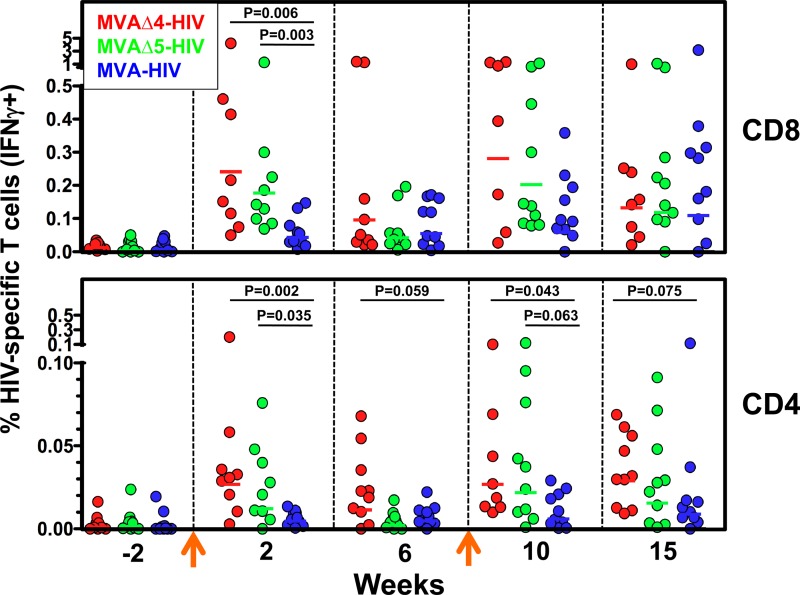
Improved vaccines that are more immunogenic than current alternatives could potentially elicit higher levels of immunity than otherwise achievable and/or allow for the use of lower doses of vaccines during immunization (i.e., dose sparing). With regard to MVA-based vaccines, dose-sparing effects would also likely be advantageous toward offsetting the difficulties anticipated in the scale-up of vaccine production. In order to investigate the immunogenicity and dose-sparing capacity of the MVAΔ4-HIV and MVAΔ5-HIV vaccines, we immunized six groups of rhesus macaques (n = 10 per group) with 2 × 108 PFU (high dose) or 1 × 107 PFU (low dose) of MVAΔ4-HIV, MVAΔ5-HIV, or MVA-HIV at 0 and 9 weeks, according to a homologous prime-boost experimental design. At various times preceding and following immunizations, we determined the frequencies of Env- and Gag-specific CD8 and CD4 T cells in PBMCs via intracellular cytokine assays for the production of IFN-γ in response to ex vivo stimulation with pools of matched overlapping Env and Gag peptides. At 2 weeks following primary immunization, macaques immunized with high-dose MVAΔ4-HIV or MVAΔ5-HIV exhibited significantly higher frequencies of total HIV-specific CD8 T cells (reported as HIV Env and Gag responses) than did macaques immunized with the control MVA-HIV vector (range of 0.05 to 4.23 and geometric mean [GM] of 0.24 for MVAΔ4-HIV, range of 0.07 to 1.29 and GM of 0.18 for MVAΔ5-HIV, and range of 0.01 to 0.15 and GM of 0.04 for MVA-HIV) (Fig. 1). When GM frequencies were compared between groups, the MVAΔ4-HIV and MVAΔ5-HIV vectors elicited 6- and 4-fold-higher frequencies of HIV-specific CD8 T cells, respectively, than did MVA-HIV. Similarly, at this time point, MVAΔ4-HIV and MVAΔ5-HIV elicited significantly higher frequencies of total HIV-specific CD4 T cell responses than did the control vector MVA-HIV (range of 0.003 to 0.20 and GM of 0.027 for MVAΔ4-HIV, range of 0 to 0.076 and GM of 0.012 for MVAΔ5-HIV, and range of 0 to 0.013 and GM of 0.005 for MVA-HIV) (Fig. 1). Group GM frequencies of total HIV-specific CD4 T cell responses elicited by the MVAΔ4-HIV and MVAΔ5-HIV vectors were 6- and 3-fold higher than that exhibited by the control vector MVA-HIV. In general, the frequencies of HIV-specific CD8 T cells that were elicited by MVA vectors were, on average, 9- to 15-fold higher than the corresponding frequencies of HIV-specific CD4 T cells.
Fig 1.
Modified MVA vectors elicit enhanced HIV-specific T cell responses in immunized macaques. Frequencies of total (Gag plus Env) HIV-specific CD8 (top) and CD4 (bottom) T cell responses were determined by IFN-γ ICS assays of PBMCs from rhesus macaques at the indicated times. Symbols represent individual macaques. Arrows indicate immunizations with 2 × 108 PFU of the MVAΔ4-HIV, MVAΔ5-HIV, or MVA-HIV vector at weeks 0 and 9. Horizontal lines denote group geometric means. Statistical comparisons of groups were performed via a Kruskal-Wallis test. Note the different ordinate scales.
At 1 week following homologous booster immunization (week 10), macaques in the MVAΔ4-HIV and MVAΔ5-HIV cohorts exhibited significantly higher frequencies of HIV-specific CD4 T cells than did those boosted with MVA-HIV (range of 0.01 to 0.10 and GM of 0.027 for MVAΔ4-HIV, range of 0.001 to 0.12 and GM of 0.022 for MVAΔ5-HIV, and range of 0 to 0.03 and GM of 0.006 for MVA-HIV). The frequencies of total HIV-specific CD8 responses at week 10 in macaques immunized with MVAΔ4-HIV or MVAΔ5-HIV also exhibited higher trends than MVA-HIV, with group GM frequencies 5- and 3-fold higher, respectively, but these values were not significant at a ≥95% level (Fig. 1). Thus, homologous prime-boost immunization with modified MVA vaccine vectors harboring 4- and 5-gene deletions demonstrated the augmented generation of primary HIV-specific CD8 and CD4 T cell responses as well as enhanced HIV-specific CD4 T cell responses following booster immunization compared to the control MVA vector.
Recombinant MVA vectors elicit HIV Env-biased CD8 and CD4 T cell responses.
We sought to determine whether the antigenic specificity of HIV-specific T cell responses elicited by modified MVA vaccine vectors differed substantially from those elicited by the control vaccine derived from the parental MVA backbone. Toward this end, we analyzed the frequencies of HIV-specific T cell responses, determined via intracellular cytokine staining (ICS) assays of PBMCs from macaques immunized with high-dose MVAΔ4-HIV, MVAΔ5-HIV, or MVA-HIV, and stratified these responses according to both the vaccine vector and peptide pool (Env-1, Env-2, or Gag) used for the stimulation of PBMCs (Fig. 2). Within both the CD8 and CD4 T cell compartments, Env-specific responses predominated among the HIV-specific responses elicited by all vaccines, including both modified and control vectors, at all postimmunization time points (Fig. 2). Thus, the magnitudes of the CD8 T cell responses elicited by both modified and control vaccines were similarly skewed in favor of HIV Env rather than Gag.
Fig 2.
Immunization of macaques with recombinant MVA vectors elicits insert-specific CD8 and CD4 T cell responses that are biased toward HIV Env. Frequencies of CD8 (left) and CD4 (right) T cells stimulated by Env-1, Env-2, and Gag peptide pools were determined by IFN-γ ICS assays of PBMCs from rhesus macaques immunized with 2 × 108 PFU of the MVAΔ4-HIV, MVAΔ5-HIV, or MVA-HIV vector at weeks 0 and 9 (arrows). Symbols represent individual macaques. Horizontal lines denote group geometric mean responses. Note the different ordinate scales between the CD8 and CD4 plots.
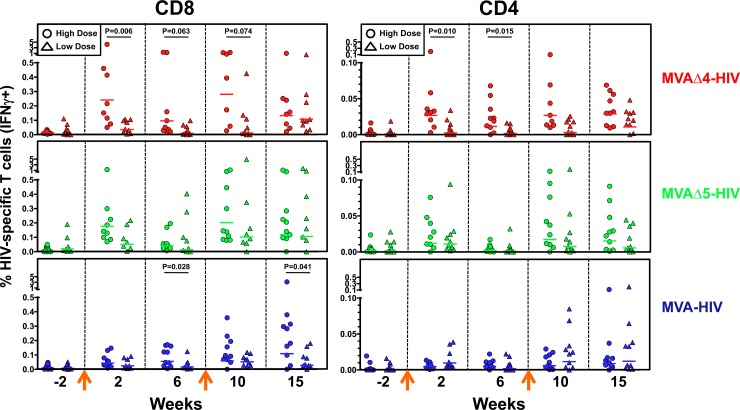
Dose-response effects on T cell responses in rhesus macaques.
The immunization of macaques with MVA vectors at a high dose (1 × 108 PFU) tended to elicit relatively higher GM frequencies of HIV-specific CD8 and CD4 T cells than did low-dose (1 × 107 PFU) immunization (Fig. 3). In particular, high-dose immunization with MVAΔ4-HIV elicited significantly higher frequencies of HIV-specific CD8 and CD4 T cells, at multiple time points, than did low-dose immunization (Fig. 3). In addition, significant increases in the frequencies of HIV-specific CD8 T cells that were elicited by MVAΔ4-HIV and MVAΔ5-HIV, compared to those elicited by MVA-HIV, were observed following low-dose immunization (see Fig. S1 in the supplemental material). Generally, the differences between the frequencies of HIV-specific T cells that were elicited following high- and low-dose immunizations with modified vaccines tended to be greater than those exhibited by high- versus low-dose immunization with the control vaccine. These results suggest that there may be a threshold dose (between 1 × 107 and 2 × 108 PFU) for the intramuscular administration of recombinant MVA vaccines, above which the modified vaccines' phenotype of T cell enhancement is fully penetrant.
Fig 3.
Effects of vaccine doses on magnitudes of elicited HIV-specific T cell responses. Frequencies of total (Gag and Env) HIV-specific CD8 (left) and CD4 (right) T cell responses were determined by IFN-γ ICS assays of PBMCs from rhesus macaques at the indicated times following high-dose (2 × 108 PFU) or low-dose (1 × 107 PFU) immunization with the MVAΔ4-HIV, MVAΔ5-HIV, or MVA-HIV vector at weeks 0 and 9 (arrows). Horizontal lines denote group geometric mean responses. Statistical comparisons of groups were performed via a Kruskal-Wallis test. High-dose data are reproduced from Fig. 1 to facilitate comparisons. Note the different ordinate scales between the CD8 and CD4 plots.
Modified MVA vectors elicit enhanced antibody responses in immunized macaques.
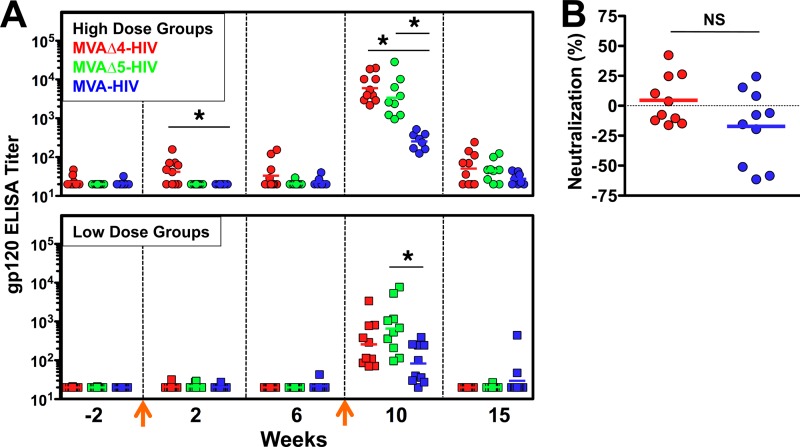
Plasma samples from macaques immunized with MVAΔ4-HIV, MVAΔ5-HIV, or MVA-HIV were assayed to determine the magnitude and quality of the Env-specific antibody responses that were elicited following immunization. Titers of HIV Env-specific binding antibodies were determined by gp120 ELISAs, utilizing recombinant gp120 that was matched to the vectored envelope as the coating antigen. Envelope (gp120) ELISA titers are shown for macaques immunized with both high (2 × 108 PFU) and low (1 × 107 PFU) doses of MVAΔ4-HIV, MVAΔ5-HIV, and MVA-HIV (Fig. 4). Levels of gp120-specific antibodies were detected beginning 2 weeks following the primary high-dose immunization and were significantly higher (2-fold) in macaques immunized with MVAΔ4-HIV than in those immunized MVA-HIV (Fig. 4A). An effective boosting of gp120 antibody titers was observed for all vectors following secondary immunizations at both high and low doses. A comparison of gp120-specific ELISA titers between immunization groups 1 week following booster immunization (week 10) revealed dramatically higher antibody titers in macaques that received MVAΔ4-HIV or MVAΔ5-HIV than in those that received MVA-HIV, with group GM titers showing 24-fold (MVAΔ4-HIV versus MVA-HIV, high dose) and 13-fold (MVAΔ5-HIV versus MVA-HIV, high dose) increases. In all high-dose groups, a significant reduction in binding titers occurred by week 15, consistent with a rapid contraction of Env-specific plasma cells during this 5-week period. Nevertheless, the antibody titers in the modified vector group continued to remain higher than those in the control vector group at this time point. Low-dose immunizations exhibited 3-fold (MVAΔ4) and 8-fold (MVAΔ5) increases over the control MVA vector at week 10 (Fig. 4A). Indeed, at week 10, low-dose immunization with both Δ4 and Δ5 induced equivalent or higher levels of anti-Env antibody than those elicited by the control at a high dose, providing evidence for a dose-sparing advantage for these modified vectors, although again, these increases were lost at week 15 (Fig. 4A).
Fig 4.
Modified MVA vectors elicit enhanced titers of HIV Env-specific antibodies in immunized macaques. (A) Titers of antibodies against vaccine-encoded gp120 were determined by ELISAs, at the indicated times, in macaques that were immunized with either the high-dose (2 × 108 PFU) or low-dose (1 × 107 PFU) MVAΔ4-HIV, MVAΔ5-HIV, or MVA-HIV vector. Symbols represent individual macaques, horizontal lines denote geometric mean titers, arrows indicate immunizations at weeks 0 and 9, and an asterisk denotes a P value of ≤0.05 (determined by a Kruskal-Wallis test). (B) Week 10 sera from macaques immunized with high-dose MVAΔ4-HIV (red) or MVA-HIV (blue) were assayed at a 1:20 dilution for ex vivo neutralization activity against an HIV Con-C Env-pseudotyped lentivirus reporter, as described in the supplemental material. Data are normalized to individual macaques' preimmune values. Horizontal lines denote group means. NS indicates “not significant.”
Because we observed such a significant increase in levels of binding antibody at week 10, we also determined the capacity of plasma from macaques immunized with MVAΔ4-HIV or MVA-HIV to cross-neutralize a nonidentical HIV subtype C Env (pCon-C gp160)-pseudotyped lentivirus reporter (Fig. 4B). Plasma samples obtained prior to immunization (preimmune) and at 1 week following booster immunization (week 10) from macaques immunized with the high-dose MVAΔ4-HIV or MVA-HIV vector were assayed ex vivo at a 1:20 dilution by an ex vivo TZM-bl cell-based infectivity reduction assay (25). Additional assays in which the plasma samples were diluted from 1:20 to 1:500 yielded similar results (data not shown). For each macaque, a percent neutralization value was calculated as the difference (preimmune minus week 10) in infectivities of the pseudotyped reporter in the assay (Fig. 4B). These data show that neither the antibody responses elicited by MVAΔ4-HIV nor those elicited by MVA-HIV demonstrated a significant ex vivo cross-neutralization of HIV subtype C Env-pseudotyped virions (Fig. 4B), and no correlation was observed between gp120-specific ELISA titers and the ex vivo neutralization activity of the consensus subtype C (Con-C) envelope (see Fig. S2 in the supplemental material).
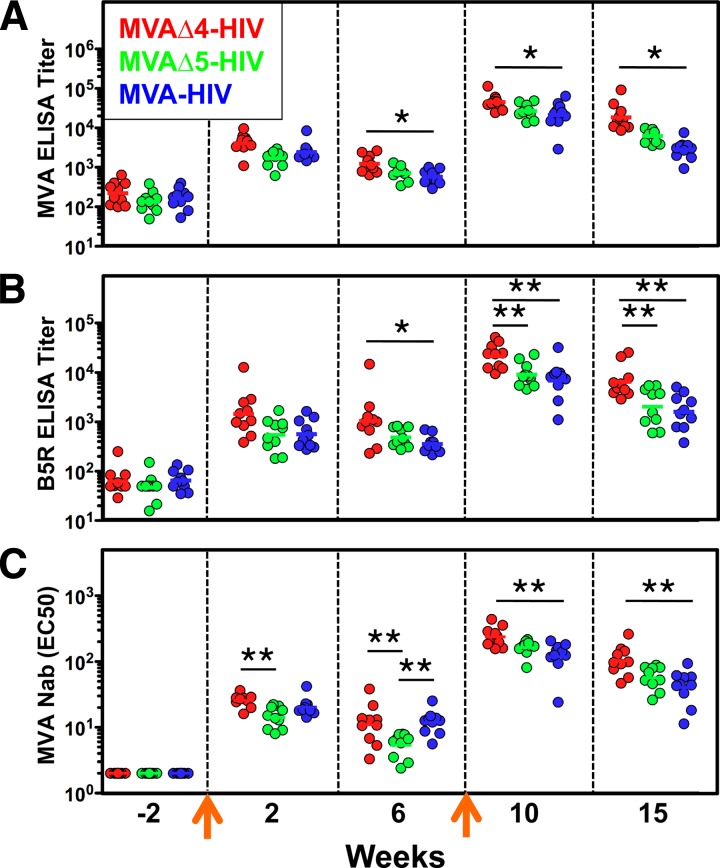
We also assessed the effects that the vector modifications had on the generation of vector-specific antibody responses, since this could be relevant for future poxvirus vaccines. Macaques immunized with MVAΔ4-HIV exhibited modest enhancements (2- to 7-fold) in the titers of binding antibodies directed against both MVA virion structural proteins (determined via MVA virion ELISAs) and the nonstructural protein B5R (Fig. 5A and B). Statistically significant differences in these antibody titers elicited by modified versus control vaccines were observed at multiple time points following both primary and secondary immunizations (Fig. 5A and B). Furthermore, the titers of vector-specific neutralizing antibodies elicited by MVAΔ4-HIV were similarly enhanced (2-fold) compared to those elicited by the control MVA-HIV vector at numerous time points following immunization (Fig. 5C). Interestingly, the titers of B5R-specific antibodies that were elicited by the MVAΔ5-HIV vector, which harbors a udg deletion that attenuates the expression of the B5R protein in infected cells, more closely resembled those of the control MVA-HIV vector and were significantly lower (2- to 3-fold) than those elicited by MVAΔ4-HIV following secondary immunization (Fig. 5B). Thus, the deletion of udg in MVAΔ5-HIV offset the enhancement of vector-specific antibody titers that otherwise resulted from immunization with a vector (MVAΔ4-HIV) harboring deletions of immune-modulatory genes only.
Fig 5.
MVAΔ4-HIV elicits high titers of vector-specific antibody responses in immunized macaques. Titers of binding antibodies against MVA virions (A) or a recombinant B5R protein (B) were determined by ELISAs, and titers of neutralizing antibodies against MVA (C) were determined by the ex vivo neutralization of an MVA-lacZ reporter virus, at the indicated times, in macaques that were immunized with the high-dose (2 × 108 PFU) MVAΔ4-HIV, MVAΔ5-HIV, or MVA-HIV vector. Symbols represent individual macaques, horizontal lines denote geometric mean titers, and arrows indicate immunizations at weeks 0 and 9. * denotes a P value of ≤0.05, and ** denotes a P value of ≤0.005 (determined by a Kruskal-Wallis test). Nab, neutralizing antibody; EC50, 50% effective concentration.
DISCUSSION
The purpose of this study was to determine whether the targeted deletion of viral genes from recombinant MVA-based HIV vaccines confers enhanced vaccine immunogenicity compared to that conferred by a control vaccine derived from parental (unmodified) MVA in rhesus macaques. The experimental vaccines (MVAΔ4-HIV and MVAΔ5-HIV) embody two different genetic modifications to the MVA backbone, which are comprised of deletions of different combinations of MVA genes. MVAΔ4-HIV harbors simultaneous deletions of four poxvirus immune-modulatory genes (MVA008L, MVA153L, MVA159R, and MVA184R), which encode an IL-18 binding protein (75, 80), a CC-chemokine binding protein (7, 62), a dominant negative Toll/IL-1 signaling adapter, and a soluble IL-1β receptor (3, 76), respectively. MVAΔ5-HIV harbors a deletion of the viral uracil-DNA glycosylase gene (MVA101R) (30), which reduces late gene expression, in addition to the deletions of the above-described four viral immune-modulatory genes. Both modified vaccines and the nondeleted parental MVA vaccine express an identical set of synthetic, codon-optimized HIV genes encoding antigenic consensus subtype C Env and Gag proteins under the control of early poxvirus promoters (modified H5) from the MVA deletion III locus. In summary, we observed that our modified vaccines elicited enhanced levels of both HIV-specific T cell and antibody (binding and nonneutralizing) immune responses in rhesus macaques but that these enhanced responses were transient in nature and not durable.
The immunization of macaques with modified vaccines resulted in the induction of higher levels of both HIV-specific T cell and antibody responses following both primary and secondary immunizations. With respect to primary T cell responses, modified vaccines induced levels of HIV-specific CD8 and CD4 T cells that were, on average, up to 5- to 6-fold higher than those induced by the control. Interestingly, the greatest differences between frequencies of HIV-specific T cells elicited by modified and control vaccines were observed acutely (2 weeks) following primary immunization. These results indicate that the modified vaccines are better able to elicit primary antigen-specific CD8 and CD4 T cell responses than the parental control and suggest that the modified vaccines may have a relatively greater impact on the generation of HIV-specific effector rather than memory T cells. Similarly, the MVAΔ4-HIV and MVAΔ5-HIV vaccines elicited 4- to 5-fold-higher frequencies of HIV-specific CD4 T cells than those elicited by the control vaccine at 1 week following homologous booster immunization. However, these augmented responses were transient in nature, as only trends toward higher-level CD4 responses were observed 5 weeks later in macaques that had been boosted with the modified vaccines.
Because of limited nonhuman primate resources, this study was not designed to assess the relative contributions of the deletions of individual MVA immune-modulatory genes on vaccine immunogenicity. However, immunogenicity studies in mice of MVA recombinants that harbor a single-gene deletion of MVA184R (78) or MVA153L (18, 21) or a double deletion of these two immune-modulatory genes (32) reported modest effects (2- to 3-fold enhancement) of these gene deletions on the magnitudes of the ensuing T cell responses. The relatively greater effects observed here for our modified vaccines in nonhuman primates may reflect positive contributions of additional gene deletions, species-specific differences between rhesus macaques and mice, or a combination of these factors with regard to the modified vaccines' ability to induce a more favorable environment for antigen presentation and/or the expansion of antigen-specific T cells in vivo.
While the deletion of udg from recombinant MVA vaccines was shown previously to effect modest enhancements of vaccine-elicited T cell responses in rhesus macaques (30), its deletion in combination with the deletion of four viral immune-modulatory genes (MVAΔ5-HIV) was not observed to confer any obvious additive enhancement of the magnitude of induced HIV-specific T cell responses beyond that similarly observed for MVAΔ4-HIV. In contrast, with regard to vector-specific antibody responses, the deletion of udg in MVAΔ5-HIV offset the enhancement otherwise conferred by the Δ4 modification alone. With regard to dose-sparing effects, both modified vectors elicited peak titers of Env-specific antibody responses following low-dose booster immunizations that were comparable to those elicited by the control vaccine at a high dose, whereas dose-sparing effects pertaining to levels of HIV-specific T cell responses were less pronounced.
Finally, our analysis of vaccine-elicited T cell breadth, at the level of peptide pools, clearly demonstrates a major bias in favor of HIV Env, compared to Gag, for both CD8 and CD4 T cell responses. Moreover, this Env bias was similarly observed following the immunization of macaques with both modified and control vaccines. Analogous observations of Env immunodominance within both CD8 and CD4 T cell populations were reported by a number of studies of other poxvirus-based HIV vaccines, including ones derived from MVA (58) and NYVAC (57, 58), in preclinical macaque studies (57, 58) and a human clinical trial evaluation (38). Because an increased breadth of HIV-specific T cell responses may be reasoned to potentially confer increased protection by HIV vaccines, it will be important, going forward, to better understand how to broaden vaccine-elicited T cell responses beyond those directed primarily toward HIV envelope antigens.
The deletion of immune-modulatory genes from MVA vectors engendered an enhanced ability to elicit antibody responses in vivo, although compared to the control, the relative magnitude and kinetics of these responses appear to be antigen specific. The immunization of macaques with both MVAΔ4-HIV and MVAΔ5-HIV induced HIV Env-specific antibody responses in plasma that were transiently of a higher magnitude than those elicited by the MVA-HIV control. Env-specific antibodies were detected as early as 2 weeks following immunization with MVAΔ4-HIV but not the control vaccine. More dramatically, MVAΔ4-HIV and MVAΔ5-HIV elicited significantly higher (up to 25-fold) titers of gp120 binding antibodies at 1 week following homologous booster immunization, which were nonneutralizing against an HIV Con-C-pseudotyped reporter vector, than the control vector. However, these augmented antibody responses were transient in nature, as similar titers of gp120 binding antibodies were observed 5 weeks later for macaques immunized with either the modified or control vaccines. In contrast, MVAΔ4-HIV elicited 2- to 7-fold-higher titers of vector-specific antibodies (both neutralizing and nonneutralizing) following booster immunization, which remained persistently elevated through at least 6 weeks postboosting, suggesting that the Δ4 modification may be relevant to the improvement of MVA as a smallpox virus vaccine per se. As anticipated, MVAΔ5-HIV, which harbors a deletion of udg in addition to deletions of the four immune-modulatory genes deleted from MVAΔ4-HIV, induced titers of vector-specific antibodies that were at best comparable to, but frequently lower than, those elicited by the control vaccine. Thus, when combined with the Δ4 modification, the deletion of udg specifically offset the enhancement of vector-specific antibody responses that was otherwise observed, presumably because the deletion of udg abrogates the expression of MVA late genes (30, 45), which encode many virion structural proteins that are antibody targets (24).
It is striking that following homologous booster immunization, with MVAΔ4-HIV or MVAΔ5-HIV in particular, the relative peak titer and ensuing rate of the postpeak decline of Env-specific antibody titers were higher than those of vector-specific antibody responses. At present, the precise mechanism by which the modified vaccines elicit transiently enhanced Env-specific antibody titers is not known and could be due to direct effects of the immune-modulatory gene deletions on responding Env-specific B cells or to indirect effects on the development of Env-specific B cell responses that are mediated by the enhanced levels of Env-specific CD4 T-helper cells that are elicited by the modified vectors. Regardless, these kinetics are consistent with the preferential development of short-lived, rather than long-lived, Env-specific antibody-secreting plasma cells following booster immunization (71, 87) and may imply that the modified vaccines magnify the expansion of short-lived Env-specific plasma cells above that otherwise achieved by the control vector.
Alternatively, the phenotype of relatively impermanent Env-specific antibodies in plasma, which we observed here in the context of immunization with recombinant MVA vaccines, may have a component that is intrinsic to the HIV envelope antigen itself. Previous analyses of HIV Env antibody responses that were elicited following either the vaccination of humans with recombinant gp120 (33) or the suppression of HIV viremia, via antiretroviral treatment (ART), in chronically infected individuals (12) indicated that HIV Env induces plasma antibody responses that are of a selectively short duration compared to those of other conventional T-dependent vaccine antigens (5). Additionally, it is possible that mechanisms governing immunological tolerance, which have been hypothesized to select against Env-specific B cell responses that are cross-reactive with self-antigens (40), could also contribute to Env-specific plasma antibody responses that are relatively less durable than those elicited by other (non-cross-reactive) antigens. Because an effective prophylactic HIV vaccine will need to elicit high levels of protective antibodies that are durable, prior to HIV exposure, in order to prevent HIV infection or to circumvent the establishment of viral latency in CD4 T cells (36, 86), it is imperative to better understand the mechanisms that govern the generation of serological memory, specifically as they pertain to HIV envelope antigens. Thus, HIV Env may not be the best indicator for assessing the benefits of specific MVA modifications toward enhancing the humoral immunogenicity (for non-Env antigens) of such modified vaccines. Indeed, the more modest, but significant, increases in the levels of antivector antibody responses induced by the Δ4 vector were sustained throughout the 15-week period of the study. Future investigations that compare the kinetics and quality of vaccine-elicited antibody responses directed against HIV Env versus a model antigen(s) should be informative in this regard.
Finally, because the higher levels of HIV-specific cellular and humoral responses that are engendered by our modified vaccines were relatively transient, it is unlikely that these vaccines would be able to mediate protection against infection which is significantly more durable than that achievable with the control. However, our results appear salient in light of the field's renewed interest in poxvirus-based HIV vaccines following the demonstration of modest efficacy against HIV acquisition (31.2%; 95% confidence interval [CI], 1.1 to 52.1%) by a canarypox virus (ALVAC-HIV)/recombinant gp120 (AIDSVAX-B/E) vaccine in the RV144 trial (68). The observation from the RV144 trial that, among vaccinees, much higher response rates were observed for gp120 binding antibodies (>98%) than for Gag-specific (8.3%) or Env-specific (15.9%) T cells supports a potential role for nonneutralizing antibodies in mediating protection against HIV infection. In addition, analyses to identify correlates of risk in the RV144 trial have shown a significant association between high V1/V2 IgG responses and lower rates of HIV infection (41; B. F. Haynes, presented at the AIDS Vaccine 2011 Meeting, Bangkok, Thailand, 12 to 15 September 2011). Results from a recent preclinical study in rhesus macaques to assess protection against the acquisition of neutralization-resistant simian immunodeficiency virus (SIV) following repeated low-dose rectal exposure further emphasize the importance of the inclusion of Env immunogens in HIV vaccines as well as the relevance of MVA-based vaccine vectors (8). As such, our identification of combinations of genes that can be deleted from MVA vectors to positively affect these vaccines' cellular and humoral immunogenicity profiles in nonhuman primates provides a platform for the further development of poxvirus-based HIV vaccines.
Supplementary Material
ACKNOWLEDGMENTS
We thank B. Moss (NIH) for providing the MVA seed stock (MVA-1974 clone 1 LVDP6) from which all recombinant vectors were derived. We also thank the veterinary and animal care staff, including Elizabeth Strobert, Sissy Chikazawa, Stephanie Ehnert, Chris Souder, and other members of the Yerkes National Primate Research Center's Research Resources staff, for providing expert care and handling of the nonhuman primates utilized in this study. HIV peptide reagents were sourced, in part, from the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH. Recombinant vaccinia virus B5R protein was obtained via the Biodefense and Emerging Infections Research Resources Repository (BEI Resources).
This work was supported by the following sources: NIH U19 AI061728 (IPCAVD), the Immunology Core of the Emory Center for AIDS Research (NIH P30 AI050409), and the Yerkes National Primate Research Center's base grant (NIH P51 RR000165).
We declare that we have no conflicts of interest with regard to the work described in the manuscript.
Footnotes
Published ahead of print 12 September 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Alcami A, Khanna A, Paul NL, Smith GL. 1999. Vaccinia virus strains Lister, USSR and Evans express soluble and cell-surface tumour necrosis factor receptors. J. Gen. Virol. 80(Pt 4):949–959 [DOI] [PubMed] [Google Scholar]
- 2. Alcami A, Smith GL. 1996. A mechanism for the inhibition of fever by a virus. Proc. Natl. Acad. Sci. U. S. A. 93:11029–11034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alcami A, Smith GL. 1992. A soluble receptor for interleukin-1 beta encoded by vaccinia virus: a novel mechanism of virus modulation of the host response to infection. Cell 71:153–167 [DOI] [PubMed] [Google Scholar]
- 4. Alcami A, Symons JA, Smith GL. 2000. The vaccinia virus soluble alpha/beta interferon (IFN) receptor binds to the cell surface and protects cells from the antiviral effects of IFN. J. Virol. 74:11230–11239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Amanna IJ, Carlson NE, Slifka MK. 2007. Duration of humoral immunity to common viral and vaccine antigens. N. Engl. J. Med. 357:1903–1915 [DOI] [PubMed] [Google Scholar]
- 6. Antoine G, Scheiflinger F, Dorner F, Falkner FG. 1998. The complete genomic sequence of the modified vaccinia Ankara strain: comparison with other orthopoxviruses. Virology 244:365–396 [DOI] [PubMed] [Google Scholar]
- 7. Bahar MW, et al. 2008. Structure and function of A41, a vaccinia virus chemokine binding protein. PLoS Pathog. 4:e5 doi:10.1371/journal.ppat.0040005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Barouch DH, et al. 2012. Vaccine protection against acquisition of neutralization-resistant SIV challenges in rhesus monkeys. Nature 482:89–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bejon P, et al. 2007. Extended follow-up following a phase 2b randomized trial of the candidate malaria vaccines FP9 ME-TRAP and MVA ME-TRAP among children in Kenya. PLoS One 2:e707 doi:10.1371/journal.pone.0000707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Beveridge NE, et al. 2007. Immunisation with BCG and recombinant MVA85A induces long-lasting, polyfunctional Mycobacterium tuberculosis-specific CD4+ memory T lymphocyte populations. Eur. J. Immunol. 37:3089–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Blanchard TJ, Alcami A, Andrea P, Smith GL. 1998. Modified vaccinia virus Ankara undergoes limited replication in human cells and lacks several immunomodulatory proteins: implications for use as a human vaccine. J. Gen. Virol. 79:1159–1167 [DOI] [PubMed] [Google Scholar]
- 12. Bonsignori M, et al. 2009. HIV-1 envelope induces memory B cell responses that correlate with plasma antibody levels after envelope gp120 protein vaccination or HIV-1 infection. J. Immunol. 183:2708–2717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bowie A, et al. 2000. A46R and A52R from vaccinia virus are antagonists of host IL-1 and Toll-like receptor signaling. Proc. Natl. Acad. Sci. U. S. A. 97:10162–10167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Buchbinder SP, et al. 2008. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet 372:1881–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Carroll MW, Moss B. 1997. Host range and cytopathogenicity of the highly attenuated MVA strain of vaccinia virus: propagation and generation of recombinant viruses in a nonhuman mammalian cell line. Virology 238:198–211 [DOI] [PubMed] [Google Scholar]
- 16. Casimiro DR, et al. 2004. Heterologous human immunodeficiency virus type 1 priming-boosting immunization strategies involving replication-defective adenovirus and poxvirus vaccine vectors. J. Virol. 78:11434–11438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cebere I, et al. 2006. Phase I clinical trial safety of DNA- and modified virus Ankara-vectored human immunodeficiency virus type 1 (HIV-1) vaccines administered alone and in a prime-boost regime to healthy HIV-1-uninfected volunteers. Vaccine 24:417–425 [DOI] [PubMed] [Google Scholar]
- 18. Clark RH, Kenyon JC, Bartlett NW, Tscharke DC, Smith GL. 2006. Deletion of gene A41L enhances vaccinia virus immunogenicity and vaccine efficacy. J. Gen. Virol. 87:29–38 [DOI] [PubMed] [Google Scholar]
- 19. Corey L, McElrath MJ, Kublin JG. 2009. Post-step modifications for research on HIV vaccines. AIDS 23:3–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Corona Gutierrez CM, et al. 2004. Therapeutic vaccination with MVA E2 can eliminate precancerous lesions (CIN 1, CIN 2, and CIN 3) associated with infection by oncogenic human papillomavirus. Hum. Gene Ther. 15:421–431 [DOI] [PubMed] [Google Scholar]
- 21. Cottingham MG, et al. 2008. Recombination-mediated genetic engineering of a bacterial artificial chromosome clone of modified vaccinia virus Ankara (MVA). PLoS One 3:e1638 doi:10.1371/journal.pone.0001638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Currier JR, et al. 2010. Phase I safety and immunogenicity evaluation of MVA-CMDR, a multigenic, recombinant modified vaccinia Ankara-HIV-1 vaccine candidate. PLoS One 5:e13983 doi:10.1371/journal.pone.0013983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dangoor A, et al. 2010. Clinical and immunological responses in metastatic melanoma patients vaccinated with a high-dose poly-epitope vaccine. Cancer Immunol. Immunother. 59:863–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Davies DH, et al. 2008. Antibody profiling by proteome microarray reveals the immunogenicity of the attenuated smallpox vaccine modified vaccinia virus Ankara is comparable to that of Dryvax. J. Virol. 82:652–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Derdeyn CA, et al. 2004. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science 303:2019–2022 [DOI] [PubMed] [Google Scholar]
- 26. Dorrell L, et al. 2007. Safety and tolerability of recombinant modified vaccinia virus Ankara expressing an HIV-1 gag/multiepitope immunogen (MVA.HIVA) in HIV-1-infected persons receiving combination antiretroviral therapy. Vaccine 25:3277–3283 [DOI] [PubMed] [Google Scholar]
- 27. Drexler I, Heller K, Wahren B, Erfle V, Sutter G. 1998. Highly attenuated modified vaccinia virus Ankara replicates in baby hamster kidney cells, a potential host for virus propagation, but not in various human transformed and primary cells. J. Gen. Virol. 79(Pt 2):347–352 [DOI] [PubMed] [Google Scholar]
- 28. Dunachie SJ, et al. 2006. A DNA prime-modified vaccinia virus Ankara boost vaccine encoding thrombospondin-related adhesion protein but not circumsporozoite protein partially protects healthy malaria-naive adults against Plasmodium falciparum sporozoite challenge. Infect. Immun. 74:5933–5942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Garber DA, Feinberg MB. 2006. Probing the possibilities for T-cell-based AIDS vaccines. Curr. Opin. HIV AIDS 1:314–322 [DOI] [PubMed] [Google Scholar]
- 30. Garber DA, et al. 2009. Expanding the repertoire of modified vaccinia Ankara-based vaccine vectors via genetic complementation strategies. PLoS One 4:e5445 doi:10.1371/journal.pone.0005445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Garber DA, Silvestri G, Feinberg MB. 2004. Prospects for an AIDS vaccine: three big questions, no easy answers. Lancet Infect. Dis. 4:397–413 [DOI] [PubMed] [Google Scholar]
- 32. Garcia-Arriaza J, Najera JL, Gomez CE, Sorzano CO, Esteban M. 2010. Immunogenic profiling in mice of a HIV/AIDS vaccine candidate (MVA-B) expressing four HIV-1 antigens and potentiation by specific gene deletions. PLoS One 5:e12395 doi:10.1371/journal.pone.0012395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gilbert PB, et al. 2005. Correlation between immunologic responses to a recombinant glycoprotein 120 vaccine and incidence of HIV-1 infection in a phase 3 HIV-1 preventive vaccine trial. J. Infect. Dis. 191:666–677 [DOI] [PubMed] [Google Scholar]
- 34. Goepfert PA, et al. 2011. Phase 1 safety and immunogenicity testing of DNA and recombinant modified vaccinia Ankara vaccines expressing HIV-1 virus-like particles. J. Infect. Dis. 203:610–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Guimaraes-Walker A, et al. 2008. Lessons from IAVI-006, a phase I clinical trial to evaluate the safety and immunogenicity of the pTHr.HIVA DNA and MVA.HIVA vaccines in a prime-boost strategy to induce HIV-1 specific T-cell responses in healthy volunteers. Vaccine 26:6671–6677 [DOI] [PubMed] [Google Scholar]
- 36. Haase AT. 2010. Targeting early infection to prevent HIV-1 mucosal transmission. Nature 464:217–223 [DOI] [PubMed] [Google Scholar]
- 37. Hansen SG, et al. 2009. Effector memory T cell responses are associated with protection of rhesus monkeys from mucosal simian immunodeficiency virus challenge. Nat. Med. 15:293–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Harari A, et al. 2008. An HIV-1 clade C DNA prime, NYVAC boost vaccine regimen induces reliable, polyfunctional, and long-lasting T cell responses. J. Exp. Med. 205:63–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hawkridge T, et al. 2008. Safety and immunogenicity of a new tuberculosis vaccine, MVA85A, in healthy adults in South Africa. J. Infect. Dis. 198:544–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Haynes BF, et al. 2005. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science 308:1906–1908 [DOI] [PubMed] [Google Scholar]
- 41. Haynes BF, et al. 2012. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N. Engl. J. Med. 366:1275–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hill AV, et al. 2010. Prime-boost vectored malaria vaccines: progress and prospects. Hum. Vaccin. 6:78–83 [DOI] [PubMed] [Google Scholar]
- 43. Hirao LA, et al. 2010. Comparative analysis of immune responses induced by vaccination with SIV antigens by recombinant Ad5 vector or plasmid DNA in rhesus macaques. Mol. Ther. 18:1568–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hirsch VM, et al. 1996. Patterns of viral replication correlate with outcome in simian immunodeficiency virus (SIV)-infected macaques: effect of prior immunization with a trivalent SIV vaccine in modified vaccinia virus Ankara. J. Virol. 70:3741–3752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Holzer GW, Falkner FG. 1997. Construction of a vaccinia virus deficient in the essential DNA repair enzyme uracil DNA glycosylase by a complementing cell line. J. Virol. 71:4997–5002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jaoko W, et al. 2008. Safety and immunogenicity of recombinant low-dosage HIV-1 A vaccine candidates vectored by plasmid pTHr DNA or modified vaccinia virus Ankara (MVA) in humans in East Africa. Vaccine 26:2788–2795 [DOI] [PubMed] [Google Scholar]
- 47. Keefer MC, et al. 2011. A phase I trial of preventive HIV vaccination with heterologous poxviral-vectors containing matching HIV-1 inserts in healthy HIV-uninfected subjects. Vaccine 29:1948–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kim DW, Krishnamurthy V, Bines SD, Kaufman HL. 2010. TroVax, a recombinant modified vaccinia Ankara virus encoding 5T4: lessons learned and future development. Hum. Vaccin. 6:784–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kim JH, Rerks-Ngarm S, Excler JL, Michael NL. 2010. HIV vaccines: lessons learned and the way forward. Curr. Opin. HIV AIDS 5:428–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Letvin NL, et al. 2006. Preserved CD4+ central memory T cells and survival in vaccinated SIV-challenged monkeys. Science 312:1530–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Liu J, et al. 2009. Immune control of an SIV challenge by a T-cell-based vaccine in rhesus monkeys. Nature 457:87–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mayr A, Hochstein-Mintzel V, Stickl H. 1975. Abstammung, eigenschaften und verwendung des attenuierten vaccinia-stammes MVA. Infection 3:6–14 [Google Scholar]
- 53. McElrath MJ, et al. 2008. HIV-1 vaccine-induced immunity in the test-of-concept Step Study: a case-cohort analysis. Lancet 372:1894–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. McShane H, et al. 2005. Boosting BCG with MVA85A: the first candidate subunit vaccine for tuberculosis in clinical trials. Tuberculosis (Edinb.) 85:47–52 [DOI] [PubMed] [Google Scholar]
- 55. McShane H, et al. 2004. Recombinant modified vaccinia virus Ankara expressing antigen 85A boosts BCG-primed and naturally acquired antimycobacterial immunity in humans. Nat. Med. 10:1240–1244 [DOI] [PubMed] [Google Scholar]
- 56. Meyer H, Sutter G, Mayr A. 1991. Mapping of deletions in the genome of the highly attenuated vaccinia virus MVA and their influence on virulence. J. Gen. Virol. 72:1031–1038 [DOI] [PubMed] [Google Scholar]
- 57. Mooij P, et al. 2009. Comparison of human and rhesus macaque T-cell responses elicited by boosting with NYVAC encoding human immunodeficiency virus type 1 clade C immunogens. J. Virol. 83:5881–5889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mooij P, et al. 2008. Differential CD4+ versus CD8+ T-cell responses elicited by different poxvirus-based human immunodeficiency virus type 1 vaccine candidates provide comparable efficacies in primates. J. Virol. 82:2975–2988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Moorthy VS, et al. 2004. Phase 1 evaluation of 3 highly immunogenic prime-boost regimens, including a 12-month reboosting vaccination, for malaria vaccination in Gambian men. J. Infect. Dis. 189:2213–2219 [DOI] [PubMed] [Google Scholar]
- 60. Moorthy VS, et al. 2004. A randomised, double-blind, controlled vaccine efficacy trial of DNA/MVA ME-TRAP against malaria infection in Gambian adults. PLoS Med. 1:e33 doi:10.1371/journal.pmed.0010033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mwau M, et al. 2004. A human immunodeficiency virus 1 (HIV-1) clade A vaccine in clinical trials: stimulation of HIV-specific T-cell responses by DNA and recombinant modified vaccinia virus Ankara (MVA) vaccines in humans. J. Gen. Virol. 85:911–919 [DOI] [PubMed] [Google Scholar]
- 62. Ng A, Tscharke DC, Reading PC, Smith GL. 2001. The vaccinia virus A41L protein is a soluble 30 kDa glycoprotein that affects virus virulence. J. Gen. Virol. 82:2095–2105 [DOI] [PubMed] [Google Scholar]
- 63. Nicol MP, Grobler LA. 2010. MVA-85A, a novel candidate booster vaccine for the prevention of tuberculosis in children and adults. Curr. Opin. Mol. Ther. 12:124–134 [PubMed] [Google Scholar]
- 64. Parrino J, et al. 2007. Safety, immunogenicity and efficacy of modified vaccinia Ankara (MVA) against Dryvax challenge in vaccinia-naive and vaccinia-immune individuals. Vaccine 25:1513–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Pathan AA, et al. 2007. Boosting BCG with recombinant modified vaccinia Ankara expressing antigen 85A: different boosting intervals and implications for efficacy trials. PLoS One 2:e1052 doi:10.1371/journal.pone.0001052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Peters BS, et al. 2007. Studies of a prophylactic HIV-1 vaccine candidate based on modified vaccinia virus Ankara (MVA) with and without DNA priming: effects of dosage and route on safety and immunogenicity. Vaccine 25:2120–2127 [DOI] [PubMed] [Google Scholar]
- 67. Pitisuttithum P, et al. 2006. Randomized, double-blind, placebo-controlled efficacy trial of a bivalent recombinant glycoprotein 120 HIV-1 vaccine among injection drug users in Bangkok, Thailand. J. Infect. Dis. 194:1661–1671 [DOI] [PubMed] [Google Scholar]
- 68. Rerks-Ngarm S, et al. 2009. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med. 361:2209–2220 [DOI] [PubMed] [Google Scholar]
- 69. Sandstrom E, et al. 2008. Broad immunogenicity of a multigene, multiclade HIV-1 DNA vaccine boosted with heterologous HIV-1 recombinant modified vaccinia virus Ankara. J. Infect. Dis. 198:1482–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Shiver JW, et al. 2002. Replication-incompetent adenoviral vaccine vector elicits effective anti-immunodeficiency-virus immunity. Nature 415:331–335 [DOI] [PubMed] [Google Scholar]
- 71. Slifka MK, Ahmed R. 1998. Long-lived plasma cells: a mechanism for maintaining persistent antibody production. Curr. Opin. Immunol. 10:252–258 [DOI] [PubMed] [Google Scholar]
- 72. Smith CL, et al. 2005. Recombinant modified vaccinia Ankara primes functionally activated CTL specific for a melanoma tumor antigen epitope in melanoma patients with a high risk of disease recurrence. Int. J. Cancer 113:259–266 [DOI] [PubMed] [Google Scholar]
- 73. Smith CL, et al. 2005. Immunodominance of poxviral-specific CTL in a human trial of recombinant-modified vaccinia Ankara. J. Immunol. 175:8431–8437 [DOI] [PubMed] [Google Scholar]
- 74. Smith GL, Symons JA, Alcami A. 1999. Immune modulation by proteins secreted from cells infected by vaccinia virus. Arch. Virol. Suppl. 15:111–129 [DOI] [PubMed] [Google Scholar]
- 75. Smith VP, Bryant NA, Alcami A. 2000. Ectromelia, vaccinia and cowpox viruses encode secreted interleukin-18-binding proteins. J. Gen. Virol. 81:1223–1230 [DOI] [PubMed] [Google Scholar]
- 76. Spriggs MK, et al. 1992. Vaccinia and cowpox viruses encode a novel secreted interleukin-1-binding protein. Cell 71:145–152 [DOI] [PubMed] [Google Scholar]
- 77. Stack J, et al. 2005. Vaccinia virus protein A46R targets multiple Toll-like-interleukin-1 receptor adaptors and contributes to virulence. J. Exp. Med. 201:1007–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Staib C, Kisling S, Erfle V, Sutter G. 2005. Inactivation of the viral interleukin 1beta receptor improves CD8+ T-cell memory responses elicited upon immunization with modified vaccinia virus Ankara. J. Gen. Virol. 86:1997–2006 [DOI] [PubMed] [Google Scholar]
- 79. Stittelaar KJ, et al. 2001. Safety of modified vaccinia virus Ankara (MVA) in immune-suppressed macaques. Vaccine 19:3700–3709 [DOI] [PubMed] [Google Scholar]
- 80. Symons JA, et al. 2002. The vaccinia virus C12L protein inhibits mouse IL-18 and promotes virus virulence in the murine intranasal model. J. Gen. Virol. 83:2833–2844 [DOI] [PubMed] [Google Scholar]
- 81. Symons JA, Tscharke DC, Price N, Smith GL. 2002. A study of the vaccinia virus interferon-gamma receptor and its contribution to virus virulence. J. Gen. Virol. 83:1953–1964 [DOI] [PubMed] [Google Scholar]
- 82. UNAIDS 2009. AIDS epidemic update. UNAIDS, Geneva, Switzerland [Google Scholar]
- 83. Vasan S, et al. 2010. Phase 1 safety and immunogenicity evaluation of ADMVA, a multigenic, modified vaccinia Ankara-HIV-1 B′/C candidate vaccine. PLoS One 5:e8816 doi:10.1371/journal.pone.0008816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Walker LM, Burton DR. 2010. Rational antibody-based HIV-1 vaccine design: current approaches and future directions. Curr. Opin. Immunol. 22:358–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Wilson NA, et al. 2006. Vaccine-induced cellular immune responses reduce plasma viral concentrations after repeated low-dose challenge with pathogenic simian immunodeficiency virus SIVmac239. J. Virol. 80:5875–5885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Wong S, Siliciano R. 2007. Biology of early infection and impact on vaccine design, p 17–22 In Koff W, Kahn P, Gust I. (ed), AIDS vaccine development: challenges and opportunities. Caister Academic Press, Wymondham, United Kingdom [Google Scholar]
- 87. Wrammert J, Ahmed R. 2008. Maintenance of serological memory. Biol. Chem. 389:537–539 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.